-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaOral Mycobiome Analysis of HIV-Infected Patients: Identification of as an Antagonist of Opportunistic Fungi
Oral microbiota contribute to health and disease, and their disruption may influence the course of oral diseases like oral candidiasis. Here we identify the core oral mycobiome (COM) and core oral bacteriome (COB) in HIV-infected and uninfected individuals, and demonstrate that the COM differs between these two groups. Decrease in abundance of Pichia (a resident oral fungus) in uninfected individuals coincided with increase in abundance of Candida, suggesting an antagonistic relationship. In vitro testing showed that Pichia spent medium (PSM) inhibits growth of pathogenic fungi; these findings were validated in an experimental mouse modal of oral candidiasis. The mechanism by which Pichia antagonizes Candida involves nutrient competition and secretory factor/s that inhibit the latter's ability to adhere, germinate, and form biofilms. This study is the first to characterize the mycobiome and the bacteriome in the oral cavity of HIV infected patients, and provides the first evidence that a fungus present in the same host microenvironment antagonizes Candida and identifies potential novel antifungal approach.
Published in the journal: . PLoS Pathog 10(3): e32767. doi:10.1371/journal.ppat.1003996
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1003996Summary
Oral microbiota contribute to health and disease, and their disruption may influence the course of oral diseases like oral candidiasis. Here we identify the core oral mycobiome (COM) and core oral bacteriome (COB) in HIV-infected and uninfected individuals, and demonstrate that the COM differs between these two groups. Decrease in abundance of Pichia (a resident oral fungus) in uninfected individuals coincided with increase in abundance of Candida, suggesting an antagonistic relationship. In vitro testing showed that Pichia spent medium (PSM) inhibits growth of pathogenic fungi; these findings were validated in an experimental mouse modal of oral candidiasis. The mechanism by which Pichia antagonizes Candida involves nutrient competition and secretory factor/s that inhibit the latter's ability to adhere, germinate, and form biofilms. This study is the first to characterize the mycobiome and the bacteriome in the oral cavity of HIV infected patients, and provides the first evidence that a fungus present in the same host microenvironment antagonizes Candida and identifies potential novel antifungal approach.
Introduction
Organisms residing in the oral cavity (oral microbiota) contribute to health and disease, and influence diseases like oral candidiasis, the most common oral complication of HIV-infection [1], [2]. Pathogenesis of oral candidiasis is linked to variables like changes in the CD4+ cell count and antiretroviral therapy (ART) in HIV-1-infected patients [3]. Although the introduction of ART has reduced mortality and morbidity as well as the incidence of opportunistic infections among HIV-infected patients, oral candidiasis remains a significant disease, even in the era of ART. In this regard, recent studies indicate that the decline of oral candidiasis among ART-experienced HIV-infected patients is transient in some HIV-infected individuals [4]. In addition, preliminary results reported by Thompson et al. [5] showed that symptomatic oral Candida infection occurred in one-third of patients with advanced AIDS (n = 122), even in the setting of ART. More recently, Patel et al. [6] reported symptomatic oral candidiasis in 27% (59/215) HIV-infected patients. Therefore, even in the era of ART, oral candidiasis remains a significant problem.
Characterization of the microbiota (bacteriome and mycobiome) in health and disease is expected to expedite the discovery, testing and validation of novel drugs [7]. Most studies that characterized the human microbiome in health and disease have focused on the bacteriome, in both oral and non-oral body sites [8]–[12]. Recently, Iliev et. al. [13] showed that while no significant differences in major phyla of commensal bacteria was observed in the intestinal microflora between wild-type and mice lacking Dectin-1, members of the mycobiome interacted with the intestinal immune system to influence inflammatory bowel disease (IBD) highlighting the role of the fungal community in disease. Earlier, we characterized the oral mycobiome in healthy individuals using high-throughput multitag pyrosequencing (MTPS), and reported that humans are colonized with up to 85 fungal genera [14]. Although these studies demonstrated the complexity of the human oral microbiome, the specific contribution of the mycobiome to oral diseases was not investigated.
Previous studies have shown that alteration in the bacterial population has direct impact on the development of Candida infections [for reviews, see 15]. However, the interactions between members of the oral microbiota and Candida in HIV disease setting have not been investigated. In the current study, we identified the core oral mycobiome (COM) and bacteriome (COB) [defined as those organisms present in ≥20% of the subjects] in HIV-infected and uninfected individuals, and demonstrated that the COM undergoes a change in HIV disease. Furthermore, we noted that a decrease in abundance of the yeast Pichia coincided with an increase in Candida colonization, suggesting an antagonistic relation between these two fungi. We also found that nutrient competition as well as Candida growth and modulation of its virulence factors by Pichia is a mechanism underlying this interaction. In addition, treatment with Pichia Spent Medium (PSM) was efficacious against oral candidiasis when tested in an experimental murine model. Our results provide the first evidence of interaction among members of the oral mycobiome community, particularly between Pichia and pathogenic fungi. These findings could lead to the development of novel antifungals to prevent and treat fungal infections including mucosal Candida infections, thereby impacting the management of oral candidiasis and other fungal infections. They also highlight the need to characterize the mycobiome at different body sites which may lead to novel discoveries of how fungi can be exploited to control health and disease.
Materials and Methods
Ethics Statement
Written informed consent was obtained from all study participants. Study participants were recruited according to protocol (#20070413) approved by the Human Subjects Institutional Review Board (IRB) at University Hospitals Case Medical Center. Oral rinse samples were obtained from 12 HIV-infected and 12 uninfected individuals (matched for age, sex, and ethnicity). Inclusion criteria for these participants were: >18 years of age and no clinical signs of oral mucosal disease including oral candidiasis, while the exclusion criteria were: recent use of antimicrobial or antifungal agents (within a month), use of topical or systemic steroids, pregnancy, and insulin-dependent diabetes mellitus. Oral wash samples were collected using a standardized Standard Operating Procedure developed by the Oral HIV/AIDS Research Alliance (OHARA) as described earlier [14]. All animal experimentation was performed in strict accordance with the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol for animal infection was approved by the Institutional Animal Care and Use Committee (IACUC) at Case Western Reserve University School of Medicine, Cleveland, Ohio (protocol approval number 2013-0020). All procedures were performed under general anesthesia and all efforts were made to minimize animal suffering.
Microbiome Analysis
Oral rinse samples were processed individually using the Fast DNA Spin Kit following manufacturer's instructions (BIO 101; Vista, CA). Each extraction tube was agitated three times using a Fast Prep FP120 instrument at a speed setting of 5 for 30 s. Tubes were cooled on ice between agitations. Fungi and bacteria present in these samples were identified with ITS-based and 16S probes, respectively. The ITS1 region from DNA sample extracts was amplified in triplicate using primers with high specificity for ascomycete fungi (fluorescently labeled forward primer ITS1F (CTTGGTCATTTAGAGGAAGTAA) and unlabeled reverse primer ITS2 (GCTGCGTTCTTCATCGATGC). The ITS primers were selected in this study to detect the presence of various fungi since these primers are able to detect consensus sequences present in a broad range of fungi [16], [17]. For bacterial identification, extracted DNA was amplified by PCR using routinely employed universal primers [fluorescently labeled forward primer 27F (5′-6FAM - AGAGTTTGATCCTGGCTCAG-3′) and unlabeled reverse primer 355R5′ (5′ - GCTGCCTCCCGTAGGAGT-3′)] [18], which amplify the first two hyper-variable regions of 16S rRNA [19] and are commonly used for microbiome analysis [10], [20]. Microbiome analysis was performed using multitag 454 pyrosequencing (MTPS) technique, which was used for characterization of nucleic acids [21] (for details, please see Method S1).
Strains
Candida albicans [strains SC5312, 10341, GDH2346), Pichia (MRL81)], Penicillium (MRL22345) and Cladosporium (MRL1458) strains tested in this study were obtained from the OHARA Repository at Case and the culture collection of the Center for Medical Mycology. Fungal strains were maintained on Sabouraud dextrose agar (SDA, [yeast extract, peptone, and dextrose at 1∶2∶1]) (Difco Laboratories, Detroit, MI) medium. Since species-level identification of Pichia based upon morphological or physiological features alone is usually not possible, we used a molecular approach (based on sequence analysis of the internal transcribed spacer and D1/D2 ribosomal DNA regions) [22] to confirm the identity of Pichia MRL81 strain. Our analysis revealed that this strain was P. farinosa. All strains were kept at −80°C for long-term storage.
Role of Nutrient Limitation
To determine whether inhibition of pathogenic fungi by Pichia spent medium (PSM) is due to nutrient competition between Pichia and Candida, we assessed Candida growth when mixed with Pichia at different ratios. P. farinosa and a GFP-tagged C. albicans strain (generous gift from Dr. B. Cormack) [23] were mixed together at equivalent cell densities (103, 104, or 105 cells each) in yeast nitrogen base (YNB) medium supplemented with glucose (0.9% w/v) and uridine (100 µg/mL). These mixed cell suspensions were incubated at 37°C and allowed to grow (as “mixed cultures”). Candida or Pichia cells alone were grown in the same growth medium (“mono cultures”) serving as controls. At specific time intervals (24, 48, 72 h), 1-mL aliquots were withdrawn from the mixed - and mono-cultures, centrifuged to collect the cells in pellet, and re-suspended in 250 µL phosphate buffered saline. Fifty µL of the resuspended cell mixture was placed on a glass slide and observed under phase-contrast and fluorescence microscope. The remaining cell suspension (200 µL) was transferred to a 96-well microtiter plate, and fluorescence was measured using a spectrofluorimeter (excitation and emission wavelength of 485 and 435 nm, respectively).
Biochemical Characterization of Pichia Spent Medium (PSM)
The effect of Pichia supernatant on Candida growth, germination, adherence, biofilm formation, and its biochemical properties was determined as described below.
Effect of Pichia Supernatant on the Growth of Candida, Aspergillus, and Fusarium
To evaluate the effect of Pichia supernatant on the growth of various fungi, Pichia spent (“conditioned”) medium (PSM) was obtained by centrifuging 100-mL culture of Pichia grown in Sabouraud dextrose broth (SDB) for 48 h, and filter sterilizing it. Next, fungal cells/conidia (1×105 cells/mL) were incubated with PSM at 35°C and growth was followed for 48 h. Aliquots were collected at 2 h intervals and fungal growth was measured spectrophotometrically at 600 nm.
Germination Assay
Effect of Pichia spent media (PSM) on Candida germination was determined using C. albicans strain SC5314, as described previously [24]. Candida cells were grown planktonically in the absence or presence of PSM. Germination rate was compared with that of cells grown in SDB media containing fetal bovine serum (FBS) (Hyclone, Thermo Fisher Scientific, Rockford, IL), a known inducer of germination. Briefly, C. albicans cells were grown and the cell density was adjusted to 1×107 cells/mL in Hanks balanced salt solution (HBSS) (Mediatech). In separate 1.5-mL tubes, 50 mL of these cells were diluted to a density of 5×105 cells/mL in HBSS (blank control), FBS, or PSM, and incubated on a rocker at 37°C for up to 4 h. At 15-min intervals, 10-µL samples from each media type were microscopically examined using a hemacytometer. Total cell count and germination (defined as a germ-tube length greater than or equal to the blastospore diameter) was determined from an average of 4 observations. One hundred to 200 cells were counted per observation. The assays were discontinued when cells clumped together, due to germination, which made it difficult to count individual cells.
Adhesion Assay
The effect of Pichia or Penicillium (used as a control) cells or supernatant on Candida adherence (using strain C. albicans SC5314, a clinical isolate used conventionally in Candida adhesion and germination assays) was determined as described earlier [24], [25]. Briefly, standardized suspensions of 50 to 200 cells/mL were added onto silicone elastomer disks for 90 min. Disks were then washed in phosphate-buffered saline (PBS) to remove non-adherent cells and placed in wells of 12-well tissue culture plates (Becton Dickinson, Franklin Lakes, NJ). Two milliliters of warm (55°C) liquefied SDA was added per well to completely cover the SE disks and allowed to solidify. Plates were incubated overnight (37°C), and the number of colonies adhering per disk was counted using a dissecting microscope.
Biofilm Evaluation
The effect of oral fungi (Pichia, Cladosporium or Penicillium, the latter fungi were used as controls since Cladosporium was present only in the uninfected subjects like Pichia, while Penicillium was present in both infected and uninfected subjects to the same abundance) or their supernatant on the ability of Candida to form biofilms was evaluated using metabolic activity assay and confocal microscopy, as described earlier [26]–[28]. Briefly, Candida cells were incubated in the presence or absence of Pichia cells or spent medium (supernatant, PSM) at different relative ratios (1∶3, 1∶1, 3∶1), and allowed to form biofilms for 48 h on silicone elastomer catheter discs. The amount of biofilm formed was assayed colorimetrically using the XTT (2,3-bis (2-methoxy-4-nitro-5-sulfophenyl)-5-[(phenylamino) carbonyl]-2H-tetrazolium hydroxide, Sigma-Aldrich) metabolic activity assay in which XTT is converted by metabolically active cells to a red formazan product [26]. In addition, the effect of fungal supernatants on the morphology and architecture of the formed biofilms was evaluated using confocal scanning laser microscopy (CSLM) [26]. Briefly, biofilms were stained with the fluorescently labeled polysaccharide-indicating lectin Concanavalin Alexa Fluor 488 conjugate (CON-A, 25 µg/mL; Invitrogen) and metabolic activity indicator dye FUN1™ (10 µM; Invitrogen). After staining, discs containing biofilms were flipped and placed on a 35-mm-diameter glass-bottom petri dish (MatTek Corp., Ashland, Mass.). Stained biofilms were observed with a Zeiss LSM510 confocal scanning laser microscope equipped with argon and HeNe lasers and mounted on a Zeiss Axiovert100 M microscope (Carl Zeis, Inc.). The objective used was a water immersion C-apochromat lens (40×; numerical aperture, 1.2).
In Vivo Model of Oral Candidiasis
Wild-type C57BL/6 mice (purchased from Charles River Laboratories, Wilmington, MA) were immunosuppressed with 4 mg of cortisone acetate (Sigma Chemical Co., St. Louis, Mo.) administered subcutaneously on the day before and 1 and 3 days after challenge with Candida cells. Mice were given tetracycline hydrochloride (Sigma Chemical Co., St. Louis, Mo.) in their drinking water (0.5 mg/ml), starting the day before infection. On the day of inoculation, mice were anesthetized and light scratches made on the dorsum of the tongue followed by the introduction of C. albicans GDH (108 blastospores). The scratches were superficial, limited to the outermost stratum corneum, and did not cause trauma or bleeding. Mice were divided into groups (n = 4); treated with Pichia supernatant, 100 µl in the oral cavity twice a day, a “mock” vehicle control, and untreated control. Topical nystatin (widely used clinically to treat oral candidiasis [29]) was used as a comparator. Treatment began on day 4 post inoculation, mice were sacrificed on day 7 and the tongues harvested for enumeration of tissue fungal burden or histopathology with Periodic acid-Schiff stain. Additionally, tongues were visually assessed daily beginning day 1 post infections to assess severity of the infection using a previously described scoring system [30]: A score of 0 indicates the appearance of a normal tongue, with intact light reflection and no visible signs of infection, a score of 1 denotes isolated patches of fungus, a score of 2 when confluent patches of fungus are observed throughout the oral cavity, and a score of 3 indicates the presence of wide-spread fungal plaques and erosive mucosal lesions. The histology slides were assessed by an independent pathologist who was blinded to the study arms. The animal studies were repeated on different days to ensure reproducibility.
Identification of PSM as a Protein
Since the antifungal activity of PSM was secretory in nature, we determined whether this activity was due to a protein, carbohydrate or small molecule (metabolite). We exposed PSM to proteinase K (which digests most proteins), NaOH (which denatures carbohydrates) [31], or acetonitrile extraction (that isolates metabolites) [32]. We also determined the effect of heat on PSM activity by exposing it to 90°C temperature in a water bath for 10 min. The ability of these differently treated PSM to inhibit Candida biofilms was evaluated as above.
Statistical Analysis
Microbiome data were analyzed using the QIIME and R platforms [33], [34]. Wilcoxon–Mann–Whitney rank sum test was used for comparison between the two groups, with P-value of <0.05 considered as a significant difference. For comparison of several groups, Kruskal–Wallis one-way analysis of variance on ranks was used, and pairwise multiple comparison procedures (Dunns method) post-hoc test was used for multiple pairwise comparisons. Univariate analyses was used to compare the prevalence of specific fungi and bacteria between the two groups using a Pearson chi-squared test or two-sample t-test (assuming unequal variances). For correlation analysis, microbiome abundance data was divided into independent data matrices (disease and no disease) and correlation analyses was conducted using the “psych” package (corr.test function) in R statistical platform (pairwise Spearman's correlation and two-tailed probability of t for each correlation) [34], [35]. The function “Circle.corr” was used to graphically illustrate the correlation coefficients and significant correlations (P<.05) in circle graph, where red and blue circle indicate positive and negative correlations, respectively. Numerical variables (e.g. metabolic activity, dry biomass, thickness) were all assessed using paired or unpaired t-test, or ANOVA as appropriate. Comparison of clinical scores were performed using box-plots and non-parametric independent samples Kruskal-Wallis test, while mean fungal burden (log CFU/g) were compared using ANOVA. All statistical analyses were performed using R [34] or SPSS (ver. 13) statistical software packages.
Results
Participant Demographics
A total of 24 individuals were enrolled in the study, with 12 HIV-infected patients and 12 uninfected individuals (11 males and one female in both study groups, Table 1). The mean age was 38.7 and 38.8 years in HIV-infected (age range: 22–56) and uninfected (age range: 22–59) groups, respectively. Among the 12 HIV-infected patients, eight had initiated antiretroviral therapy. In both study groups, self-reported ethnicities were: six African-Americans, two Hispanics, and four Caucasians. While all samples were analyzed for fungal microbiota, one of the samples did not provide robust signals for the bacterial microbiome, and hence was excluded from the analysis. In addition, the corresponding matched uninfected control sample was also excluded. As a result, there were 12 uninfected-HIV-infected sample pairs for mycobiome analysis but only 11 sample pairs for bacteriome analysis.
Tab. 1. Demographic information of study participants. 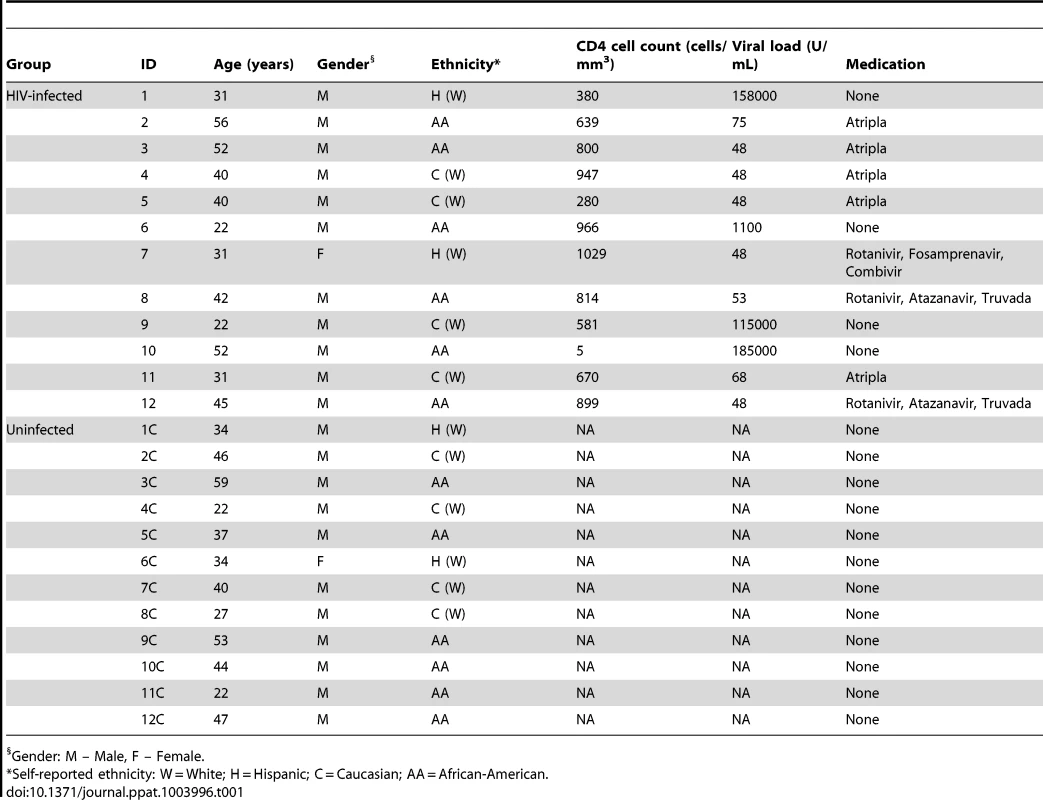
Gender: M – Male, F – Female. Oral Bacteriome of HIV-infected Participants Were Similar to That of Uninfected Individuals
Our results showed that the number of bacterial genera in the oral microbiota of study participants ranged between 8–14 per person among HIV-infected and uninfected individuals. Among HIV-infected patients, Prevotella, Streptococcus and Rothia were the most common genera; while in controls the most abundant bacteria were Prevotella, Streptococcus and Fusobacterium (Fig. 1A, and Table S1). The core oral bacteriome (COB) consisted of 14 genera in both HIV-infected and uninfected individuals, of which 13 (Actinomyces, Granulicatella, Fusobacterium, Leptotrichia, Rothia, Neisseria, Haemophilus, Pasteurella, Porphyromonas, Prevotella, Gemella, Streptococcus, and Veillonella) were common to both groups (Fig. 1B). We found that Capnocytophaga was present only in HIV-infected patients while Aggregatibacter was present in uninfected individuals only (Fig. 1B). These results suggest that the COB of HIV-infected patients was similar to that of uninfected individuals with minimal difference.
Fig. 1. Bacterial microbiome (bacteriome) of HIV-infected patients and uninfected individuals. 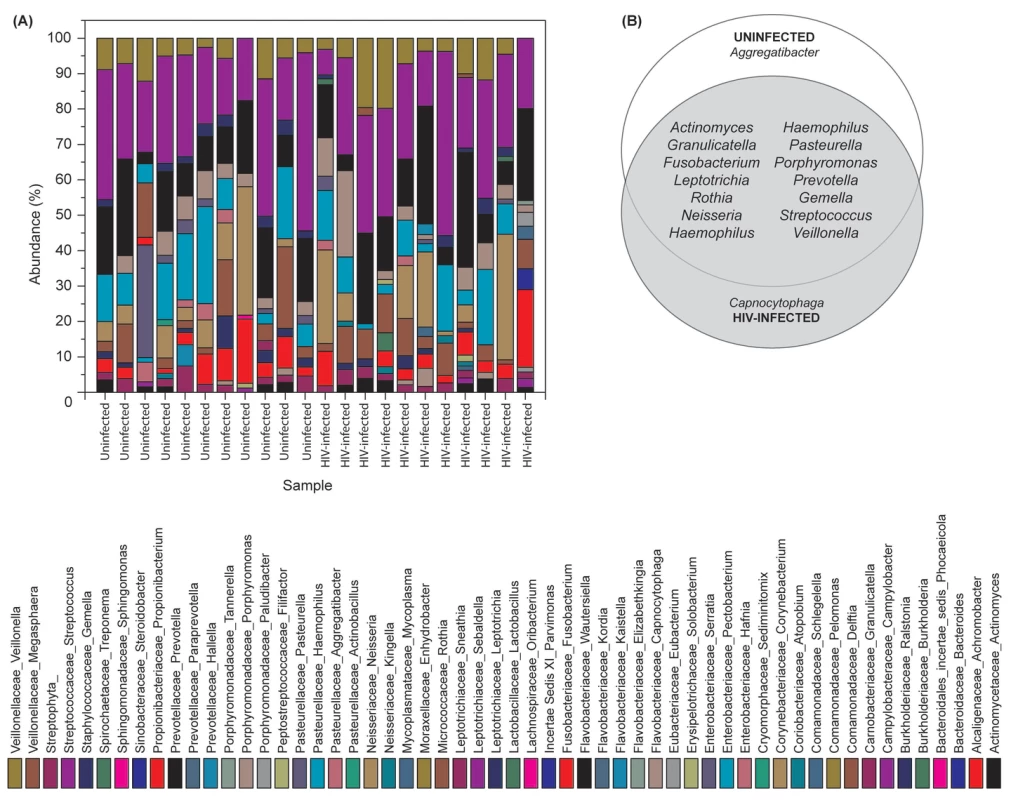
(A) Relative abundance bacteria in uninfected and HIV-infected patients (N = 11 for each group), (B) Bacteria present in the core oral bacteriome of uninfected and HIV-infected individuals. Oral Mycobiome of HIV-infected Patients Exhibits Differences from Uninfected Individuals
Our results showed that the number of fungal genera present in oral wash samples ranged between 1–9 per person among uninfected and HIV-infected individuals (Fig. 2A and Table S2). Among HIV-infected patients, Candida, Epicoccum, and Alternaria were the most common genera (present in 92%, 33%, and 25%, respectively), while in uninfected participants, the most abundant fungi were Candida, Pichia, and Fusarium (58%, 33%, and 33%, respectively; Fig. 2A). The COM of HIV-infected and uninfected individuals consisted of five genera (Fig. 2B); of these, Candida and Penicillium were common between the two groups, while differing in the remaining genera demonstrating that the COM of HIV-infected patients differs from that of age - and sex-matched uninfected controls. Among the Candida species detected, C. albicans was the most common (58% in uninfected and 83% in HIV-infected patients), followed by C. dubliniensis (17% in both groups). Interestingly, C. intermedia and C. sake were present only in uninfected (n = 1) and HIV-infected (n = 1) groups, respectively.
Fig. 2. Fungal microbiome (mycobiome) of HIV-infected patients and uninfected individuals. 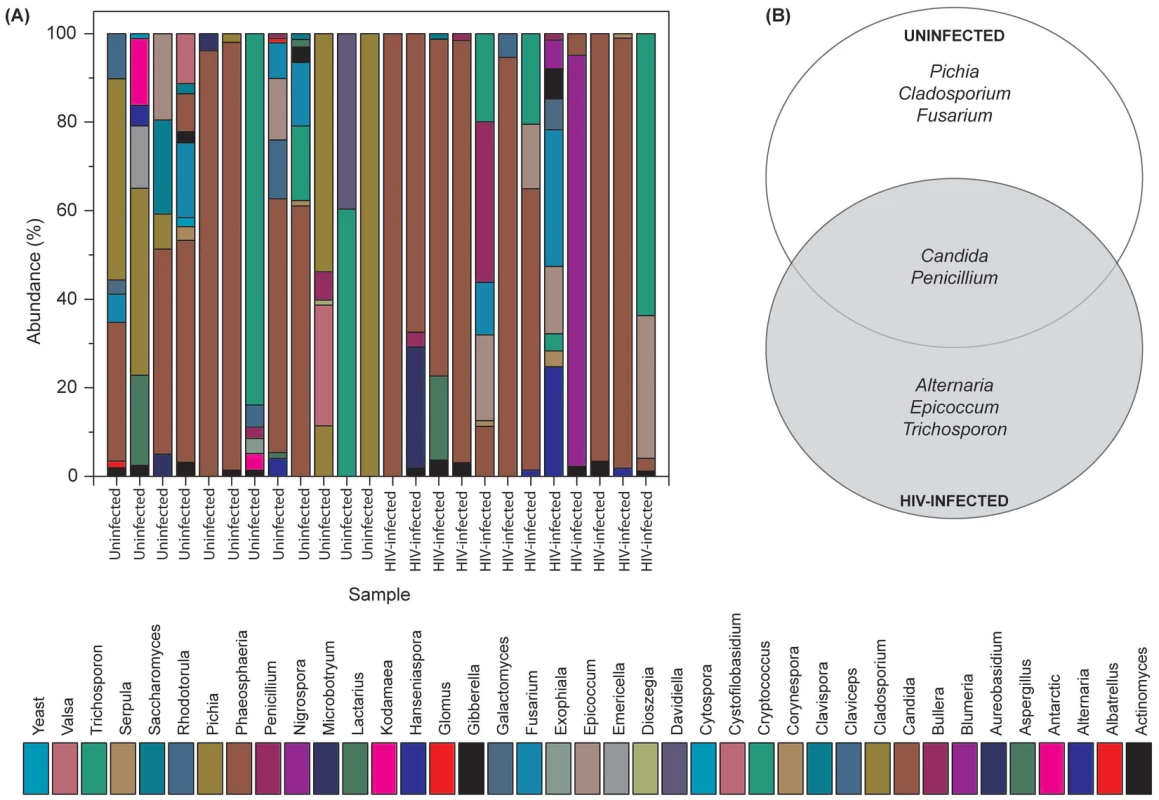
(A) Relative abundance of fungi in uninfected and HIV-infected patients (N = 12 for each group), (B) Fungi present in the core oral mycobiome of uninfected and HIV-infected individuals. Correlation between Members of the Oral Bacteriome and Mycobiome in HIV-Infected Patients
Next, we determined how the individual members of the oral bacteriome and mycobiome are correlated within their respective communities, and also across the two communities. We grouped the microbiome abundance data into independent mycobiome and bacteriome data matrices and conducted correlation analysis using R statistical computing software. We found 15 bacteria-fungi pairs that were correlated significantly in samples from non-infected study participants (Fig. 3 A, Table 2). Among these significant correlation pairs, two pairs (Rothia-Cladosporium and Granulicatella-Cryptococcus) were negatively correlated (coefficient −0.61 and −0.65, respectively). The remaining 13 pairs of significantly correlated pairs exhibited positive correlation with coefficients ranging from 0.64 (Aggregatibacter-Lactarius) to 0.86 (Capnocytophaga-Cladosporium). In comparison, there were 12 statistically significant bacteria-fungi pairs in HIV-infected patients, with 11 positive (coefficient of 0.64 for 8 pairs, 0.74 for 2 pairs, Fig. 3 B, Table 2) and one with negative correlation (Campylobacter-Candida, coefficient −0.67).
Fig. 3. Correlation coefficients of abundance of microbes in uninfected and HIV-infected patients. 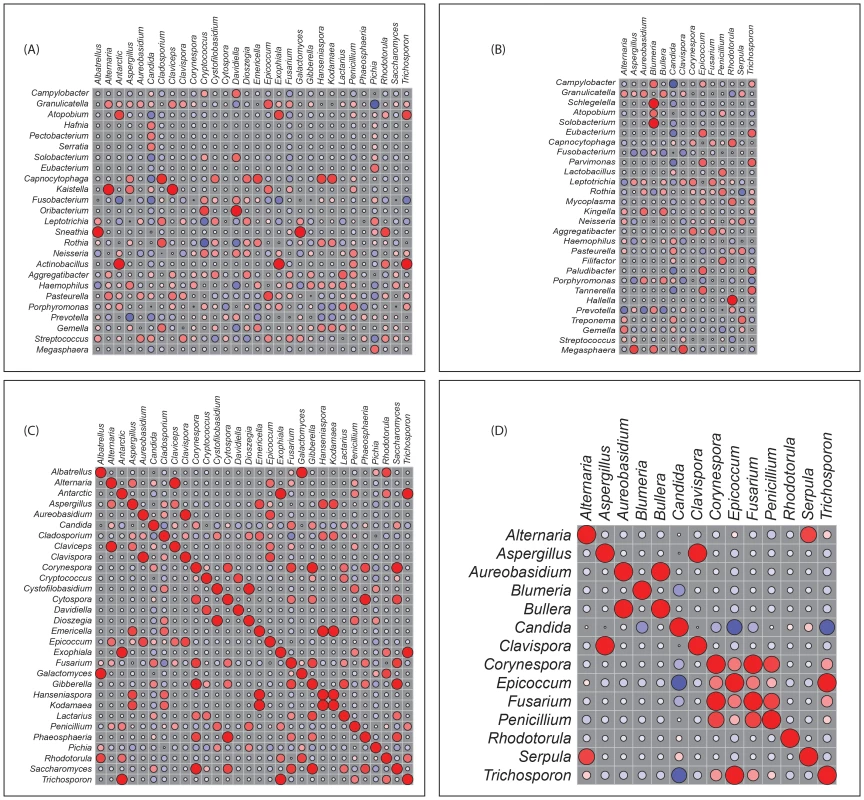
Correlation of bacteriome and mycobiome was determined for (A) uninfected and (B) HIV-infected study participants using R statistical computing software (Spearman's correlation and two-tailed probability of t for each correlation) for the two groups. Interactions among different fungi in the mycobiome of (C) uninfected and (D) HIV-infected study participants was also determined using similar approach. Red: Positive correlation; Blue: negative correlation; diameter of circles represent the absolute value of correlation for each pair of the microbe-microbe matrix. Tab. 2. Correlation between bacteriome and mycobiome in uninfected and HIV-infected study participants. 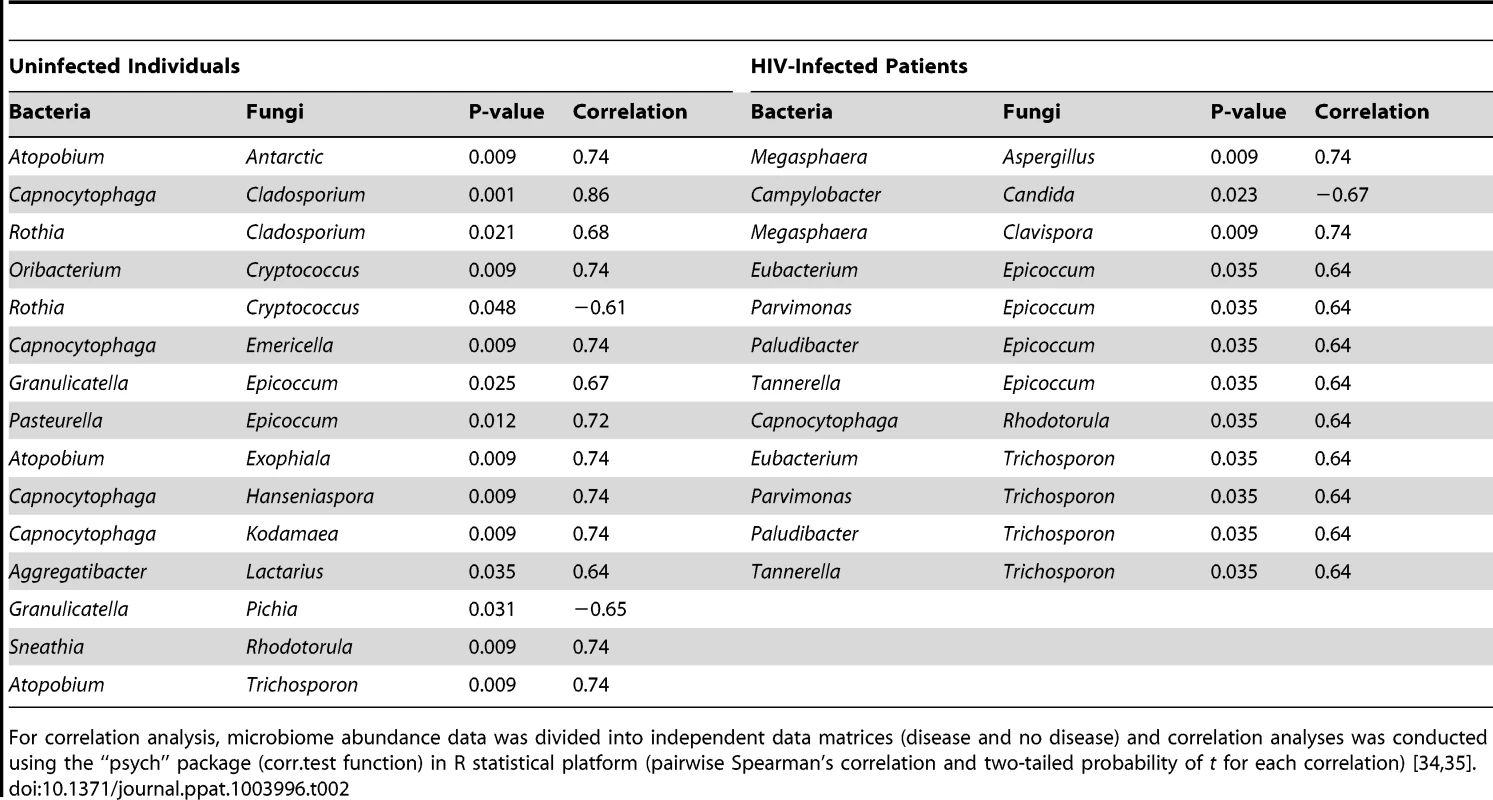
For correlation analysis, microbiome abundance data was divided into independent data matrices (disease and no disease) and correlation analyses was conducted using the “psych” package (corr.test function) in R statistical platform (pairwise Spearman's correlation and two-tailed probability of t for each correlation) [34], [35]. We also evaluated the correlation between different members of the mycobiome in the uninfected and HIV-infected groups. Our analyses revealed that in the uninfected group, 23 fungal-fungal interactions were statistically significant (P≤0.035, Fig. 3C, Table 3), while in the HIV-infected group, 6 fungus-fungus pairs were significantly correlated (P-value of ≤0.03, Fig. 3D), which included Candida-Epicoccum, Candida-Trichosporon, Epicoccum-Trichosporon, Penicillium-Corynespora, Penicillium-Fusarium, and Alternaria-Serpula.
Tab. 3. Correlation among oral fungi in uninfected study participants. 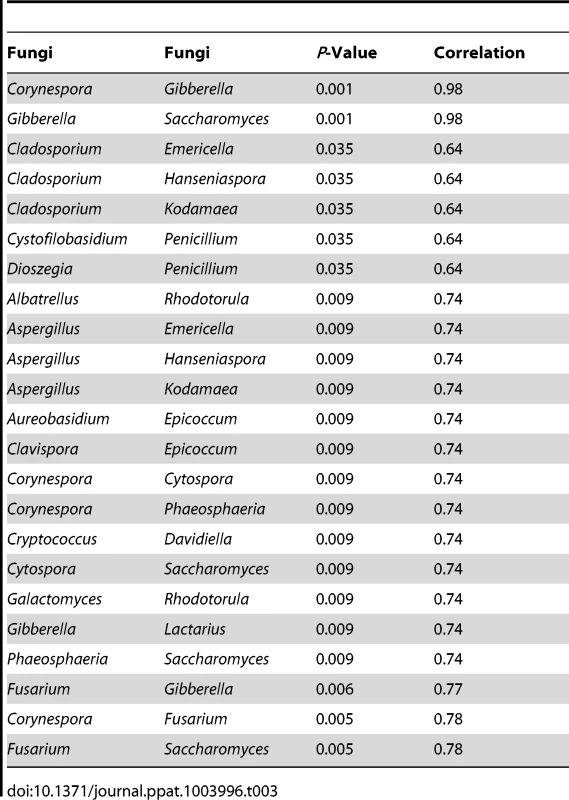
Pichia, a Member of the Core Oral Mycobiome, Exhibits Antagonism against Candida
Having defined the core mycobiome, next we investigated whether members of the core oral mycobiome are associated with Candida, the most common oral fungal pathogen of HIV-infection [36]. Our sequencing data indicated the presence of 3 Pichia species, including P. guillermondii, P. burtonii, and P. jadinii in the tested samples. We found that decrease in Pichia abundance coincided with an increase in Candida colonization (Fig. 4A), suggesting antagonism between Pichia and Candida. Furthermore, we found that among the 4 uninfected subjects where Pichia was present, 24 fungal genera were absent, including Aspergillus and Cryptococcus (Table 4). In addition, in these 4 uninfected individuals, 9 fungal genera (including Fusarium) were present as co-colonizers (Table 4). Analysis of the abundance profile of Fusarium in all uninfected individuals (n = 12), revealed that its abundance was 3-fold lower when Pichia was present (0.016%, n = 4) compared to where Pichia was absent (0.048%, n = 8). Taken together, these results show that Pichia interacts with other fungi, with an antagonistic interaction with known pathogens including Candida, Cryptococcus, Aspergillus and Fusarium.
Fig. 4. Antagonistic relation between Pichia and other fungi. 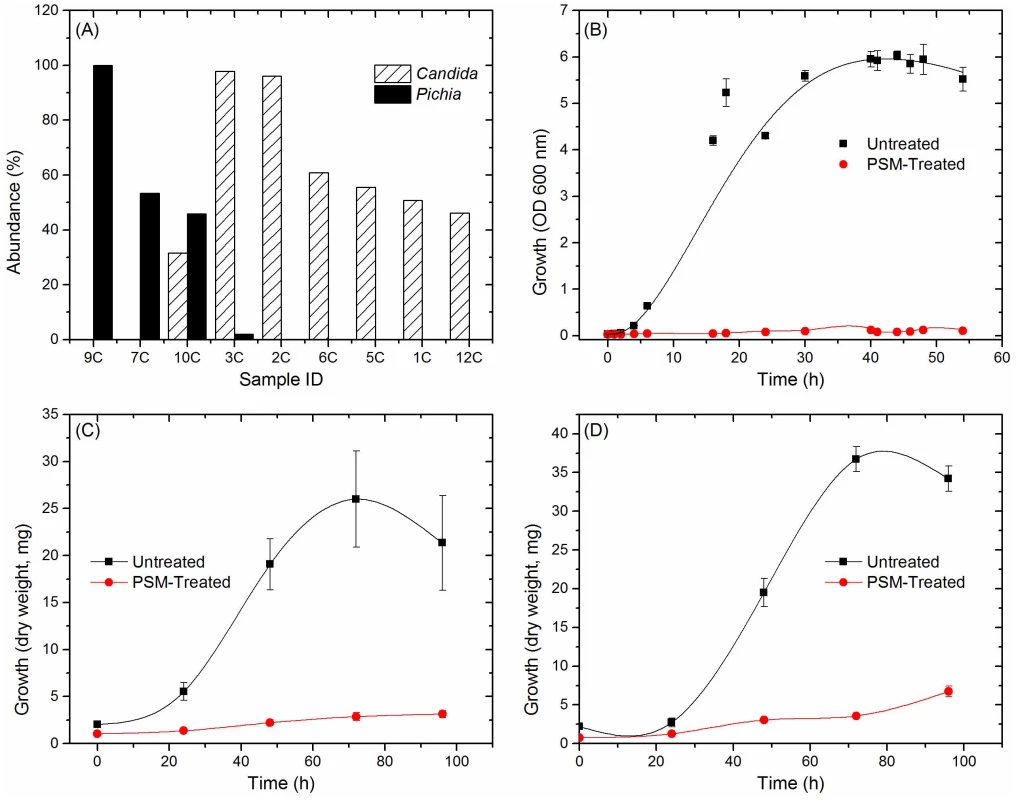
(A) Relative abundance of Pichia and Candida in uninfected individuals. (B) Effect of PSM on growth of Candida was determined by measuring optical density (OD). Effect of PSM on (C) Aspergillus, and (D) Fusarium was determined by measuring dry weight of fungi. Pichia spent (“conditioned”) medium (PSM) was obtained by centrifuging 100-mL culture of Pichia grown in Sabouraud dextrose broth (SDB). Tab. 4. Relationship between Pichia and other fungi in oral wash of uninfected patients. 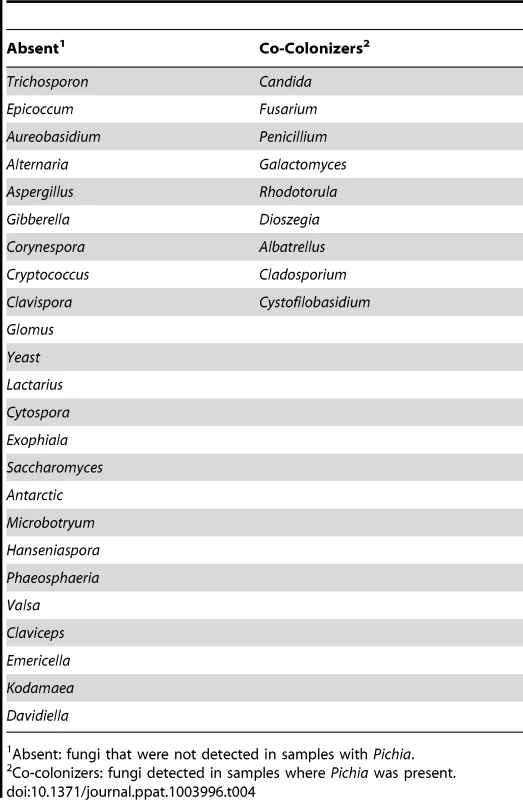
Absent: fungi that were not detected in samples with Pichia. Pichia Inhibits Growth of Candida, Aspergillus and Fusarium
Next, we investigated the ability of Pichia to inhibit growth of C. albicans, by allowing blastospores to grow in the presence or absence of Pichia spent medium (PSM). As mentioned above, our results revealed three Pichia species (P. guillermondii, P. burtonii, and P. jadinii) in the tested samples. These Pichia spp, along with P. farinosa, are common biocontrol agents used against plant pathogens [37]–[40]. Moreover, literature search showed that P. farinosa exhibits more biocontrol activity than P. guilliermondii [41]. Since P. farinosa and P. guilliermondii are very closely related to each other based on their whole genome [38] and mitochondrial DNA [42] sequences, we selected P. farinosa to investigate the interactions between this species and pathogenic fungi including Candida. As shown in Figure 4B, PSM completely inhibited Candida growth, demonstrating a direct inhibitory effect of Pichia against Candida. We also assessed the effect of PSM on growth of Aspergillus and Fusarium by determining their dry weight. As shown in Figure 4C and D, Aspergillus and Fusarium were unable to exhibit growth in presence of PSM. These studies demonstrated that PSM exhibits broad-spectrum activity against pathogenic fungi.
Next, we initiated studies to gain insight into the underlying mechanism for the allowing Pichia to inhibit Candida. Since Pichia is commonly used as a post-harvest biocontrol agent against plant pathogens [43]–[47], and several studies have suggested that this inhibitory activity involves nutrient competition, biofilm formation, germination, metabolites, secretory proteins [48]–[57], we explored whether Pichia-mediated inhibition of Candida involves these mechanisms.
Pichia Overgrows Candida in Mixed Cultures
To determine whether inhibition of Candida by Pichia is due to nutrient competition, we assessed Candida growth when mixed with Pichia at different ratios. We found that control Candida or Pichia cells exhibited robust, confluent growth when incubated by themselves (Fig. 5A–C, G). In contrast, mixing of Candida and Pichia (at 103 cells each) resulted in noticeably reduced cell density (Fig. 5D–F). Quantitative measurement of fluorescence signal confirmed reduced Candida growth in the presence of Pichia cells, with up to 58% decrease in fluorescence (Fig. 5H). Similar trends were observed when Candida and Pichia cells were mixed at 104 or 105 cells each (data not shown). These results suggested that Pichia outcompetes/overgrows Candida.
Fig. 5. Pichia overgrows Candida in mixed cultures. 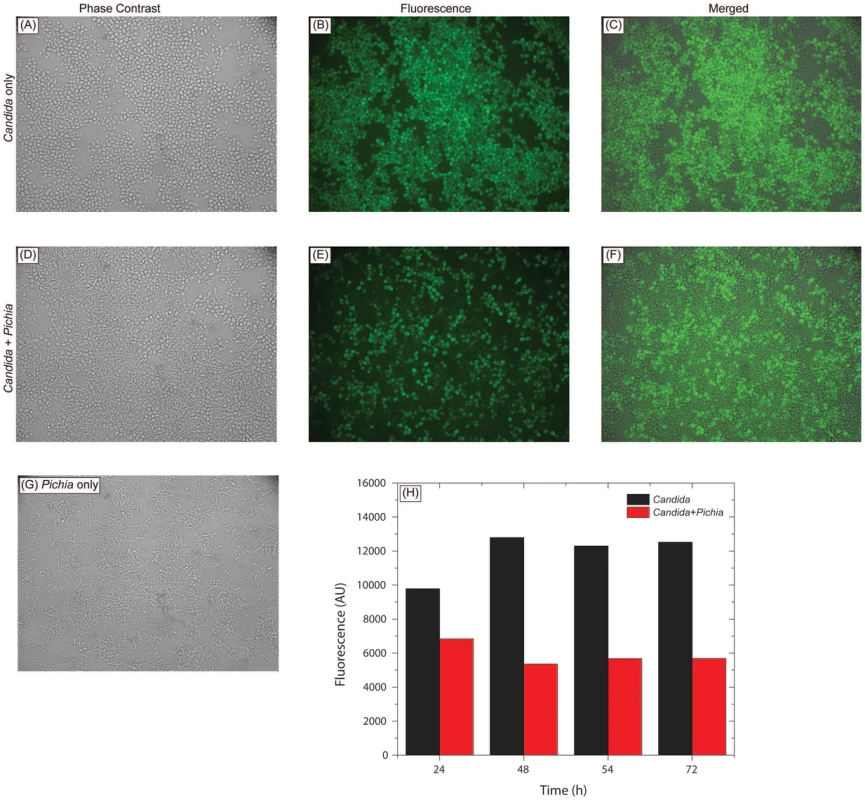
Pichia farinosa was mixed with GFP-tagged C. albicans strain at 103 cells each in yeast nitrogen base (YNB) growth medium and incubated at 37°C for 54 h. Growth of Candida cells was monitored by phase-contrast and fluorescence microscopy. Panels A, D, G: phase-contrast images of Candida alone, Candida+Pichia, and Pichia alone, respectively. Panels B, E: fluorescence images of Candida alone and Candida+Pichia mixture. Panels C, F: Composite figures with phase-contrast images merged with fluorescence image, for Candida only and Candida+Pichia mixture of cells. Panel H: quantitative measurement of fluorescence (excitation and emission wavelength = 485 and 435 nm, respectively). Pichia Inhibits the Ability of C. albicans to Form Biofilms
Our results showed that when both Candida and Pichia were present together, their abundance ratio was close to 1∶1. Therefore, to determine whether a threshold number of Pichia cells was necessary to inhibit Candida biofilms, we used mixtures of Pichia and Candida combined at different ratios (1∶1, 1∶3, 3∶1). Co-incubation of C. albicans with Pichia cells at different ratios resulted in significant inhibition of biofilm formation at all ratios tested (Fig. 6A, P<.05). Moreover, there was no significant difference in the extent of biofilm inhibition between the different cells densities of Pichia examined.
Fig. 6. Activity of Pichia spent medium (PSM) against fungal biofilms. 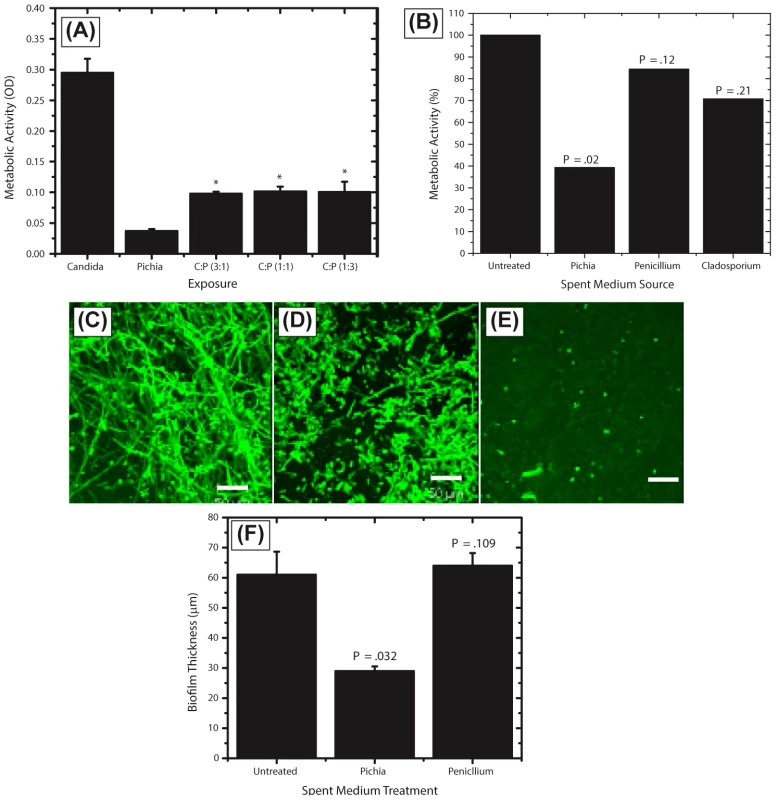
(A) Effect of Pichia cells on the ability of Candida to form biofilms. Candida and Pichia were co-incubated [Candida∶Pichia (C∶P) = 3∶1, 1∶1, or 1∶3] and biofilm formation was monitored (*P≤.002, compared to Candida or Pichia controls). (B) Effect of media supernatant obtained from Pichia, Penicillium, or Cladosporium on Candida biofilms. Mean ± SD of ≥3 separate experiments. (C–E) Confocal microscopy images of Candida biofilms formed in presence of (C) no media supernatant, (D) Penicillium supernatant or (E) Pichia supernatant. (F) Thickness of biofilms formed in presence of media supernatant of Pichia or Penicillium. Next, to determine whether the biofilm-inhibitory activity of P. farinosa was mediated by secretory factor/s, we determined the effect of Pichia spent medium (PSM) on C. albicans biofilms using a metabolic activity (XTT) assay described earlier [26]. Spent medium from Penicillium or Cladosporium (both prepared similarly as PSM) was used as a control since Penicillium had the same abundance in HIV-infected and uninfected individuals (present in 25% of samples in each group), while Cladosporium was present only in uninfected participants. Our results showed that when exposed to PSM, metabolic activity of Candida biofilms was significantly reduced (39% compared to untreated control, P = .02, Fig. 6B). Moreover, exposure to equivalently prepared spent media from Penicillium or Cladosporium did not have a significant effect on Candida biofilms (metabolic activity = 84% and 71%, respectively, P≥.12 for both comparisons, Fig. 6B). Next, we used confocal laser scanning microscopy (CLSM) to determine the effect of PSM on C. albicans biofilm architecture. While untreated and Penicillium-treated Candida formed robust biofilms (Fig. 6C–D, respectively), exposure to PSM resulted in biofilm disruption, with sparse yeast cells and no extracellular matrix or hyphae observed (Fig. 6E). Moreover, thickness of Candida biofilms exposed to PSM was significantly reduced compared to that of controls (Fig. 6F, P<.05). Finally, to determine whether the activity of PSM is dose-dependent, we compared the ability of 50% and 100% PSM to inhibit Candida biofilms. Metabolic activity and thickness of Candida biofilms exposed to 100% PSM was significantly lower than those exposed to 50% PSM (Fig. 7A, B; P = .007 and .006, respectively). Taken together, these results demonstrate that the Candida-inhibitory activity of Pichia is specific and dose-dependent.
Fig. 7. Dose dependent activity of PSM, and its effect on Candida germination and adhesion. 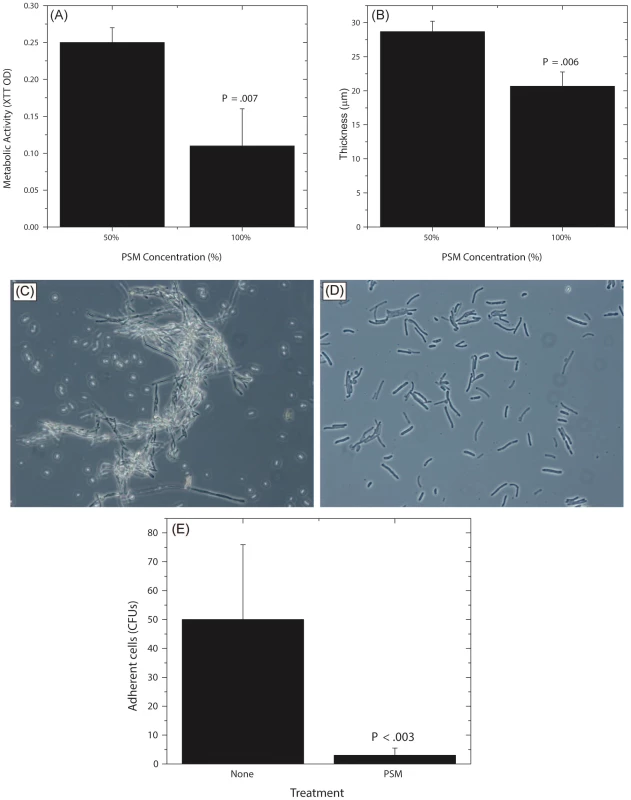
Effect of undiluted and diluted (50%) PSM on (A) metabolic activity and (B) thickness of Candida biofilms was assessed. (C) Germination in Candida grown in SDB, exposed to fetal bovine serum, (D) Stunted germ tubes formed by Candida exposed to Pichia supernatant (Magnification 20X), (E) Effect of PSM on the ability of Candida (grown in SDB) to adhere to solid substrates. (SDB - Sabouraud dextrose broth.) Pichia Inhibits Candida Germination and Adhesion
Since adhesion and germination are key steps in mature Candida biofilm formation [58]–[60] and are known Candida virulence factors, we examined whether P. farinosa spent medium affects these processes. Our data showed that while untreated Candida formed robust hyphae (Fig. 7C), exposure to PSM resulted in stunted Candida germ tubes (Fig. 7D), indicating that a secreted component of Pichia inhibits Candida germination. We also found that the number of Candida colony forming units (CFUs) adhering to silicone elastomer catheter substrate when treated with Pichia supernatant was significantly lower than untreated Candida cells (Fig. 7E, P<.003). These results showed that Pichia inhibits the ability of Candida to germinate and adhere to catheter substrate.
Active Ingredient of PSM Is a Protein
To determine whether the active ingredient of PSM is a metabolite or a protein, we evaluated Candida growth in presence or absence of metabolites extracted [32] from PSM. Extracted Pichia metabolites had no effect on Candida growth (Fig. 8A). Next, we evaluated the influence of proteinase-, alkali-, or heat (90°C for 10 min)-treated PSM on C. albicans biofilms. Our data showed that proteinase-K treatment abrogated the ability of PSM to inhibit biofilms, while alkali - and heat-treatment did not have any effect, as determined by analysis of biofilm thickness (Fig. 8B) and architecture (Fig. 8C–G). These studies indicate that the active component in PSM is proteinaceous, heat-stable, and non-glycosylated.
Fig. 8. Biochemical characterization of Pichia spent medium. 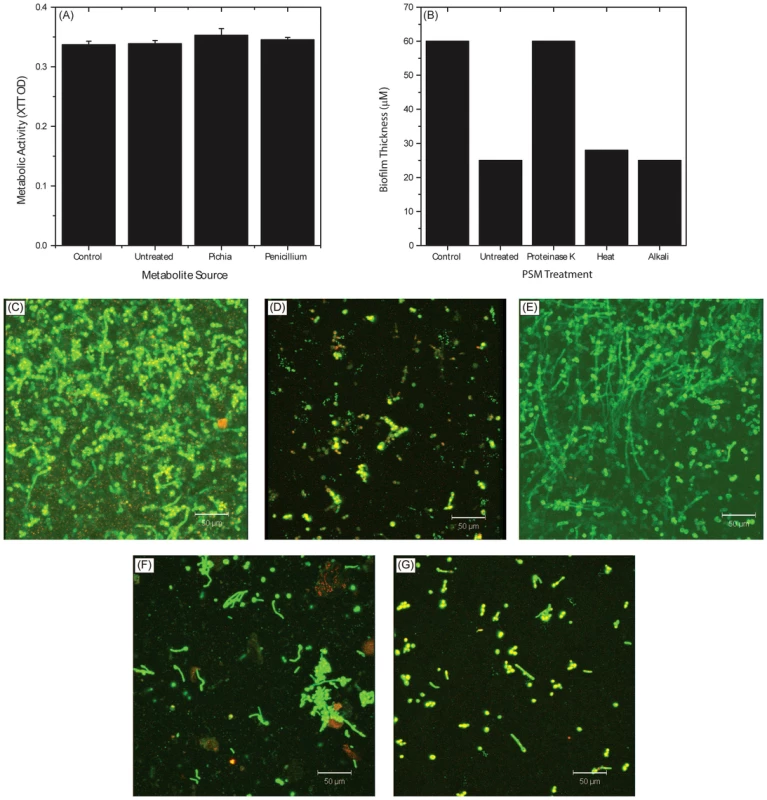
(A) Metabolic activity of Candida biofilms exposed to metabolites extracted from spent media of Pichia, Penicillium, or media control. (B) Effect of PSM exposed to proteinase, heat, or alkali on Candida biofilms. Confocal images show architecture of Candida biofilms exposed to (C) no PSM (control), (D) untreated PSM, (E) proteinase-K treated PSM, (F) heat, or (G) alkali. PSM Is Effective against Oral Candidiasis in an Experimental Murine Model
To determine whether the in vitro activity of PSM against Candida is also exhibited in vivo, we evaluated the efficacy of PSM in an experimental murine model of oral candidiasis. We found that at the end of treatment (Day 7), clinical score of PSM-treated mice was significant reduced compared to untreated, vehicle-treated, or nystatin-treated mice (Fig. 9A, P = .002 by Kruskal-Wallis test). The fungal burden of tongue from PSM-treated mice was also significantly reduced compared to untreated, vehicle-treated, or nystatin-treated controls (P≤.029 for all comparisons, Fig. 9B). Additionally, histological examination showed extensive tissue invasion by fungal hyphae and destruction of the epithelium in untreated or vehicle-treated controls or nystatin-treated mice (Fig. 9C–E). In contrast, tongue epithelium in PSM-treated mice revealed only superficial hyphal invasion and intact tissue structures (Fig. 9F). These results demonstrated that PSM was efficacious in treating oral candidiasis in vivo when tested in an experimental oral model of candidiasis.
Fig. 9. Efficacy of Pichia spent medium (PSM) in an experimental murine morel of oral candidiasis. 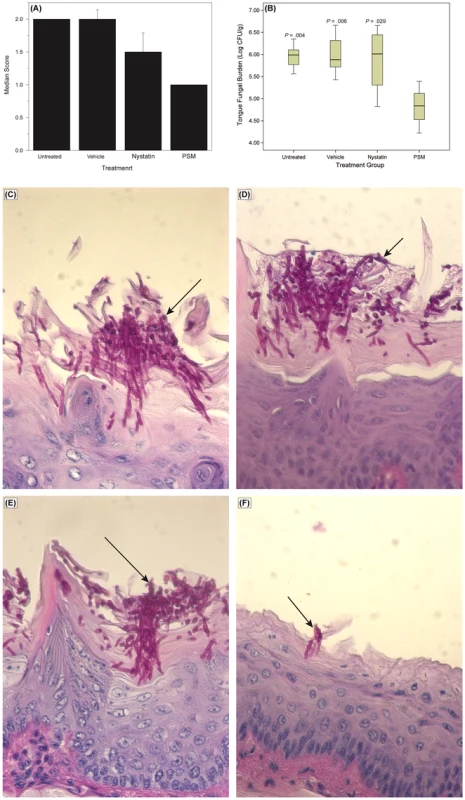
Assessment of oral candidiasis in mice infected with Candida was performed by (A) clinical score and (B) tongue fungal burden. (A) Median clinical scores of oral candidiasis in mice after treatment with PSM and nystatin. (B) Box-plot of tissue fungal burden (log CFUs/g) in different groups of mice infected with Candida (P-values, compared to PSM treatment). Histology analyses of tissue section of tongue from mouse infected with Candida, followed by (C) no treatment or treated with (D) vehicle control, (E) nystatin, or (F) PSM. Arrows – fungal hyphae. Discussion
In the current study, we identified the core oral mycobiome and core oral bacteriome in HIV-infected and uninfected individuals, and demonstrated that the oral mycobiome in HIV infection differs from non-diseased controls. In contrast to previous studies that characterized the bacterial component of the oral microbiome [61]–[63], our study defines both the bacterial and fungal components of the core oral microbiome in the same sample in HIV setting. Our findings revealed that the oral bacteriome of HIV-infected individuals was similar to that of uninfected individuals, indicating the presence of a shared bacteriome in these individuals. The core oral bacteriome of uninfected individuals in our study is in agreement with results reported earlier by Zaura et al. [64] who showed that 15 bacterial genera were present in the core oral bacteriome of healthy individuals.
Correlation analyses of the relationship between bacteriome and mycobiome revealed that 15 and 12 bacteria-fungi pairs were correlated significantly in samples from uninfected and HIV-infected patients, respectively. It is possible that these correlations indicate mutually dependent relationships within the oral microbiome, in which bacteria may be assisting or scavenging their fungal neighbors. Alternatively, fungal members of the microbiome may impact bacterial growth and drug susceptibility. Such interactions could influence the course and extent of oral diseases in the HIV setting. In this regard, interactions between bacteria and Candida have been investigated previously by Hogan and Kolter [65], who showed that P. aeruginosa kills hyphal form of C. albicans via biofilm formation. Furthermore, Candida-bacterial interactions are also associated with diseases like ventilator-associated pneumonia [66], [67] and bloodstream infections [68]. Our analyses revealed no correlation between Candida and bacteria in uninfected individuals, while in HIV-infected patients Candida and Campylobacter were negatively correlated. This correlation is in agreement with the findings of Navazesh et al. [69], who showed that antiretroviral therapy increased the risk for recovering bacteria (including Campylobacter species) with a concomitant decrease in the recovery rate of Candida, in HIV-infected women. Moreover, Workman et al. [70] reported that proteins secreted by Campylobacter inhibit the growth of C. albicans. The clinical relevance of this correlation remains to be investigated.
In the current study, we also defined the oral mycobiome of HIV-infected and uninfected study participants, and showed this fungal community to comprise up to nine different genera in both groups. To our knowledge, the only other study to have used a sequencing-based approach to identify oral fungi in HIV setting was that performed by Aas et al. [12], who analyzed sub-gingival plaque of HIV-infected patients, and reported the presence of only two fungal species (Saccharomyces cerevisiae and C. albicans in 4 and 2 patients, respectively). The difference in fungal profile between these investigators and our study may be due to differences in sample types (oral wash vs. sub-gingival plaque), detection probe (pan-fungal ITS probe vs. 18S rDNA), and sequencing technique (real-time pyrosequencing vs. rDNA sequencing).
Our study showed that the core oral mycobiome of HIV-infected patients was different from that of uninfected individuals. In a recent study, Iliev et al. [13] characterized the gut microbiome in wild type mice and isogenic Clec7a−/− mice that lack Dectin-1 (the innate immune receptor) and exhibited increased levels of chemically induced colitis. These investigators reported that there were no significant differences in bacteriome between wild type and Clec7a−/− mice, while the mycobiome profile differed between these isogenic mice, with an increase in opportunistic pathogenic fungi (Candida and Trichosporon) during colitis in Clec7a−/− mice, and a decrease in the nonpathogenic fungi (e.g. Saccharomyces). Thus, severe colitis in Dectin-1 knockout mice was associated with alterations in the gut mycobiome (but not the bacteriome). This study is similar to our findings, showing that HIV disease is associated with changes in the oral mycobiome, and highlight the importance of characterizing the mycobiome and its role in disease.
Our analyses showed that increase in Candida colonization was associated with a concomitant decrease in the abundance of Pichia, suggesting an antagonistic relation between these two fungi. In addition, these analyses also suggested that Pichia exhibits antagonistic interaction with other known pathogens including Cryptococcus, Aspergillus and Fusarium. These interactions were confirmed using growth assays that demonstrated broad-spectrum inhibitory activity of Pichia. The anti-Candida activity of Pichia was validated in vivo using an experimental murine model of oral candidiasis.
The biocontrol activity of Pichia against fungal plant pathogens has been shown to involve multiple mechanisms, including biofilm formation and germination [48]–[53]. The results of the current study are in agreement with these findings, where we showed that the anti-Candida activity of Pichia is mediated by inhibition of Candida growth and virulence factors like germination, adherence, and biofilm formation. It is well known that interfering with virulence factors decreases the ability of Candida to cause infection including oral candidiasis [71]–[79]. Therefore, Pichia, by inhibiting Candida virulence factors may limit the ability of this pathogenic fungus to cause infection. Such involvement of virulence factors is a common theme in interactions among microorganisms in the context of human infection [65].
Pichia biocontrol activity has also been attributed to nutrient limitation [39], [41], [51], [80]. Our results showed that Pichia outcompetes Candida when the two fungi are mixed together and allowed to grow. The possible reason for this phenomenon could be due to the ability of Pichia to consume nutrients more efficiently than Candida. Alternatively, it is possible that Pichia secretes factor/s that attenuate the ability of Candida to grow. Data in support of the latter possibility can be derived from our findings that Pichia spent medium inhibits pathogenic fungi including Candida, Aspergillus, Fusarium, and Cryptococcus. Furthermore, our results also demonstrate that the inhibitory activity of PSM is proteinaceous in nature, and not a metabolite. Earlier studies have shown that biocontrol activity of Pichia against fungal plant pathogens is mediated by secretory metabolite or proteins [48], [53], [81], [82].
In conclusion, we identified the core bacteriome and mycobiome in HIV setting, and identified HIV-specific changes in the mycobiome. We also identified a critical antagonistic interaction between Pichia and fungal pathogens including Candida. This interaction was demonstrated using in vitro and in vivo models. We also defined the mechanisms underlying the antagonistic interaction between Pichia and Candida. Our findings show for the first time that normal fungal community interacts with Candida in the oral cavity. Detailed investigations are warranted to purify and characterize the secretory factor/s mediating such interactions, and their mechanism/s of action at the molecular level. Our findings have wide implications regarding the discovery of novel antifungal agents that new therapeutic approaches for the management of fungal infections.
Supporting Information
Zdroje
1. JenkinsonHF, LamontRJ (2005) Oral microbial communities in sickness and in health. Trends Microbiol 13 : 589–595.
2. PattonLL, PhelanJA, Ramos-GomezFJ, NittayanantaW, ShiboskiCH, et al. (2002) Prevalence and classification of HIV-associated oral lesions. Oral Dis 8 : 98–109.
3. ChattopadhyayA, CaplanDJ, SladeGD, ShugarsDC, TienHC, et al. (2005) Risk indicators for oral candidiasis and oral hairy leukoplakia in HIV-infected adults. Community Dent Oral Epidemiol 33 : 35–44.
4. Koletar SL, Smurzynski M, Wu K, Collier AC, Bosch RJ, et al.. AIDS-defining illnesses occurring in treatment-experienced HIV-1-infected persons followed in the ACTG Longitudinal Linked Randomized Trials (ALLRT) study [Abstract 1017]. In Proceedings of the 15th Conference on Retroviruses & Opportunistic Infections; February 3–6, 2008; Boston, MA. Available: http://retroconference.org/. Accessed 2/3/13.
5. ThompsonGR3rd, PatelPK, KirkpatrickWR, WestbrookSD, BergD, et al. (2010) Oropharyngeal candidiasis in the era of antiretroviral therapy. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 109 : 488–495.
6. PatelPK, ErlandsenJE, KirkpatrickWR, BergDK, WestbrookSD, et al. (2012) The Changing Epidemiology of Oropharyngeal Candidiasis in Patients with HIV/AIDS in the Era of Antiretroviral Therapy. AIDS Res Treat 2012 : 262471.
7. KinrossJM, DarziAW, NicholsonJK (2011) Gut microbiome-host interactions in health and disease. Genome Med 3 : 14.
8. NelsonKE, WeinstockGM, HighlanderSK, WorleyKC, CreasyHH, et al. (2010) A catalog of reference genomes from the human microbiome. Science 328 : 994–999.
9. GriceEA, KongHH, ConlanS, DemingCB, DavisJ, et al. (2009) Topographical and temporal diversity of the human skin microbiome. Science 324 : 1190–1192.
10. HuseSM, YeY, ZhouY, FodorAA (2012) A Core Human Microbiome as Viewed through 16S rRNA Sequence Clusters. PLoS ONE 7: e34242.
11. ClementeJC, UrsellLK, ParfreyLW, KnightR (2012) The Impact of the Gut Microbiota on Human Health: An Integrative View. Cell 148 : 1258–1270.
12. AasJA, BarbutoSM, AlpagotT, OlsenI, DewhirstFE, et al. (2007) Subgingival plaque microbiota in HIV positive patients. Journal of Clinical Periodontology 34 : 189–195.
13. IlievID, FunariVA, TaylorKD, NguyenQ, ReyesCN, et al. (2012) Interactions Between Commensal Fungi and the C-Type Lectin Receptor Dectin-1 Influence Colitis. Science 336 : 1314–1317.
14. GhannoumMA, JurevicRJ, MukherjeePK, CuiF, SikaroodiM, et al. (2010) Characterization of the Oral Fungal Microbiome (Mycobiome) in Healthy Individuals. PLoS Pathogens 6: e1000713.
15. PelegAY, HoganDA, MylonakisE (2010) Medically important bacterial-fungal interactions. Nat Rev Microbiol 8 : 340–349.
16. LandlingerC, BaskovaL, PreunerS, WillingerB, BuchtaV, et al. (2008) Identification of fungal species by fragment length analysis of the internally transcribed spacer 2 region. Eur J Clin Microbiol Infect Dis 28 (6) 613–22.
17. BormanAM, LintonCJ, MilesSJ, JohnsonEM (2008) Molecular identification of pathogenic fungi. Journal of Antimicrobial Chemotherapy 61: i7–12.
18. GillevetP, SikaroodiM, KeshavarzianA, MutluEA (2010) Quantitative assessment of the human gut microbiome using multitag pyrosequencing. Chem Biodivers 7 : 1065–1075.
19. ChakravortyS, HelbD, BurdayM, ConnellN, AllandD (2007) A detailed analysis of 16S ribosomal RNA gene segments for the diagnosis of pathogenic bacteria. Journal of Microbiological Methods 69 : 330–339.
20. SpearGT, SikaroodiM, ZariffardMR, LandayAL, FrenchAL, et al. (2008) Comparison of the diversity of the vaginal microbiota in HIV-infected and HIV-uninfected women with or without bacterial vaginosis. J Infect Dis 198 : 1131–1140.
21. Gillevet PM, inventor; BioSpherex LLC, assignee (2013 December 10) Multitag Sequencing and Ecogenomic Analysis. United States Patent 8,603,749.
22. RomanelliAM, SuttonDA, ThompsonEH, RinaldiMG, WickesBL (2010) Sequence-based identification of filamentous basidiomycetous fungi from clinical specimens: a cautionary note. J Clin Microbiol 48 : 741–752.
23. CormackBP, BertramG, EgertonM, GowNA, FalkowS, et al. (1997) Yeast-enhanced green fluorescent protein (yEGFP) a reporter of gene expression in Candida albicans. Microbiology 143 : 303–311.
24. SwindellK, LattifAA, ChandraJ, MukherjeePK, GhannoumMA (2009) Parenteral Lipid Emulsion Induces Germination of Candida albicans and Increases Biofilm Formation on Medical Catheter Surfaces. J Infect Dis 200 : 473–480.
25. KuhnDM, MukherjeePK, ClarkeTA, PujolC, ChandraJ, et al. (2004) Candida parapsilosis characterization in an outbreak setting. Emerging Infectious Diseases 10 : 1074–1081.
26. ChandraJ, MukherjeePK, GhannoumMA (2008) In vitro growth and analysis of Candida biofilms. Nature Protocols 3 : 1909–1924.
27. ChandraJ, KuhnDM, MukherjeePK, HoyerLL, McCormickT, et al. (2001) Biofilm formation by the fungal pathogen Candida albicans - development, architecture and drug resistance. Journal of Bacteriology 183 : 5385–5394.
28. ChandraJ, MukherjeePK, LeidichSD, FaddoulFF, HoyerLL, et al. (2001) Antifungal resistance of candidal biofilms formed on denture acrylic in vitro. Journal of Dental Research 80 : 903–908.
29. PienaarED, YoungT, HolmesH (2010) Interventions for the prevention and management of oropharyngeal candidiasis associated with HIV infection in adults and children. Cochrane Database of Systematic Reviews 11: CD003940.
30. HiseAG, TomalkaJ, GanesanS, PatelK, HallBA, et al. (2009) An essential role for the NLRP3 inflammasome in host defense against the human fungal pathogen Candida albicans. Cell Host Microbe 5 : 487–497.
31. TangN, LiuL, KangK, MukherjeePK, TakaharaM, et al. (2004) Inhibition of monocytic interleukin-12 production by Candida albicans via selective activation of ERK mitogen-activated protein kinase. Infection and Immunity 72 : 2513–2520.
32. GhannoumMA, MukherjeePK, JurevicRJ, RetuertoM, BrownRE, et al. (2011) Metabolomics Reveals Differential Levels of Oral Metabolites in HIV-Infected Patients: Toward Novel Diagnostic Targets. Omics 17 : 5–15.
33. CaporasoJG, KuczynskiJ, StombaughJ, BittingerK, BushmanFD, et al. (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7 : 335–336.
34. R Core Team (2013) R: A language and environment for statistical computing. Available http://www.R-project.org. Accessed 2 February 2013.
35. Revelle W (2013) psych: procedures for personality and psychological research. Available http://CRAN.R-project.org/package=psych Version = 1.3.10. Accessed 20 December 2013.
36. NoktaM (2008) Oral manifestations associated with HIV infection. Curr HIV/AIDS Rep 5 : 5–12.
37. SchislerD, KurtzmanC, BothastR, SliningerP (1995) Evaluation of yeasts for biological control of Fusarium dry rot of potatoes. American Potato Journal 72 : 339–353.
38. StarmerWT, GanterPF, PhaffHJ (1986) Quantum and continuous evolution of DNA base composition in the yeast genus Pichia. Evolution 40 : 1263–1274.
39. Golubev W (2006) Antagonistic interactions among yeasts. Biodiversity and Ecophysiology of Yeasts: Springer. pp. 197–219.
40. AntunesJ, AguiarC (2012) Search for killer phenotypes with potential for biological control. Annals of microbiology 62 : 427–433.
41. DruveforsUA, SchnurerJ (2005) Mold-inhibitory activity of different yeast species during airtight storage of wheat grain. FEMS Yeast Res 5 : 373–378.
42. JungPP, FriedrichA, SoucietJL, LouisV, PotierS, et al. (2010) Complete mitochondrial genome sequence of the yeast Pichia farinosa and comparative analysis of closely related species. Curr Genet 56 : 507–515.
43. WalkerGM (2010) Pichia anomala: cell physiology and biotechnology relative to other yeasts. Antonie Van Leeuwenhoek 99 (1) 25–34.
44. EladY (2003) Biocontrol of foliar pathogens: mechanisms and application. Commun Agric Appl Biol Sci 68 : 17–24.
45. CaoS, YuanY, HuZ, ZhengY (2010) Combination of Pichia membranifaciens and ammonium molybdate for controlling blue mould caused by Penicillium expansum in peach fruit. Int J Food Microbiol 141 : 173–176.
46. SantosA, San MauroM, BravoE, MarquinaD (2009) PMKT2, a new killer toxin from Pichia membranifaciens, and its promising biotechnological properties for control of the spoilage yeast Brettanomyces bruxellensis. Microbiology 155 : 624–634.
47. OlstorpeM, PassothV (2010) Pichia anomala in grain biopreservation. Antonie Van Leeuwenhoek 99 (1) 57–62.
48. KaganBL (1983) Mode of action of yeast killer toxins: channel formation in lipid bilayer membranes. Nature 302 : 709–711.
49. İzgüF, AltınbayD, AcunT (2006) Killer toxin of Pichia anomala NCYC 432; purification, characterization and its exo-β-1,3-glucanase activity. Enzyme and Microbial Technology 39 : 669–676.
50. CaoS, ZhengY, TangS, WangK (2008) Improved control of anthracnose rot in loquat fruit by a combination treatment of Pichia membranifaciens with CaCl(2). Int J Food Microbiol 126 : 216–220.
51. DruveforsUA, PassothV, SchnurerJ (2005) Nutrient effects on biocontrol of Penicillium roqueforti by Pichia anomala J121 during airtight storage of wheat. Appl Environ Microbiol 71 : 1865–1869.
52. XuXB, TianSP (2008) Reducing oxidative stress in sweet cherry fruit by Pichia membranaefaciens: a possible mode of action against Penicillium expansum. J Appl Microbiol 105 : 1170–1177.
53. ZhaoJ, MouY, ShanT, LiY, ZhouL, et al. (2010) Antimicrobial metabolites from the endophytic fungus Pichia guilliermondii isolated from Paris polyphylla var. yunnanensis. Molecules 15 : 7961–7970.
54. JijakliMH, LepoivreP (1998) Characterization of an Exo-beta-1,3-Glucanase Produced by Pichia anomala Strain K, Antagonist of Botrytis cinerea on Apples. Phytopathology 88 : 335–343.
55. DrobyS, HofsteinR, WilsonCL, WisniewskiM, FridlenderB, et al. (1993) Pilot Testing of Pichia guilliermondii: A Biocontrol Agent of Postharvest Diseases of Citrus Fruit. Biological Control 3 : 47–52.
56. DrobyS, WisniewskiME, CohenL, WeissB, TouitouD, et al. (1997) Influence of CaCl(2) on Penicillium digitatum, Grapefruit Peel Tissue, and Biocontrol Activity of Pichia guilliermondii. Phytopathology 87 : 310–315.
57. GiobbeS, MarcedduS, SchermB, ZaraG, MazzarelloVL, et al. (2007) The strange case of a biofilm-forming strain of Pichia fermentans, which controls Monilinia brown rot on apple but is pathogenic on peach fruit. FEMS Yeast Res 7 : 1389–1398.
58. KumamotoCA, VincesMD (2005) Alternative Candida albicans lifestyles: Growth on Surfaces. Annual Review of Microbiology 59 : 113–133.
59. ChandraJ, MukherjeePK, GhannoumMA (2011) Candida Biofilms Associated with CVC and Medical Devices. Mycoses 55 : 46–57.
60. ChandraJ, MukherjeePK, GhannoumMA (2010) Fungal Biofilms in the Clinical Lab Setting. Current Reports in Fungal Infection 4 : 137–144.
61. ConlanS, KongHH, SegreJA (2012) Species-level analysis of DNA sequence data from the NIH Human Microbiome Project. PLoS One 7: e47075.
62. LiK, BihanM, YoosephS, MetheBA (2012) Analyses of the microbial diversity across the human microbiome. PLoS One 7: e32118.
63. ZarcoMF, VessTJ, GinsburgGS (2012) The oral microbiome in health and disease and the potential impact on personalized dental medicine. Oral Dis 18 : 109–120.
64. ZauraE, KeijserBJ, HuseSM, CrielaardW (2009) Defining the healthy “core microbiome” of oral microbial communities. BMC Microbiol 9 : 259.
65. HoganDA, KolterR (2002) Pseudomonas - Candida interactions: an ecological role for virulence factors. Science 296 : 2229–2232.
66. AzoulayE, TimsitJ-F, TaffletM, de LassenceA, DarmonM, et al. (2006) Candida Colonization of the Respiratory Tract and Subsequent Pseudomonas Ventilator-Associated Pneumonia. Chest 129 : 110–117.
67. NseirS, JozefowiczE, CavestriB, SendidB, Di PompeoC, et al. (2007) Impact of antifungal treatment on Candida-Pseudomonas interaction: a preliminary retrospective case-control study. Intensive Care Med 33 : 137–142.
68. DyessDL, GarrisonRN, FryDE (1985) Candida sepsis. Implications of polymicrobial blood-borne infection. ArchSurg 120 : 345–348.
69. NavazeshM, MulliganR, PogodaJ, GreenspanD, AlvesM, et al. (2005) The effect of HAART on salivary microbiota in the Women's Interagency HIV Study (WIHS). Oral Surg Oral Med Oral Pathol Oral Radiol Endod 100 : 701–708.
70. WorkmanSN, BeenFE, CrawfordSR, LavoieMC (2008) Bacteriocin-like inhibitory substances from Campylobacter spp. Antonie Van Leeuwenhoek 93 : 435–436.
71. DupontPF (1995) Candida albicans, the opportunist. A cellular and molecular perspective. JAmPodiatrMedAssoc 85 : 104–115.
72. MartinMV, CraigGT, LambDJ (1984) An investigation of the role of true hypha production in the pathogenesis of experimental oral candidosis. Sabouraudia 22 : 471–476.
73. CannonRD, HolmesAR, MasonAB, MonkBC (1995) Oral Candida: clearance, colonization, or candidiasis? JDentRes 74 : 1152–1161.
74. SamaranayakeYH, WuPC, SamaranayakeLP, SoM, YuenKY (1994) Adhesion and colonisation of Candida krusei on host surfaces. JMedMicrobiol 41 : 250–258.
75. De BernardisF, BoccaneraM, RainaldiL, GuerraCE, QuintiI, et al. (1992) The secretion of aspartyl proteinase, a virulence enzyme, by isolates of Candida albicans from the oral cavity of HIV-infected subjects. EurJEpidemiol 8 : 362–367.
76. SardiJC, DuqueC, HoflingJF, GoncalvesRB (2012) Genetic and phenotypic evaluation of Candida albicans strains isolated from subgingival biofilm of diabetic patients with chronic periodontitis. Med Mycol 50 : 467–475.
77. GhannoumMA, Abu-ElteenKH (1990) Pathogenicity determinants of Candida. Mycoses 33 : 265–282.
78. GhannoumMA (2000) Potential role of phospholipases in virulence and fungal pathogenesis. Clinical Microbiology Reviews 13 : 122–143.
79. RichardsonMD, SmithH (1981) Production of germ tubes by virulent and attenuated strains of Candida albicans. J Infect Dis 144 : 565–569.
80. RestucciaC, GiusinoF, LicciardelloF, RandazzoC, CaggiaC, et al. (2006) Biological control of peach fungal pathogens by commercial products and indigenous yeasts. J Food Prot 69 : 2465–2470.
81. SantosA, MarquinaD (2004) Ion channel activity by Pichia membranifaciens killer toxin. Yeast 21 : 151–162.
82. FredlundE, BrobergA, BoysenME, KenneL, SchnurerJ (2004) Metabolite profiles of the biocontrol yeast Pichia anomala J121 grown under oxygen limitation. Appl Microbiol Biotechnol 64 : 403–409.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek DHX36 Enhances RIG-I Signaling by Facilitating PKR-Mediated Antiviral Stress Granule FormationČlánek Oral Bacteria and CancerČlánek A Non-Coding RNA Promotes Bacterial Persistence and Decreases Virulence by Regulating a Regulator in
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2014 Číslo 3- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Conflicting Interests in the Pathogen–Host Tug of War: Fungal Micronutrient Scavenging Versus Mammalian Nutritional Immunity
- Putting Fungi to Work: Harvesting a Cornucopia of Drugs, Toxins, and Antibiotics
- Mycobacteriophages: Windows into Tuberculosis
- Human African Trypanosomiasis and Immunological Memory: Effect on Phenotypic Lymphocyte Profiles and Humoral Immunity
- Five Things to Know about Genetically Modified (GM) Insects for Vector Control
- A Missing Dimension in Measures of Vaccination Impacts
- Eosinophils Are Important for Protection, Immunoregulation and Pathology during Infection with Nematode Microfilariae
- Clonality of HTLV-2 in Natural Infection
- Production, Fate and Pathogenicity of Plasma Microparticles in Murine Cerebral Malaria
- Group B Streptococcal Infection of the Choriodecidua Induces Dysfunction of the Cytokeratin Network in Amniotic Epithelium: A Pathway to Membrane Weakening
- New Insights into How Adapts to Its Mammalian Host during Bubonic Plague
- Foodborne Transmission of Nipah Virus in Syrian Hamsters
- A Polysaccharide Virulence Factor from Elicits Anti-inflammatory Effects through Induction of Interleukin-1 Receptor Antagonist
- Structural and Functional Characterization of a Complex between the Acidic Transactivation Domain of EBNA2 and the Tfb1/p62 Subunit of TFIIH
- Adaptive Gene Amplification As an Intermediate Step in the Expansion of Virus Host Range
- DHX36 Enhances RIG-I Signaling by Facilitating PKR-Mediated Antiviral Stress Granule Formation
- Hepatitis B Virus Infection and Immunopathogenesis in a Humanized Mouse Model: Induction of Human-Specific Liver Fibrosis and M2-Like Macrophages
- Crk Adaptors Negatively Regulate Actin Polymerization in Pedestals Formed by Enteropathogenic (EPEC) by Binding to Tir Effector
- Fatty Acid Biosynthesis Contributes Significantly to Establishment of a Bioenergetically Favorable Environment for Vaccinia Virus Infection
- A Cytosolic Chaperone Complexes with Dynamic Membrane J-Proteins and Mobilizes a Nonenveloped Virus out of the Endoplasmic Reticulum
- Intracellular Promote Invasive Cell Motility through Kinase Regulation of the Host Actin Cytoskeleton
- MAVS-MKK7-JNK2 Defines a Novel Apoptotic Signaling Pathway during Viral Infection
- RON5 Is Critical for Organization and Function of the Moving Junction Complex
- Immune Suppression by Neutrophils in HIV-1 Infection: Role of PD-L1/PD-1 Pathway
- and Exhibit Metabolic Symbioses
- The Herpes Virus Fc Receptor gE-gI Mediates Antibody Bipolar Bridging to Clear Viral Antigens from the Cell Surface
- Target Cell Availability, Rather than Breast Milk Factors, Dictates Mother-to-Infant Transmission of SIV in Sooty Mangabeys and Rhesus Macaques
- Evolution of the Retroviral Restriction Gene : Inhibition of Non-MLV Retroviruses
- Infection of Adult Thymus with Murine Retrovirus Induces Virus-Specific Central Tolerance That Prevents Functional Memory CD8 T Cell Differentiation
- Fha Interaction with Phosphothreonine of TssL Activates Type VI Secretion in
- In Vivo Administration of a JAK3 Inhibitor during Acute SIV Infection Leads to Significant Increases in Viral Load during Chronic Infection
- Lack of Detectable HIV-1 Molecular Evolution during Suppressive Antiretroviral Therapy
- Activation of HIV-1 from Latent Infection via Synergy of RUNX1 Inhibitor Ro5-3335 and SAHA
- A Compact, Multifunctional Fusion Module Directs Cholesterol-Dependent Homomultimerization and Syncytiogenic Efficiency of Reovirus p10 FAST Proteins
- The Role of Host and Microbial Factors in the Pathogenesis of Pneumococcal Bacteraemia Arising from a Single Bacterial Cell Bottleneck
- Genetic Dissection of Gut Epithelial Responses to
- Two-Component System Cross-Regulation Integrates Response to Heme and Cell Envelope Stress
- Oral Mycobiome Analysis of HIV-Infected Patients: Identification of as an Antagonist of Opportunistic Fungi
- A Model System for Studying the Transcriptomic and Physiological Changes Associated with Mammalian Host-Adaptation by Serovar Copenhageni
- Inflammasome Sensor NLRP1 Controls Rat Macrophage Susceptibility to
- ChIP-Seq and RNA-Seq Reveal an AmrZ-Mediated Mechanism for Cyclic di-GMP Synthesis and Biofilm Development by
- The Hypervariable Amino-Terminus of P1 Protease Modulates Potyviral Replication and Host Defense Responses
- Caspase-1-Dependent and -Independent Cell Death Pathways in Infection of Macrophages
- The Effect of Cell Growth Phase on the Regulatory Cross-Talk between Flagellar and Spi1 Virulence Gene Expression
- Different Mutagenic Potential of HIV-1 Restriction Factors APOBEC3G and APOBEC3F Is Determined by Distinct Single-Stranded DNA Scanning Mechanisms
- Oral Bacteria and Cancer
- Identification of OmpA, a Protein Involved in Host Cell Invasion, by Multi-Phenotypic High-Content Screening
- Transovarial Transmission of a Plant Virus Is Mediated by Vitellogenin of Its Insect Vector
- VE-Cadherin Cleavage by LasB Protease from Facilitates Type III Secretion System Toxicity in Endothelial Cells
- Dimerization of VirD2 Binding Protein Is Essential for Induced Tumor Formation in Plants
- Crystal Structure of the Vaccinia Virus DNA Polymerase Holoenzyme Subunit D4 in Complex with the A20 N-Terminal Domain
- Post-Translational Regulation via Clp Protease Is Critical for Survival of
- Modulation of Phagosomal pH by Promotes Hyphal Morphogenesis and Requires Stp2p, a Regulator of Amino Acid Transport
- Rotavirus Activates Lymphocytes from Non-Obese Diabetic Mice by Triggering Toll-Like Receptor 7 Signaling and Interferon Production in Plasmacytoid Dendritic Cells
- Cytomegalovirus m154 Hinders CD48 Cell-Surface Expression and Promotes Viral Escape from Host Natural Killer Cell Control
- Interferon Regulatory Factor-1 Protects from Fatal Neurotropic Infection with Vesicular Stomatitis Virus by Specific Inhibition of Viral Replication in Neurons
- HMGB1-Promoted and TLR2/4-Dependent NK Cell Maturation and Activation Take Part in Rotavirus-Induced Murine Biliary Atresia
- An Immunomics Approach to Schistosome Antigen Discovery: Antibody Signatures of Naturally Resistant and Chronically Infected Individuals from Endemic Areas
- PPARγ Agonists Improve Survival and Neurocognitive Outcomes in Experimental Cerebral Malaria and Induce Neuroprotective Pathways in Human Malaria
- A Non-Coding RNA Promotes Bacterial Persistence and Decreases Virulence by Regulating a Regulator in
- Viral OTU Deubiquitinases: A Structural and Functional Comparison
- Heterogeneity and Breadth of Host Antibody Response to KSHV Infection Demonstrated by Systematic Analysis of the KSHV Proteome
- Influenza A Virus Assembly Intermediates Fuse in the Cytoplasm
- Broadly Reactive Human CD8 T Cells that Recognize an Epitope Conserved between VZV, HSV and EBV
- Oncogenic Human Papillomaviruses Activate the Tumor-Associated Lens Epithelial-Derived Growth Factor (LEDGF) Gene
- Erythrocyte Invasion: Combining Function with Immune Evasion
- IL-1α and Complement Cooperate in Triggering Local Neutrophilic Inflammation in Response to Adenovirus and Eliminating Virus-Containing Cells
- Chronic Exposure to Type-I IFN under Lymphopenic Conditions Alters CD4 T Cell Homeostasis
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Cytomegalovirus m154 Hinders CD48 Cell-Surface Expression and Promotes Viral Escape from Host Natural Killer Cell Control
- Human African Trypanosomiasis and Immunological Memory: Effect on Phenotypic Lymphocyte Profiles and Humoral Immunity
- DHX36 Enhances RIG-I Signaling by Facilitating PKR-Mediated Antiviral Stress Granule Formation
- Conflicting Interests in the Pathogen–Host Tug of War: Fungal Micronutrient Scavenging Versus Mammalian Nutritional Immunity
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



