-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaInfluenza A Virus Assembly Intermediates Fuse in the Cytoplasm
Influenza A viruses, containing eight single stranded RNA segments, cause seasonal epidemics and occasional pandemics. Reassortment of the influenza viral genome in co-infected cells confers an evolutionary advantage for the virus, and can result in viruses with pandemic potential like the 2009 pandemic H1N1 and 2013 H7N9 virus. Replication of the viral genome occurs in the nucleus of the host cell and the progeny viral RNA (vRNA) segments must be transported to the plasma membrane for budding. The dynamics of vRNA assembly into progeny virions remains unknown. We used novel techniques to visualize the 3D-localization of four distinct vRNA segments in an infected cell and a fluorescent virus to visualize vRNA transport during a productive infection to determine where, when and how assembly occurs. Our data suggest that vRNA segments are exported from the nucleus as subcomplexes that undergo additional assembly en route to the plasma membrane through dynamic fusion events of vRNA-containing cytoplasmic foci. These observations have broad implications for understanding the intracellular requirements behind reassortment of influenza viruses and may lead to the development of new antiviral targets.
Published in the journal: . PLoS Pathog 10(3): e32767. doi:10.1371/journal.ppat.1003971
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1003971Summary
Influenza A viruses, containing eight single stranded RNA segments, cause seasonal epidemics and occasional pandemics. Reassortment of the influenza viral genome in co-infected cells confers an evolutionary advantage for the virus, and can result in viruses with pandemic potential like the 2009 pandemic H1N1 and 2013 H7N9 virus. Replication of the viral genome occurs in the nucleus of the host cell and the progeny viral RNA (vRNA) segments must be transported to the plasma membrane for budding. The dynamics of vRNA assembly into progeny virions remains unknown. We used novel techniques to visualize the 3D-localization of four distinct vRNA segments in an infected cell and a fluorescent virus to visualize vRNA transport during a productive infection to determine where, when and how assembly occurs. Our data suggest that vRNA segments are exported from the nucleus as subcomplexes that undergo additional assembly en route to the plasma membrane through dynamic fusion events of vRNA-containing cytoplasmic foci. These observations have broad implications for understanding the intracellular requirements behind reassortment of influenza viruses and may lead to the development of new antiviral targets.
Introduction
Influenza viruses cause severe annual morbidity and mortality [1]. The genome of influenza A viruses is composed of 8 negative-sense single stranded RNA gene segments (PB2, PB1, PA, HA, NP, NA, M and NS) that encode 10 major proteins and several auxiliary peptides. Production of infectious progeny virions requires incorporation of all 8 vRNA segments and occurs at the apical membrane of infected cells [2]. The segmented nature of the viral genome allows for the generation of novel reassortant viruses containing genes from distinct parental viruses in co-infected cells. The 2009 pandemic H1N1 virus arose from a reassortment event between two swine origin viruses [3]. Additionally, the 2013 H7N9 virus responsible for the outbreak in Mainland China and Taiwan is a reassortant virus deriving gene segments from avian H9N2 and H7N9 viruses [4]. Reassortment provides an evolutionary advantage for influenza viruses and thus reassortant viruses pose a major public health risk. Understanding how vRNA segments are assembled and packaged into progeny virions is key for unlocking how reassortant viruses are generated, yet little is known about this process.
Replication of the influenza virus genome occurs in the nucleus by a virally encoded heterotrimeric RNA-dependent RNA polymerase composed of PB1, PB2, and PA proteins [2]. Each newly synthesized vRNA segment is coated with the viral nucleoprotein (NP) and the heterotrimeric polymerase complex associates with each vRNA segment via the 5′ and 3′ ends. This resultant vRNP complex is exported from the nucleus and travels to the plasma membrane for packaging into a budding virion [2], [5]. Recent studies have suggested a role for Rab11a-containing recycling endosomes in the transport of vRNA segments to the plasma membrane for packaging [6]–[9]. However, the dynamics of vRNA transport, including whether vRNA segments are transported individually or as a complex, remain largely unclear.
Studies using electron tomography or fluorescent in situ hybridization (FISH) on purified influenza viral particles suggest that the majority of progeny virions incorporate a single copy of all 8 vRNA segments [10]–[13]. Selective packaging of all 8 segments into progeny virions is thought to occur via RNA-RNA interactions between the vRNA segments identified through in vitro studies, which suggests the formation of an interaction network between vRNA segments [5], [11], [13]–[16]. However, the organization and mechanics of vRNA assembly are poorly understood, due mostly to the lack of adequate tools for observing the transport and spatial arrangement of vRNA segments in situ.
Visualization of intracellular vRNA segments has been limited to two-color FISH [12], [17], hindering assessment of the localization and interactions between more than two vRNA segments within a single cell. Additionally, tracking vRNA movement during a productive infection requires a virus that expresses a fluorescent tag fused to a viral protein involved in vRNA trafficking, which is packaged into progeny virions and is maintained in subsequent generations. Although some fluorescent influenza viruses have been generated, they either contain GFP fused with a viral protein not involved in vRNA transport or as a separate polypeptide used as a marker for infection that fails to highlight any specific process of the viral lifecycle [18]–[20]. An influenza virus with PB2 tagged with split-GFP was recently used to assess the movement of vRNA [21]. However, while split-GFP is useful in studying protein-protein interactions, it is a poor choice for visualizing protein dynamics because 1) visualization is limited to cells expressing the remainder of the GFP molecule; 2) visualization is not possible prior to formation of the complete GFP; 3) split-GFP fragments are unstable compared to intact GFP; and 4) split-GFP is prone to aggregation [22]–[24].
To explore the mechanics of vRNA assembly we developed two novel imaging tools: a system to visualize four different vRNA segments within an infected cell and a fluorescent influenza virus to track vRNA movement in live cells during a productive infection. Using the four-color FISH assay we were able to visualize all 8 vRNA segments and quantify the number of cytoplasmic foci and found that the majority of the foci at the external nuclear periphery contain more than one vRNA segment. To assess the dynamics of vRNA assembly in the cytoplasm we generated an influenza virus, A/WSN/1933 (H1N1) (WSN) expressing full-length GFP fused to the viral polymerase PA protein (WSN PA-GFP) and observed that PA-GFP foci colocalize and travel together in the cytoplasm by an inverted selective-plane illumination microscope. Our data provide evidence that vRNA segments form subcomplexes throughout the cytoplasm during infection, that these subcomplexes contain multiple distinct vRNA segments, and that vRNA-containing cytoplasmic foci can fuse together during transport.
Results
Visualization of multiple vRNA segments within a single infected cell
In order to explore where vRNA assembly occurs during a viral infection we established a system to simultaneously visualize four different vRNA segments within an infected cell (Figure 1). We visualized four distinct fluorophores conjugated to vRNA probes with a tunable white light laser confocal microscope, using a sequential scanning program to ensure minimal cross talk between channels (Figure S1). We designed probes against all 8 vRNA segments of A/WSN/1933 (H1N1) virus to enable visualization of all 8 segments in relation to each other in multiple four-color FISH experiments. Figure 1 depicts two representative cells displaying all 8 vRNA segments; one cell probed for vRNA segments PB2, PB1, PA, and NP and the other probed for HA, NA, M and NS vRNA segments. We confirmed the specificity of each FISH probe set by testing each of the probes on cells expressing a single vRNA transcript from a PolI expression plasmid (Figure S2). To our knowledge, our study is the first to visualize four distinct vRNA species within a single cell.
Fig. 1. Visualization of four distinct influenza viral gene segments within a single cell. 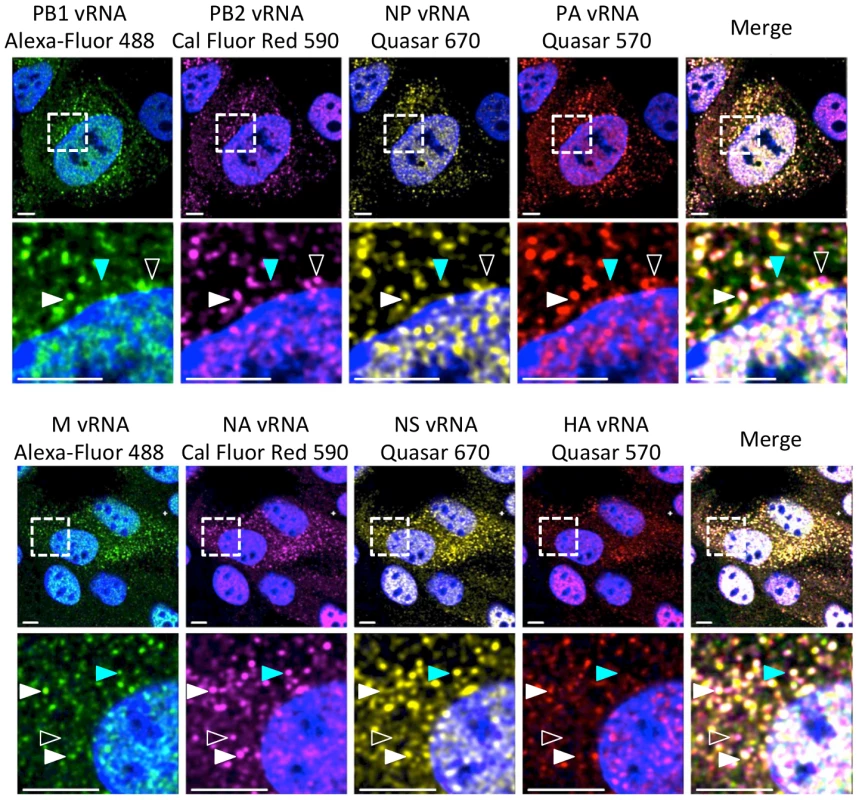
MDCK cells were infected with wild type (WT) WSN at a multiplicity of infection (MOI) of 3 for 8 hours post infection (hpi) and then imaged with FISH probes directed against four distinct vRNA species. The first row displays a cell probed for influenza vRNA segments PB2, PB1, PA, and NP. Row 3 contains images of a cell probed for the other 4 influenza vRNA segments: HA, NA, M and NS. The second and fourth row of images show an enlargement of the area defined by the dashed boxes in the first and third rows. Solid white arrowheads identify a cytoplasmic focus with all four distinct vRNA segments, turquoise arrowheads indicate a focus with only three vRNA species, and open arrowheads show a focus with only two vRNA species. All scale bars are 10 µm. DAPI marks the cellular nucleus. Staining of vRNA in WT WSN infected Madin-Darby canine kidney (MDCK) cells using four-color FISH displayed diffuse nuclear staining and distinct cytoplasmic foci. Closer inspection of the cytoplasmic foci in the enlarged areas of Figure 1 revealed foci containing combinations of 4, 3, 2, or a single vRNA segment (various arrowheads). Staining of vRNA segments in the nucleus indicated a high degree of colocalization between the vRNA foci (white pixels in the merged nucleus image); however, the high density of the vRNA segments and their diffuse appearance prevented accurate assessment of potential colocalization between vRNA segments in the nucleus. Therefore, we focused on the fate of vRNA segments after nuclear export. To explore the composition of each cytoplasmic focus we used the imaging software Imaris to mask the nucleus, thus excluding the vRNA nuclear staining from our analysis. Figure 2A shows a 3D rendering of all the cytoplasmic foci in a MDCK cell infected with WT WSN and probed for PB2 (red), PB1 (green), PA (orange), and NP (yellow), which encode proteins that form the viral RNP complex. This image was rotated to provide an axial view of the cell and to highlight the cytoplasmic foci in relationship with the nucleus. To quantify the total number of foci containing a single labeled vRNA segment, 2 distinct vRNA segments, 3 different segments, or all 4 vRNA segments, we determined the signal intensity for each fluorophore in each cytoplasmic focus from three independent cells. We found that a significantly larger number of cytoplasmic foci contained all 4 labeled vRNA segments (Figure 2B). This observation was consistent for each set of four vRNA segments that we tested (Figure S3).
Fig. 2. Viral RNA composition and spatial location of cytoplasmic foci. 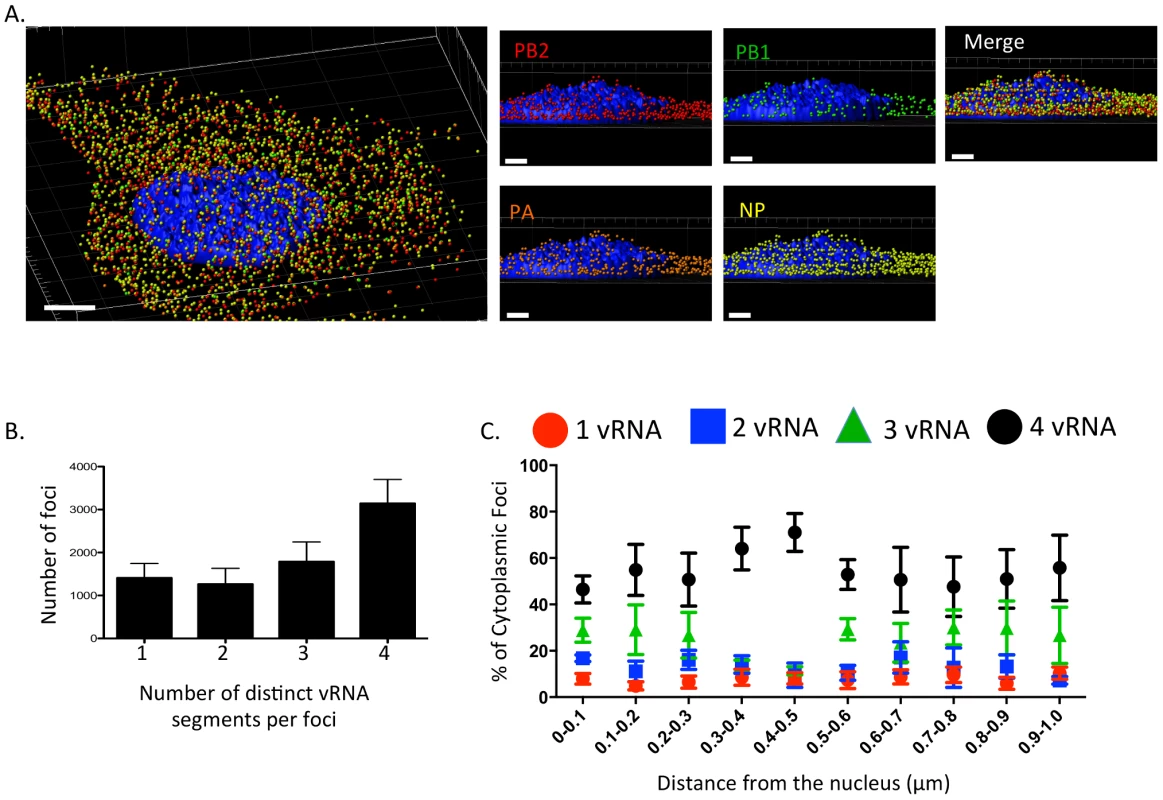
3D rendering of an MDCK cell infected with WT WSN (MOI = 3) for 8 hpi were stained with probe B from Table S1: WSN PB2 Quasar 570 (red spots), PB1 Alexa-Fluor 488 (green spots), PA Cal Fluor Red 590 (orange spots), and NP Quasar 670 (yellow spots) (A). The DAPI-stained nucleus is labeled in blue. An enlarged region of the 3D image was tilted 90° in the z-direction to provide an axial-view of the cytoplasmic foci and nucleus. Scale bars are 4 µm in the whole cell image and 2 µm in the rotated images. The total number of foci containing 1, 2, 3, or 4 vRNA segments was quantified within a given cell (B). Each bar represents the average from 3 independently analyzed cells with standard error indicated. The distance from the nucleus for each focus from four independent cells was calculated (C). The proportion of foci containing 4,3,2,or 1 distinct vRNA segment with a given range from the nucleus is represented graphically as a scatter plot. Each spot is an average from 4 independent cells and the standard error is indicated. To determine whether the proportion of cytoplasmic foci containing 4, 3, 2, or 1 vRNA segments changed spatially in relationship to the nucleus, we determined the 3D distance from the nucleus for each focus using the DAPI signal to define the nuclear boundary. We found that the majority of the foci near or at the nucleus (within 0.1 µm) contained more than one vRNA segment (Figure 2C), suggesting that vRNA segments are not exported from the nucleus as individual segments. In general, independent of distance from the nucleus the proportion of foci containing 3 or 4 vRNA segments was much higher than those containing 1 or 2 vRNA segments (Figure 2C). Interestingly, we observed an increase in the proportion of foci containing four vRNA as the distance from the nucleus increased to 0.5 µm (Figure 2C). Based on the axial view of the cell in Figure 2A, the top of nucleus is very close to the apical cellular membrane, as defined by FISH staining. There may be a bias at 0.5 µm from the nucleus for foci that are at the plasma membrane being incorporated into progeny virions preparing to bud. The predominance of foci containing all four labeled vRNA segments close to the apical surface of the cell is consistent with the notion that vRNA assembly is selective and that there is a mechanism to ensure packaging of all 8 vRNA segments. However, since the distance from the nucleus to the cellular membrane is not uniform in all directions, we observed many foci located at distances greater than 0.5 µm from the nucleus that did not contain all 4 of the labeled vRNA segments (Figure 2C).
Colocalization of vRNA segments
In vitro studies have shown that vRNA segments associate with each other [13], [15], [16], suggesting that selective packaging of vRNA segments is mediated via vRNA-vRNA interactions. A proposed mechanism for assembly includes the formation of a single linear interaction network between vRNA segments for packaging [5]. We multiplexed FISH probes (Table S1) to compare each vRNA segment to each other in an attempt to identify a linear interaction network between vRNA segments, and calculated the Pearson correlation coefficient between each vRNA segment in the cytoplasm (Figure 3). The Pearson correlation coefficient has been used extensively for estimating the colocalization between two channels [25] as it suggests the degree of colocalization: values of 0.5–1 are considered high, 0.3–0.5 moderate, and 0–0.3 low. As a positive control, we probed the same segment with two FISH probes conjugated to different fluorophores. The Pearson coefficient for the positive controls was in the high range, with the exception of the NS gene segment (Figure 3). In general we observed colocalization coefficient values between vRNA segments in the high to moderate range, making it difficult to ascribe a linear order of assembly between vRNA segments. Interestingly, we observed a low correlation of ∼0.2 between PB2 and HA vRNA segments (Figure 3). This observation is of particular interest since these two gene segments encode the most well characterized virulence motifs of influenza viruses [26]–[29].
Fig. 3. Colocalization coefficients between influenza vRNA segments. 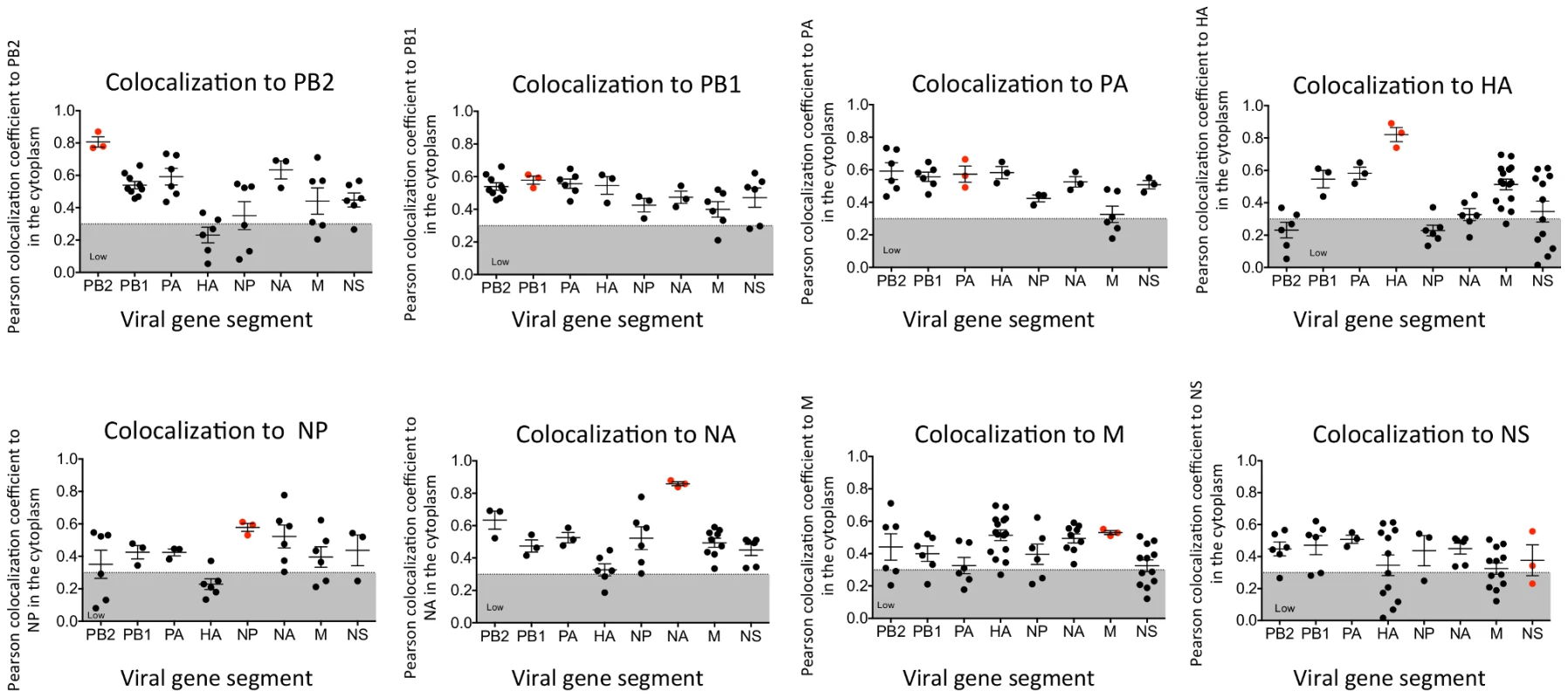
Graphical representation of the Pearson correlation coefficient values between each vRNA segment and all other vRNA segments from exclusively cytoplasmic staining. Dual staining of infected cells with FISH probes against the same target served as a positive control and is indicated in red. Each spot represents a single analyzed infected cell. Gray shading highlights the region of low colocalization based on the Pearson correlation coefficient. Generation of WSN PA-GFP virus
The four-color FISH studies suggest that vRNA segments are exported from the nucleus as subcomplexes that assemble en route to the plasma membrane. In order to investigate this further, we sought to observe cytoplasmic vRNA trafficking in live cells. Since it is technically challenging to label vRNA segments for live cell imaging, we developed a fluorescent virus that could be used to study viral replication during a productive infection. We generated a WSN virus encoding a full length GFP fused to the viral PA protein, a component of the heterotrimeric viral polymerase complex with endonuclease activity that is associated with the 3′ and 5′ ends of vRNA segments [2], [30]–[32]. Creation of the WSN PA-GFP virus built upon two previously published observations. First, the WSN PA protein, fused to GFP at its C-terminus, functions like wild-type (WT) PA protein in in vitro replication experiments [33]. Second, rescue of viruses with a vRNA segment containing a large fragment of foreign RNA requires duplication of approximately 150 base pairs (bp) of the coding region prior to the 3′ non-coding region [34]. Thus, we generated a WSN PA-GFP virus with a portion of the C-terminal coding region duplicated after the GFP stop codon and prior to the non-coding region of the PA gene segment (Figure 4A).
Fig. 4. Generation of WSN PA-GFP influenza virus. 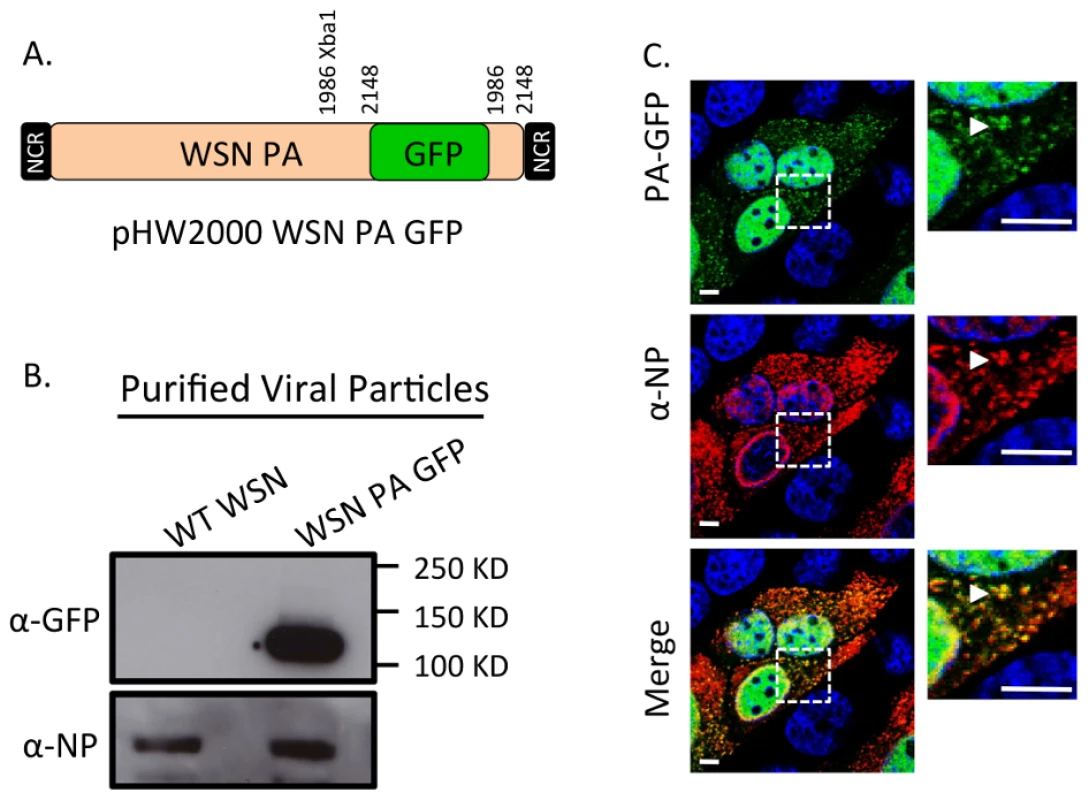
Schematic of the reverse genetics pHW2000 plasmid created by inserting the coding region of full length GFP into the C-terminal domain of the WSN PA open reading frame (A). This insertion included a duplication of 162 nucleotides of WSN PA prior to the 5′ non-coding region (NCR) depicted in the black rectangle. Western blot of purified recombinant WT WSN or WSN PA-GFP virus grown in embryonated eggs (B). The GFP signal corresponds to the appropriate size of a PA-GFP fusion protein. Viral NP protein was used as a control to ensure loading of equivalent amounts of virus. Immunofluorescence of MDCK cells infected with WSN PA-GFP (MOI = 3) for 16 hpi with anti-influenza NP antibody (C). Images to the right are enlarged regions identified by the dashed square. White arrows show areas of co-localization. All scale bars are 5 µm. Western blot analysis of purified WT WSN and WSN PA-GFP virions demonstrate that GFP is present in mature virions and runs slightly above the 100 kDa marker, which is appropriate for a PA-GFP fusion since PA is 82 kDa and GFP is about 35 kDa (Figure 4B). Presence of the viral NP protein was used to ensure equal loading of both viruses. The localization pattern of WSN PA-GFP in MDCK cells 16 hours post-infection was diffusely nuclear with discrete cytoplasmic foci that colocalized with the viral NP protein (Figure 4C). We confirmed that PA-GFP colocalized with vRNA segments by using FISH against vRNA segments PB2 and HA in both MDCK (Figure 5A) and human lung adenocarcinoma epithelial (A549) cells (Figure 5B) 16 hours post-infection. Additionally, treatment of infected cells with leptomycin B (LMB) 4 hours post-infection restricted the localization of PA-GFP, NP, and vRNA segments to the nucleus 24 hours post-infection (Figure S4). Nuclear retention of these vRNP components in the presence of LMB demonstrates that nuclear export depends upon exportin 1 (Crm1), as previously suggested [35]–[37]. Taken together, our data demonstrate that PA-GFP can serve as a surrogate for vRNA gene segments in cells infected with the WSN PA-GFP virus.
Fig. 5. Colocalization of WSN PA-GFP and influenza vRNA segments. 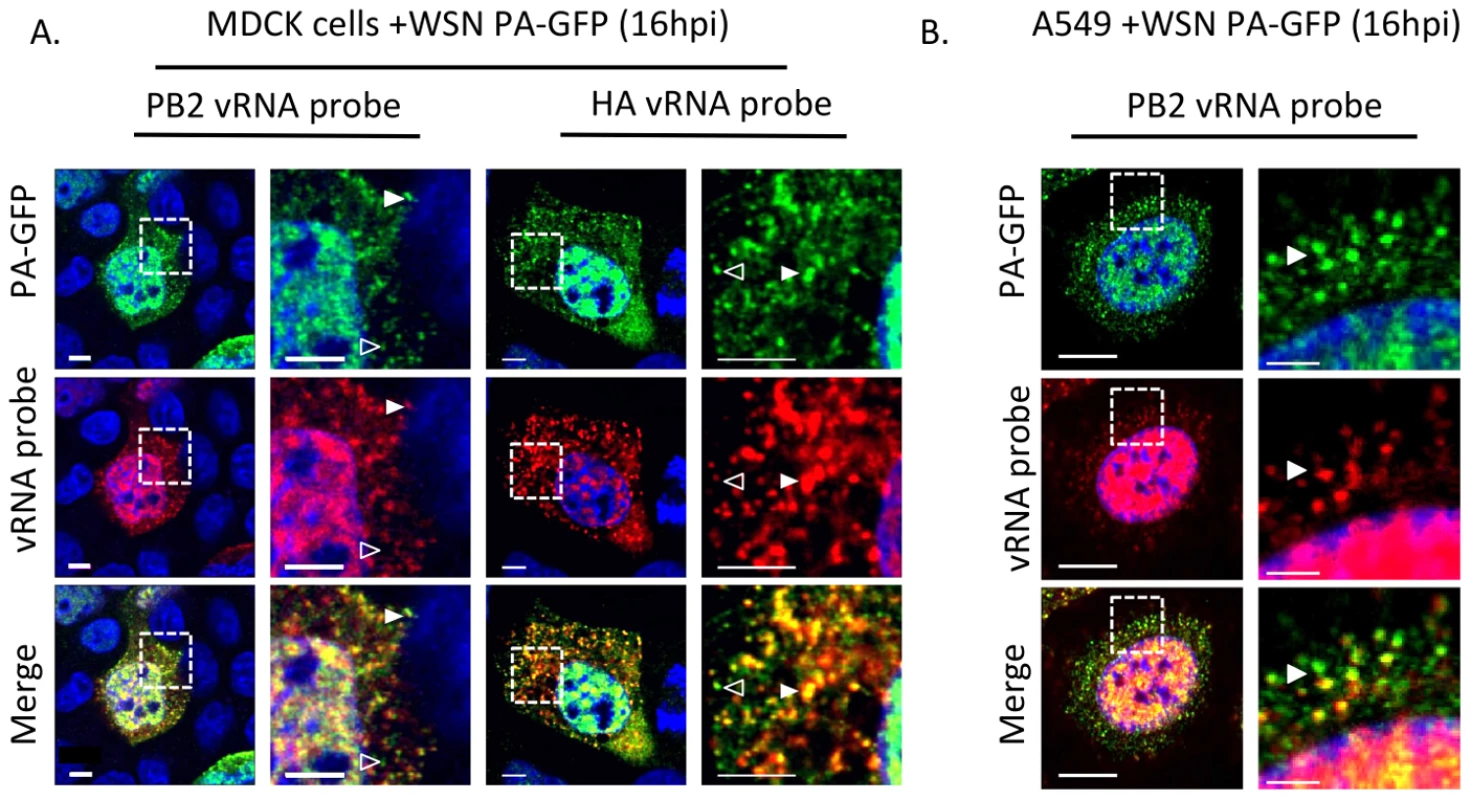
Fluorescent in situ hybridization (FISH) for viral RNA segments 1 (PB2) and 4 (HA) on MDCK (A) or A549 cells (B) cells 16 hpi with WSN PA-GFP (MOI = 0.5). Images to the right are enlarged regions identified by the dashed box. White arrowheads show colocalization of PA-GFP and vRNA, and open arrowheads indicate GFP foci not colocalized with viral RNA signal. DAPI marks the cellular nucleus. Scale bars are 5 µm in all panels of part A. In part B scale bars are 10 µm in the whole cell images and 2 µm in length in the enlarged region. Characterization of PA-GFP movements during productive viral infection
Previous studies exploring the dynamics of vRNA transport using transient transfection systems or the PB2 split-GFP virus have described both fast (∼1 µm/s) and slow (∼0.12 µm/s) movements [7], [21]. In order to capture the 3D dynamics spanning a large temporal range, while minimizing photobleaching and photodamage, we used the recently developed inverted selective-plane illumination microscopy (iSPIM) [38] to image the movement of PA-GFP foci every 2 seconds for 30 minutes in MDCK and A549 cells starting 16 hours post-infection (Movies S1 and S2). Multiple WSN PA-GFP trajectories were analyzed from each cell. In order to define the movement of the foci we determined the mean squared displacement (MSD) of each PA-GFP focus during a trajectory [39]. We found that many foci displayed diffusive movement, based on the characteristic linear dependence of their MSDs on time, while a quarter of the foci appeared to undergo active transport, as their MSDs displayed nonlinear time dependence (Figure S5). PA-GFP trajectories with active transport had average speeds of 0.23+/−0.06 µm/s and 0.27+/−0.11 µm/s in MDCK and A549 cells respectively. These speeds are similar to the maximum speeds of microtubule-mediated endosomal transport in a HeLa-derived cell line (0.25–1.5 µm/s) [40].
Closer inspection of PA-GFP tracks revealed that many foci colocalized during their trajectories. Strikingly, we observed that foci displayed distinct fates after colocalization. Some foci appeared to come together and persist as a single diffraction-limited spot for the remainder of the trajectory (herein referred to as ‘fusion’) while other foci transiently colocalized, then separated and continued on as distinct foci. We quantified the number of fusion and transient colocalization events in WSN PA-GFP infected MDCK and A549 cells (Tables S2 and S3). Most trajectories displayed transient colocalization events (69% and 88% in MDCK and A549 cells, respectively), whereas fusion events were relatively rare (9% and 16% for MDCK and A549 cells, respectively).
Figure 6 depicts an example of a fusion event involving two cytoplasmic PA-GFP foci within an infected MDCK cell (Figure 6A). A single focus is tracked from near the nuclear edge into the cytoplasm and towards the cell periphery (Figure 6B). After 36 s, the PA-GFP focus (Figure 6C, red arrow) colocalized and fused with another focus (blue arrow). We confirmed that these two foci persisted as a single focus after colocalization by observing 1) a single peak in a fluorescence intensity vs. depth plot at the site of fusion and 2) an increase in fluorescence intensity of the fused focus at the time of fusion compared to a stationary, unfused adjacent focus (Figure 6D and E). While previous studies reported similar average speeds of vRNA trafficking [21] they did not observe dynamic fusion and colocalization events in the cytoplasm. The WSN PA-GFP virus enabled us to extend these previous studies and describe a novel aspect of vRNA transport. The fusion of multiple PA-GFP foci in the cytoplasm may reflect the coalescence of polymerase complexes attached to different vRNA segments, suggesting that the subcomplexes we identified using four-color FISH dynamically associate with each other in the cytoplasm en route to the plasma membrane.
Fig. 6. Fusion event of an WSN PA-GFP focus in the cytoplasm of MDCK cells. 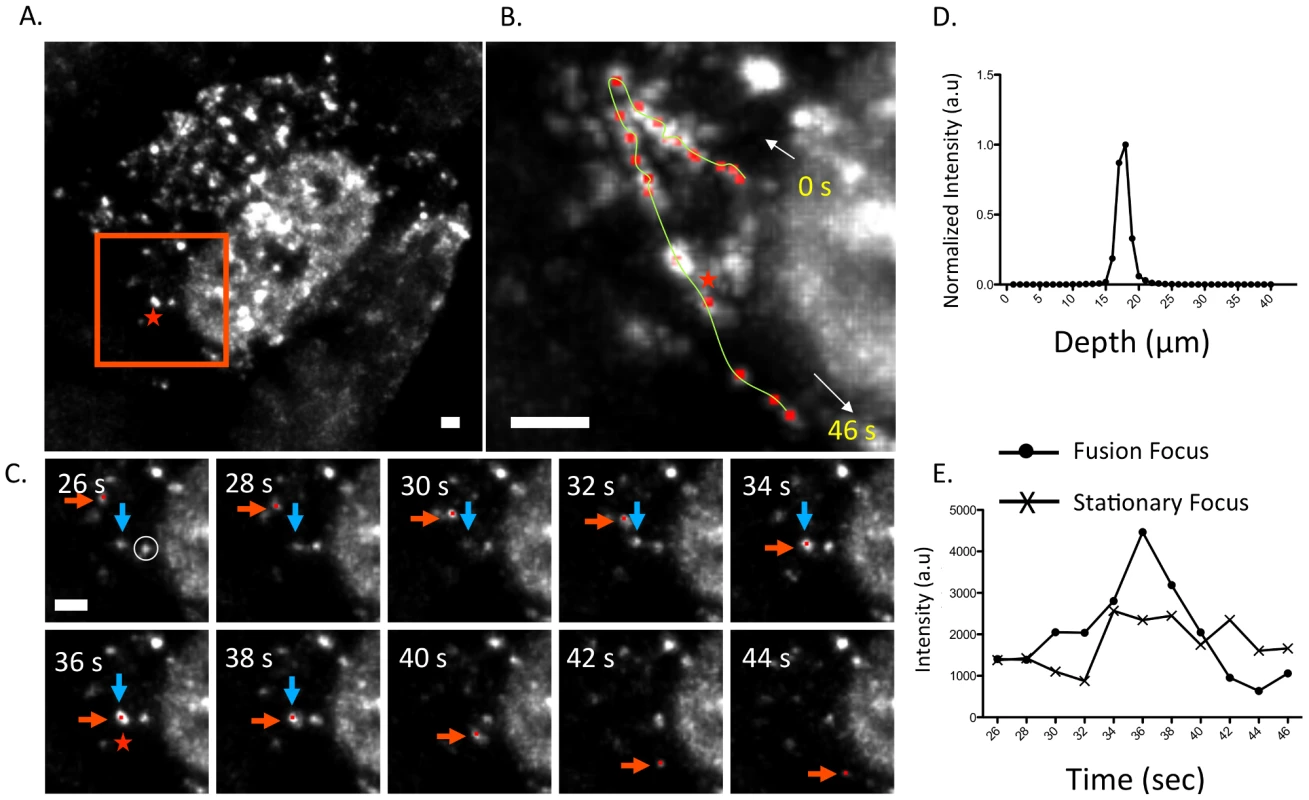
Maximum intensity projection from inverted selective-plane illumination microscopy (iSPIM) movie of MDCK cells infected with WSN PA-GFP (MOI = 5) at 16 hpi, 36 seconds (s) after onset of tracking (A). The red-boxed region is highlighted in (B and C), and the red star indicates the region where colocalization of two GFP foci (marked in red and blue arrows) occurred. A single WSN PA-GFP focus trajectory; each red dot is a spot tracked in time, the yellow line is the overall trace, and white arrows indicate the direction of the PA-GFP movement (B). The total track duration is 46 s. Part C contains still frames of the red-boxed region in part A from 26–44 s, and illustrates colocalization and subsequent fusion of two foci. The red arrow indicates the PA-GFP focus that is being tracked and the blue arrow indicates the PA-GFP focus with which it fuses. All scale bars are 2 µm. An intensity versus depth plot of the colocalized foci, indicated by the red star in part A, demonstrates a single peak indicating that the fusion occurs in a single diffraction-limited spot (D). An intensity vs. time plot of the fused focus compared to an adjacent stationary focus, identified by the white circle in part C, during the same time frame (E). The asterisk marks the time corresponding to the colocalization event. Discussion
Reassortment of influenza virus gene segments can lead to the emergence of novel influenza viruses with pandemic potential. The data from this study provide insight into vRNA assembly, which is important for understanding the intracellular mechanism of vRNA reassortment. First, our data suggest that vRNA segments are not exported as individual segments since the majority of foci at the external nuclear periphery contain more than one vRNA segment (Figure 2C). Second, we observed many cytoplasmic foci with fewer than 4 vRNA segments (Figure 1, 2A, 2C and Figure S3), implying that all 8 vRNA segments are not exported from the nucleus together. Third, individual vRNA segments do not reach the plasma membrane separately since many foci contain more than a single vRNA segment and foci can fuse together in the cytoplasm (Figure 1, 2B, and 6C). Therefore, we believe that vRNA assembly is an active process that includes the formation of flexible subcomplexes that export from the nucleus and then undergo further assembly en route to the plasma membrane via dynamic colocalization events (Figure 7).
Fig. 7. Proposed model for vRNA assembly. 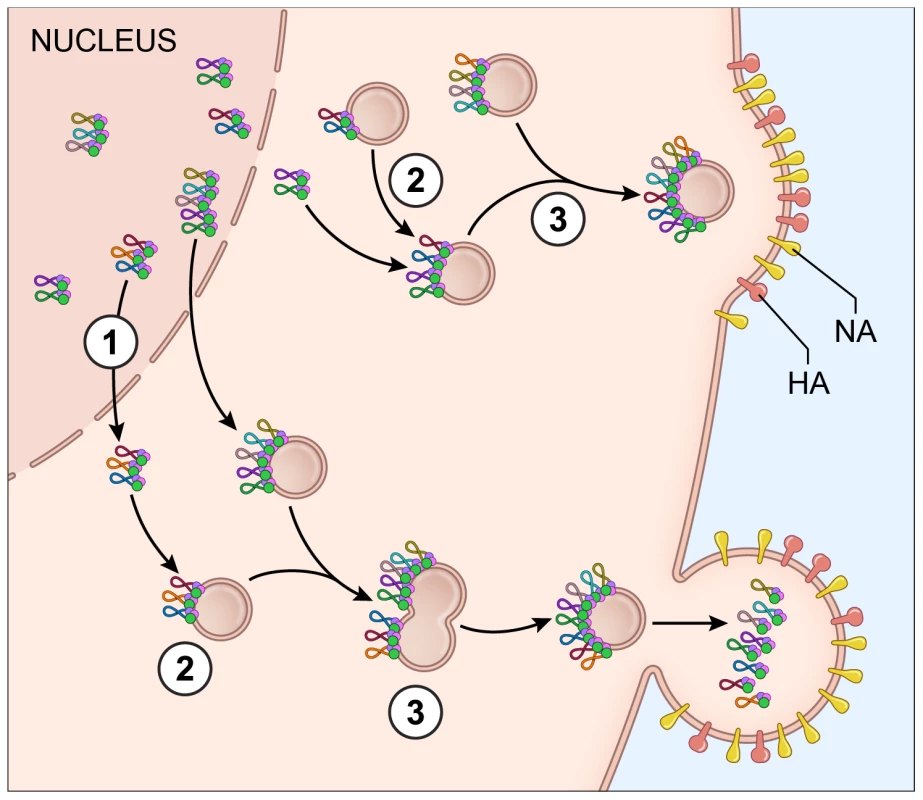
The data presented in this paper supports a model of vRNA assembly whereby 1) vRNA segments export from the nucleus as complexes containing multiple vRNA segments, 2) cytoplasmic foci containing vRNA segments transport through the cytoplasm and 3) these cytoplasmic foci fuse together in the cytoplasm prior to budding. Each vRNA segment is shown as a loop where the ends are associated with a heterotrimeric polymerase complex identified as 3 circles. The PA protein of the polymerase complex is shown in green since this was the protein used as a surrogate for vRNA transport. The cellular Rab11a-containing endosomes that colocalize with vRNA-containing cytoplasmic foci are indicated. Electron tomography has shown that vRNA segments within progeny virions have a conserved ‘7+1’ structure, where 7 segments surrounding a center segment [11], [13]. Selective assembly of vRNA segments into this type of conformation would seem to require either a linear ordering of vRNA-vRNA associations or a the existence of a master segment in the center that can interact with all other segments [5]. In vitro studies exploring the RNA-RNA interactions between all 8 segments have suggested that indeed a linear interaction network between vRNA segments is used for vRNA assembly [13], [16], but these interaction networks are not conserved between different subtypes of influenza A viruses [15]. In this study, we attempted to identify preferences between vRNA segments using the four-color FISH assay and found that most vRNA segments colocalize with each other with a high confidence value; we did not observe a distinct linear preference between vRNA segments (Figure 3). We believe that the high specificity of each FISH probe set (Figure S2) limits the likelihood of false positive results in our colocalization studies. In addition, our data are based on the colocalization profiles of vRNA segments throughout the entire cytoplasm, including the plasma membrane where vRNA segments are localized in high concentration prior to budding. Therefore, our data neither support nor refute the hypothesis that vRNA segments associate in an ordered fashion. Further analysis that carefully examines the colocalization profiles of vRNA segments and the vRNA composition of the cytoplasmic foci at different distances from the nucleus would provide spatially defined colocalization coefficients and interaction networks, which may help to clarify the nature of the selective arrangement between vRNA segments.
Interestingly, we observed a low correlation coefficient between the HA and PB2 vRNA segments, which encode known viral host range determinants [29]. HA mediates attachment and entry into host cells by binding to terminally attached sialic acid moieties on cell-surface glycoproteins; avian influenza viruses predominantly associate with α2,3-linked sialic acids while human viruses bind α2,6-linked sialic acids [41], [42]. Multiple residues of the PB2 protein, a component of the viral heterotrimeric polymerase complex, restrict replication of the virus in mammalian cells; for example the residue at position 627 typically encodes glutamic acid in avian influenza viruses and lysine in most human influenza viruses [26], [43], [44]. Interestingly, the recent studies generating airborne transmissible avian H5N1 viruses described viruses where the HA protein was modified to associate with α2,6 sialic acid moieties and either contained a human PB2 vRNA segment or the PB2 627K residue [45]–[47], demonstrating the importance of these two gene segments in emerging avian influenza pandemic viruses. Based on our data, a low correlation coefficient between the HA and PB2 vRNA segments may indicate that these segments are less likely to reassort together than two segments with a high correlation coefficient like PB1 and HA vRNA, which reassorted together in the 1957 H2N2 pandemic virus [48].
The observations we present here extend and refine previous assumptions regarding vRNA assembly. A recent study [12] proposed a scheme for vRNA assembly based on visualization of only 2 vRNA segments (PB2 and NA), in which they suggest that vRNA segments are exported from the nucleus individually. By visualizing four vRNA segments simultaneously we demonstrate that most cytoplasmic foci contain more than one vRNA segment. While our data support the overall conclusion proposed by Chou et al that vRNA assembly occurs en route to the PM, we not only provide much stronger support for this model but we also extend the observations made in fixed cells by visualizing vRNA transport in live cells during a productive infection. We demonstrate that cytoplasmic foci dynamically colocalize with each other, potentially mediating further assembly of vRNA subcomplexes. In addition, previous studies have shown that influenza vRNA segments colocalize with recycling endosomes containing the small GTPase Rab11a, and that these endosomes facilitate vRNA trafficking in the cytoplasm [7], [9], [36]. We confirmed colocalization of Rab11a and influenza PB2 vRNA segment in A549 cells transfected with Rab11a-GFP (Figure S6). Since WSN PA-GFP foci colocalize with PB2 vRNA segment (Figure 5) it is likely that WSN PA-GFP and Rab11a also associate, suggesting that the dynamic colocalization events observed during viral infection might also involve Rab11a-containing recycling endosomes. Further studies characterizing the importance of the transient or sustained colocalization events in viral replication may reveal novel targets for antiviral therapeutics.
The results we present here provide insight into the dynamics of vRNA transport and assembly, but our methods have applications beyond vRNA trafficking. For example, the WSN PA-GFP virus can be used to observe the kinetics of spread of influenza virus in cells (Movie S3), because the fluorescence can be tracked for extended periods of time. Development of fluorescent HIV, coronavirus and measles viruses have greatly enhanced our understanding of the biology of these pathogens, from production of progeny virions to viral spread [49]–[51]. We believe the observations and tools described in this study will enable similar progress in understanding of influenza virus biology.
Materials and Methods
Cells and viruses
Madin-Darby Canine kidney (MDCK) epithelial cells and adenocarcinoma human alveolar basal epithelial cells (A549) obtained from ATCC were maintained in modified Eagle's medium (MEM) with 10% fetal calf serum (FCS) and L-glutamine or Dulbecco's MEM (DMEM) with 10% FCS respectively. 293T cells (from ATCC) were used to rescue recombinant viruses and were maintained in 10% FCS DMEM.
Recombinant A/WSN/1933 (H1N1) viruses were rescued using an eight-plasmid (pHW2000) reverse genetics system, a kind gift from Dr. Richard Webby of St. Jude Children's Research Hospital. WSN PA-GFP was constructed by engineering a silent Xba1 site in PA using site-directed mutagenesis (Stratagene Santa Clara, CA) into both pHW2000 WSN PA and pCDNA WSN PA-GFP [33] at position 1986 (WT PA 1984 - TCA AGA – 1989; PA XBA1 1984 - TCT AGA – 1989). The pCDNA WSN PA-GFP plasmid contained an Xba1 site just after the GFP stop codon, so this plasmid was cut with Xba1, and the C-terminal PA-GFP fusion was inserted into the pHW2000 WSN PA Xba1 digested vector. This strategy will result in a duplication of 165 base pairs of the WSN PA coding region after the GFP stop codon as depicted in Figure 4A. WT WSN and WSN PA-GFP viruses were rescued by transfection in 293T cells with TransIT transfection reagent (Mirus Bio Madison, WI) and then plaque purified on MDCK cells. GFP positive plaques were used to inoculate 10-day old embryonated chicken eggs. Infectivity titers for all virus stocks were calculated the 50% tissue culture infectious dose (TCID50) per mL as described in [52].
Western blot analysis for GFP (mouse anti-GFP antibody, Living Colors, Invitrogen 1∶1,000) or viral NP (mouse anti-Influenza A NP antibody, Millipore 1∶5,000) was performed on purified viruses amplified in embryonated egg allantoic fluid.
Immunofluorescence
MDCK or A549 cells grown on circular coverslips (12Cir-1.5, Fisher Scientific Pittsburgh, PA) were infected with virus for the time and multiplicity of infection (MOI) specified in the figure legends. The cells were fixed with 4% paraformaldehyde (PFA) and probed with anti-Influenza A NP antibody (Millipore 1∶2,000) for one hour followed by Alexa Fluor-594 goat anti-mouse Fab' fragment (Invitrogen 1∶2,000) for visualization of viral NP protein. To determine the role of Crm1 on the export of WSN PA-GFP protein, cells were infected with WSN PA-GFP virus and treated 4 hours post infection (hpi) with leptomycin B (2.5 mg/mL) that was kept on for the remainder of the infection. Cells were fixed 16 hpi and stained as described above.
Four color fluorescent in situ hybridization (FISH)
Probes for FISH were designed and purchased from BioSearch Technologies (Novato, CA) and were directed against each vRNA segment. Each probe is ∼20 base pairs and ∼30 probes cover the length of a given vRNA target. The sequence of each probe was manually compared against the entire WSN influenza genome to ensure specificity to the targeted vRNA segment; any probes with greater than 10 base pairs complementarity to another vRNA target or to a mRNA sequence were excluded prior to purchase. This rigorous evaluation of the FISH probe sequences allowed for high specificity of the FISH probes to the specific vRNA segment.
The fluorophores used in our four-color FISH experiments are: Quasar 570, CAL Fluor Red 590, Quasar 670, and Alexa-Fluor 488. FISH probes conjugated to the Alexa-Fluor 488 fluorophore were produced manually to amine-terminal oligos synthesized by BioSearch Technologies and conjugated using the Invitrogen Alexa-Fluor 488 Oligonucleotide Amine Labeling kit per the manufacturer's recommendations.
Infected cells were stained with FISH probes from BioSearch using a standard protocol previously described in [53]. Briefly, cells grown on circular coverslips were infected at an MOI = 3 and fixed 8 hpi with 4% PFA stored in ice cold 70% EtOH overnight. The next day the coverslips were rehydrated with the wash buffer (10% formamide and 2× SSC in DEPC-treated water). Cells were incubated overnight in 200 µl of hybridization buffer (10% dextran sulfate, 2 mM vanadyl-ribonucleoside complex, 0.02% RNA-free BSA, 1 mg/ml e.Coli tRNA, 2× SSC, and 10% formamide in DEPC treated water) with 2 µl of labeled probes (probe concentrations ranged from 2.5 µM–10 µM) and incubated overnight in a 28°C incubator. The following day, the hybridization buffer was removed and cells were incubated in wash buffer containing DAPI for 10 minutes, washed then mounted. All incubations were performed in the dark.
Spectral separation of the four fluorophores was achieved using a Leica SP5 white-light laser confocal microscope. A sequential scanning program with a line average of 4 between frames was established based on the manufacturer's excitation and emission spectra for each fluorophore. The following parameters were used: PMT1 (DAPI) UV laser (power = 4%) wavelength range 415–470 nm, PMT2 (Alexa-Fluor 488) 488 nm excitation wavelength (λex) (power = 10%) range 500–540 nm, PMT3 (Quasar 570) 545 nm λex (12%) range 553–585 nm, PMT4 (Cal Fluor Red 590) 575 nm λex (12%) range 590–635 nm, and PMT5 (Quasar 670) 647 nm λex (12%) range 675–750 nm. Spectral separation was confirmed in each experiment with single color controls (infected cells stained with only a single vRNA FISH probe set).
FISH colocalization
WT WSN virus infected cells were probed with different combinations of FISH probes (Table S1) to assess colocalization between individual segments. Cells were imaged using the sequential scanning program on the Leica SP5 white light laser confocal microscope. To obtain close to Nyquist sampling, stacks of each cell were taken with 0.17 µm separation between slices and the height of the stacks was determined for each cell based on the FISH staining. Images were acquired at 1024×1024 pixel resolution with a 63× Leica objective with a numerical aperture (NA) of 1.4 and a zoom of 4; the resulting pixel size was ∼50×50×168 nm. Background subtraction and subsequent deconvolution of each stack was performed manually for each channel using Huygens Essential software (Version 4.3.1p3, Scientific Volume Imaging BV, Hilversum, Netherlands).
Imaris software (version 7.6.3, Bitplane AG, Zurich, Switzerland) was used for 3D reconstruction, particle tracking, colocalization and statistical analysis. Pearson coefficients in colocalized volumes were obtained between each vRNA probe set from the deconvolved images using Imaris colocalization analysis. The DAPI stain was used to create a channel mask (smoothing = 0.9) to exclude the vRNA nuclear stain from the colocalization quantification. Thus, we were able to compare the staining of different vRNA segments in the cytoplasm of an infected cell. At least 3 different cell volumes were analyzed for each vRNA pair.
To determine the vRNA composition and distance from the nucleus of each cytoplasmic focus we used the Imaris software to create ‘Spots’ from the signal of each channel in the cytoplasm (using the nuclear mask created earlier). All spots from each channel were merged into one ‘Spots’ channel, which created a single spot for every cytoplasmic focus within an infected cell, with intensity values for each channel. This information was used to determine the number of spots positive for all 4 channels versus 3 channels, 2 channels, or a single channel. The frequency of spots from three independent cells was recorded; the numbers were compared using column statistical comparison and a Chi-Squared test using the graphing software, Prism (version 5.0, GraphPad, San Diego, CA), to determine if there was a significant difference between any of the distributions. A separate surface to define the boundary of the cell was created manually based on the vRNA staining pattern in every z-plane. The distance from the nucleus, defined by DAPI staining, was calculated for each spot within this surface and the frequency of spots in 100 nm distance increments was recorded from four independent cells.
FISH probe specificity
To confirm the specificity of each FISH probe set, two coverslips of A549 cells were transfected with a pPol1 plasmid expressing each vRNA segment of the WSN virus using Fugene HD (Promega, Madison, WI). Cells were fixed 24 hours post-transfection and processed for FISH staining as described above. Transfected cells were stained with FISH probe reactions B and E (Table S1) to ensure coverage of all 8 vRNA segments. FISH stained slides were visualized on a Leica SP5 white light laser microscope using the sequential parameters described above for four color FISH.
Visualization of Rab11a
Plasmids expressing Rab11a GFP were a kind gift from Dr. Julie Donaldson, National Heart Lung and Blood Institute (NHLBI) and were transfected into A549 cells using Fugene HD transfection reagent (Promega, Madison, WI). 24 hours post-transfection the cells were infected with WT WSN virus (MOI = 3) for subsequent FISH analysis as described above. Images of cells transfected with Rab11a-GFP and stained for vRNA segments were taken on a Leica SP5 confocal microscope using a 63× Leica objective with NA of 1.4.
Live cell imaging
iSPIM
MDCK or A549 cells were seeded onto rectangular coverslips (VWR, 48393-241) and then placed into an imaging chamber (Applied Scientific Instrumentation, I-3078-2450) that fit into a custom-built inverted selective plane illumination microscope (iSPIM). Cells were infected in the chamber at an MOI = 5 and then placed in a 37°C incubator for 16 hours prior to imaging. The iSPIM system was modified from our original design [38], equipped with a 0.3 NA excitation objective (Nikon 10×, 3.5 mm working distance) and a higher, 1.0 NA detection objective (Zeiss, W Plan-Apochromat 63×, 2.1 mm working distance) to increase spatial resolution and fluorescence signal collection. The 488 nm excitation power was set at 50 µW (as measured before the excitation objective), and an electron-multiplying charge coupled device (EM-CCD, Andor, iXon DU-897T) was used to capture fluorescence. We recorded 40 planes per volume for each cell, spacing planes every 1 µm. Volumes were acquired every 2 seconds for a total of 30 minutes.
Point scanning confocal microscopy
The spread of influenza was captured on a Leica SP5 point scanning confocal microscope using a 1.4 NA 63× objective. MDCK cells were seeded into an 8-well chambered coverglass slide (Nunc, Rochester, NY) and infected with an MOI = 0.1 with WSN PA-GFP virus. Four hours post-infection, the slides were placed onto the microscope inside a temperature (37°C) and 5% CO2 controlled incubator (Microscope incubator S-2, PeCon GmbH, Erbach, Germany). We captured 5 slices per field of view, with ∼1 µm separation between slices, every 10 min for 16 hours. The DIC channel was used to determine stack height.
Tracking cytoplasmic PA-GFP
3D image stacks captured using the iSPIM were background subtracted and deconvolved using the Huygens software package (SVI, Huygens Professional) using the ‘Classic MLE’ setting with 40 iterations per deconvolution. The resulting datasets were analyzed in ImageJ, generating maximum intensity projections (as displayed in Figure 6A) and axial intensity profiles (Figure 6D). In order to track the motion of cytoplasmic foci, deconvolved datasets were loaded into Imaris for tracking. Manual tracking was performed using the ‘Spots’ function under the Imaris Surpass toolbox and the location of a PA-GFP focus was determined over all frames. A minimum of 30 tracks were collected for each cell type. Statistics for tracks, including the mean track speed, track duration, intensity of the focus over the length of the track, and the xyz position of each focus in each frame throughout the trace were calculated using Imaris. The trajectories for each focus were next analyzed in MATLAB using custom software in order to calculate the mean squared displacements (MSD) vs. time lag (Figure S4). Fusion and transient colocalization events were tallied. Fusions were classified as events wherein two foci came together in a diffraction limited spot and subsequently moved together as a single particle, and confirmed by assessing the axial intensity profile of the particle (Figure 6D). Transient colocalizations were scored as events where two foci came together within a diffraction limited spot for a limited period of time, and did not move together afterwards.
Supporting Information
Zdroje
1. Estimates of deaths associated with seasonal influenza — United States, 1976–2007. MMWR Morbidity and mortality weekly report 59 : 1057–1062.
2. Fields BN, Knipe DM, Howley PM (2013) Fields virology. Philadelphia: Wolters Kluwer/Lippincott Williams & Wilkins Health. p. p.
3. GartenRJ, DavisCT, RussellCA, ShuB, LindstromS, et al. (2009) Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science 325 : 197–201.
4. LiuD, ShiW, ShiY, WangD, XiaoH, et al. (2013) Origin and diversity of novel avian influenza A H7N9 viruses causing human infection: phylogenetic, structural, and coalescent analyses. Lancet 381 : 1926–1932.
5. HutchinsonEC, von KirchbachJC, GogJR, DigardP (2010) Genome packaging in influenza A virus. The Journal of general virology 91 : 313–328.
6. BruceEA, StuartA, McCaffreyMW, DigardP (2012) Role of the Rab11 pathway in negative-strand virus assembly. Biochemical Society transactions 40 : 1409–1415.
7. AmorimMJ, BruceEA, ReadEK, FoegleinA, MahenR, et al. (2011) A Rab11 - and microtubule-dependent mechanism for cytoplasmic transport of influenza A virus viral RNA. Journal of virology 85 : 4143–4156.
8. BruceEA, DigardP, StuartAD (2010) The Rab11 pathway is required for influenza A virus budding and filament formation. Journal of virology 84 : 5848–5859.
9. EisfeldAJ, KawakamiE, WatanabeT, NeumannG, KawaokaY (2011) RAB11A is essential for transport of the influenza virus genome to the plasma membrane. Journal of virology 85 : 6117–6126.
10. NodaT, SagaraH, YenA, TakadaA, KidaH, et al. (2006) Architecture of ribonucleoprotein complexes in influenza A virus particles. Nature 439 : 490–492.
11. NodaT, SugitaY, AoyamaK, HiraseA, KawakamiE, et al. (2012) Three-dimensional analysis of ribonucleoprotein complexes in influenza A virus. Nature communications 3 : 639.
12. ChouYY, HeatonNS, GaoQ, PaleseP, SingerR, et al. (2013) Colocalization of Different Influenza Viral RNA Segments in the Cytoplasm before Viral Budding as Shown by Single-molecule Sensitivity FISH Analysis. PLoS pathogens 9: e1003358.
13. FournierE, MoulesV, EssereB, PaillartJC, SirbatJD, et al. (2012) A supramolecular assembly formed by influenza A virus genomic RNA segments. Nucleic acids research 40 : 2197–2209.
14. DuhautSD, DimmockNJ (2002) Defective segment 1 RNAs that interfere with production of infectious influenza A virus require at least 150 nucleotides of 5′ sequence: evidence from a plasmid-driven system. The Journal of general virology 83 : 403–411.
15. GavazziC, IselC, FournierE, MoulesV, CavalierA, et al. (2013) An in vitro network of intermolecular interactions between viral RNA segments of an avian H5N2 influenza A virus: comparison with a human H3N2 virus. Nucleic acids research 41 : 1241–1254.
16. FournierE, MoulesV, EssereB, PaillartJC, SirbatJD, et al. (2012) Interaction network linking the human H3N2 influenza A virus genomic RNA segments. Vaccine 30 : 7359–7367.
17. ChouYY, VafabakhshR, DoganayS, GaoQ, HaT, et al. (2012) One influenza virus particle packages eight unique viral RNAs as shown by FISH analysis. Proceedings of the National Academy of Sciences of the United States of America 109 : 9101–9106.
18. GaoQ, LowenAC, WangTT, PaleseP (2010) A nine-segment influenza a virus carrying subtype H1 and H3 hemagglutinins. Journal of virology 84 : 8062–8071.
19. LiF, FengL, PanW, DongZ, LiC, et al. (2010) Generation of replication-competent recombinant influenza A viruses carrying a reporter gene harbored in the neuraminidase segment. Journal of virology 84 : 12075–12081.
20. ManicassamyB, ManicassamyS, Belicha-VillanuevaA, PisanelliG, PulendranB, et al. (2010) Analysis of in vivo dynamics of influenza virus infection in mice using a GFP reporter virus. Proceedings of the National Academy of Sciences of the United States of America 107 : 11531–11536.
21. AvilovSV, MoisyD, MunierS, SchraidtO, NaffakhN, et al. (2012) Replication-competent influenza A virus that encodes a split-green fluorescent protein-tagged PB2 polymerase subunit allows live-cell imaging of the virus life cycle. Journal of virology 86 : 1433–1448.
22. BlakeleyBD, ChapmanAM, McNaughtonBR (2012) Split-superpositive GFP reassembly is a fast, efficient, and robust method for detecting protein-protein interactions in vivo. Molecular bioSystems 8 : 2036–2040.
23. LalondeS, EhrhardtDW, FrommerWB (2005) Shining light on signaling and metabolic networks by genetically encoded biosensors. Current opinion in plant biology 8 : 574–581.
24. MaglieryTJ, WilsonCG, PanW, MishlerD, GhoshI, et al. (2005) Detecting protein-protein interactions with a green fluorescent protein fragment reassembly trap: scope and mechanism. Journal of the American Chemical Society 127 : 146–157.
25. ZinchukV, ZinchukO, OkadaT (2007) Quantitative colocalization analysis of multicolor confocal immunofluorescence microscopy images: pushing pixels to explore biological phenomena. Acta histochemica et cytochemica 40 : 101–111.
26. SubbaraoEK, LondonW, MurphyBR (1993) A single amino acid in the PB2 gene of influenza A virus is a determinant of host range. Journal of virology 67 : 1761–1764.
27. FukuyamaS, KawaokaY (2011) The pathogenesis of influenza virus infections: the contributions of virus and host factors. Current opinion in immunology 23 : 481–486.
28. NaffakhN, TomoiuA, Rameix-WeltiMA, van der WerfS (2008) Host restriction of avian influenza viruses at the level of the ribonucleoproteins. Annual review of microbiology 62 : 403–424.
29. NeumannG, KawaokaY (2006) Host range restriction and pathogenicity in the context of influenza pandemic. Emerging infectious diseases 12 : 881–886.
30. FodorE, CrowM, MingayLJ, DengT, SharpsJ, et al. (2002) A single amino acid mutation in the PA subunit of the influenza virus RNA polymerase inhibits endonucleolytic cleavage of capped RNAs. Journal of virology 76 : 8989–9001.
31. YuanP, BartlamM, LouZ, ChenS, ZhouJ, et al. (2009) Crystal structure of an avian influenza polymerase PA(N) reveals an endonuclease active site. Nature 458 : 909–913.
32. DiasA, BouvierD, CrepinT, McCarthyAA, HartDJ, et al. (2009) The cap-snatching endonuclease of influenza virus polymerase resides in the PA subunit. Nature 458 : 914–918.
33. FodorE, SmithM (2004) The PA subunit is required for efficient nuclear accumulation of the PB1 subunit of the influenza A virus RNA polymerase complex. Journal of virology 78 : 9144–9153.
34. Dos Santos AfonsoE, EscriouN, LeclercqI, van der WerfS, NaffakhN (2005) The generation of recombinant influenza A viruses expressing a PB2 fusion protein requires the conservation of a packaging signal overlapping the coding and noncoding regions at the 5′ end of the PB2 segment. Virology 341 : 34–46.
35. EltonD, Simpson-HolleyM, ArcherK, MedcalfL, HallamR, et al. (2001) Interaction of the influenza virus nucleoprotein with the cellular CRM1-mediated nuclear export pathway. Journal of virology 75 : 408–419.
36. MomoseF, KikuchiY, KomaseK, MorikawaY (2007) Visualization of microtubule-mediated transport of influenza viral progeny ribonucleoprotein. Microbes and infection/Institut Pasteur 9 : 1422–1433.
37. WatanabeK, TakizawaN, KatohM, HoshidaK, KobayashiN, et al. (2001) Inhibition of nuclear export of ribonucleoprotein complexes of influenza virus by leptomycin B. Virus research 77 : 31–42.
38. WuY, GhitaniA, ChristensenR, SantellaA, DuZ, et al. (2011) Inverted selective plane illumination microscopy (iSPIM) enables coupled cell identity lineaging and neurodevelopmental imaging in Caenorhabditis elegans. Proceedings of the National Academy of Sciences of the United States of America 108 : 17708–17713.
39. HuangF, WatsonE, DempseyC, SuhJ (2013) Real-time particle tracking for studying intracellular trafficking of pharmaceutical nanocarriers. Methods in molecular biology 991 : 211–223.
40. ChenH, YangJ, LowPS, ChengJX (2008) Cholesterol level regulates endosome motility via Rab proteins. Biophysical journal 94 : 1508–1520.
41. ShinyaK, EbinaM, YamadaS, OnoM, KasaiN, et al. (2006) Avian flu: influenza virus receptors in the human airway. Nature 440 : 435–436.
42. van RielD, MunsterVJ, de WitE, RimmelzwaanGF, FouchierRA, et al. (2006) H5N1 Virus Attachment to Lower Respiratory Tract. Science 312 : 399.
43. GabrielG, HerwigA, KlenkHD (2008) Interaction of polymerase subunit PB2 and NP with importin alpha1 is a determinant of host range of influenza A virus. PLoS pathogens 4: e11.
44. LiZ, ChenH, JiaoP, DengG, TianG, et al. (2005) Molecular basis of replication of duck H5N1 influenza viruses in a mammalian mouse model. Journal of virology 79 : 12058–12064.
45. HerfstS, SchrauwenEJ, LinsterM, ChutinimitkulS, de WitE, et al. (2012) Airborne transmission of influenza A/H5N1 virus between ferrets. Science 336 : 1534–1541.
46. ImaiM, WatanabeT, HattaM, DasSC, OzawaM, et al. (2012) Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature 486 : 420–428.
47. LuX, ShiY, ZhangW, ZhangY, QiJ, et al. (2013) Structure and receptor-binding properties of an airborne transmissible avian influenza A virus hemagglutinin H5 (VN1203mut). Protein & cell 4 : 502–511.
48. KawaokaY, KraussS, WebsterRG (1989) Avian-to-human transmission of the PB1 gene of influenza A viruses in the 1957 and 1968 pandemics. Journal of virology 63 : 4603–4608.
49. LemonK, de VriesRD, MesmanAW, McQuaidS, van AmerongenG, et al. (2011) Early target cells of measles virus after aerosol infection of non-human primates. PLoS pathogens 7: e1001263.
50. McDonaldD, VodickaMA, LuceroG, SvitkinaTM, BorisyGG, et al. (2002) Visualization of the intracellular behavior of HIV in living cells. The Journal of cell biology 159 : 441–452.
51. SimsAC, BaricRS, YountB, BurkettSE, CollinsPL, et al. (2005) Severe acute respiratory syndrome coronavirus infection of human ciliated airway epithelia: role of ciliated cells in viral spread in the conducting airways of the lungs. Journal of virology 79 : 15511–15524.
52. LakdawalaSS, LamirandeEW, SuguitanALJr, WangW, SantosCP, et al. (2011) Eurasian-origin gene segments contribute to the transmissibility, aerosol release, and morphology of the 2009 pandemic H1N1 influenza virus. PLoS pathogens 7: e1002443.
53. ShihJD, WaksZ, KedershaN, SilverPA (2011) Visualization of single mRNAs reveals temporal association of proteins with microRNA-regulated mRNA. Nucleic acids research 39 : 7740–7749.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek DHX36 Enhances RIG-I Signaling by Facilitating PKR-Mediated Antiviral Stress Granule FormationČlánek Oral Bacteria and CancerČlánek A Non-Coding RNA Promotes Bacterial Persistence and Decreases Virulence by Regulating a Regulator in
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2014 Číslo 3- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Conflicting Interests in the Pathogen–Host Tug of War: Fungal Micronutrient Scavenging Versus Mammalian Nutritional Immunity
- Putting Fungi to Work: Harvesting a Cornucopia of Drugs, Toxins, and Antibiotics
- Mycobacteriophages: Windows into Tuberculosis
- Human African Trypanosomiasis and Immunological Memory: Effect on Phenotypic Lymphocyte Profiles and Humoral Immunity
- Five Things to Know about Genetically Modified (GM) Insects for Vector Control
- A Missing Dimension in Measures of Vaccination Impacts
- Eosinophils Are Important for Protection, Immunoregulation and Pathology during Infection with Nematode Microfilariae
- Clonality of HTLV-2 in Natural Infection
- Production, Fate and Pathogenicity of Plasma Microparticles in Murine Cerebral Malaria
- Group B Streptococcal Infection of the Choriodecidua Induces Dysfunction of the Cytokeratin Network in Amniotic Epithelium: A Pathway to Membrane Weakening
- New Insights into How Adapts to Its Mammalian Host during Bubonic Plague
- Foodborne Transmission of Nipah Virus in Syrian Hamsters
- A Polysaccharide Virulence Factor from Elicits Anti-inflammatory Effects through Induction of Interleukin-1 Receptor Antagonist
- Structural and Functional Characterization of a Complex between the Acidic Transactivation Domain of EBNA2 and the Tfb1/p62 Subunit of TFIIH
- Adaptive Gene Amplification As an Intermediate Step in the Expansion of Virus Host Range
- DHX36 Enhances RIG-I Signaling by Facilitating PKR-Mediated Antiviral Stress Granule Formation
- Hepatitis B Virus Infection and Immunopathogenesis in a Humanized Mouse Model: Induction of Human-Specific Liver Fibrosis and M2-Like Macrophages
- Crk Adaptors Negatively Regulate Actin Polymerization in Pedestals Formed by Enteropathogenic (EPEC) by Binding to Tir Effector
- Fatty Acid Biosynthesis Contributes Significantly to Establishment of a Bioenergetically Favorable Environment for Vaccinia Virus Infection
- A Cytosolic Chaperone Complexes with Dynamic Membrane J-Proteins and Mobilizes a Nonenveloped Virus out of the Endoplasmic Reticulum
- Intracellular Promote Invasive Cell Motility through Kinase Regulation of the Host Actin Cytoskeleton
- MAVS-MKK7-JNK2 Defines a Novel Apoptotic Signaling Pathway during Viral Infection
- RON5 Is Critical for Organization and Function of the Moving Junction Complex
- Immune Suppression by Neutrophils in HIV-1 Infection: Role of PD-L1/PD-1 Pathway
- and Exhibit Metabolic Symbioses
- The Herpes Virus Fc Receptor gE-gI Mediates Antibody Bipolar Bridging to Clear Viral Antigens from the Cell Surface
- Target Cell Availability, Rather than Breast Milk Factors, Dictates Mother-to-Infant Transmission of SIV in Sooty Mangabeys and Rhesus Macaques
- Evolution of the Retroviral Restriction Gene : Inhibition of Non-MLV Retroviruses
- Infection of Adult Thymus with Murine Retrovirus Induces Virus-Specific Central Tolerance That Prevents Functional Memory CD8 T Cell Differentiation
- Fha Interaction with Phosphothreonine of TssL Activates Type VI Secretion in
- In Vivo Administration of a JAK3 Inhibitor during Acute SIV Infection Leads to Significant Increases in Viral Load during Chronic Infection
- Lack of Detectable HIV-1 Molecular Evolution during Suppressive Antiretroviral Therapy
- Activation of HIV-1 from Latent Infection via Synergy of RUNX1 Inhibitor Ro5-3335 and SAHA
- A Compact, Multifunctional Fusion Module Directs Cholesterol-Dependent Homomultimerization and Syncytiogenic Efficiency of Reovirus p10 FAST Proteins
- The Role of Host and Microbial Factors in the Pathogenesis of Pneumococcal Bacteraemia Arising from a Single Bacterial Cell Bottleneck
- Genetic Dissection of Gut Epithelial Responses to
- Two-Component System Cross-Regulation Integrates Response to Heme and Cell Envelope Stress
- Oral Mycobiome Analysis of HIV-Infected Patients: Identification of as an Antagonist of Opportunistic Fungi
- A Model System for Studying the Transcriptomic and Physiological Changes Associated with Mammalian Host-Adaptation by Serovar Copenhageni
- Inflammasome Sensor NLRP1 Controls Rat Macrophage Susceptibility to
- ChIP-Seq and RNA-Seq Reveal an AmrZ-Mediated Mechanism for Cyclic di-GMP Synthesis and Biofilm Development by
- The Hypervariable Amino-Terminus of P1 Protease Modulates Potyviral Replication and Host Defense Responses
- Caspase-1-Dependent and -Independent Cell Death Pathways in Infection of Macrophages
- The Effect of Cell Growth Phase on the Regulatory Cross-Talk between Flagellar and Spi1 Virulence Gene Expression
- Different Mutagenic Potential of HIV-1 Restriction Factors APOBEC3G and APOBEC3F Is Determined by Distinct Single-Stranded DNA Scanning Mechanisms
- Oral Bacteria and Cancer
- Identification of OmpA, a Protein Involved in Host Cell Invasion, by Multi-Phenotypic High-Content Screening
- Transovarial Transmission of a Plant Virus Is Mediated by Vitellogenin of Its Insect Vector
- VE-Cadherin Cleavage by LasB Protease from Facilitates Type III Secretion System Toxicity in Endothelial Cells
- Dimerization of VirD2 Binding Protein Is Essential for Induced Tumor Formation in Plants
- Crystal Structure of the Vaccinia Virus DNA Polymerase Holoenzyme Subunit D4 in Complex with the A20 N-Terminal Domain
- Post-Translational Regulation via Clp Protease Is Critical for Survival of
- Modulation of Phagosomal pH by Promotes Hyphal Morphogenesis and Requires Stp2p, a Regulator of Amino Acid Transport
- Rotavirus Activates Lymphocytes from Non-Obese Diabetic Mice by Triggering Toll-Like Receptor 7 Signaling and Interferon Production in Plasmacytoid Dendritic Cells
- Cytomegalovirus m154 Hinders CD48 Cell-Surface Expression and Promotes Viral Escape from Host Natural Killer Cell Control
- Interferon Regulatory Factor-1 Protects from Fatal Neurotropic Infection with Vesicular Stomatitis Virus by Specific Inhibition of Viral Replication in Neurons
- HMGB1-Promoted and TLR2/4-Dependent NK Cell Maturation and Activation Take Part in Rotavirus-Induced Murine Biliary Atresia
- An Immunomics Approach to Schistosome Antigen Discovery: Antibody Signatures of Naturally Resistant and Chronically Infected Individuals from Endemic Areas
- PPARγ Agonists Improve Survival and Neurocognitive Outcomes in Experimental Cerebral Malaria and Induce Neuroprotective Pathways in Human Malaria
- A Non-Coding RNA Promotes Bacterial Persistence and Decreases Virulence by Regulating a Regulator in
- Viral OTU Deubiquitinases: A Structural and Functional Comparison
- Heterogeneity and Breadth of Host Antibody Response to KSHV Infection Demonstrated by Systematic Analysis of the KSHV Proteome
- Influenza A Virus Assembly Intermediates Fuse in the Cytoplasm
- Broadly Reactive Human CD8 T Cells that Recognize an Epitope Conserved between VZV, HSV and EBV
- Oncogenic Human Papillomaviruses Activate the Tumor-Associated Lens Epithelial-Derived Growth Factor (LEDGF) Gene
- Erythrocyte Invasion: Combining Function with Immune Evasion
- IL-1α and Complement Cooperate in Triggering Local Neutrophilic Inflammation in Response to Adenovirus and Eliminating Virus-Containing Cells
- Chronic Exposure to Type-I IFN under Lymphopenic Conditions Alters CD4 T Cell Homeostasis
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Cytomegalovirus m154 Hinders CD48 Cell-Surface Expression and Promotes Viral Escape from Host Natural Killer Cell Control
- Human African Trypanosomiasis and Immunological Memory: Effect on Phenotypic Lymphocyte Profiles and Humoral Immunity
- DHX36 Enhances RIG-I Signaling by Facilitating PKR-Mediated Antiviral Stress Granule Formation
- Conflicting Interests in the Pathogen–Host Tug of War: Fungal Micronutrient Scavenging Versus Mammalian Nutritional Immunity
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



