-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaDimerization of VirD2 Binding Protein Is Essential for Induced Tumor Formation in Plants
Agrobacterium tumefaciens causes crown gall disease (tumors) in agriculturally important plant species. It initiates infection through its Ti plasmid, which integrates a portion of its own DNA (T-DNA) into that of the host genome. The T-DNA is bound to VirD2 relaxase, and this complex is required for the efficient translocation and integration of the T-DNA into the plant genome for tumor formation. Two additional proteins, among others, are also required for Agrobacterium tumorigenesis: VirD4-coupling protein (CP) and VirD2-binding protein (VBP). VBP is responsible for recruiting VirD2–T-DNA to VirD4 CP to help localize T-DNA to the Type IV Secretion System apparatus for transfer. However, it is still unclear how VBP recruits the complex to VirD4 CP. Here, we report the crystal structure and associated functional studies of the C-terminal domain of VBP. We show that the C-terminal domain is the dimerization domain of VBP and only dimeric VBP is functional and essential for the induction of tumor in plants. This study enhances the understanding of the role of VBP in recruiting VirD2–T-DNA in A. tumefaciens prior to its transfer into the host plant. This mode of action can be extended to other pathogenic bacteria employing similar secretion systems.
Published in the journal: . PLoS Pathog 10(3): e32767. doi:10.1371/journal.ppat.1003948
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1003948Summary
Agrobacterium tumefaciens causes crown gall disease (tumors) in agriculturally important plant species. It initiates infection through its Ti plasmid, which integrates a portion of its own DNA (T-DNA) into that of the host genome. The T-DNA is bound to VirD2 relaxase, and this complex is required for the efficient translocation and integration of the T-DNA into the plant genome for tumor formation. Two additional proteins, among others, are also required for Agrobacterium tumorigenesis: VirD4-coupling protein (CP) and VirD2-binding protein (VBP). VBP is responsible for recruiting VirD2–T-DNA to VirD4 CP to help localize T-DNA to the Type IV Secretion System apparatus for transfer. However, it is still unclear how VBP recruits the complex to VirD4 CP. Here, we report the crystal structure and associated functional studies of the C-terminal domain of VBP. We show that the C-terminal domain is the dimerization domain of VBP and only dimeric VBP is functional and essential for the induction of tumor in plants. This study enhances the understanding of the role of VBP in recruiting VirD2–T-DNA in A. tumefaciens prior to its transfer into the host plant. This mode of action can be extended to other pathogenic bacteria employing similar secretion systems.
Introduction
The Type IV Secretion System (T4SS) has an unmatched versatility among the seven different secretion systems known in bacteria. T4SS can translocate not only proteins but also DNA to phylogenetically diverse taxa, including many bacterial species and various eukaryotic cells, as well as import and export DNA from the extracellular milieu. The T4SS shares a common ancestry with bacterial conjugation systems [1], with three types of T4SS described to date: (1) conjugation systems, defined as machines that translocate DNA substrates to recipient cells by a contact-dependent process; (2) effector translocation systems, functioning to deliver proteins or other effector molecules to eukaryotic target cells; and (3) DNA release or uptake systems, which translocate DNA to or from the extracellular milieu [2].
The VirB/D4 secretion system of Agrobacterium tumefaciens is a typical example of a T4SS, which serves as both a conjugation and an effector translocation system. Conjugation systems within the T4SS facilitate the translocation of protein-DNA complexes from the bacterium into the cytoplasm of the host cell. For instance, A. tumefaciens translocates to the host cell a segment of the Ti (tumor inducing) plasmid between the right and left borders (T-DNA) in complex with a cytoplasmic relaxase protein (VirD2) [3]. Proteins involved in T-DNA processing and translocation are classified into three functionally distinct classes [4]. Class I constitutes proteins involved in the processing of the DNA intermediates, such as VirC1, VirC2, VirD1 and VirD2 relaxase [5]. VirC1 and VirC2 proteins assemble the relaxosomal complex at the border sequences of the Ti plasmid and initiate T-DNA processing [6], whereas VirD2 relaxase, when present in the relaxosomal complex, cleaves the bottom strand of the T-DNA, where it remains covalently attached to the 5′ end of the single-stranded T-DNA [7]. Class II comprises 11 VirB proteins that form the T4SS apparatus which is responsible for the translocation of the VirD2–T-DNA complex and the effector proteins into the host cell cytoplasm [4], [8]. Class III constitutes the coupling proteins (CP), which mediate the interaction between the substrate (VirD2–T-DNA complex) and the transport apparatus [9]. VirD4 CP, the coupling protein in the A. tumefaciens VirB/D4 system, is an inner membrane protein with a large cytoplasmic domain essential for the transfer of both the T-DNA strand and VirE2 to host cells [10]–[12]. However, the VirD2–T-DNA complex and VirE2 are translocated separately [7], [13], [14]. The translocated effector proteins — VirE2, VirE3 and VirF which are involved in nuclear targeting, import and integration of the T-DNA into the host genome—mediate the interactions between the VirD2–T-DNA complex and host cellular factors [1]. In particular, VirE2 coats single-stranded T-DNA and protects it from host cell nucleases [15], whereas both VirE2 and VirD2 carry the nuclear localization signals that aid in the nuclear import of the T-DNA. VirE2 also interacts with host cell proteins, such as VIP1 and VIP2 [16]. VIP1 and VIP2 localize to plant nuclei and probably facilitate delivery of the T-DNA complex to its site of integration [16] VirF, on the other hand, localizes in the host nucleus and targets VIP1 and VirE2 for proteolysis and thus uncoats the T-DNA from its cognate proteins. This uncoating mechanism is a crucial step that must occur prior to integration of the T-DNA in the host genome [17].
The VirD2–T-DNA complex forms in the bacterial cytoplasm, but there is no evidence to suggest that VirD4 CP can recruit the bulky substrate complex to the T4SS apparatus [18]. In 2007, Guo et al. reported the existence of a subset of proteins defined as ‘recruiting proteins’ that are involved in bringing the nucleoprotein substrate complex formed in the bacterial cytoplasm to the VirD4 CP. The VirD2-binding protein (VBP) was subsequently identified as a key protein belonging to this subset and was shown to recruit the VirD2–T-DNA complex to the VirD4 CP [8]. Site-directed mutagenesis experiments have indeed shown that the interaction between VBP and VirD2 is important for T-DNA transfer [8], with VBP interacting with both the VirD2–T-DNA complex and with several of the T4SS components independently, including VirD4 CP, VirB4 and VirB11. However, the molecular mechanism(s) by which VBP recruits the complex to VirD4 CP remains unknown.
A. tumefaciens is a gram-negative bacteria that causes crown gall disease (tumor formation) in over 140 species of dicots [19].Despite this negative role, researchers have started to exploit the ability of Agrobacterium to infiltrate and infect other organisms to create transgenic plants. Thus, given the importance of this bacterium in both crop infection and as a research tool, we sought to better understand how VBP recruits the protein-DNA complex to VirD4 CP by exploring the structure of VBP.
Here we report the crystal structure of the C-terminal domain of VBP along with its functional studies. We show that the two monomers of the asymmetric unit form a tight anti-parallel dimer, with the dimerization confirmed by solution studies. Moreover, we demonstrate that this C-terminal domain is the dimerization domain of full-length VBP. Pull-down analyses and plant virulence assays showed that only Agrobacterium harboring dimeric VBP can interact with the key proteins to induce tumor formation in plants. These studies broaden our understanding about the role of VBP in the VirD2–T-DNA transfer from A. tumefaciens to the plant cell.
Results
Overall structure
We initially attempted to crystallize full-length VBP with intact N - and C-terminal domains (Figure 1A). Peptide-mass finger printing analysis on the crystals showed that only the C-terminal domain of VBP had crystallized. The boundaries of the crystallized protein were determined using N-terminal sequencing and mass spectrometric analysis. Subsequently, we generated a new construct that consisted of the C-terminal domain, and obtained crystals that diffracted up to 2.7 Å resolution. The structure was determined using the single wavelength anomalous diffraction (SAD) method (Table 1).The asymmetric unit consists of two monomers forming a tight dimer. Each monomer contains a six-helix bundle with three long anti-parallel α helices (α1, α2, and α6) that forms the major part of this domain, and three shorter helices (α3, α4 and α5) that stack at an angle to the long helices (Figure 1B, Figure S1A, S1B). The monomers are arranged as anti-parallel dimers with a large buried surface area of 1203 Å2 (or 14% of the total surface area of each monomer). The dimer is held together by tight interactions between α1 and α2 helices from both monomers (Figure 1C).
Fig. 1. Overall structure. 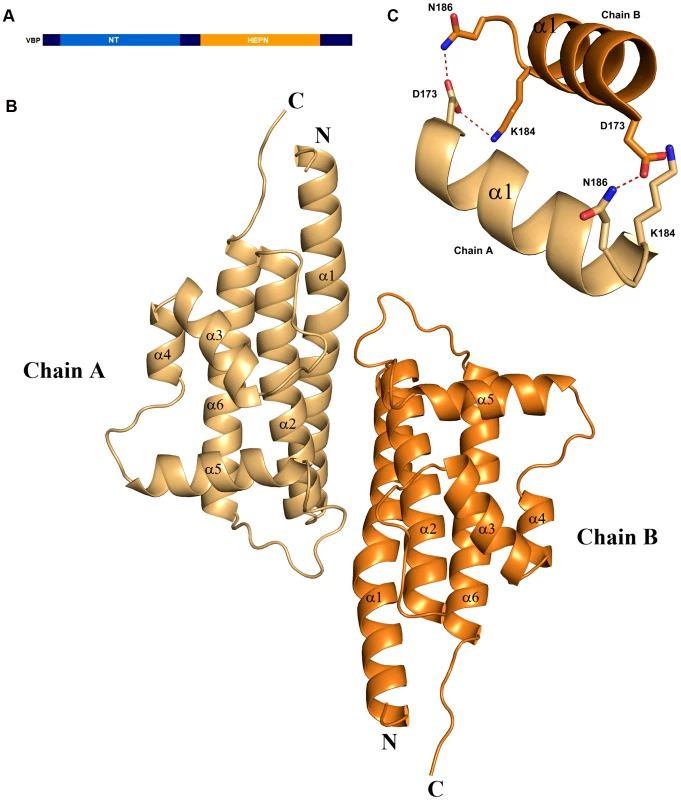
(A) Schematic representation of VirD2-binding protein (VBP) and its domains. Based on the sequence analysis, the N-terminal (aa 10–133) is predicted to be a nucleotidyltransferase (NT) domain and the C-terminal (aa 159–279) is predicted to be higher eukaryotes and prokaryotes nucleotide (HEPN) binding domain. (B) Structure of the HEPN domain of VBP. The asymmetric unit contains two monomers that form a tight dimer. Chain A is shown in light orange and Chain B is shown in deep orange. (C) Dimer interface of HEPN dimer. The contacts at the core of the HEPN dimer are shown. Tab. 1. Crystallographic statistics and refinement details. 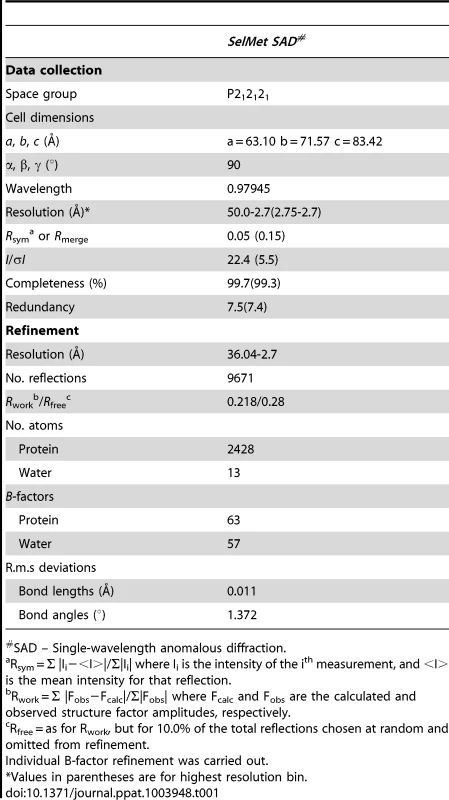
SAD – Single-wavelength anomalous diffraction. Sequence and structural homology of VBP
PSI–BLAST searches of the non-redundant protein database using the full length VBP (gi|159141484) resulted in several hits of nucleotidyltransferase proteins from Rhizobium, Sinorhizobium and other species of Agrobacterium. Sequence-based predictions revealed that VBP has two domains: an NT_KNTase -like domain at the N-terminus and a HEPN (higher eukaryotes and prokaryotes nucleotide-binding) domain at the C-terminus. The NT (nucleotidyltransferase) domain is implicated in nucleotidyl transfer function, whereas the HEPN domain is implicated in nucleotide binding[20]. A DALI [21] search for the structural homologs of VBP C-terminal domain identified several proteins with a HEPN domain (Table S1); this confirmed that the C-terminal domain of VBP adopts a HEPN fold (hereafter referred to as HEPN domain). Notably, the HEPN domain of VBP aligns with the C-terminal domain of kanamycin nucleotidyltransferase from Staphylococcus aureus (PDB code: 1KNY) with an rmsd of 2.9 Å for the 104 Cα atoms. Although VBP has very low sequence identity (14 to 16%) with its structural homologs, it might have similar nucleotide binding and transfer function.
VBP is a dimer
The crystal structure of the HEPN domain shows the presence of a tight dimer in the asymmetric unit. The molecular mass based on the sequence of VBP is 37.5 kDa (including the 6His tag). However, size-exclusion chromatography showed that VBP elutes as a single peak at an elution volume corresponding to an apparent molecular mass of 75 kDa (Figure 2A). Further, the analytical ultracentrifugation (AUC) analysis showed that VBP sediments as a single species corresponding to an apparent molecular mass of dimeric VBP (75 kDa) (Figure 2B). Taken together, these results show that VBP forms a homodimer in solution and likely functions as a dimer in the cell.
Fig. 2. VBP is a dimer. 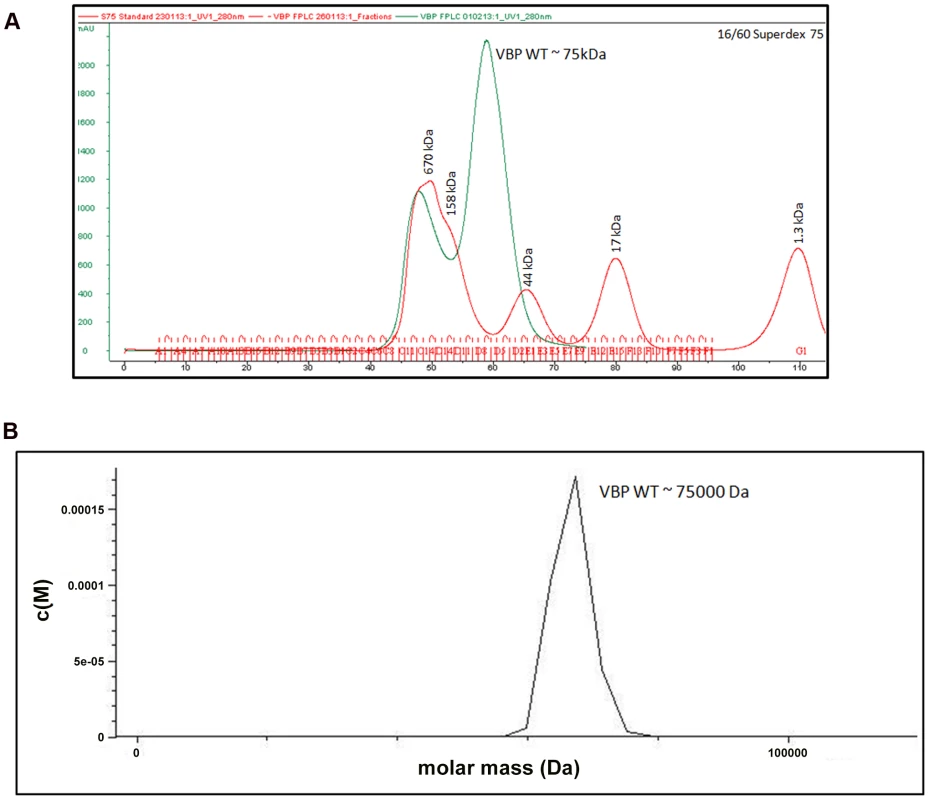
(A) Gel filtration profile of VBP. Full-length VBP elutes as a single peak (in green) at an elution volume corresponding to an apparent molecular mass of 75 kDa. The molecular weight standard is shown in red. The peak at 670 kDa corresponds to aggregated VBP that elutes in the void. (B) Analytical ultra-centrifugation profile of VBP. The full-length VBP sediments as a single species at an apparent molecular mass of 75 kDa. HEPN domain of VBP is the dimerization domain
We examined the role of the HEPN domain in VBP oligomerization by generating individual constructs for the N-terminal (NT) and C-terminal (HEPN) domains of VBP. The size-exclusion chromatography of the purified proteins showed that the NT domain elutes as a single peak at an elution volume corresponding to an apparent molecular mass of 16.8 kDa (NT as monomer), whereas the HEPN domain elutes as a single peak at an elution volume corresponding to an apparent molecular mass of 37 kDa (HEPN as a dimer) (Figure S2). These results were further confirmed using analytical ultracentrifugation experiments (Figure 3A, 3B), and are consistent with the crystal structure findings that show the presence of tight dimeric HEPN domains in the asymmetric unit. Taken together, these results suggest that HEPN domain is responsible for the dimerization of the full-length VBP.
Fig. 3. The HEPN domain of VBP is the dimerization domain. 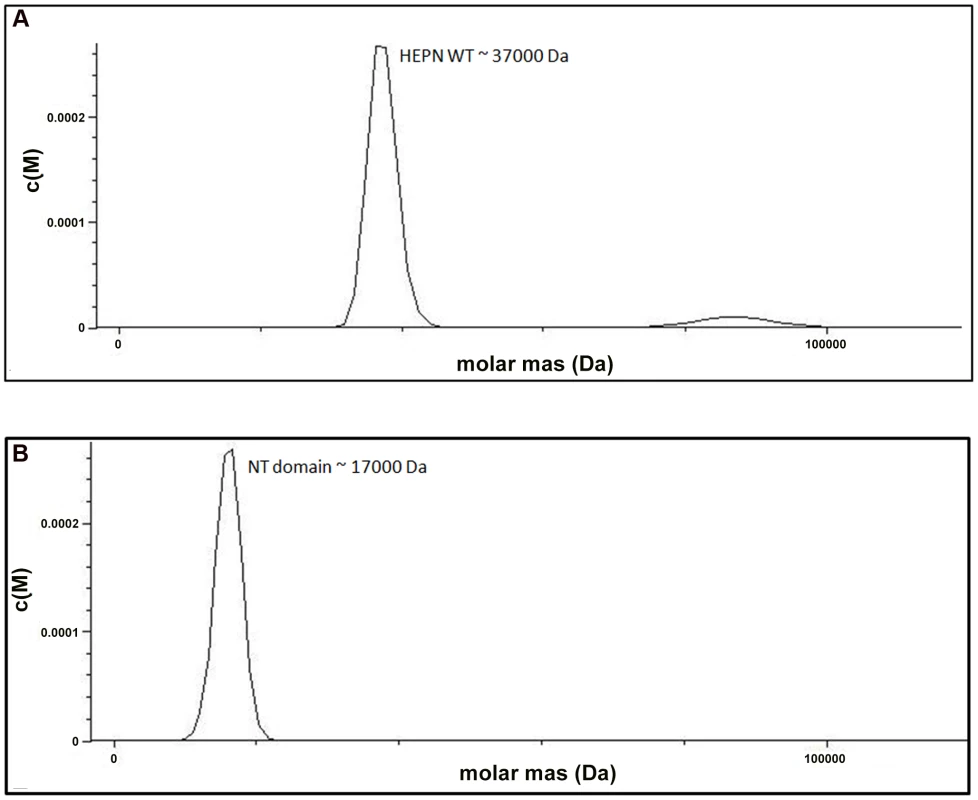
(A) Analytical ultra-centrifugation profile of the HEPN domain of VBP. The HEPN domain sediments as a major species at an apparent molecular mass of 37 kDa. (B) Analytical ultra-centrifugation profile of the NT domain of VBP. The NT domain sediments as a major species at an apparent molecular mass of 17 kDa. Abbreviations: HEPN, higher eukaryotes and prokaryotes nucleotide binding domain; NT, Nucleotidyltransferase domain. Substitution of key residues disrupts the dimerization
The structural analysis indicated that the HEPN dimer is held together by contacts throughout helices α1 and α2; in particular, residues Asp173, Lys184 and Asn186 of the HEPN domain play important roles in maintaining the dimer. Asn186 is located at the edge of a loop connecting the α1 and α2 helices of the dimer interface (Figure 1C). This residue, along with Lys184 and Asp173 of one monomer, is involved in hydrogen bonding contacts with Asp173 and Asn186, Lys184 of the second monomer. We found that a single substitution of Asp173Asn, Lys184Asp or Asn186Asp in the HEPN domain is sufficient to disrupt dimer formation, as verified by size-exclusion chromatography and analytical ultracentrifugation experiments (Figure 4A, Figure S3A, S3B and Figure S4).
Fig. 4. Substitution of key residues disrupts dimerization. 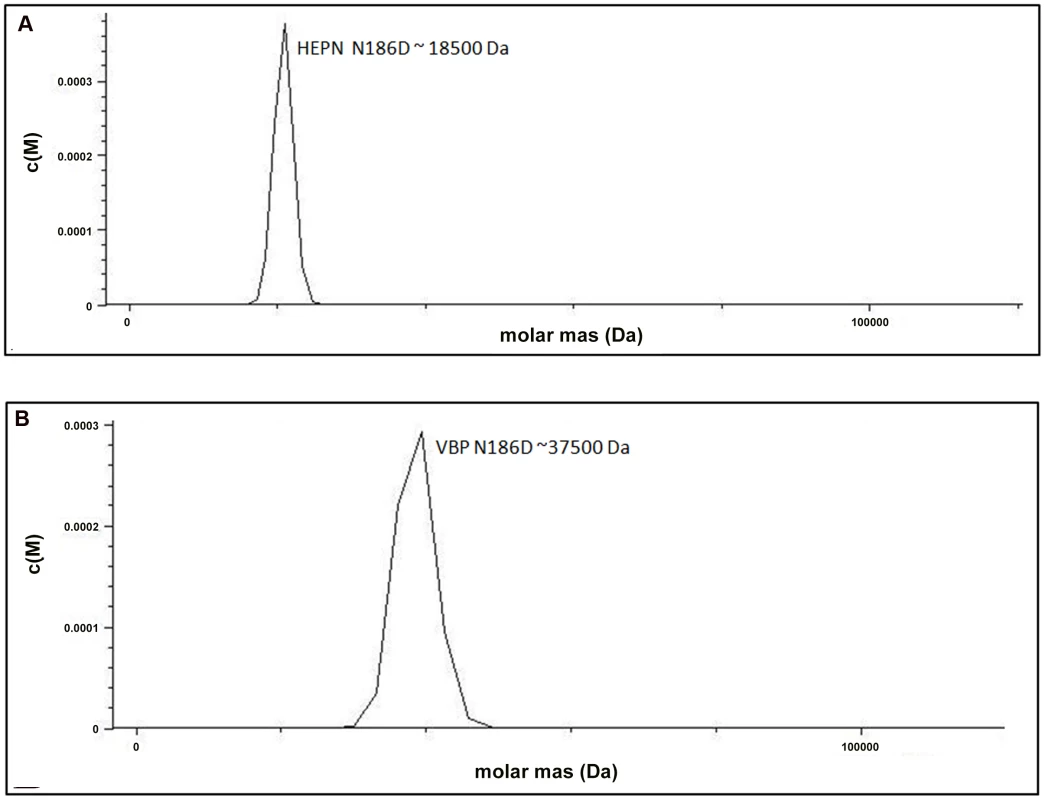
(A) Analytical ultra-centrifugation profile of HEPN Asn186Asp domain of VBP. HEPN domain with Asn186Asp substitution sediments as a single species at an apparent molecular mass of 18.5 kDa. (B) Analytical ultra-centrifugation profile of VBP Asn186Asp. VBP with Asn186Asp substitution sediments as a single species at an apparent molecular mass of 37.5 kDa. Abbreviations: HEPN, higher eukaryotes and prokaryotes nucleotide (domain). Next, we verified the role of these key residues using full-length VBP. Single point substitutions of Asp173Asn, Lys184Asp or Asn186Asp in full-length VBP caused the dimer to break and the protein to elute as a single peak at an elution volume corresponding to an apparent molecular mass of 37.5 kDa (monomeric VBP). Further AUC analyses of full-length VBP bearing one of these point substitutions showed that the protein sediments as a single species at an apparent molecular mass corresponding to monomeric VBP (37.5 kDa) (Figure 4B, Figure S5A, S5B and Figure S6). The circular dichroism (CD) spectrum of the wild-type VBP, mutated VBP and HEPN domain suggested that these mutants have the same structural fold as the wild-type proteins (Figures S7A, S7B). These results indicate that the hydrogen bonds at the dimeric interface play a key role in maintaining an intact dimeric HEPN domain and reiterate the involvement of the HEPN domain in VBP dimerization.
VBP binds to VirD2 and VirD4 CP as a dimer
VBP is the key recruiting protein that binds to the VirD2–T-DNA complex and brings it into contact with VirD4 CP for subsequent translocation to the host cell [8]. Thus, we sought to verify the binding property of VBP with VirD2 and VirD4 CP using pull-down assays. Using an in vitro pull-down assay with recombinant proteins (6His-VBP and MBP-VirD2), we showed that wild-type VBP binds to VirD2, whereas VBP with one of these aforementioned single point substitutions—Asp173Asn, Lys184Asp or Asn186Asp (which results in monomeric VBP)—does not bind VirD2 (Figure 5A). To confirm these results, we used His-VBP and substituted His-VBP (with (Asp173Asn/Lys184Asp/Asn186Asp mutations) as bait and pulled down the VBP-interacting proteins from a vir gene-induced Agrobacterium null mutant that lacks VBP: GMV123 (A triple vbp null mutant strain GMI9017Δvbp2Δvbp3 (for which all three existing vbp genes were knocked out)). WT His-VBP was able to pull down VirD2 and VirD4 CP, whereas substituted VBP could not pull down either of these proteins. These results indicate that the dimeric nature of VBP is important for its interaction with VirD2 and VirD4 CP (Figure 5B).
Fig. 5. Pull down assays. 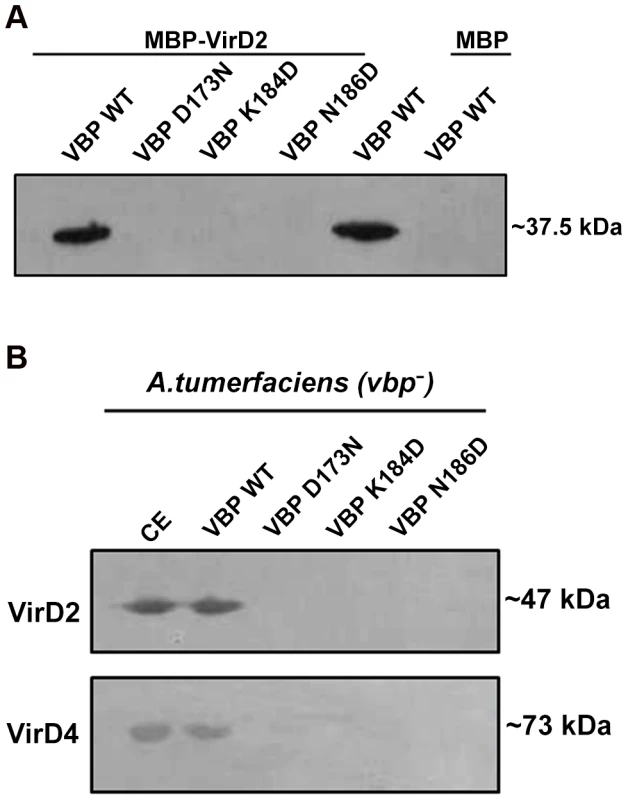
(A) MBP/MBP-VirD2 bound to amylose resin was incubated overnight with 6His-VBP, followed by washes. The final beads were resolved on a 12.5% SDS gel, transferred to a PVDF membrane and treated with anti-His monoclonal antibody (1∶10,000). 6His-VBP was loaded into the lane 5 as a reference. The VBP species used include: lane 1. Wild-type (WT) VBP; lane 2.VBP D173N; lane 3.VBP K184D; lane 4. VBP N186D; lane 5. VBP wild-type loaded on to the gel for reference; .lane 6. VBP passed through MBP bound to amylose beads. (B) 6His-VBP/substituted 6His-VBP bound to Ni-NTA metal affinity resin was incubated with freshly prepared A. tumefaciens crude extracts. After incubation at 4°C for 1 h, the resin was washed four times. The bound complex was eluted with 250 mM imidazole. The eluted protein was resolved on SDS-Gel, transferred to a PVDF membrane and the protein was detected using protein (VirD2 and VirD4 CP) specific monoclonal antibodies. The VBP species used include: lane 1. Crude extract loaded for reference; lane 2. WT VBP; lane 3.VBP D173N; lane 4.VBP K184D; lane 5. VBP N186D. VBP functions as a dimer in vivo
The observed dimeric nature of VBP in solution and in the crystal structure prompted us to verify the functional state of VBP inside the cell using a plant virulence assay. GMV123 (vbp KO strain of A. tumefaciens) was complemented with pQH300 plasmid harboring substituted constructs for a functional vbp gene, a gene expressing the HEPN domain, or a gene expressing the NT domain. We found that null mutants transformed with a plasmid expressing VBP were able to cause a tumor-like phenotype in the wild-type plants (Figure 6A and 6B and Figure S8). Strains expressing a substituted VBP (Asp173Asn/Lys184Asp/Asn186Asp) or one of the other deletion mutants (pQH-NTD or pQH-HEPN) did not cause tumors (Figure 6A and 6B and Figure S8). These results indicate that VBP functions as a dimer in the cells and that full-length VBP is required for tumor formation in plants.
Fig. 6. VBP is a functional dimer. 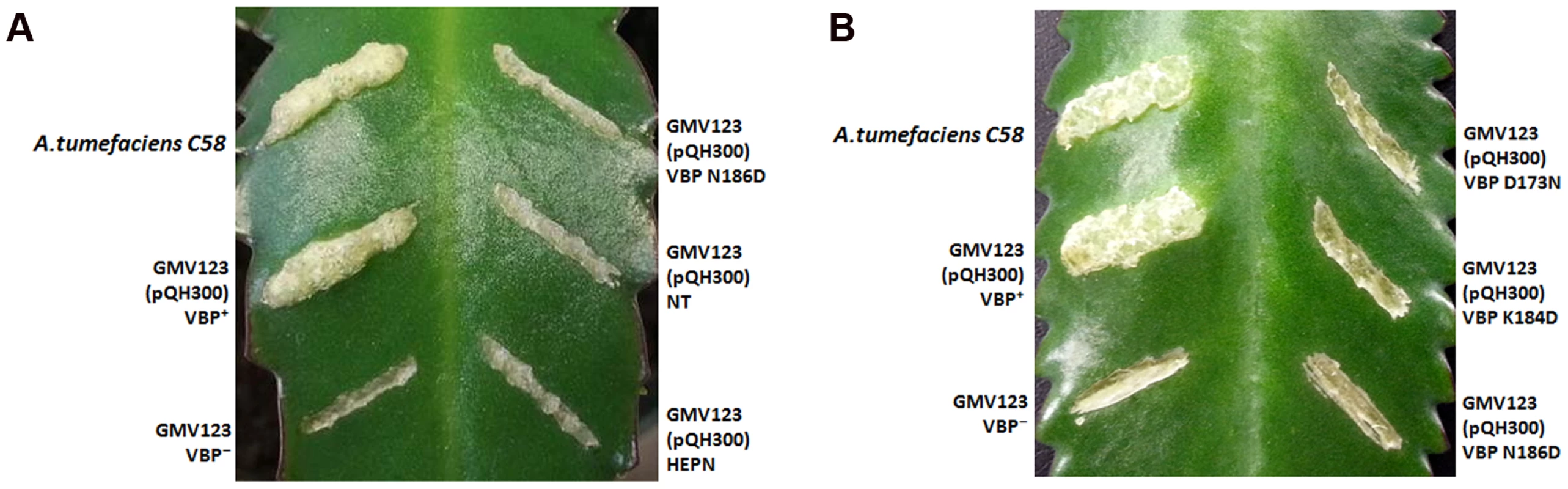
The effect of VBP mutations on tumorigenesis. A. tumefaciens strains were grown in MG/L medium at 28°C overnight. The cell density was adjusted to 108cells/ml. This cell suspension (5 µl) was inoculated into each wound site on the leaves of Kalanchoe plants. The tumors were photographed at 25 days (A) The wounds on the Kalanchoe leaf were inoculated with A.tumefaciens WT strain or GMV123 strain complemented with plasmid expressing VBP WT, VBP N186D, NT domain or HEPN domain. Only wounds inoculated with A.tumefaciens WT strain or GMV123 strain complemented with plasmid expressing VBP WT showed tumor, other wounds showed no tumor clearly indicating that only Agrobacterium harboring full length VBP can induce tumor. (B) The wounds on the Kalanchoe leaf were inoculated with A.tumefaciens WT strain or GMV123 strain complemented with plasmid expressing VBP WT, VBP N186D, VBP D173N or K184D. Only wounds inoculated with A.tumefaciens WT strain or GMV123 strain complemented with plasmid expressing VBP WT showed tumor, other wounds showed no tumor clearly indicating that only Agrobacterium harboring full length dimeric VBP can induce tumor. The particular mutants are labeled at the respective scars. Interaction of VBP with ATP
Proteins homologous to VBP are predicted to bind ATP [22]. A previously reported structure of kanamycin nucleotidyltransferase (PDB code: 1KNY), a structural homolog of VBP, in complex with an ATP analog and kanamycin, shows that the nucleotide binding pocket involves residues from the N-terminal domain of one monomer and the C-terminal domain of the second monomer. We verified the ATP binding property of VBP using isothermal titration calorimetry (ITC) (Figure 7A and Table S2). The ITC analysis showed that VBP binds to the ATP analog (AMPPNP) (Kd = 2.0 µM), whereas VBP with the three point substitutions described above does not bind to AMPPNP (Figure 7B, Figure S9A and S9B), indicating that only dimeric VBP binds to ATP. Further, using ITC experiments, we sought to verify whether the HEPN domain alone can bind nucleotides (Figure 7C). Our results indicated that the HEPN domain alone cannot bind nucleotides. We infer that, similar to the kanamycin nucleotidyltransferase, the ATP binding in VBP might involve both N-terminal and C-terminal domains. Notably, the structure of the kanamycin nucleotidyltransferase (PDB code: 1KNY) complexed with a nucleotide analog and kanamycin shows that the two monomers of the dimer interact in an anti-parallel fashion to form the ATP binding pocket. Similarly, the structure of the HEPN domain from VBP shows that the two HEPN monomers form a tight dimer in which the monomers run in anti-parallel. Although the relative orientation of monomers in the dimers of both proteins is not the same, both proteins might have a similar ATP-binding mechanism.
Fig. 7. Interaction of VBP with AMPPNP (ATP analog) by isothermal titration calorimetry (ITC). 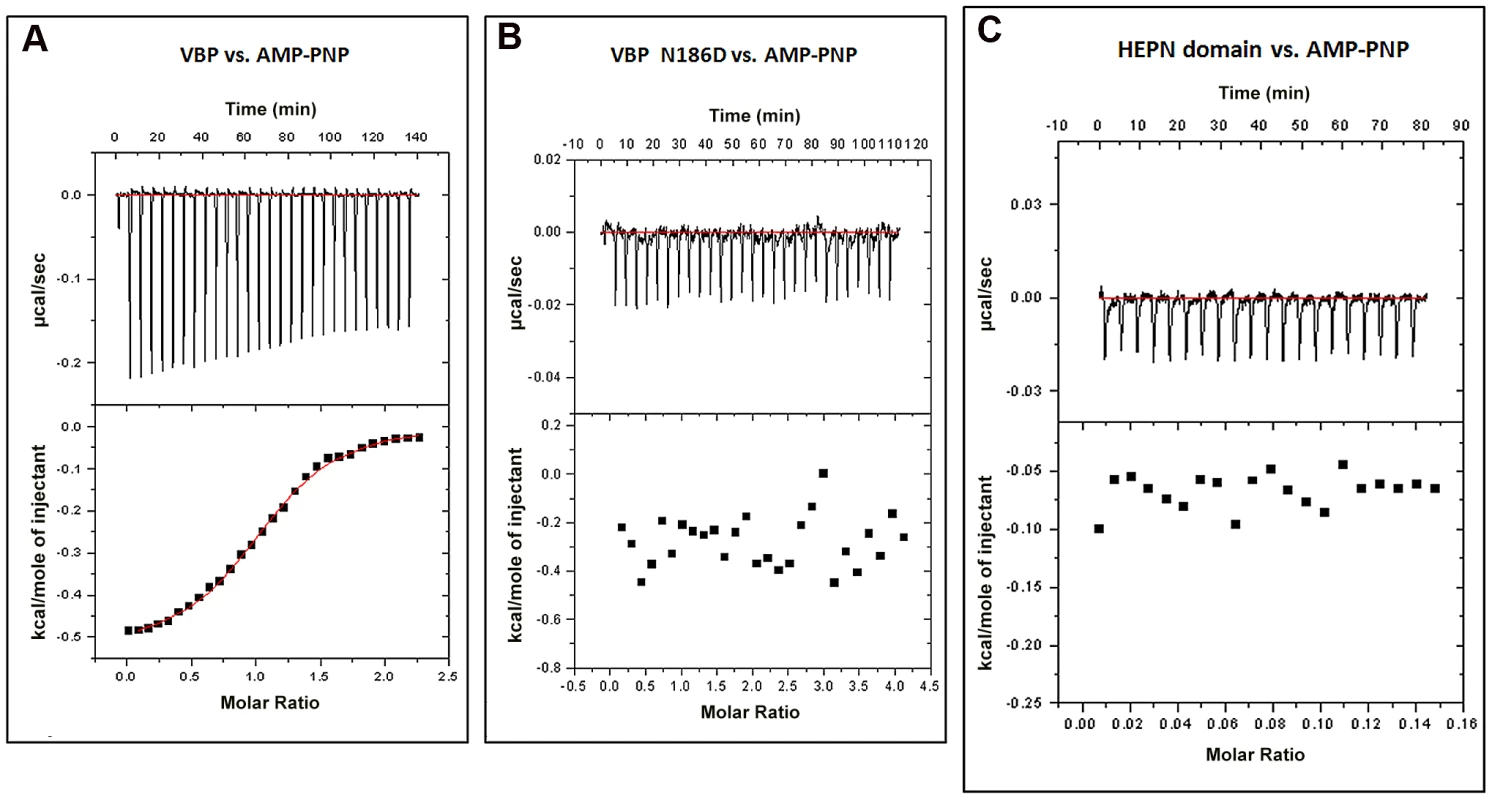
Representative ITC assays are shown. The upper part of each panel shows the thermogram (thermal power vs. time) after baseline correction and the bottom part of each panel shows the binding isotherm (normalized heat vs. molar ratio of reactants). (A) Calorimetric titration for VBP. (B) Calorimetric titration for VBP N186D. (C) Calorimetric titration for HEPN. Discussion
A. tumefaciens affects more than 140 species of dicots [19], instigating infection through the efficient translocation of the VirD2–T-DNA complex [2], a prerequisite for the integration of T-DNA into the plant genome and eventual tumor formation in plants [23]. The T-DNA is a segment of the Ti plasmid that encodes most of the proteins that are involved in VirD2–T-DNA complex translocation and T-DNA integration into the host genome, with each protein catering to a particular stage of the translocation process. The VirD4 CP is known to couple the VirD2–T-DNA complex mediated by VBP as a recruiting complex (VBP: VirD2–T-DNA) to the T4SS secretion apparatus. VBP is thus a key component of the recruiting complex [8].
Based on the sequence analysis, it has been predicted that the NT domain of VBP belongs to the DNA polymerase β superfamily of proteins [21]. This superfamily includes nucleotidyltransferases that catalyze nucleotidylation of proteins in yet unidentified pathways [21]. Similarly the C-terminal of VBP is predicted to have the HEPN domain [24]. VBP interaction with VirD2, and several other energizing components of the T4SS (VirD4, VirB4 and VirB11) [13], makes it difficult to predict the exact nucleotidylation site of the protein.
In this study, we sought to analyze the structural and functional aspects of the C-terminal domain of VBP. We show that this domain adopts a HEPN fold and facilitates the dimerization of VBP, forming tight anti-parallel dimers. The structural similarity observed between the HEPN domain of VBP and kanamycin nucleotidyltransferase, as well as the results from our ITC experiments, suggest that a nucleotide binding pocket is formed by the dimeric interface in this protein. Furthermore, our pull-down assays show that only dimeric VBP can bind VirD2 and VirD4 CP and that dimerization of VBP is essential for A. tumefaciens-induced tumor formation in plants (Figures 5, 6 and 8).
Fig. 8. Schematic representation shows the induction of tumors in plants by Agrobacterium tumefaciens and the role of VBP. 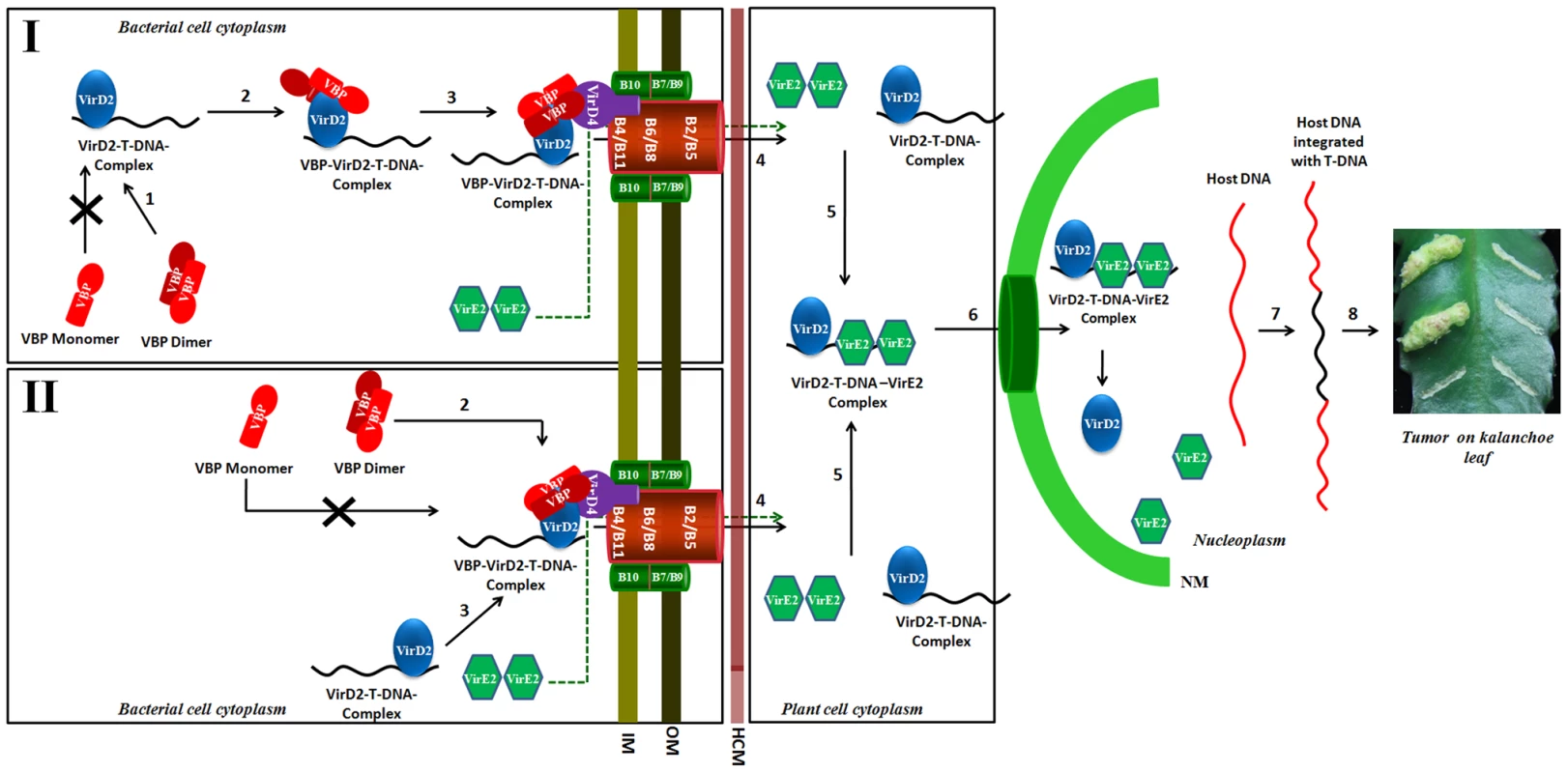
Our experiments show that only dimeric VBP can bind to VirD2 or VirD4 CP. We propose that the VirD2–T-DNA complex could possibly be recruited to the T4SS apparatus by two different mechanisms. Mechanism I: Dimeric VBP binds to VirD2 (steps 1 and 2), it recruits the VirD2–T-DNA complex to the (VirD4 CP) T4SS apparatus which constitutes the 11 VirB proteins (step 3). Mechanism II: Dimeric VBP binds to VirD4 CP (step 2). VBP acts as a docking station to recruit the VirD2–T-DNA complex (step 3). Once recruited to the T4SS apparatus, the VirD2–T-DNA complex is translocated into the host cell cytoplasm (step 4). It is yet unclear whether VBP is translocated or not [8]. VirE2 is one among the several effector proteins that are translocated through the T4SS. Inside the host cell, VirE2 coats the single-stranded T-DNA to form the VirD2–T-DNA-VirE2 complex (step 5). Certain host cytoplasmic proteins recognize and bind to the nuclear localization signals on VirD2 and VirE2 and translocate the VirD2–T-DNA:VirE2 complex to the nucleus (step 6) [35]. Inside the nucleus, VirD2 and VirE2, along with a plethora of host proteins, help the T-DNA to integrate with the host DNA (step 7) [36]. The integrated T-DNA modulates the host cell process to enable the bacterial colonization and growth, which leads to the formation of tumor (step 8). IM: Inner membrane; OM: Outer membrane; HCM: Host cell membrane; NM: Nuclear membrane. Previously, we showed that VBP has independent interactions with VirD4 CP, VirB4 and VirB11 ATPases [8]. VirD4 CP has a prominent cytoplasmic domain, besides its membrane embedded domain [1], whereas VirB4 and VirB11 have both membrane and the cytoplasmic regions [25]. Our previous studies have shown that VBP is a cytoplasmic protein that localizes at the poles only in the presence of T4SS [8]. This is probably because of the interaction between VBP and the cytoplasmic regions of the T4SS proteins, particularly VirD4 CP, VirB4 and VirB11. Furthermore, the polar localization of VBP does not depend on the presence of VirD2 [8].
Although VirD2 is a cytoplasmic protein, it localizes to the poles in the presence of VBP and T4SS apparatus, but remains in the cytoplasm in the absence of VBP, irrespective of the presence of T4SS apparatus [8]. This indicates that when T4SS is present, VBP binds to the cytoplasmic region of VirD4 CP either independently or as a complex with VirD2, when it is available. In light of this finding, we propose that either VBP binds to the VirD2–T-DNA complex and recruits it to the VirD4 CP, or VBP initially binds to VirD4 CP proteins at the cytoplasmic region and serves as a docking station for the VirD2–T-DNA complex (Figure 8). Using a transfer DNA immunoprecipitation (TrIP) assay, Cascales et al. elegantly showed that T-DNA recruited to VirD4 CP is transferred to VirB6 through VirB11 [7] and that VirB4 and VirB11 ATPases interact to drive the export of T-DNA [7].
The present study sheds light on the role of VBP in the VirD2–T-DNA complex translocation in VirB/D4 T4SS of A. tumefaciens and other similar bacterial system that use the Type IV secretion conjugation systems.
Methods
Plasmid and strain construction
The strains and plasmids used are given in Table S3. Intact vbp and vird2 genes were amplified from A. tumefaciens C58 plasmid and Ti plasmid, respectively. These genes were then cloned to pET32a (Novagen; Madison, WI, USA) and pRSET (Invitrogen, Carlsbad, CA, USA) vectors, respectively. N-terminal nucleotidyltransferase (NT) domain and C-terminal HEPN domain constructs were created using specific primers that amplify these regions and were cloned into pGEX-6p1 (GE Healthcare; Buckinghamshire, UK). Site-specific mutations in vbp were introduced by overlapping PCR, as described previously [26]. Each construct was verified by DNA sequencing. A fragment of virF cassette cloned from pTiBo542 (GenBank: DQ058764.1) was inserted into the SphI–ApaI site on pCB301 [27]. The virF coding sequence was substituted with a multiple cloning site, resulting in pQH300.
Protein expression and purification
The plasmid pET32a-vbp was transformed into E. coli BL21 (DE3) cells and was grown in LB broth at 37°C overnight. The overnight culture was transferred into 1 L of LB broth and the protein expression was induced at an absorbance of 0.6 with 350 µM IPTG for 20 h at 20°C.Cells were harvested and lysed in lysis buffer (20 mM Tris-HCl, pH 8.0, 200 mM NaCl and 1 mM PMSF). Cell lysates were centrifuged and the supernatants transferred to affinity columns containing Ni-NTA agarose (Qiagen; Valencia, CA, USA), pre-equilibrated with the lysis buffer. The 6His-VBP bound to Ni-NTA was eluted with 400 mM imidazole following three wash steps to remove non-specific proteins. The eluted protein was purified through size-exclusion chromatography using a HiLoad 16/12075 Superdex75 gel filtration column (AKTA FPLC UPC-900 system, GE Healthcare) containing buffer (20 mMTris-HCl, pH 8.0, 200 mM NaCl, and 5% glycerol). The GST fusion proteins (GST-HEPN and GST-NTD) were expressed as described above using M9 media [28]. The fusion proteins were purified by affinity chromatography on GST-Sepharose resin, and the tags were removed by cleavage with PreScission proteases (GE Healthcare; Buckinghamshire, UK). The HEPN domain was additionally purified by size-exclusion chromatography in gel-filtration buffer (30 mM CHES pH 9.0, NaCl 200 mM, 5% glycerol). The NT domain was purified in the same way but using a buffer containing 30 mM Tris-HCl, pH 7.5 and 200 mM NaCl buffer.
Crystallization and data collection
Initial crystallization conditions were identified by hanging drop vapor diffusion method using an index screen (Hampton Research, Aliso Viejo, CA, USA). Diffraction-quality crystals were obtained by equilibrating l.0 µl drop of protein (4 mg/ml) in 30 mM CHES, pH 9.0, 200 mM NaCl and 5% glycerol mixed with 1.0 µl of reservoir solution (8% (w/v) PEG 3350, 2% v/v tacsimate, 5% v/v 2-proponal, and 0.1 M imidazole) suspended over 1 ml of reservoir solution. Crystals grew in 1–3 days at 16°C. For data collection, 15% glycerol was added as a cryo-protectant and the crystals were flash-cooled in an N2 cold stream.
A complete single wavelength anomalous diffraction (SAD) [29] dataset was collected to 2.7 Å resolution at the synchrotron beamline X6A (National Synchrotron Light Source, Brookhaven National Laboratory, Upton, NY) using a Quantum4-CCD detector (Area Detector Systems Corp., Poway, CA). The datasets were processed and scaled using HKL2000 [30]. The crystals belonged to a P212121 space group. There were two monomers in the asymmetric unit corresponding to Vm = 2.49 Å3 Da−1 with a solvent content of 50.6%. The position of the selenium atoms were determined using the program Phenix-Autosol [31]. The obtained phases were further improved by density modification using RESOLVE [31]. Over 50% of the backbone atoms of the model were built by RESOLVE. The remaining residues were manually built using Coot [32] and subsequently refined using Refmac [33]. Refinement was continued until the R-value converged to 0.22 (Rfree = 0.28) for reflections I>σ (I) to 2.7 Å resolution (Table 1). The model had good stereochemistry, with 99.3% residues within the allowed regions of the Ramachandran plot. Subsequently, the importance of the key residues at the dimeric interface was validated by structure-based in vitro studies, such as analytical ultracentrifugation and pull down assays, and in vivo plant virulence studies. Coordinates of HEPN domain of VBP have been deposited in the Protein Data Bank (http://www.pdb.org) under accession code 4NQF.
Analytical ultracentrifugation
The oligomeric state of full-length VBP, HEPN and NTD domain of VBP and their mutants was investigated by monitoring the sedimentation properties of each protein in sedimentation velocity experiments. For these experiments, 400 µl of samples at 1 mg/ml in appropriate buffer were used, with the experiments carried out in duplicate. The sedimentation velocity profiles were collected by monitoring the absorbance at 280 nm. The samples were centrifuged at 40,000 rpm at 24°C in a Beckman Optima XL-I centrifuge fitted with a four-hole AN-60 rotor and double-sector aluminum centerpieces and equipped with absorbance optics. Eighty scans were collected and analyzed using the Sedfit program [34].
Pull down assay
MBP-VirD2 bound to amylose resin (New England Biolabs, Ipswich, MA, USA) was incubated with purified 6His-VBP, with or without substitution at Asp173Asn/Lys184Asp/Asn186Asp. The beads were washed several times before resolving on 12.5% SDS-PAGE. For western blot analysis, the proteins were transferred to a PVDF membrane. 6His-VBP was detected by the addition of diluted anti-His antibody (Santa Cruz Biotechnology; Santa Cruz, CA, USA). The signal was detected using the Super Signal WestPico Chemiluminescent substrate (Pierce Biotechnology; Rockford, IL, USA) under the conditions recommended by the manufacturer. For pull down assay from A. tumefaciens crude extracts, E. coli strain BL21 was used to produce 6His-VBP/substituted 6His-VBP as described above.6His-VBP/substituted 6His-VBP bound to Ni-NTA metal affinity resin was incubated with freshly prepared A. tumefaciens (vbp KO strain) crude extracts (2.5 mM MgCl2, 50 mM NaCl, 2 mM PMSF, 50 mM Tris-HCl, pH 7.4, 0.5% Triton X-100). After incubation at 4°C for 1 h, the resin was washed four times. The bound proteins were eluted with 250 mM imidazole. The eluted proteins were resolved on 12.5% SDS-PAGE. For western blot analysis, the proteins were transferred to a PVDF membrane. VirD2 and VirD4 CP in the eluent were detected by western blot using anti-VirD2 (1∶5000) and anti-VirD4 (1∶4000) antibodies. The signal was detected using the Super Signal WestPico Chemiluminescent substrate (Pierce Biotechnology; Rockford, IL, USA) under the conditions recommended by the manufacturer.
Isothermal titration calorimetry
6His-VBP, with or without substitution of key residues, were purified in gel filtration buffer containing 30 mM Tris-HCl, pH 8.0 and 200 mM NaCl. ITC experiments were carried out using a VP-ITC calorimeter (MicroCal, LLC, Northampton, MA, USA) at 25°C using 0.01 mM VBP protein in the sample cell and 0.25 mM AMP-PNP in the injecting syringe. All samples were thoroughly degassed and then centrifuged to remove precipitates. With the exception of the first injection, 10 µl volumes per injection were used for different experiments. Consecutive injections were separated by 5 min to allow the peak to return to baseline levels. ITC data were analyzed with a single-site binding model using Origin 7.0 (Origin Lab Corp., Northampton, MA, USA) software.
Circular dichroism spectrometry
Far UV spectra (260–190 nm) of VBP, HEPN, NTD domains and their mutants were measured using a Jasco J-810 spectropolarimeter (Jasco Europe, MI, Italy) in phosphate buffer (pH 7.5) at room temperature using a 0.1-cm path length-stoppered cuvette. Six scans were recorded, averaged and then baseline-subtracted.
Plant virulence assay
A. tumefaciens strains were grown in MG/L liquid medium overnight at 28°C supplemented with appropriate antibiotics. The bacterial cells were collected by centrifugation and re-suspended in a buffer solution consisting of 10 mM MgCl2 and 10 mM MES, pH 5.5. Cell concentrations were adjusted to OD600 = 0.1. The leaves of Kalanchoe plants were wounded with a hypodermic needle and 5 µl of bacterial cell suspension was inoculated onto each wound area. The tumors were photographed at different time points after inoculation.
Supporting Information
Zdroje
1. CascalesE, ChristiePJ (2003) The versatile bacterial type IV secretion systems. Nat Rev Microbiol 1 : 137–149.
2. Alvarez-MartinezCE, ChristiePJ (2009) Biological diversity of prokaryotic type IV secretion systems. Microbiol Mol Biol Rev 73 : 775–808.
3. LesslM, LankaE (1994) Common mechanisms in bacterial conjugation and Ti-mediated T-DNA transfer to plant cells. Cell 77 : 321–324.
4. ChristiePJ, AtmakuriK, KrishnamoorthyV, JakubowskiS, CascalesE (2005) Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu Rev Microbiol 59 : 451–485.
5. TzfiraT, CitovskyV (2000) From host recognition to T-DNA integration: the function of bacterial and plant genes in the Agrobacterium-plant cell interaction. Mol Plant Pathol 1 : 201–212.
6. AtmakuriK, CascalesE, BurtonOT, BantaLM, ChristiePJ (2007) Agrobacterium ParA/MinD-like VirC1 spatially coordinates early conjugative DNA transfer reactions. EMBO J 26 : 2540–2551.
7. CascalesE, ChristiePJ (2004) Definition of a bacterial type IV secretion pathway for a DNA substrate. Science 304 : 1170–1173.
8. GuoM, JinS, SunD, HewCL, PanSQ (2007) Recruitment of conjugative DNA transfer substrate to Agrobacterium type IV secretion apparatus. Proc Natl Acad Sci U S A 104 : 20019–20024.
9. ChristiePJ (1997) Agrobacterium tumefaciens T-complex transport apparatus: a paradigm for a new family of multifunctional transporters in eubacteria. J Bacteriol 179 : 3085–3094.
10. OkamotoS, Toyoda-YamamotoA, ItoK, TakebeI, MachidaY (1991) Localization and orientation of the VirD4 protein of Agrobacterium tumefaciens in the cell membrane. Mol Gen Genet 228 : 24–32.
11. HamiltonCM, LeeH, LiPL, CookDM, PiperKR, et al. (2000) TraG from RP4 and TraG and VirD4 from Ti plasmids confer relaxosome specificity to the conjugal transfer system of pTiC58. J Bacteriol 182 : 1541–1548.
12. KumarRB, DasA (2002) Polar location and functional domains of the Agrobacterium tumefaciens DNA transfer protein VirD4. Mol Microbiol 43 : 1523–1532.
13. VeluthambiK, ReamW, GelvinSB (1988) Virulence genes, borders, and overdrive generate single-stranded T-DNA molecules from the A6 Ti plasmid of Agrobacterium tumefaciens. J Bacteriol 170 : 1523–1532.
14. AtmakuriK, DingZ, ChristiePJ (2003) VirE2, a type IV secretion substrate, interacts with the VirD4 transfer protein at cell poles of Agrobacterium tumefaciens. Mol Microbiol 49 : 1699–1713.
15. CitovskyV, GDEV, ZambryskiP (1988) Single-Stranded DNA Binding Protein Encoded by the virE Locus of Agrobacterium tumefaciens. Science 240 : 501–504.
16. TzfiraT, CitovskyV (2002) Partners-in-infection: host proteins involved in the transformation of plant cells by Agrobacterium. Trends Cell Biol 12 : 121–129.
17. TzfiraT, VaidyaM, CitovskyV (2004) Involvement of targeted proteolysis in plant genetic transformation by Agrobacterium. Nature 431 : 87–92.
18. GuoM, HouQ, HewCL, PanSQ (2007) Agrobacterium VirD2-binding protein is involved in tumorigenesis and redundantly encoded in conjugative transfer gene clusters. Mol Plant Microbe Interact 20 : 1201–1212.
19. MooreLW, ChiltonWS, CanfieldML (1997) Diversity of opines and opine-catabolizing bacteria isolated from naturally occurring crown gall tumors. Appl Environ Microbiol 63 : 201–207.
20. AltschulSF, MaddenTL, SchafferAA, ZhangJ, ZhangZ, et al. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25 : 3389–3402.
21. HolmL, RosenstromP (2010) Dali server: conservation mapping in 3D. Nucleic Acids Res 38: W545–549.
22. KozlovG, DenisovAY, GirardM, DicaireMJ, HamlinJ, et al. (2011) Structural basis of defects in the sacsin HEPN domain responsible for autosomal recessive spastic ataxia of Charlevoix-Saguenay (ARSACS). J Biol Chem 286 : 20407–20412.
23. PitzschkeA, HirtH (2010) New insights into an old story: Agrobacterium-induced tumour formation in plants by plant transformation. EMBO J 29 : 1021–1032.
24. GrynbergM, ErlandsenH, GodzikA (2003) HEPN: a common domain in bacterial drug resistance and human neurodegenerative proteins. Trends Biochem Sci 28 : 224–226.
25. DangTA, ZhouXR, GrafB, ChristiePJ (1999) Dimerization of the Agrobacterium tumefaciens VirB4 ATPase and the effect of ATP-binding cassette mutations on the assembly and function of the T-DNA transporter. Mol Microbiol 32 : 1239–1253.
26. HoSN, HuntHD, HortonRM, PullenJK, PeaseLR (1989) Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77 : 51–59.
27. OliverDJ, XiangCB, HanP, LutzigerI, WangK (1999) A mini binary vector series for plant transformation. Plant molecular biology 40 : 711–717.
28. DoubliéS (1997) Preparation of selenomethionyl proteins for phase determination. Methods Enzymol 276 : 523–530.
29. TerwilligerTC, BerendzenJ (1997) Bayesian correlated MAD phasing. Acta Crystallogr D Biol Crystallogr 53 : 571–579.
30. Otwinowski Z, Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. In: Carter Jr, CW, editor. Methods in enzymology. London: Elsevier Academic press. pp. 307–326.
31. TerwilligerT (2003) SOLVE and RESOLVE: automated structure solution and density modification. Methods Enzymol 374 : 22–37.
32. EmsleyP, CowtanK (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60 : 2126–2132.
33. MurshudovGN, SkubakP, LebedevAA, PannuNS, SteinerRA, et al. (2011) REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr 67 : 355–367.
34. BrownPH, SchuckP (2006) Macromolecular size-and-shape distributions by sedimentation velocity analytical ultracentrifugation. Biophys J 90 : 4651–4661.
35. ZupanJR, ZambryskiP (1995) Transfer of T-DNA from Agrobacterium to the plant cell. Plant Physiol 107 : 1041–1047.
36. CitovskyV, KozlovskySV, LacroixB, ZaltsmanA, Dafny-YelinM, et al. (2007) Biological systems of the host cell involved in Agrobacterium infection. Cell Microbiol 9 : 9–20.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek DHX36 Enhances RIG-I Signaling by Facilitating PKR-Mediated Antiviral Stress Granule FormationČlánek Oral Bacteria and CancerČlánek A Non-Coding RNA Promotes Bacterial Persistence and Decreases Virulence by Regulating a Regulator in
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2014 Číslo 3- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Conflicting Interests in the Pathogen–Host Tug of War: Fungal Micronutrient Scavenging Versus Mammalian Nutritional Immunity
- Putting Fungi to Work: Harvesting a Cornucopia of Drugs, Toxins, and Antibiotics
- Mycobacteriophages: Windows into Tuberculosis
- Human African Trypanosomiasis and Immunological Memory: Effect on Phenotypic Lymphocyte Profiles and Humoral Immunity
- Five Things to Know about Genetically Modified (GM) Insects for Vector Control
- A Missing Dimension in Measures of Vaccination Impacts
- Eosinophils Are Important for Protection, Immunoregulation and Pathology during Infection with Nematode Microfilariae
- Clonality of HTLV-2 in Natural Infection
- Production, Fate and Pathogenicity of Plasma Microparticles in Murine Cerebral Malaria
- Group B Streptococcal Infection of the Choriodecidua Induces Dysfunction of the Cytokeratin Network in Amniotic Epithelium: A Pathway to Membrane Weakening
- New Insights into How Adapts to Its Mammalian Host during Bubonic Plague
- Foodborne Transmission of Nipah Virus in Syrian Hamsters
- A Polysaccharide Virulence Factor from Elicits Anti-inflammatory Effects through Induction of Interleukin-1 Receptor Antagonist
- Structural and Functional Characterization of a Complex between the Acidic Transactivation Domain of EBNA2 and the Tfb1/p62 Subunit of TFIIH
- Adaptive Gene Amplification As an Intermediate Step in the Expansion of Virus Host Range
- DHX36 Enhances RIG-I Signaling by Facilitating PKR-Mediated Antiviral Stress Granule Formation
- Hepatitis B Virus Infection and Immunopathogenesis in a Humanized Mouse Model: Induction of Human-Specific Liver Fibrosis and M2-Like Macrophages
- Crk Adaptors Negatively Regulate Actin Polymerization in Pedestals Formed by Enteropathogenic (EPEC) by Binding to Tir Effector
- Fatty Acid Biosynthesis Contributes Significantly to Establishment of a Bioenergetically Favorable Environment for Vaccinia Virus Infection
- A Cytosolic Chaperone Complexes with Dynamic Membrane J-Proteins and Mobilizes a Nonenveloped Virus out of the Endoplasmic Reticulum
- Intracellular Promote Invasive Cell Motility through Kinase Regulation of the Host Actin Cytoskeleton
- MAVS-MKK7-JNK2 Defines a Novel Apoptotic Signaling Pathway during Viral Infection
- RON5 Is Critical for Organization and Function of the Moving Junction Complex
- Immune Suppression by Neutrophils in HIV-1 Infection: Role of PD-L1/PD-1 Pathway
- and Exhibit Metabolic Symbioses
- The Herpes Virus Fc Receptor gE-gI Mediates Antibody Bipolar Bridging to Clear Viral Antigens from the Cell Surface
- Target Cell Availability, Rather than Breast Milk Factors, Dictates Mother-to-Infant Transmission of SIV in Sooty Mangabeys and Rhesus Macaques
- Evolution of the Retroviral Restriction Gene : Inhibition of Non-MLV Retroviruses
- Infection of Adult Thymus with Murine Retrovirus Induces Virus-Specific Central Tolerance That Prevents Functional Memory CD8 T Cell Differentiation
- Fha Interaction with Phosphothreonine of TssL Activates Type VI Secretion in
- In Vivo Administration of a JAK3 Inhibitor during Acute SIV Infection Leads to Significant Increases in Viral Load during Chronic Infection
- Lack of Detectable HIV-1 Molecular Evolution during Suppressive Antiretroviral Therapy
- Activation of HIV-1 from Latent Infection via Synergy of RUNX1 Inhibitor Ro5-3335 and SAHA
- A Compact, Multifunctional Fusion Module Directs Cholesterol-Dependent Homomultimerization and Syncytiogenic Efficiency of Reovirus p10 FAST Proteins
- The Role of Host and Microbial Factors in the Pathogenesis of Pneumococcal Bacteraemia Arising from a Single Bacterial Cell Bottleneck
- Genetic Dissection of Gut Epithelial Responses to
- Two-Component System Cross-Regulation Integrates Response to Heme and Cell Envelope Stress
- Oral Mycobiome Analysis of HIV-Infected Patients: Identification of as an Antagonist of Opportunistic Fungi
- A Model System for Studying the Transcriptomic and Physiological Changes Associated with Mammalian Host-Adaptation by Serovar Copenhageni
- Inflammasome Sensor NLRP1 Controls Rat Macrophage Susceptibility to
- ChIP-Seq and RNA-Seq Reveal an AmrZ-Mediated Mechanism for Cyclic di-GMP Synthesis and Biofilm Development by
- The Hypervariable Amino-Terminus of P1 Protease Modulates Potyviral Replication and Host Defense Responses
- Caspase-1-Dependent and -Independent Cell Death Pathways in Infection of Macrophages
- The Effect of Cell Growth Phase on the Regulatory Cross-Talk between Flagellar and Spi1 Virulence Gene Expression
- Different Mutagenic Potential of HIV-1 Restriction Factors APOBEC3G and APOBEC3F Is Determined by Distinct Single-Stranded DNA Scanning Mechanisms
- Oral Bacteria and Cancer
- Identification of OmpA, a Protein Involved in Host Cell Invasion, by Multi-Phenotypic High-Content Screening
- Transovarial Transmission of a Plant Virus Is Mediated by Vitellogenin of Its Insect Vector
- VE-Cadherin Cleavage by LasB Protease from Facilitates Type III Secretion System Toxicity in Endothelial Cells
- Dimerization of VirD2 Binding Protein Is Essential for Induced Tumor Formation in Plants
- Crystal Structure of the Vaccinia Virus DNA Polymerase Holoenzyme Subunit D4 in Complex with the A20 N-Terminal Domain
- Post-Translational Regulation via Clp Protease Is Critical for Survival of
- Modulation of Phagosomal pH by Promotes Hyphal Morphogenesis and Requires Stp2p, a Regulator of Amino Acid Transport
- Rotavirus Activates Lymphocytes from Non-Obese Diabetic Mice by Triggering Toll-Like Receptor 7 Signaling and Interferon Production in Plasmacytoid Dendritic Cells
- Cytomegalovirus m154 Hinders CD48 Cell-Surface Expression and Promotes Viral Escape from Host Natural Killer Cell Control
- Interferon Regulatory Factor-1 Protects from Fatal Neurotropic Infection with Vesicular Stomatitis Virus by Specific Inhibition of Viral Replication in Neurons
- HMGB1-Promoted and TLR2/4-Dependent NK Cell Maturation and Activation Take Part in Rotavirus-Induced Murine Biliary Atresia
- An Immunomics Approach to Schistosome Antigen Discovery: Antibody Signatures of Naturally Resistant and Chronically Infected Individuals from Endemic Areas
- PPARγ Agonists Improve Survival and Neurocognitive Outcomes in Experimental Cerebral Malaria and Induce Neuroprotective Pathways in Human Malaria
- A Non-Coding RNA Promotes Bacterial Persistence and Decreases Virulence by Regulating a Regulator in
- Viral OTU Deubiquitinases: A Structural and Functional Comparison
- Heterogeneity and Breadth of Host Antibody Response to KSHV Infection Demonstrated by Systematic Analysis of the KSHV Proteome
- Influenza A Virus Assembly Intermediates Fuse in the Cytoplasm
- Broadly Reactive Human CD8 T Cells that Recognize an Epitope Conserved between VZV, HSV and EBV
- Oncogenic Human Papillomaviruses Activate the Tumor-Associated Lens Epithelial-Derived Growth Factor (LEDGF) Gene
- Erythrocyte Invasion: Combining Function with Immune Evasion
- IL-1α and Complement Cooperate in Triggering Local Neutrophilic Inflammation in Response to Adenovirus and Eliminating Virus-Containing Cells
- Chronic Exposure to Type-I IFN under Lymphopenic Conditions Alters CD4 T Cell Homeostasis
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Cytomegalovirus m154 Hinders CD48 Cell-Surface Expression and Promotes Viral Escape from Host Natural Killer Cell Control
- Human African Trypanosomiasis and Immunological Memory: Effect on Phenotypic Lymphocyte Profiles and Humoral Immunity
- DHX36 Enhances RIG-I Signaling by Facilitating PKR-Mediated Antiviral Stress Granule Formation
- Conflicting Interests in the Pathogen–Host Tug of War: Fungal Micronutrient Scavenging Versus Mammalian Nutritional Immunity
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



