-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaNsrR, GadE, and GadX Interplay in Repressing Expression of the O157:H7 LEE Pathogenicity Island in Response to Nitric Oxide
Expression of genes of the locus of enterocyte effacement (LEE) is essential for adherence of enterohemorrhagic Escherichia coli (EHEC) to intestinal epithelial cells. Gut factors that may modulate LEE gene expression may therefore influence the outcome of the infection. Because nitric oxide (NO) is a critical effector of the intestinal immune response that may induce transcriptional regulation in enterobacteria, we investigated its influence on LEE expression in EHEC O157:H7. We demonstrate that NO inhibits the expression of genes belonging to LEE1, LEE4, and LEE5 operons, and that the NO sensor nitrite-sensitive repressor (NsrR) is a positive regulator of these operons by interacting directly with the RNA polymerase complex. In the presence of NO, NsrR detaches from the LEE1/4/5 promoter regions and does not activate transcription. In parallel, two regulators of the acid resistance pathway, GadE and GadX, are induced by NO through an indirect NsrR-dependent mechanism. In this context, we show that the NO-dependent LEE1 down-regulation is due to absence of NsrR-mediated activation and to the repressor effect of GadX. Moreover, the inhibition of expression of LEE4 and LEE5 by NO is due to loss of NsrR-mediated activation, to LEE1 down-regulation and to GadE up-regulation. Lastly, we establish that chemical or cellular sources of NO inhibit the adherence of EHEC to human intestinal epithelial cells. These results highlight the critical effect of NsrR in the regulation of the LEE pathogenicity island and the potential role of NO in the limitation of colonization by EHEC.
Published in the journal: . PLoS Pathog 10(1): e32767. doi:10.1371/journal.ppat.1003874
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1003874Summary
Expression of genes of the locus of enterocyte effacement (LEE) is essential for adherence of enterohemorrhagic Escherichia coli (EHEC) to intestinal epithelial cells. Gut factors that may modulate LEE gene expression may therefore influence the outcome of the infection. Because nitric oxide (NO) is a critical effector of the intestinal immune response that may induce transcriptional regulation in enterobacteria, we investigated its influence on LEE expression in EHEC O157:H7. We demonstrate that NO inhibits the expression of genes belonging to LEE1, LEE4, and LEE5 operons, and that the NO sensor nitrite-sensitive repressor (NsrR) is a positive regulator of these operons by interacting directly with the RNA polymerase complex. In the presence of NO, NsrR detaches from the LEE1/4/5 promoter regions and does not activate transcription. In parallel, two regulators of the acid resistance pathway, GadE and GadX, are induced by NO through an indirect NsrR-dependent mechanism. In this context, we show that the NO-dependent LEE1 down-regulation is due to absence of NsrR-mediated activation and to the repressor effect of GadX. Moreover, the inhibition of expression of LEE4 and LEE5 by NO is due to loss of NsrR-mediated activation, to LEE1 down-regulation and to GadE up-regulation. Lastly, we establish that chemical or cellular sources of NO inhibit the adherence of EHEC to human intestinal epithelial cells. These results highlight the critical effect of NsrR in the regulation of the LEE pathogenicity island and the potential role of NO in the limitation of colonization by EHEC.
Introduction
Enterohemorrhagic Escherichia coli (EHEC), especially those belonging to the O157:H7 serotype, are foodborne pathogens and healthy rearing animals are the main reservoir. Human infection occurs through the ingestion of contaminated food. This primary infection yields to the development of intestinal disorders, including aqueous or bloody diarrhea. Moreover, EHEC express a cardinal and well-defined virulence factor, the Shiga-toxin (Stx) encoded by genes located in lysogenic lambdoid bacteriophages. Stx is produced in the gut lumen and crosses the epithelial barrier to reach the blood and the target organs including the kidneys. In this context, infected patients may develop life-threatening complications such as the hemolytic and uremic syndrome (HUS), the main cause of renal failure in children in developed countries [1].
EHEC genes carried by the locus of enterocyte effacement (LEE), a chromosomal pathogenicity island organized in 5 operons, encode bacterial factors implicated in the intimate adherence of these bacteria to intestinal epithelial cells [2]. These genes encode a type 3 secretion system (T3SS; LEE1, LEE2, LEE3), a translocon and a syringe (LEE4) that allows bacteria to inject effectors in epithelial cells, such as the LEE5-encoded intimin receptor Tir; moreover, other proteins not carried by the LEE can be translocated by the T3SS into enterocytes [3], [4]. The injected effectors and/or protein of the translocon itself interact with the host signal transduction, leading to actin polymerization and to microvilli effacement [2], to regulation of the innate immune response [5], [6], and to increased electrolyte transport [7]. Regulation of gene expression within the LEE is known to be complex and governed by a large number of influences, including environmental cues or quorum sensing, and involves several specific or global regulators [8], [9]. The first gene of the LEE1 operon, ler, encodes a transcriptional regulator that positively regulates the expression of all the other operons [9]–[11]. However a variety of extra-transcriptional mechanisms have also been involved in the regulation of LEE expression, though little detailed mechanistic information is available [12]. GadE (YhiE) and GadX (YhiX) are two main regulators of the acid fitness island involved in acid-resistance (AR) in E. coli K12 [13]–[15]. At acidic pH values, GadE and GadX positively regulate the gadA and gadBC genes, encoding the components of the glutamate-dependent AR. In E. coli O157:H7, GadE has acquired additional functions and inversely coordinates expression of AR and LEE genes [16]: It has been proposed that, during passage through the human stomach, GadE protects EHEC by inducing the glutamate-dependent AR system and inhibits the unnecessary expression of the LEE genes, while environmental cues in the intestine lead to downregulation of gadE and upregulation of the LEE genes [16]. GadE has been shown to directly bind the ler (LEE1) and sepZ (LEE2) promoters in vitro [8], but in vivo binding of GadE and the role of GadX have never been investigated.
We have previously shown that nitric oxide (NO) decreases Stx2 synthesis by EHEC O157:H7 at the transcriptional level [17]. This occurs through the inhibition of the SOS response by the NO sensor nitrite-sensitive repressor (NsrR) [17], the key regulator of the nitrosative stress in enterobacteria [18]. In this context, our aim was to investigate whether NO also modulates LEE gene transcription and therefore EHEC adhesion to epithelial cells. Here we show that NsrR is a direct positive regulator of the transcription of LEE1, LEE4 and LEE5 genes and an indirect repressor of gadE and gadX genes. In the presence of NO, LEE1/4/5 activation is abrogated, GadE is induced and yields to gadX expression. Finally, we identify GadE and GadX as repressors of LEE4/5 and of LEE1, respectively. Using a human intestinal epithelial cells/EHEC co-culture model we demonstrate that bacterial adhesion is inhibited in NO producing cells.
Results
The adhesion of EHEC to intestinal epithelial cells is reduced by NO
We first examined adhesion of the E. coli O157:H7 strain EDL933 to cultured Hct-8 intestinal epithelial cells in the presence of the NO donor NOR-4. Exposure to NOR-4 at 200 µM or 500 µM did not cause any significant difference in the growth rate of EDL933, as described [17]. However, EHEC adhesion to Hct-8 cells was dramatically inhibited when NOR-4 was added to the co-cultures (Figs. 1A and 1B). The number of EHEC fixed to the cells was significantly decreased by 41±5% and 89±2% in the presence of 200 µM and 500 µM NOR-4, respectively (Fig. 1B).
Fig. 1. Adhesion of EDL933 to intestinal epithelial cells. 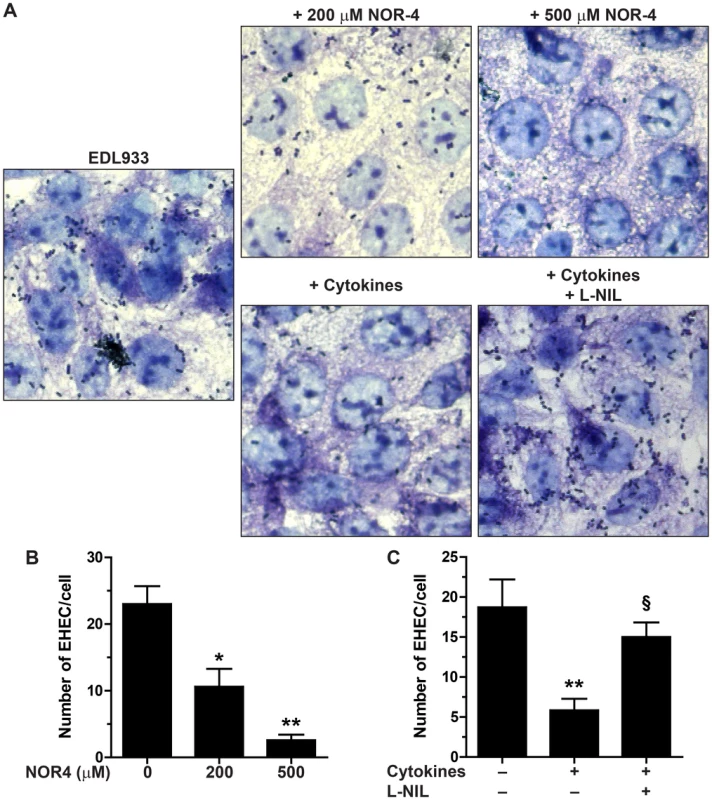
Hct-8 cells, pre-treated or not with a cytokine cocktail for 24 h, were co-cultured for 6 h with the EHEC strain EDL933 ± NOR-4 or L-NIL. A: Cells and bacteria were visualized after Giemsa staining; magnification, ×63. B and C: The number of bacteria adherent per Hct-8 cell was counted on 15 microscopic fields. For B, * P<0.05, ** P<0.01 compared to the co-cultures without NOR-4; n = 6. For C, ** P<0.01 vs. cells not stimulated with cytokines; § P<0.05 vs. cells treated with cytokines; n = 6. To further confirm this result, we analyzed the effect of endogenous NO released by enterocytes. Hct-8 cells were first treated for 24 h with a cytokine cocktail known to stimulate the inducible NO synthase (iNOS) expression [19], washed, and then infected with the strain EDL933 in the presence or absence of the iNOS inhibitor N6-(1-iminoethyl)-l-lysine (l-NIL). There was less EHEC fixed to NO-producing epithelial cells than to control cells (Figs. 1A and 1C). The inhibition of EHEC adherence to Hct-8 cells treated with cytokines was abolished by the use of l-NIL (Figs. 1A and 1C).
NO inhibits LEE1/4/5 gene expression and stimulates the Gad system
The expression of genes that represent the five operons of the LEE (Fig. 2A) was analyzed after treatment with NOR-4 for 6 h. NO was consistently generated in the bacteria culture medium and reached a plateau after 6 h (Fig. S1). The expression of ler (LEE1), espA (LEE4), tir and eae (LEE5) was down-regulated by NO, while the transcription of sepZ (LEE2) was induced by 2.4-fold (Fig. 2B). The expression of the gene escV (LEE3) was not modulated by NOR-4 (Fig. 2B). Because GadE and GadX modulates LEE expression in EHEC and EPEC, respectively, [16], [20], we investigated the effect of NOR-4 on gadE and gadX transcription. As shown in Figure 2C, the expression of gadE and gadX was significantly induced by 2.4 - and 2.7-fold in bacteria exposed to NOR-4, respectively. Thereby, these data prompted us to wonder whether NO-dependent down-regulation of LEE1, LEE4 and LEE5 requires GadE and/or GadX.
Fig. 2. Influence of NO on LEE, gadX, and gadE gene expression. 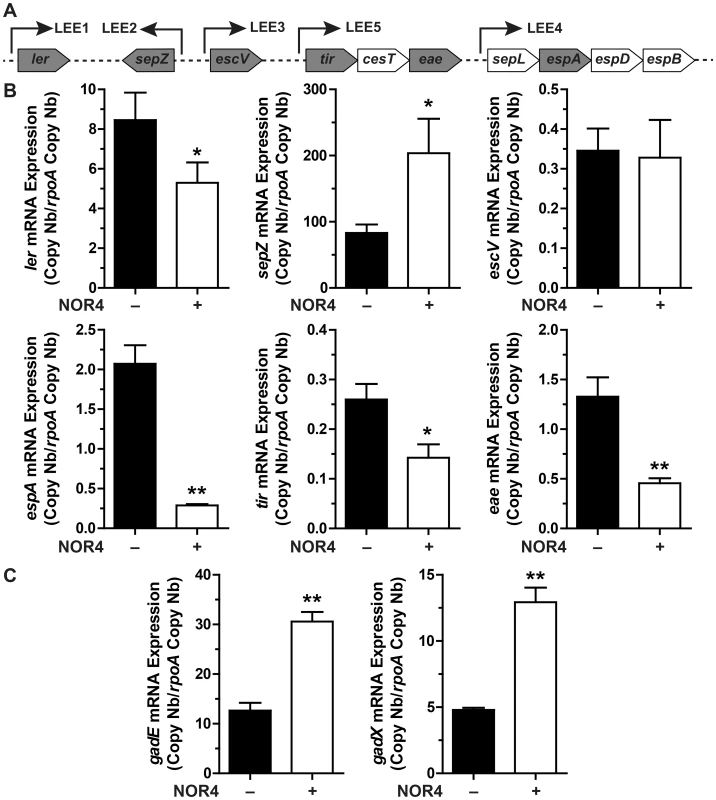
A: schematic representation of the LEE showing the structural organization of the main operons. Arrows indicate the orientation of the transcription. The genes analyzed in this study are in grey boxes. B and C: EDL933 was grown for 6 h with or without NOR-4. The expression of the LEE genes (B) and of gadE and gadX (C) was analyzed by RT-qPCR. * P<0.05, ** P<0.01 compared to the strain grown in the absence of NOR-4; n = 3–6. GadE and GadX modulate the expression of LEE genes
Since the role of GadX and GadE on LEE expression is not well defined and is strongly dependent on the growth conditions [16], [20], [21], we first analyzed the expression of ler, espA, and tir in EDL933 ΔgadE and ΔgadX mutants. When compared to the EDL933 strain, the mRNA levels of ler, espA and tir were increased by ∼1.4-, 2.3-, and 2-fold in the ΔgadE strain, respectively (Fig. 3A); these effects were reversed when the gadE mutant was trans-complemented with the gadE gene in a low copy number plasmid vector (Fig. 3A). The gadX mutation was associated with a spontaneous increase of ler transcription and with a significant reduction of espA and tir gene expression (Fig. 3A). The transcription of ler was repressed while the expression of espA was activated and that of tir was restored to the same level as the WT in the trans-complemented strain (EDL933 ΔgadX-c; Fig. 3A). These data suggest that GadE represses the expression of LEE4 and LEE5 genes independently of Ler, and that GadX represses LEE1 but activates LEE4 and LEE5 gene expression. Interestingly, the NOR-4-dependent down-regulation of ler, espA, and tir was still observed in the ΔgadE, ΔgadX and ΔgadE/gadX mutants (Fig. 3A), suggesting that another factor is implicated in the inhibition of LEE1/4/5 by NO.
Fig. 3. Regulation of LEE1/4/5 by GadE and GadX. 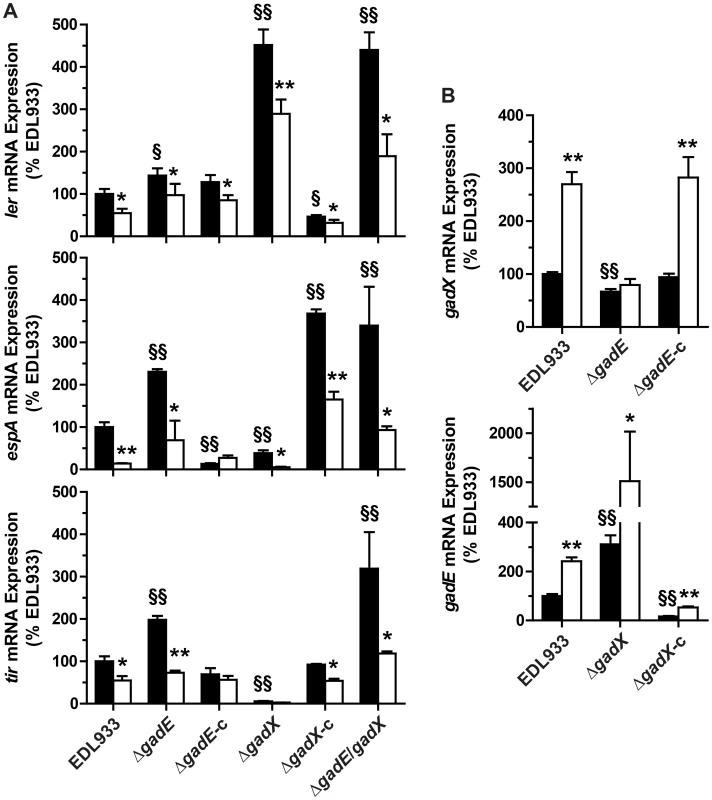
The mRNA levels of ler (LEE1), espA (LEE4), and tir (LEE5) (A) and of gadE and gadX (B) were assessed in the strain EDL933, in the ΔgadE, ΔgadX and ΔgadE/gadX isogenic mutants, and in the complemented strains ΔgadE-c and ΔgadX-c. Bacteria were grown in the absence (black bars) or presence (white bars) of NOR-4 for 6 h. * P<0.05, ** P<0.01 compared to the same strain without NOR-4; § P<0.05, §§ P<0.01 vs. EDL933; n = 3–7. We next wonder whether GadE and GadX repressed the LEE independently from each other or whether GadX is epistatic to GadE as in E. coli K12 [22]. The expression of ler was similar in a ΔgadE/gadX double mutant and in the EDL933 ΔgadX strain (Fig. 3A), indicating that GadX is epistatic to GadE in controlling LEE1. Conversely, espA and tir mRNA levels were increased in EDL933 ΔgadE/gadX when compared to the WT strain, as in the ΔgadE strain, demonstrating that GadE is epistatic to GadX for the regulation of LEE4 and LEE5. Therefore we investigated whether GadE controls gadX expression. Figure 3B shows a 33% decrease in gadX mRNA levels in the gadE mutant, indicating that GadE activates gadX expression. In addition, we observed 3.1-fold more gadE mRNA copies in the ΔgadX strain than in the WT strain (Fig. 3B) and gadE mRNA levels were dramatically reduced in the complemented strain (Fig. 3E), suggesting that GadX is a repressor of gadE expression. Therefore, the moderate increase in ler expression observed in the ΔgadE strain (Fig. 3A) is likely due to the lower level of GadX in this strain and not to a direct effect of GadE on ler transcription. Lastly, the activation of gadX transcription by NOR-4 was suppressed in the gadE mutant, but not in the EDL933 ΔgadE-c strain (Fig. 3B), while the NO-dependent induction of gadE mRNA expression was still observed in the ΔgadX strain (Fig. 3B). These data indicate that NO activates gadX expression through GadE.
The repression of LEE1/4/5 genes is mediated by NsrR
NsrR is a transcriptional regulator that regulates gene expression in response to NO [18]. Therefore we investigated whether NsrR regulates gadE, gadX, and the LEE genes. In the absence of NO, the mRNA levels of ler, espA, and tir were 6.8, 7.1, and 14.3-fold lower in the ΔnsrR mutant than in the WT strain, respectively (Fig. 4A). The expression of these genes was similar in the strains EDL933 and EDL933 ΔnsrR-c (Fig. 4A). Moreover, the NO-dependent regulation of these LEE genes was abrogated in EDL933 ΔnsrR and was restored in the complemented strain (Fig. 4A). Inversely, the transcription of gadE and gadX was significantly increased in the ΔnsrR mutant, but not in the complemented strain. The expression of these two genes was not affected by NOR-4 in the nsrR-deficient strain (Fig. 4B).
Fig. 4. Effect of NsrR on LEE, gadX, and gadE gene expression. 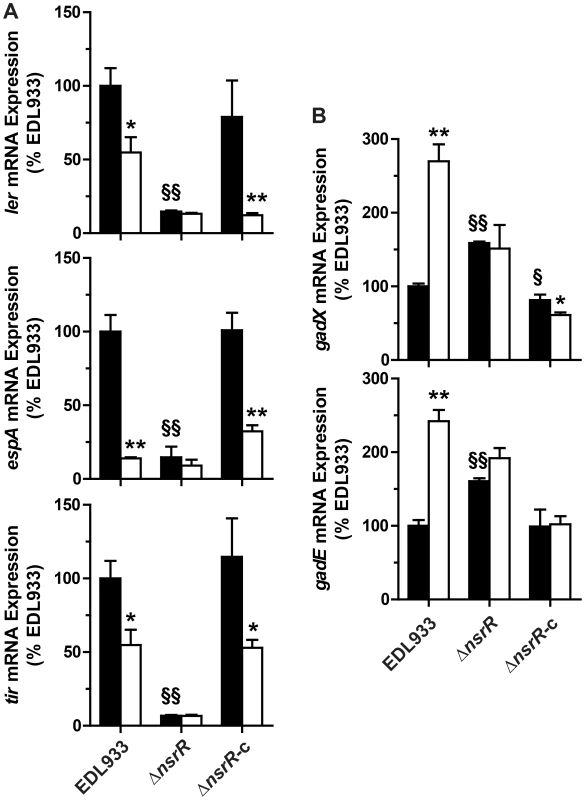
The strains EDL933, ΔnsrR, or ΔnsrR-c were grown with (white bars) or without (black bars) NOR-4. The expression of the genes ler, espA, tir (A) and gadX and gadE (B) was analyzed. * P<0.05, ** P<0.01 compared to the same strain without NOR-4; § P<0.05, §§ P<0.01 vs. EDL933; n = 3. These data suggest that NsrR is a transcriptional activator of LEE1, LEE4, and LEE5 and a repressor of gadE, which in turn modulates gadX expression. NsrR loses its ability to regulate the expression of LEE and gad genes in the presence of NO.
The NsrR binding on the promoter regions of LEE 1/4/5 is inhibited by NO
The investigation of GadE, GadX and/or NsrR direct binding to the gadE, gadX, and LEE promoter regions was performed by chromatin immunoprecipitation (ChIP) experiments using the EDL933 ΔgadE, ΔgadX, and ΔnsrR mutants expressing the 6-His-GadE, the 6-His-GadX, and the 6-His-NsrR fusion proteins, respectively.
We first analyzed the gadX promoter described by Hommais et al. [15], the three promoters described for gadE in E. coli K12 [22] (Fig. S2), and the gadA promoter as a positive control for GadE and GadX binding [23]. Surprisingly, we found that GadE and GadX did not bind to the gadX and gadE promoters, respectively (Figs. 5A and 5B), indicating that activation of gadX by GadE and repression of gadE by GadX occur through indirect regulations. As expected, the binding of GadX and GadE to the gadA promoter region was observed (Figs. 5A and 5B).
Fig. 5. Binding of GadE, GadX and NsrR on various promoter regions. 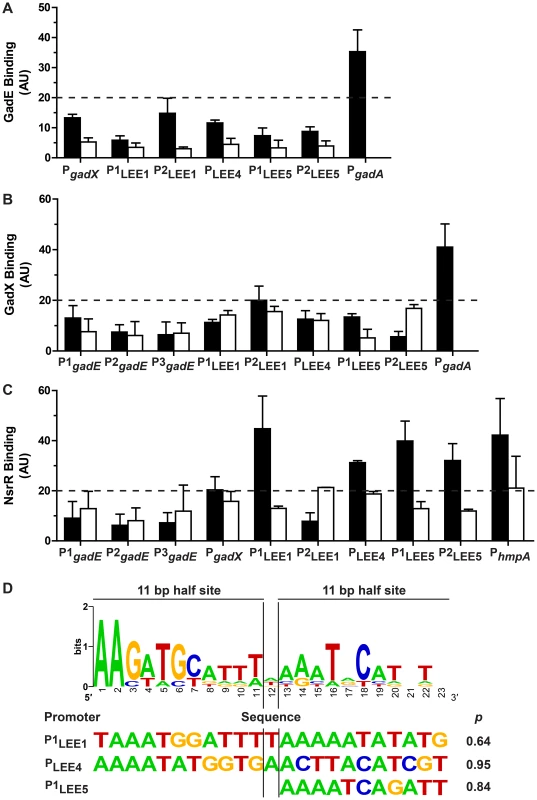
The strains EDL933 ΔgadE-pBADMycHisA::gadE, ΔgadX-pBADHisA::gadX, and ΔnsrR-pBADMycHisA::nsrR were grown in the absence (black bars) or in the presence (white bars) of NOR-4. ChIP assays followed by qPCR were performed to determine the relative enrichment in DNA molecules bound to GadE (A), GadX (B), NsrR (C). Values higher than 20 (twice the values obtained for the strain EDL933 containing the empty pBADmycHisA vector) indicate protein binding to the promoters of interest. D: Bio-informatics analyses of NsrR-binding sites. Sequence logo determined from seven putative NsrR-binding sites in EDL933 (upper panel), and sequences with the best matches for the entire or one of the half sites are shown with their statistical scores (lower panel). Two ler promoters have been described in EHEC, the distal P1 promoter and a putative proximal P2 promoter (Fig. S2). The P1 promoter is common to EHEC and EPEC, while the P2 promoter is present only in EHEC [24]–[26]. Neither GadE (Fig. 5A) nor GadX (Fig. 5B) bound to either of these promoters (Figs. 5A and 5B). These data indicate that GadE and GadX do not repress ler expression directly. The LEE4 promoter has been identified in EHEC upstream of sepL [27], espA being the second gene of the operon. In EPEC, it has been shown that Ler-mediated activation of the LEE5 operon requires sequences between positions -198 and -75 relative to the transcriptional start site [28]. Two primer pairs overlapping this region have been designed for ChIP experiments, amplifying a LEE5 distal (P1LEE5) and a LEE5 proximal (P2LEE5) region (Fig. S2). ChIP experiments showed that neither GadE nor GadX bound to the LEE4 and LEE5 promoters (Figs. 5A and 5B). Lastly, the binding of GadE and GadX to the LEE1/4/5 promoter regions was not modulated by NOR-4. These data indicate that control of LEE4 and LEE5 expression by GadE and GadX is due to indirect effects.
In contrast, NsrR bound to the distal LEE1 promoter (P1LEE1), to the LEE4 and LEE5 promoters, and to the promoter of hmpA, a well-know NsrR target gene (Fig. 5C). Furthermore, NsrR binding to these promoter regions was inhibited when the bacteria were grown in the presence of NOR-4 (Fig. 5C). We did not observed NsrR binding to the gadE and gadX promoters (Fig. 5C).
We thus performed bio-informatics analysis to identify putative NsrR-binding sites in the LEE1, LEE4 LEE5, gadE, and gadX promoters in the strain EDL933. We used the homologous sequences of seven NsrR-binding sites described in E. coli K12 [29] to generate the sequence logo of the NsrR box in the strain EDL933 (Fig. 5D). We then performed bioinformatics analysis on the LEE1, LEE4 and LEE5 promoter sequences by the Gibbs Sampler Motif Software, using the matrix of the seven putative NsrR-binding sites of EDL933. In agreement with the ChIP data, bioinformatics analysis identified sequences presenting high identity with the NsrR consensus binding site in the LEE1 (P1), LEE4 and LEE5 promoter regions (Fig. 5D and Fig. S2), but not in the promoters of gadE and gadX. The analysis indicated a 23 bp putative NsrR-binding site in the promoters of LEE1 (86.9% identity) and LEE4 (78.2% identity), but only a second half-site NsrR-binding site in the LEE5 promoter (90.9% identity for the half site; Fig. 5D). In silico analyses performed using the BLAST program indicated that these putative binding sites are conserved in a number of EHEC and EPEC strains, but not in Citrobacter rodentium (Fig. S3), an attaching/effacing pathogen that infects rodents.
NsrR interacts with the RNA polymerase complex
Since NsrR has been exclusively described as a transcriptional repressor, we investigated the molecular mechanism underlying the direct activation of LEE gene expression by NsrR. For many transcriptional activators, increase of the transcription level results from the recruitment of RNA polymerase through direct interaction between the regulatory protein and one or several subunits of the polymerase [30]. We therefore examined if NsrR can interact with α and σ RNA polymerase subunits. To this end, His-tagged NsrR and hemagglutinin (HA)-tagged polymerase subunits α (RpoA) or σ38 (RpoS) were co-expressed in bacteria. His-NsrR was purified under native conditions using a nickel affinity resin and the different fractions were analyzed by western-blot. As positive controls, RpoA and RpoS were also co-expressed with His-Crp or His-Crl, respectively, two well-known interacting partners [31], [32]. All tagged proteins were properly expressed as revealed by their immunodetection in the whole extract samples (Fig. 6). As expected, HA-RpoA and HA-RpoS co-eluted with His-Crp or His-Crl, respectively. No HA-tagged protein was detected in the His eluates of the negative controls, i.e., bacteria expressing only HA-tagged proteins (Fig. 6). Importantly, HA-RpoA and HA-RpoS were also specifically recovered in the eluted fractions from the His-NsrR purifications. This finding demonstrates that NsrR can interact with the RNA polymerase complex and suggests that NsrR activates LEE gene expression through the recruitment of RNA polymerase.
Fig. 6. NsrR interacts with RNA polymerase complex. 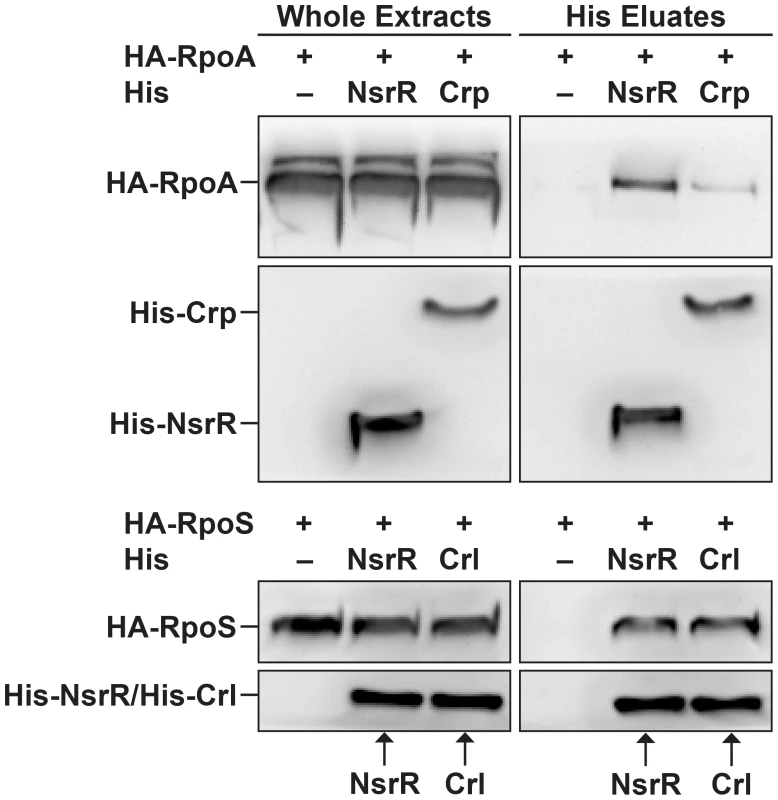
His-NsrR, His-Crp or His-Crl proteins were co-expressed in bacteria with HA-RpoA or HA-RpoS as indicated. His tagged proteins were then purified from bacterial lysates using nickel affinity. Whole extracts (1 µg) and His eluted fractions (5 µl) were probed with anti-His or anti-HA Abs. Adhesion of the regulatory mutants to HeLa cells
In order to confirm the role of NO, NsrR, GadE, and GadX in regulating LEE expression, we investigated the attachment of the regulatory mutants to HeLa cells after 6 h of infection in the presence or absence of NOR-4. As expected, EDL933 adhered to HeLa cells and when NOR-4 was added to the co-culture the level of adhesion was dramatically reduced to that of the ΔescN mutant that lacks a functional T3SS (Figs. 7A and 7B). The adhesion of the ΔgadE and ΔgadX strains was higher than that of the parent strain, correlating with the repressive effect of AR regulatory proteins on LEE gene expression (Figs. 7A and 7B). Conversely, the nsrR mutant was less adherent than the WT strain (Figs. 7A and 7B). The complementation of these three mutants restored the adhesion phenotype of the parental strain (Figs. 7A and 7B). Under NO exposure, adherence properties were affected for the ΔgadE and ΔgadX mutants but not for the ΔnsrR mutant (Figs. 7A and 7B), demonstrating that NsrR is the key regulator controlling the T3SS-dependent adhesion of EHEC in response to NO.
Fig. 7. Regulation of adhesion of EDL933 to human epithelial cells. 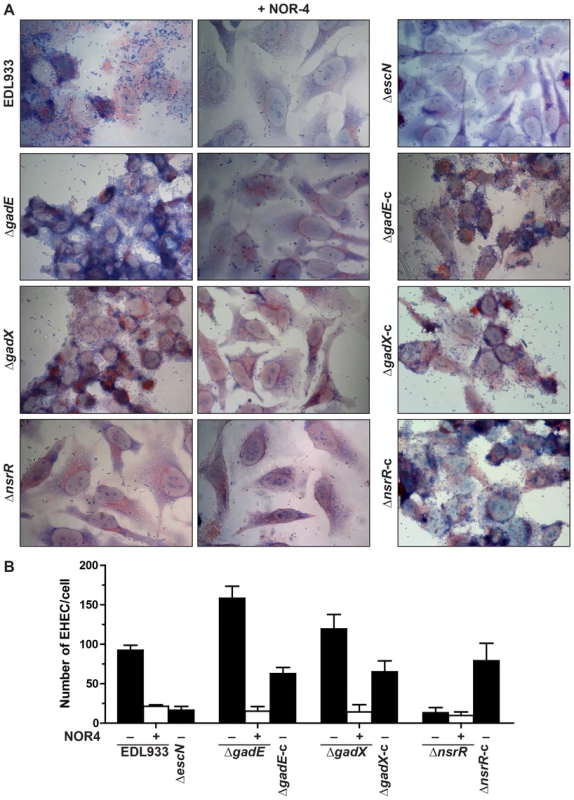
HeLa cells were infected with EDL933, ΔgadE, ΔgadX, ΔnsrR, or with the corresponding trans-complemented strains, in the presence or absence of NOR-4. After 6 h, cells were washed and colored with May-Grünwald Giemsa (A). The number of adherent bacteria per Hela cell was determined from 50 cells (B). Discussion
In the present report, we show that NO, a critical mediator of the host innate immune response, is a potent inhibitor of LEE gene expression in EHEC O157:H7 and consequently inhibits the adhesion of these pathogens to intestinal epithelial cells. We identified NsrR as an unrecognized regulator that controls the expression of LEE genes in response to NO, and we propose a regulatory model presenting the role of NsrR, GadE and GadX in LEE expression (Fig. 8). In the absence of NO (Fig. 8A), NsrR directly activates LEE1, LEE4, and LEE5 gene expression, and indirectly represses gadE and therefore gadX expression. We also show that GadE indirectly activates gadX expression and represses LEE4 and LEE5 expression independently of Ler, while GadX inhibits gadE and LEE1 expression. When NsrR binds NO (Fig. 8B), it is released from its target DNA, leading to loss of induction of LEE1/4/5 genes and to the up-regulation of gadE and, consequently, gadX. In this context, the NO-dependent LEE1 down-regulation is due to absence of NsrR-mediated activation and to the inhibitory effect of GadX. In parallel, the inhibition of LEE4 and LEE5 gene expression is due to absence of NsrR - and Ler-dependent activation and to increase of GadE level. This model assumes that repression of gadX expression by NsrR is mediated by GadE, which is consistent with the observation that the NO-dependent activation of gadX is abrogated in the ΔgadE and ΔnsrR mutants.
Fig. 8. A model for the NO-dependent regulation of LEE1/4/5. 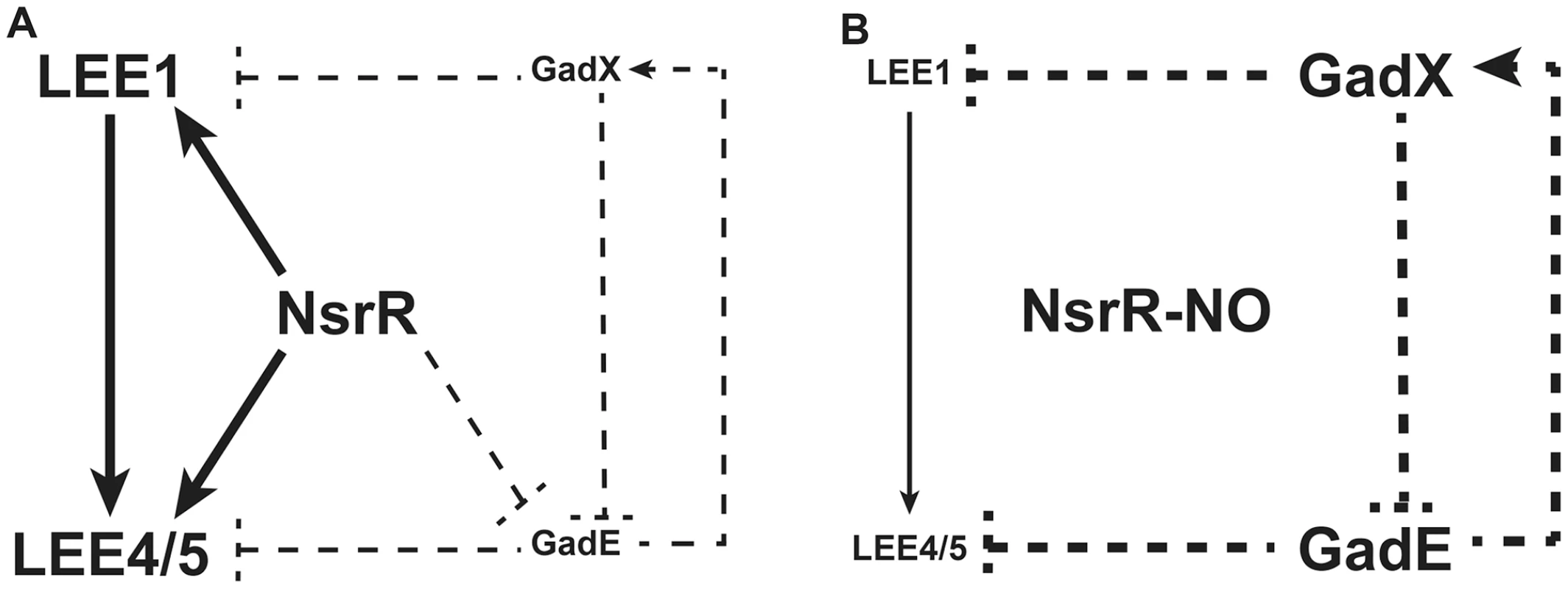
Solid lines indicate physical interaction between the regulator and the promoter as demonstrated by ChIP; dotted lines indicate that no physical interaction has been demonstrated between the regulator and the promoter. A: In the absence of NO, NsrR directly activates LEE1, LEE4, and LEE5 expression and indirectly represses gadE and therefore gadX expression. GadE activates gadX expression and acts as an indirect repressor of LEE4 and LEE5. GadX is an indirect repressor of gadE and LEE1 expression. B: Under NO exposure, NO binds to NsrR, which is consequently released from its target DNA. Thus, the activation of LEE1/4/5 genes by NsrR is suppressed. In parallel, gadE expression is restored and induces gadX up-regulation. In this context, GadE strongly represses LEE4 and LEE5 genes while GadX inhibits LEE1 expression. NsrR is a key negative regulator of the nitrosative stress in enterobacteria [18], [33]. NsrR has always been described as a transcriptional repressor. In addition, its DNA-binding activity is suppressed in the presence of NO, yielding to the expression of various genes mainly involved in NO detoxification [18], [33]. In non-pathogenic E. coli, NsrR also regulates expression of genes involved in metabolism, motility, protein degradation, surface attachment, stress response and transmembrane transport [29], [34]. Our data indicate that NsrR is also a repressor of the genes gadE and gadX. Nonetheless, the NsrR-dependent repression of gadX is probably mediated by GadE since the NO-dependent up-regulation of gadX is abrogated in the ΔgadE mutant. We did not find a sequence matching the NsrR consensus binding site in the gadE promoter, and ChIP experiments failed to demonstrate physical interaction between NsrR and the gadE promoter. Therefore, the effect of NsrR on gadE transcription is probably indirect and mediated by an unknown regulatory cascade controlled by NsrR.
Here we provide compelling evidence that NsrR is a direct positive regulator of LEE1, LEE4, and LEE5 operons in EHEC by binding to their own promoters. Moreover, our data also suggest that NsrR acts as a transcriptional activator by recruiting RNA polymerase to promoter regions since NsrR is able to pull-down the α and σ38 subunits of the RNA polymerase. Supporting the concept that it may also be a transcriptional activator, it has been reported that NsrR activates virulence gene expression in Salmonella Typhimurium, in particular expression of genes important for eukaryotic cell adherence, invasion and intestinal translocation, and that an nsrR mutant is impaired in invasion of HeLa cells [35]. However, in silico analysis failed to identify an NsrR consensus binding site in the promoter regions of these genes, indicating that the positive regulatory effect of NsrR is probably indirect in this pathogen [35]. Moreover, using an E. coli K12 strain harboring a multicopy plasmid that titrates out NsrR, Filenko et al. have identified by a microarray analysis 22 transcripts that could be directly or indirectly activated by NsrR [34].
The NsrR binding site is a 23 bp palindrome sequence composed of two 11 bp half sites separated by a single nucleotide, and NsrR binds to DNA as a dimer [36]. However, a number of NsrR target promoters contain only a single half site [29]. Potential NsrR consensus sequence were identified in the LEE1, LEE4 and LEE5 promoters, with a 23 pb putative NsrR-binding site in the LEE1 and LEE4 promoters, and a putative second half-site in the LEE5 promoter. It has been suggested that, when the NsrR binding site contains only a single half site, one NsrR monomer makes specific contact to the consensus half site and the other monomer forms nonspecific contact [37]. Alternatively, it has been suggested that NsrR binds as a tetramer to the complete binding motif and as a dimer when only one half site is conserved [29]. It is noteworthy that the putative NsrR binding sites identified in the LEE1, LEE4 and LEE5 promoters are conserved in a number of other EHEC and EPEC strains, but not in C. rodentium, suggesting that NO also influences cell adhesion via NsrR in other E. coli attaching/effacing pathogenic human strains.
Influence of GadE on LEE gene expression remains controversial. While Tatsuno et al. described an increased expression of LEE2, LEE4, and LEE5 in a ΔgadE mutant, which is not correlated with enhancement of ler expression [20], KailasanVanaja et al. showed that GadE represses LEE expression by down-regulating ler transcription [16]. These discrepancies are proposed to be due to differences in growth medium and/or differences in the sensitivity of the assays used in each study. Interestingly, our data indicate that GadE may repress the expression of LEE4 and LEE5 via two regulatory cascades, mediated or not by Ler (Figure 8). On the one hand, we show that GadE inhibits LEE1 through GadX, because a decreased expression of gadX and an induction of LEE1 are observed in the gadE-deficient strain; this results in loss of Ler-dependent induction of LEE4/5. On the other hand, the deletion of gadX is associated with an increased expression of ler and gadE, and with an inhibition of LEE4/5, suggesting that GadE inhibits these operons independently of Ler. In accordance, the induction of espA and tir in the gadE mutant and in the ΔgadE/gadX strain demonstrates that GadX regulates LEE4/5 via the repression of gadE. However, although it has been shown in vitro that GadE can bind to the ler promoter in EHEC O157:H7 [8], we did not observe such an interaction in vivo in our experiments; this difference is probably due to the presence of binding competitors in live bacteria. Regarding GadX, we show herein that it negatively regulates ler transcription in EHEC. However, the effect of GadX on LEE1 expression is indirect since no physical interaction between GadX and the LEE1 promoter has been demonstrated. Interestingly, it has been described in EPEC that LEE1 is down-regulated under conditions in which GadX is induced, namely at pH 5.5 or in contact to epithelial cells [21]; this occurs through the inhibition of the transcription of the per locus by GadX [21]. Because the perC homologue in EHEC, named pch, is involved in LEE1 induction [38], it would be interesting to now determine the role of GadX on pch expression.
The biological relevance of LEE1, LEE4, and LEE5 inhibition by NO is the decreased adhesion of E. coli O157:H7 to epithelial cells. When EHEC are ingested with the contaminated food, they first reach the stomach. It has been proposed that the acidic conditions of this ecological niche favor GadE induction and therefore limit EHEC adhesion to gastric tissues [16]. There is also abundant nonenzymatically formed NO in the gastric juice caused by acidification of nitrate and nitrite. In this context, we now propose that the NO-dependent LEE4/5 inhibition is a supplementary mechanism developed by EHEC to avoid their persistence in the stomach and to favor bacterial colonization in the colon. Moreover, we have shown in the present study that, not only a chemical source of NO, but also the reactive nitrogen species released by iNOS-expressing colonic epithelial cells inhibit the adherence of O157:H7 E. coli, and our previous work has identified NO as a potent inhibitor of Stx synthesis [17]. Together, these results suggest that NO might limit the infectious process and HUS development. Nonetheless, it has been described that EHEC inhibit the inducible transcription of iNOS in human enterocytes [19], thus, by limiting NO production, EHEC might favor their own virulence by increasing the intimate adherence to the intestinal epithelium and Stx synthesis. We can therefore speculate that the issue of the crosstalk between EHEC and the host-derived NO might determine the outcome of the infection.
Materials and Methods
Bacteria, mutagenesis, and growth conditions
Strains and plasmids used in this study are listed in Table S1. The EHEC O157:H7 strain EDL933 [39] was used throughout the study. The EDL933 ΔgadE and ΔgadX mutants and the ΔgadE/gadX double mutant were constructed using the one-step PCR-based method [40], [41]. Mutants were verified by PCR to assess the loss of the gene and by RT-qPCR to confirm lack of expression of the gene of interest, using the primers listed in Table S2. The ΔnsrR mutant strain has been previously described [17]. For complementation analysis and ChIP experiments, the gadE, gadX, and nsrR genes were amplified with the high fidelity polymerase Pfx50 (Invitrogen) and cloned under the control of the araC promoter into a low-copy plasmid containing a 6-histidine tag (pBADHisA or pBADMycHisA; Invitrogen), or in pBAD33. The cloned genes were checked by nucleotide sequencing, and their expression was analyzed by RT-qPCR. The 6-His-NsrR-, 6-His-GadE-, and 6-His-GadX-encoding genes were expressed at the same level than the WT genes. To verify the mutation of the gadE and gadX genes, we analyzed the acid resistance of the mutant strains [42]: Acid-resistance of the ΔgadE and ΔgadX mutants dropped to 0 and 1.41% of the parent strain, respectively; acid resistance was restored in the complemented mutant strains (data not shown).
A single colony of EDL933 or isogenic mutants was grown overnight in DMEM Low glucose containing 10 mM HEPES. These cultures were diluted in fresh medium to an OD600 = 0.03 and grown at 37°C. The medium was supplemented with ampicillin (50 µg/ml), kanamycin (50 µg/ml), chloramphenicol (25 µg/ml), L-arabinose (0.1 mM–0.5 mM), or the NO donor NOR-4 (Enzo Life Science) when required.
Bioinformatics analysis of NsrR-binding sites
The NsrR-binding sequence logo of the strain EDL933 was generated using homologous sequence of the seven NsrR-binding sites described previously by Partridge et al. in E. coli K-12 strain MG1655 [29] and the software Weblogo (http://weblogo.berkeley.edu/logo.cgi). The probabilities of occurrence matrix from the seven homologous sequences in EHEC O157:H7 strain EDL933 served as a model for the identification of a consensus sequence in the promoter regions of the target genes using the online software Gibbs Motif Sampler (http://ccmbweb.ccv.brown.edu/gibbs/gibbs.html). The sequence alignment of the LEE1, LEE4 and LEE5 putative sites in other EHEC strains, in EPEC strains, and in C. rodentium was performed with the MEGA5 software.
ChIP assay
The pBADMycHisA::gadE, pBADHisA::gadX, and pBADMycHisA::nsrR plasmids, encoding 6His-GadE, 6His-GadX and 6His-NsrR, were electroporated into the respective mutants to avoid native protein interference. Overnight cultures of each strain in LB medium were diluted 1∶100 in 25 ml of fresh DMEM medium buffered with 10 mM HEPES, with or without NOR-4. GadE and GadX expression was induced with 0.5 mM l-arabinose and NsrR with 0.1 mM l-arabinose. After 6 h of growth with shaking, ChIP was performed as described by Lannois et al. [43] with slight modifications. First, the protein-DNA complexes were cross-linked by treating bacteria with 1% formaldehyde at room temperature for 30 min. Bacteria were then washed twice with cold PBS and incubated for 30 min at 37°C in 0.7 ml of lysis buffer (10 mM Tris pH 8, 50 mM NaCl, 10 mM EDTA, and 20% sucrose) containing 10 mg/ml lysozyme (Sigma). Then, 0.7 ml of 2X IP buffer (100 mM Tris pH 8, 300 mM NaCl, 2% Igepal CA-630, 0.5% Na deoxycholate) containing 1 mM PMSF was added and samples were incubated 15 min at 37°C, cooled down on ice, sonicated, and incubated on ice for 1 min. Sonication was repeated 11 times to obtain a solution of fragmented chromatin. A 50 µl aliquot of each sample was treated with 100 µl TE containing 36 µg proteinase K for 2 hours at 37°C, incubated 8 hours at 67°C to reverse crosslinking, and the DNA was purified with the kit Qiaquick (Qiagen); this was termed as Input fraction. The rest of the fragmented chromatin was used to generate the IP fraction. After a 2 h-incubation with an anti-Histidine monoclonal antibody (Sigma), protein G sepharose 50% (40 µl) was added to each sample and incubated 1 hour at room temperature. The beads were washed twice with IP buffer, twice with 1 ml of ChIP wash buffer (10 mM Tris HCl pH 8, 250 mM LiCl, 1 mM EDTA, 0.5% Igepal CA-630, and 0.5% Na deoxycholate) and twice with 1 ml of TE buffer. The beads were resuspended in 100 µl of elution buffer (50 mM Tris HCl pH 8, 10 mM EDTA, 1% SDS), incubated 15 min at 65°C, and centrifuged at 9500× g for 1 min. The supernatants containing the immunoprecipitated DNA were collected and incubated with 100 µl TE containing 36 µg proteinase K 2 hours at 37°C and 8 hours at 65°C. DNA was purified with the Qiaquick kit (Qiagen) and amplified by qPCR using the primers listed in Table S2 and depicted in Fig. S2.
The enrichment of DNA targets was calculated as follows for each protein: the promoters of interest as well as a non-specific rpoA intragenic region were amplified with specific primers (Table S2). For each DNA target, we calculated the ratio between the copy number in the IP fraction and the Input fraction; each value was then divided with the ratio obtained for the non-specific rpoA intragenic region. Then the same ratio was calculated from the parent strain EDL933 containing the empty pBADMycHisA vector. Values higher than 20, corresponding to twice the values obtained for the strain EDL933 containing the empty pBADmycHisA vector, indicate protein binding to the promoter of interest.
Pull-down assays
For bacterial co-expression experiments, genes encoding NsrR, Crp or Crl were cloned into the first multiple cloning site of pCDFDuet-1 vector (Novagen) allowing expression of the proteins tagged with a N-terminal hexahistidine motif. Genes encoding RpoA or RpoS were cloned into the second multiple cloning site using PCR primers allowing the insertion of a N-terminal HA motif (see Table S2 for primers). E. coli BL21(DE3) harboring the different constructs was grown at 37°C to OD600 nm of 0.7, then induced with 1 mM IPTG and grown for an additional 2 h. After resuspension of bacteria with a 1/10e volume of lysis buffer (50 mM NaH2PO4, 300 mM NaCl), samples were sonicated and centrifuged. Supernatants (whole bacterial extracts) were incubated with Ni-NTA beads at 4°C for 16 h. Beads were washed four times with lysis buffer containing 60 mM imidazole and bound proteins were eluted with lysis buffer containing 250 mM imidazole.
Bacterial mRNA analysis
Total RNA from bacteria was extracted using the TRI Reagent RNA Isolation Reagent (Sigma). Each RNA sample (1 µg) was reverse transcribed with Superscript II enzyme (Invitrogen) and random primers (Invitrogen). The cDNAs and serial dilutions of EDL933 genomic DNA, which were used for the standard curves, were amplified with gene-specific primers (Table S2) in the Eppendorf Mastercycler eprealplex (Eppendorf) apparatus. The results are presented as the ratios between the copy number of mRNA of the gene of interest and the copy number of rpoA mRNA.
Western-blot analysis
Samples were mixed with a 2X SDS-PAGE sample buffer, heated for 5 min at 100°C, resolved on 14% SDS-PAGE gels and blotted on PVDF membranes. Membranes were blocked in PBS-0.05% Tween 20 supplemented with 5% non-fat dry milk, then probed with murine monoclonal anti-HA or HRP-conjugated anti-HIS Abs (Sigma; 1/4000 for each). An HRP-conjugated goat anti-murine IgG Ab (Sigma) was also used for the HA blots. Acquisitions were performed with a G:box system (Syngene).
Cell cultures and infections
The epithelial cell lines Hct-8 and HeLa were maintained in DMEM with 10% FCS, 10 mM Hepes, 100 U/ml penicillin, 100 µg/ml streptomycin at 37°C under 5% CO2. Hct-8 cells were plated on LabTek (Nunc), cultured for 7 days, and stimulated for 24 h with human IFN-γ (50 ng/ml), TNF-α (20 ng/ml), and IL-1β (5 ng/ml). HeLa cells were seeded into LabTek and grown for 24 h. These Hct-8 and HeLa cells were washed, and infected with bacteria with an MOI of 100, in the presence or absence of NOR-4 or of the iNOS inhibitor l-NIL. After 4 washes with PBS, cells were fixed using 1 ml methanol for 15 min at −20°C and stained with Giemsa or May-Grünwald Giemsa for 30 min. The number of adherent bacteria per cell was counted using the AxioVision 4 software.
Determination of NO concentration
The concentration of the stable oxidized products of NO, NO3− and NO2−, was measured using the Nitrite/Nitrate Assay Kit (Cayman Chemical).
Statistics
All the data represent the mean ± SEM. Student's t test or ANOVA with the Newman-Keuls test were used to determine significant differences between two groups or to analyze significant differences among multiple test groups, respectively.
Supporting Information
Zdroje
1. KarmaliMA, GannonV, SargeantJM (2010) Verocytotoxin-producing Escherichia coli (VTEC). Vet Microbiol 140 : 360–370.
2. McDanielTK, JarvisKG, DonnenbergMS, KaperJB (1995) A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc Natl Acad Sci U S A 92 : 1664–1668.
3. GarmendiaJ, PhillipsAD, CarlierMF, ChongY, SchullerS, et al. (2004) TccP is an enterohaemorrhagic Escherichia coli O157:H7 type III effector protein that couples Tir to the actin-cytoskeleton. Cell Microbiol 6 : 1167–1183.
4. GruenheidS, SekirovI, ThomasNA, DengW, O'DonnellP, et al. (2004) Identification and characterization of NleA, a non-LEE-encoded type III translocated virulence factor of enterohaemorrhagic Escherichia coli O157:H7. Mol Microbiol 51 : 1233–1249.
5. HaufN, ChakrabortyT (2003) Suppression of NF-kappa B activation and proinflammatory cytokine expression by Shiga toxin-producing Escherichia coli. J Immunol 170 : 2074–2082.
6. GobertAP, CosteA, GuzmanCA, VareilleM, HindreT, et al. (2008) Modulation of chemokine gene expression by Shiga-toxin producing Escherichia coli belonging to various origins and serotypes. Microbes Infect 10 : 159–165.
7. CollingtonGK, BoothIW, DonnenbergMS, KaperJB, KnuttonS (1998) Enteropathogenic Escherichia coli virulence genes encoding secreted signalling proteins are essential for modulation of Caco-2 cell electrolyte transport. Infect Immun 66 : 6049–6053.
8. TreeJJ, RoeAJ, FlockhartA, McAteerSP, XuX, et al. (2011) Transcriptional regulators of the GAD acid stress island are carried by effector protein-encoding prophages and indirectly control type III secretion in enterohemorrhagic Escherichia coli O157:H7. Mol Microbiol 80 : 1349–1365.
9. DengW, PuenteJL, GruenheidS, LiY, VallanceBA, et al. (2004) Dissecting virulence: systematic and functional analyses of a pathogenicity island. Proc Natl Acad Sci U S A 101 : 3597–3602.
10. ElliottSJ, SperandioV, GironJA, ShinS, MelliesJL, et al. (2000) The locus of enterocyte effacement (LEE)-encoded regulator controls expression of both LEE - and non-LEE-encoded virulence factors in enteropathogenic and enterohemorrhagic Escherichia coli. Infect Immun 68 : 6115–6126.
11. SperandioV, MelliesJL, DelahayRM, FrankelG, CrawfordJA, et al. (2000) Activation of enteropathogenic Escherichia coli (EPEC) LEE2 and LEE3 operons by Ler. Mol Microbiol 38 : 781–793.
12. BhattS, RomeoT, KalmanD (2011) Honing the message: post-transcriptional and post-translational control in attaching and effacing pathogens. Trends Microbiol 19 : 217–224.
13. MaZ, RichardH, TuckerDL, ConwayT, FosterJW (2002) Collaborative regulation of Escherichia coli glutamate-dependent acid resistance by two AraC-like regulators, GadX and GadW (YhiW). J Bacteriol 184 : 7001–7012.
14. TuckerDL, TuckerN, MaZ, FosterJW, MirandaRL, et al. (2003) Genes of the GadX-GadW regulon in Escherichia coli. J Bacteriol 185 : 3190–3201.
15. HommaisF, KrinE, CoppeeJY, LacroixC, YeramianE, et al. (2004) GadE (YhiE): a novel activator involved in the response to acid environment in Escherichia coli. Microbiology 150 : 61–72.
16. Kailasan VanajaS, BergholzTM, WhittamTS (2009) Characterization of the Escherichia coli O157:H7 Sakai GadE regulon. J Bacteriol 191 : 1868–1877.
17. VareilleM, de SabletT, HindreT, MartinC, GobertAP (2007) Nitric oxide inhibits Shiga-toxin synthesis by enterohemorrhagic Escherichia coli. Proc Natl Acad Sci U S A 104 : 10199–10204.
18. BodenmillerDM, SpiroS (2006) The yjeB (nsrR) gene of Escherichia coli encodes a nitric oxide-sensitive transcriptional regulator. J Bacteriol 188 : 874–881.
19. VareilleM, RannouF, ThelierN, GlasserAL, de SabletT, et al. (2008) Heme oxygenase-1 is a critical regulator of nitric oxide production in enterohemorrhagic Escherichia coli-infected human enterocytes. J Immunol 180 : 5720–5726.
20. TatsunoI, NaganoK, TaguchiK, RongL, MoriH, et al. (2003) Increased adherence to Caco-2 cells caused by disruption of the yhiE and yhiF genes in enterohemorrhagic Escherichia coli O157:H7. Infect Immun 71 : 2598–2606.
21. ShinS, Castanie-CornetMP, FosterJW, CrawfordJA, BrinkleyC, et al. (2001) An activator of glutamate decarboxylase genes regulates the expression of enteropathogenic Escherichia coli virulence genes through control of the plasmid-encoded regulator, Per. Mol Microbiol 41 : 1133–1150.
22. SayedAK, OdomC, FosterJW (2007) The Escherichia coli AraC-family regulators GadX and GadW activate gadE, the central activator of glutamate-dependent acid resistance. Microbiology 153 : 2584–2592.
23. TramontiA, ViscaP, De CanioM, FalconiM, De BiaseD (2002) Functional characterization and regulation of gadX, a gene encoding an AraC/XylS-like transcriptional activator of the Escherichia coli glutamic acid decarboxylase system. J Bacteriol 184 : 2603–2613.
24. SperandioV, MelliesJL, NguyenW, ShinS, KaperJB (1999) Quorum sensing controls expression of the type III secretion gene transcription and protein secretion in enterohemorrhagic and enteropathogenic Escherichia coli. Proc Natl Acad Sci U S A 96 : 15196–15201.
25. SperandioV, LiCC, KaperJB (2002) Quorum-sensing Escherichia coli regulator A: a regulator of the LysR family involved in the regulation of the locus of enterocyte effacement pathogenicity island in enterohemorrhagic E. coli. Infect Immun 70 : 3085–3093.
26. PorterME, MitchellP, FreeA, SmithDG, GallyDL (2005) The LEE1 promoters from both enteropathogenic and enterohemorrhagic Escherichia coli can be activated by PerC-like proteins from either organism. J Bacteriol 187 : 458–472.
27. LodatoPB, KaperJB (2009) Post-transcriptional processing of the LEE4 operon in enterohaemorrhagic Escherichia coli. Mol Microbiol 71 : 273–290.
28. HaackKR, RobinsonCL, MillerKJ, FowlkesJW, MelliesJL (2003) Interaction of Ler at the LEE5 (tir) operon of enteropathogenic Escherichia coli. Infect Immun 71 : 384–392.
29. PartridgeJD, BodenmillerDM, HumphrysMS, SpiroS (2009) NsrR targets in the Escherichia coli genome: new insights into DNA sequence requirements for binding and a role for NsrR in the regulation of motility. Mol Microbiol 73 : 680–694.
30. LeeDJ, MinchinSD, BusbySJ (2012) Activating transcription in bacteria. Annu Rev Microbiol 66 : 125–152.
31. BusbyS, EbrightRH (1999) Transcription activation by catabolite activator protein (CAP). J Mol Biol 293 : 199–213.
32. BougdourA, LelongC, GeiselmannJ (2004) Crl, a low temperature-induced protein in Escherichia coli that binds directly to the stationary phase sigma subunit of RNA polymerase. J Biol Chem 279 : 19540–19550.
33. RodionovDA, DubchakIL, ArkinAP, AlmEJ, GelfandMS (2005) Dissimilatory metabolism of nitrogen oxides in bacteria: comparative reconstruction of transcriptional networks. PLoS Comput Biol 1: e55.
34. FilenkoN, SpiroS, BrowningDF, SquireD, OvertonTW, et al. (2007) The NsrR regulon of Escherichia coli K-12 includes genes encoding the hybrid cluster protein and the periplasmic, respiratory nitrite reductase. J Bacteriol 189 : 4410–4417.
35. KarlinseyJE, BangIS, BeckerLA, FrawleyER, PorwollikS, et al. (2012) The NsrR regulon in nitrosative stress resistance of Salmonella enterica serovar Typhimurium. Mol Microbiol 85 : 1179–1193.
36. TuckerNP, HicksMG, ClarkeTA, CrackJC, ChandraG, et al. (2008) The transcriptional repressor protein NsrR senses nitric oxide directly via a [2Fe-2S] cluster. PLoS One 3: e3623.
37. TuckerNP, Le BrunNE, DixonR, HutchingsMI (2010) There's NO stopping NsrR, a global regulator of the bacterial NO stress response. Trends Microbiol 18 : 149–156.
38. IyodaS, WatanabeH (2004) Positive effects of multiple pch genes on expression of the locus of enterocyte effacement genes and adherence of enterohaemorrhagic Escherichia coli O157:H7 to HEp-2 cells. Microbiology 150 : 2357–2571.
39. O'BrienAO, LivelyTA, ChenME, RothmanSW, FormalSB (1983) Escherichia coli O157:H7 strains associated with haemorrhagic colitis in the United States produce a Shigella dysenteriae 1 (SHIGA) like cytotoxin. Lancet 1 : 702.
40. BeloinC, ValleJ, Latour-LambertP, FaureP, KzreminskiM, et al. (2004) Global impact of mature biofilm lifestyle on Escherichia coli K-12 gene expression. Mol Microbiol 51 : 659–674.
41. DatsenkoKA, WannerBL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97 : 6640–6645.
42. LargeTM, WalkST, WhittamTS (2005) Variation in acid resistance among shiga toxin-producing clones of pathogenic Escherichia coli. Appl Environ Microbiol 71 : 2493–2500.
43. LanoisA, JubelinG, GivaudanA (2008) FliZ, a flagellar regulator, is at the crossroads between motility, haemolysin expression and virulence in the insect pathogenic bacterium Xenorhabdus. Mol Microbiol 68 : 516–533.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Serotonin Signaling in : A Serotonin–Activated G Protein-Coupled Receptor Controls Parasite MovementČlánek Regulators of Cell Cycle Progression and Differentiation Identified Using a Kinome-Wide RNAi ScreenČlánek IFNγ/IL-10 Co-producing Cells Dominate the CD4 Response to Malaria in Highly Exposed ChildrenČlánek Functions of CPSF6 for HIV-1 as Revealed by HIV-1 Capsid Evolution in HLA-B27-Positive SubjectsČlánek Decreases in Colonic and Systemic Inflammation in Chronic HIV Infection after IL-7 Administration
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2014 Číslo 1- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- HIV-1 Accessory Proteins Adapt Cellular Adaptors to Facilitate Immune Evasion
- Ranaviruses: Not Just for Frogs
- Effectors and Effector Delivery in
- Plasmacytoid Dendritic Cell Dynamics Tune Interferon-Alfa Production in SIV-Infected Cynomolgus Macaques
- Lu/BCAM Adhesion Glycoprotein Is a Receptor for Cytotoxic Necrotizing Factor 1 (CNF1)
- A Substrate-Fusion Protein Is Trapped inside the Type III Secretion System Channel in
- Parvovirus-Induced Depletion of Cyclin B1 Prevents Mitotic Entry of Infected Cells
- Red Blood Cell Invasion by : Structural Basis for DBP Engagement of DARC
- NsrR, GadE, and GadX Interplay in Repressing Expression of the O157:H7 LEE Pathogenicity Island in Response to Nitric Oxide
- Loss of Circulating CD4 T Cells with B Cell Helper Function during Chronic HIV Infection
- TREM-1 Deficiency Can Attenuate Disease Severity without Affecting Pathogen Clearance
- Origin, Migration Routes and Worldwide Population Genetic Structure of the Wheat Yellow Rust Pathogen f.sp.
- Glutamate Utilization Couples Oxidative Stress Defense and the Tricarboxylic Acid Cycle in Phagosomal Escape
- Serotonin Signaling in : A Serotonin–Activated G Protein-Coupled Receptor Controls Parasite Movement
- Recovery of an Antiviral Antibody Response following Attrition Caused by Unrelated Infection
- Regulators of Cell Cycle Progression and Differentiation Identified Using a Kinome-Wide RNAi Screen
- Absence of Intestinal PPARγ Aggravates Acute Infectious Colitis in Mice through a Lipocalin-2–Dependent Pathway
- Induction of a Stringent Metabolic Response in Intracellular Stages of Leads to Increased Dependence on Mitochondrial Metabolism
- CTCF and Rad21 Act as Host Cell Restriction Factors for Kaposi's Sarcoma-Associated Herpesvirus (KSHV) Lytic Replication by Modulating Viral Gene Transcription
- Gammaherpesviral Gene Expression and Virion Composition Are Broadly Controlled by Accelerated mRNA Degradation
- The Arabidopsis Silencing Pathway Modulates PAMP- and Effector-Triggered Immunity through the Post-transcriptional Control of Disease Resistance Genes
- Inflammatory Stimuli Reprogram Macrophage Phagocytosis to Macropinocytosis for the Rapid Elimination of Pathogens
- Alphavirus Mutator Variants Present Host-Specific Defects and Attenuation in Mammalian and Insect Models
- Phosphopyruvate Carboxylase Identified as a Key Enzyme in Erythrocytic Carbon Metabolism
- IFNγ/IL-10 Co-producing Cells Dominate the CD4 Response to Malaria in Highly Exposed Children
- Electron Tomography of HIV-1 Infection in Gut-Associated Lymphoid Tissue
- Characterisation of a Multi-ligand Binding Chemoreceptor CcmL (Tlp3) of
- Single Cell Stochastic Regulation of Pilus Phase Variation by an Attenuation-like Mechanism
- Cell Tropism Predicts Long-term Nucleotide Substitution Rates of Mammalian RNA Viruses
- Functions of CPSF6 for HIV-1 as Revealed by HIV-1 Capsid Evolution in HLA-B27-Positive Subjects
- RNA-seq Analysis of Host and Viral Gene Expression Highlights Interaction between Varicella Zoster Virus and Keratinocyte Differentiation
- Kaposi's Sarcoma Associated Herpesvirus Tegument Protein ORF75 Is Essential for Viral Lytic Replication and Plays a Critical Role in the Antagonization of ND10-Instituted Intrinsic Immunity
- DAMP Molecule S100A9 Acts as a Molecular Pattern to Enhance Inflammation during Influenza A Virus Infection: Role of DDX21-TRIF-TLR4-MyD88 Pathway
- Variable Suites of Non-effector Genes Are Co-regulated in the Type III Secretion Virulence Regulon across the Phylogeny
- Reengineering Redox Sensitive GFP to Measure Mycothiol Redox Potential of during Infection
- Preservation of Tetherin and CD4 Counter-Activities in Circulating Alleles despite Extensive Sequence Variation within HIV-1 Infected Individuals
- KSHV 2.0: A Comprehensive Annotation of the Kaposi's Sarcoma-Associated Herpesvirus Genome Using Next-Generation Sequencing Reveals Novel Genomic and Functional Features
- Nutrient Limitation Governs Metabolism and Niche Adaptation in the Human Nose
- Decreases in Colonic and Systemic Inflammation in Chronic HIV Infection after IL-7 Administration
- Investigation of Acetylcholine Receptor Diversity in a Nematode Parasite Leads to Characterization of Tribendimidine- and Derquantel-Sensitive nAChRs
- Intranasal Vaccination Promotes Detrimental Th17-Mediated Immunity against Influenza Infection
- -Mediated Inhibition of Iron Export Promotes Parasite Replication in Macrophages
- Variation in RNA Virus Mutation Rates across Host Cells
- A Single Amino Acid in the Stalk Region of the H1N1pdm Influenza Virus HA Protein Affects Viral Fusion, Stability and Infectivity
- Group B Engages an Inhibitory Siglec through Sialic Acid Mimicry to Blunt Innate Immune and Inflammatory Responses
- Synthesis and Biological Properties of Fungal Glucosylceramide
- HIV Protective KIR3DL1/S1-HLA-B Genotypes Influence NK Cell-Mediated Inhibition of HIV Replication in Autologous CD4 Targets
- Recruitment of PfSET2 by RNA Polymerase II to Variant Antigen Encoding Loci Contributes to Antigenic Variation in
- Human and Plant Fungal Pathogens: The Role of Secondary Metabolites
- Lyme Disease: Call for a “Manhattan Project” to Combat the Epidemic
- Enhancing Virus-Specific Immunity by Combining Therapeutic Vaccination and PD-L1 Blockade in Chronic Hepadnaviral Infection
- Suppression of Interferon Lambda Signaling by SOCS-1 Results in Their Excessive Production during Influenza Virus Infection
- Inflammation Fuels Colicin Ib-Dependent Competition of Serovar Typhimurium and in Blooms
- Host-Specific Enzyme-Substrate Interactions in SPM-1 Metallo-β-Lactamase Are Modulated by Second Sphere Residues
- STING-Dependent Type I IFN Production Inhibits Cell-Mediated Immunity to
- From Scourge to Cure: Tumour-Selective Viral Pathogenesis as a New Strategy against Cancer
- Lysine Acetyltransferase GCN5b Interacts with AP2 Factors and Is Required for Proliferation
- Narrow Bottlenecks Affect Populations during Vertical Seed Transmission but not during Leaf Colonization
- Targeted Cytotoxic Therapy Kills Persisting HIV Infected Cells During ART
- Murine Gammaherpesvirus M2 Protein Induction of IRF4 via the NFAT Pathway Leads to IL-10 Expression in B Cells
- iNKT Cell Production of GM-CSF Controls
- Malaria-Induced NLRP12/NLRP3-Dependent Caspase-1 Activation Mediates Inflammation and Hypersensitivity to Bacterial Superinfection
- Detection of Host-Derived Sphingosine by Is Important for Survival in the Murine Lung
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Lyme Disease: Call for a “Manhattan Project” to Combat the Epidemic
- Origin, Migration Routes and Worldwide Population Genetic Structure of the Wheat Yellow Rust Pathogen f.sp.
- IFNγ/IL-10 Co-producing Cells Dominate the CD4 Response to Malaria in Highly Exposed Children
- Human and Plant Fungal Pathogens: The Role of Secondary Metabolites
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



