-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaPopulation Genetic Analyses Reveal the African Origin and Strain Variation of var.
article has not abstract
Published in the journal: . PLoS Pathog 8(2): e32767. doi:10.1371/journal.ppat.1002495
Category: Pearls
doi: https://doi.org/10.1371/journal.ppat.1002495Summary
article has not abstract
Cryptococcus neoformans is a ubiquitous, saprobic yeast and the cause of life-threatening infections. Humans acquire the infection by inhaling airborne cells from the environment. In the lungs, these cells become encapsulated yeasts and proliferate. In people with healthy immune responses, the infection may resolve or remain latent and subsequently cause disease. However, in immunocompromised people, such as HIV/AIDS patients, and less often in healthy hosts, the yeasts can disseminate to almost any part of the body; however, they are neurotropic, and meningoencephalitis is the most frequent and deadliest clinical manifestation [1]–[3]. An estimated 1 million new infections are acquired each year, and the majority of these cases occur in sub-Saharan Africa, which has the highest prevalence of patients with HIV/AIDS [4]. In this region, C. neoformans is the most common cause of meningitis, and mortality hovers around 50%. Others who succumb to cryptococcosis are apparently immunocompetent and exhibit no evidence of underlying disease. For example, 71% of cryptococcal infections in China occur in people without pre-existing conditions [5].
There are two varieties, C. neoformans var. grubii (Cng) and C. neoformans var. neoformans (Cnn), which are distinguishable by molecular markers or their capsular serotypes, A or D, respectively. Diploid AD hybrids also occur in the environment and patients [6]–[8]. In addition, a sibling species, Cryptococcus gattii, causes similar infections. However, isolates of both serotype D and AD hybrids, as well as C. gattii, are much less common. At least 90% of human cryptococcal disease and fatalities are caused by Cng (serotype A) [9]–[11].
Non-African Global Isolates of Cng Are Highly Clonal
Strains of Cng have been isolated from all continents except Antarctica. Molecular epidemiological studies identified significant clonality among global strains, as strains with identical molecular genotypes have been isolated from different geographic areas, continents apart [5], [12], [13]. The use of reproducible and robust multilocus sequence typing (MLST) has determined that the overwhelming majority of non-African strains of Cng are represented by only a few genotypes [13]–[15].
Isolates from southeastern Asia are remarkably homogeneous. For example, all the clinical isolates from a cohort of 120 Chinese patients were infected with the same cosmopolitan MLST genotype, M5 [5]. Similarly, all seven clinical isolates from Japan [13] and 70 of 75 from South Korea possessed the M5 genotype (designated “VNIc” in the original paper) [16]. In Thailand, 183 clinical and environmental isolates were analyzed, and 96% of the isolates were represented by only three MLST genotypes, one of which was M5 (designated “ST46” in the original paper) [15].
In the United States, an analysis of over 800 isolates yielded only ten distinct genotypes, and M5 was the most prevalent among both clinical and environmental samples (designated “A5” in the original paper) [10]. In comparison, five genotypes were found in Europe, and nine genotypes among isolates from central and eastern Africa, but only two different MLST genotypes were detected from South American and Australian isolates, although these were small samples [13]. More recently, we found that M5 was the most prevalent genotype among isolates from patients with recurrent cryptococcosis in South Africa (unpublished data), even though, as described below, this region has the highest overall genetic diversity.
Southern African Isolates of Cng Are Highly Diverse
Unlike the rest of the world, southern Africa harbors a geographically restricted, genetically diverse population of Cng. In 2003, isolates from 200 HIV-seropositive patients in Botswana were shown to possess novel genotypes that differed from isolates found anywhere else. Analyses of mating indicated that 12% of these strains possess the MATa mating type, which is exceedingly rare among non-African strains, and population genetic analysis demonstrated evidence for recombination in this population [17]. Subsequent environmental sampling confirmed that two genetically isolated subpopulations are localized in southern Africa: (i) a genetically diverse, endemic population that is restricted to southern African and associated with indigenous African trees, especially the mopane tree (Colophospermum mopane), and (ii) a cosmopolitan population of strains with molecular types that are found worldwide and frequently associated with the excreta of feral pigeons (Columba livia) [14].
Population genetic analyses of the environmental strains revealed limited genetic interaction between the endemic (arboreal) and cosmopolitan (coprophilic) populations. The arboreal population was characterized by linkage equilibrium among loci and high genetic diversity, which can be explained by recombination, ancestral origin, or both. However, when putative recombinant haplotypes were removed from the analysis, significantly high indices of genetic diversity were still detected in the African population [14].
Phylogenetic Analyses Indicate That African Strains of Cng Possess Ancestral Haplotypes
Phylogenetic analysis and principal component analysis (PCA) of the MLST profiles indicate that Cng is comprised of three isolated subpopulations, VNI, VNII, and VNB (Figure 1). The global population is markedly clonal and consists of VNI and VNII strains with few unique genotypes. The arboreal, southern African population is geographically confined and comprised of almost all the known VNB strains and a large number of VNI strains that are genetically diverse. Thus, multiple features of the African strains—greatest genetic diversity, prevalence of both mating types, and association with an indigenous reservoir—suggest they represent the ancestral origin of Cng.
Fig. 1. The genetic relationships among MLST genotypes are visualized by PCA. 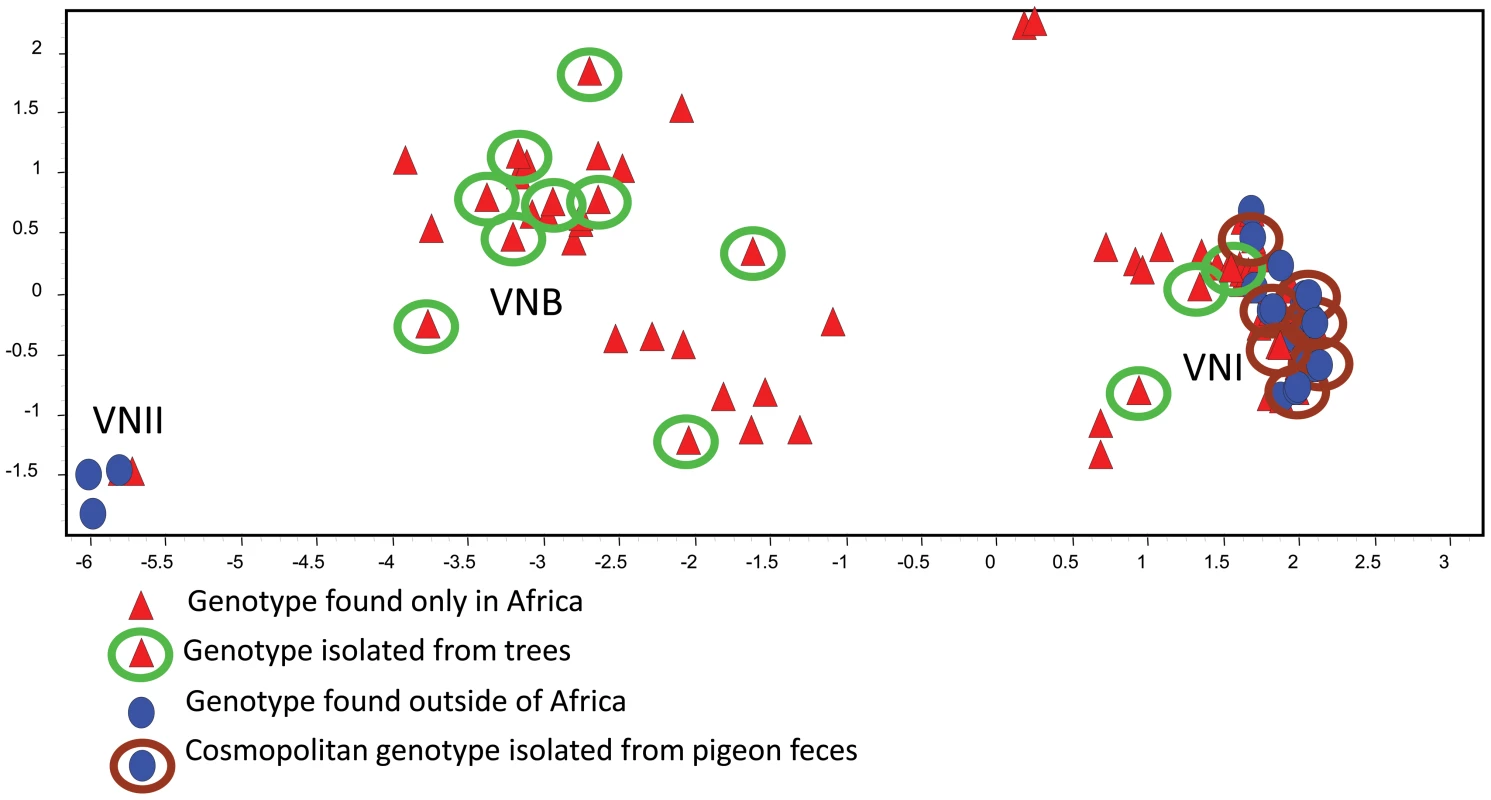
Each symbol represents a genotype with a unique eight-digit allelic profile. Red triangles represent genotypes of strains that are endemic to Africa, and blue circles represent genotypes of global strains. Genotypes associated with African trees are enclosed in green circles, and genotypes associated with pigeon excreta are enclosed in brown circles. Genotypes without circles represent clinical strains that to date have not been isolated from the environment. (From reference [14] and used with permission of the publisher.) This conclusion was supported by phylogenetic analyses of individual loci using methods of maximum likelihood and parsimony. To illustrate, haplotype networks were analyzed by statistical parsimony to infer any phylogenetic relationships among the haplotypes. The internal nodes of these networks represent ancestral haplotypes from which the distal, derived haplotypes evolved. Numerous haplotypes from the endemic African population occupied both internal (ancestral) and distal (derived) positions on the networks (Figure 2, green circles). Conversely, haplotypes that are unique to the global population were scarce and almost always in distal positions, which suggests they originated more recently. All of these genotypes were obtained from pigeon habitats (Figure 2, brown circles) [14], [15]. Furthermore, maximum likelihood analyses of three loci (TEF1, CAP59, and PLB1) indicated that the ancestral haplotypes of both the VNI and VNB populations are confined to southern Africa and associated with endemic African trees [14].
Fig. 2. Haplotype networks of the eight MLST loci. 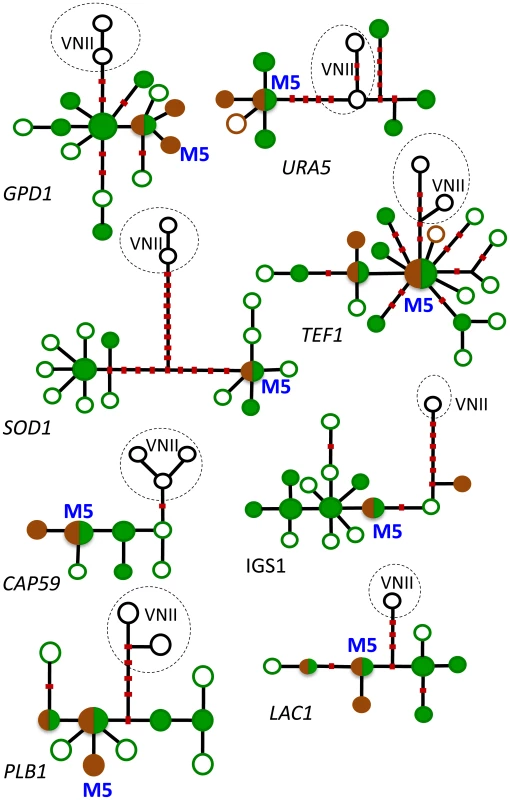
Haplotypes of strains of Cng that have never been found outside Africa are shown in green: filled green circles designate haplotypes of strains that were obtained from trees (most were also found in patients), and empty green circles signify haplotypes that were obtained only from patients. Cosmopolitan haplotypes are shown in brown: filled brown circles designate haplotypes of strains from pigeon excreta (most were also found in patients), and empty brown circles signify haplotypes that were obtained only from patients. Circles that are half green and half brown indicate haplotypes of strains found in trees and pigeon excreta. Haplotypes from the global VNII subpopulation of Cng are included as an outgroup; they are shown in black and lightly encircled. Red dots on the lines connecting the haplotypes represent the most parsimonious number of mutational steps required to generate the allelic polymorphisms. Recombinant haplotypes are excluded. The most common haplotype in Asia and elsewhere, M5, is shown in blue. (From reference [14] and used with permission of the publisher.) Evidence That Pigeons Facilitated the Global Dispersion of Southern African Strains
Environmental sampling has demonstrated that African strains of VNI and VNB are associated with native African trees, whereas cosmopolitan strains of VNI are isolated from pigeon droppings. VNI strains with identical MLST genotypes have been isolated from pigeon habitats in North and South America, Europe, Asia, and Africa [10], [13], [14]. Coalescent simulations estimated that the cosmopolitan and African populations diverged approximately 5,000 years ago, which is around the postulated time period when C. livia (rock doves or pigeons) were domesticated [15]. Although the exact origin of C. livia is unknown, historical records indicate that pigeons were probably native to the north African Mediterranean region and were spread globally over the last 400 years of European expansion [18], [19]. Thus, multiple features of the cosmopolitan population of VNI—exceptionally low genetic diversity, dominance of a single mating type, and global distribution in association with pigeon excreta—support the parsimonious conclusion that pigeons facilitated the global exportation and dispersion of African strains.
Phenotypic Diversity among Isolates of Cng
Cryptococcal virulence is complex and polygenic, involving dozens of genes and signal transduction pathways [20]. Several studies have documented variation among strains of Cng in the expression of virulence phenotypes, such as the size, composition, and biological activity of the capsule, susceptibility to antifungal drugs, resistance to phagocytes, and others [21]–[25]. This marked phenotypic variation among wild-type isolates of Cng suggests that some strains are inherently more virulent. As observed in several reports and noted above, patients may survive or succumb to cryptococcosis regardless of their immune status, which suggests that the virulence of an infecting strain may be as important as the host's defenses. Does the genotype of a strain impart any clinical relevance? A recent population genetics investigation used 11 MLST markers to genotype isolates from South African pediatric cases of cryptococcosis [26]. Seventy-one children, nearly all HIV-positive, were infected with 17 different VNI genotypes; most genotypes were equally present in boys and girls, but one genotype was significantly more prevalent in boys [26]. When the relevance of genotype was tested experimentally, clinical and environmental isolates with identical MLST profiles varied dramatically in murine virulence; however, virulence was associated with the clinical or environmental source of a strain rather than its genotype [27]. Clearly, strains with the same genotype exhibit phenotypic variation. Nevertheless, some genotypes are highly prevalent in the environment but rare or absent in patients and vice versa [10], [28]. The most important phenotype is the production of disease in people. With the advent of next-generation sequencing, it is now possible to use MLST genotypes to select strains differing in genetic diversity and clinical prevalence and to conduct comparative genomic analyses to address the question of why some strains are more pathogenic.
Zdroje
1. LinXHeitmanJ 2006 The biology of the Cryptococcus neoformans species Complex. Annu Rev Microbiol 60 69 105
2. MaHMayRC 2009 Virulence in Cryptococcus species. Adv Appl Microbiol 67 131 190
3. PerfectJRDismukesWEDromerFGoldmanDLGraybillJR 2010 Clinical practice guidelines for the management of cryptococcal disease: 2010 update by the Infectious Diseases Society of America. Clin Infect Dis 50 291 322
4. ParkBJWannemuehlerKAMarstonBJGovenderNPappasPG 2009 Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS 23 525 530
5. ChenJVarmaADiazMRLitvintsevaAPWollenbergKK 2008 Cryptococcus neoformans strains and infection in apparently immunocompetent patients, China. Emerg Infect Dis 14 755 762
6. LengelerKCoxGHeitmanJ 2001 Serotype AD strains of Cryptococcus neoformans are diploid or aneuploid and heterozygous at the mating-type locus. Infect Immun 69 115 122
7. LitvintsevaAPLinXTempletonIHeitmanJMitchellTG 2007 Many globally isolated AD hybrid strains of Cryptococcus neoformans originated in Africa. PLoS Pathog 3 e114 doi:10.1371/journal.ppat.0030114
8. XuJLuoGVilgalysRJBrandtMEMitchellTG 2002 Multiple origins of hybrid strains of Cryptococcus neoformans with serotype AD. Microbiology 148 203 212
9. GovenderNMitchellTGlitvintsevaAPMigliaKJ 2011 Cryptococcosis in Africa. HeitmanJKozelTRKwon-ChungJPerfectJRCasadevallA Cryptococcus: from human pathogen to model yeast Washington (D.C.) ASM Press 269 285
10. LitvintsevaAPKestenbaumLVilgalysRMitchellTG 2005 Comparative analysis of environmental and clinical populations of Cryptococcus neoformans. J Clin Microbiol 43 556 564
11. XuJManosuthiWBanerjeeUZhuL-PChenJ 2011 Cryptococcosis in Asia. HeitmanJKozelTRKwon-ChungJPerfectJRCasadevallA Cryptococcus from human pathogen to model yeast Washington (D.C.) ASM Press 287 298
12. BoversMHagenFKuramaeEEBoekhoutT 2008 Six monophyletic lineages identified within Cryptococcus neoformans and Cryptococcus gattii by multi-locus sequence typing. Fungal Genet Biol 45 400 421
13. LitvintsevaAPThakurRVilgalysRMitchellTG 2006 Multilocus sequence typing reveals three genetic subpopulations of Cryptococcus neoformans var. grubii (serotype A), including a unique population in Botswana. Genetics 172 2223 2238
14. LitvintsevaAPCarboneIRossouwJThakurRGovenderNP 2011 Evidence that the human pathogenic fungus Cryptococcus neoformans var. grubii may have evolved in Africa. PLoS ONE 6 e19688 doi:10.1371/journal.pone.0019688
15. SimwamiSPKhayhanKHenkDAAanensenDMBoekhoutT 2011 Low diversity Cryptococcus neoformans variety grubii multilocus sequence types from Thailand are consistent with an ancestral African origin. PLoS Pathog 7 e1001343 doi:10.1371/journal.ppat.1001343
16. ChoiYHNgamskulrungrojPVarmaASionovEHwangSM 2010 Prevalence of the VNIc genotype of Cryptococcus neoformans in non-HIV-associated cryptococcosis in the Republic of Korea. FEMS Yeast Res 10 769 778
17. LitvintsevaAPMarraRENielsenKHeitmanJVilgalysR 2003 Evidence of sexual recombination among Cryptococcus neoformans serotype A isolates in sub-Saharan Africa. Eukaryot Cell 2 1162 1168
18. LongJL 1981 Introduced birds of the world: the worldwide history, distribution and influence of birds introduced to new environments London David & Charles 208 211
19. GrzimekBSchlagerNOlendorfD 2004 Pigeons and doves. McDadeMC Grzimek's animal life encyclopedia. Volume 9: Birds II Detroit Gale 247 268
20. KronstadJWAttarianRCadieuxBChoiJD'SouzaCA 2011 Expanding fungal pathogenesis: Cryptococcus breaks out of the opportunistic box. Nat Rev Microbiol 9 193 203
21. KumarPYangMHaynesBCSkowyraMLDoeringTL 2011 Emerging themes in cryptococcal capsule synthesis. Curr Opin Struct Biol 21 597 602
22. LiawSJWuHCHsuehPR 2010 Microbiological characteristics of clinical isolates of Cryptococcus neoformans in Taiwan: serotypes, mating types, molecular types, virulence factors, and antifungal susceptibility. Clin Microbiol Infect 16 696 703
23. MitchellTGLitvintsevaAP 2010 Typing species of Cryptococcus and epidemiology of cryptococcosis. AshbeeHRBignellEM Pathogenic yeasts Berlin Heidelberg Springer-Verlag 167 190
24. PedrosoRSLavradorMAFerreiraJCCandidoRCMaffeiCM 2010 Cryptococcus neoformans var. grubii - pathogenicity of environmental isolates correlated to virulence factors, susceptibility to fluconazole and molecular profile. Mem Inst Oswaldo Cruz 105 993 1000
25. AlanioADesnos-OllivierMDromerF 2011 Dynamics of Cryptococcus neoformans-macrophage interactions reveal that fungal background influences outcome during cryptococcal meningoencephalitis in humans. mBio 2 e00158 11
26. MigliaKJGovenderNPRossouwJMeiringSMitchellTG 2011 Analyses of pediatric isolates of Cryptococcus neoformans from South Africa. J Clin Microbiol 49 307 314
27. LitvintsevaAPMitchellTG 2009 Most environmental isolates of Cryptococcus neoformans var. grubii (serotype A) are not lethal for mice. Infect Immun 77 3188 3195
28. Illnait-ZaragoziMTMartinez-MachinGFFernandez-AndreuCMBoekhoutTMeisJF 2010 Microsatellite typing of clinical and environmental Cryptococcus neoformans var. grubii isolates from Cuba shows multiple genetic lineages. PLoS ONE 5 e9124 doi:10.1371/journal.pone.0009124
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2012 Číslo 2- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Mechanisms of Pathogenesis, Infective Dose and Virulence in Human Parasites
- Structural and Functional Insights into the Pilotin-Secretin Complex of the Type II Secretion System
- Characterising the Mucosal and Systemic Immune Responses to Experimental Human Hookworm Infection
- Population Genetic Analyses Reveal the African Origin and Strain Variation of var.
- How Do Microbial Pathogens Make s?
- Substance P Causes Seizures in Neurocysticercosis
- Phagosomal Rupture by Results in Toxicity and Host Cell Death
- Absence of HIV-1 Evolution in the Gut-Associated Lymphoid Tissue from Patients on Combination Antiviral Therapy Initiated during Primary Infection
- Five Questions about Viral Trafficking in Neurons
- Selecting an Invertebrate Model Host for the Study of Fungal Pathogenesis
- A Co-Opted DEAD-Box RNA Helicase Enhances Tombusvirus Plus-Strand Synthesis
- Biochemical Properties of Highly Neuroinvasive Prion Strains
- ClpP1 and ClpP2 Function Together in Protein Degradation and Are Required for Viability and During Infection
- Discrete Cyclic di-GMP-Dependent Control of Bacterial Predation versus Axenic Growth in
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Discrete Cyclic di-GMP-Dependent Control of Bacterial Predation versus Axenic Growth in
- Characterising the Mucosal and Systemic Immune Responses to Experimental Human Hookworm Infection
- How Do Microbial Pathogens Make s?
- Substance P Causes Seizures in Neurocysticercosis
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



