-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaStructural and Functional Insights into the Pilotin-Secretin Complex of the Type II Secretion System
Gram-negative bacteria secrete virulence factors and assemble fibre structures on their cell surface using specialized secretion systems. Three of these, T2SS, T3SS and T4PS, are characterized by large outer membrane channels formed by proteins called secretins. Usually, a cognate lipoprotein pilot is essential for the assembly of the secretin in the outer membrane. The structures of the pilotins of the T3SS and T4PS have been described. However in the T2SS, the molecular mechanism of this process is poorly understood and its structural basis is unknown. Here we report the crystal structure of the pilotin of the T2SS that comprises an arrangement of four α-helices profoundly different from previously solved pilotins from the T3SS and T4P and known four α-helix bundles. The architecture can be described as the insertion of one α-helical hairpin into a second open α-helical hairpin with bent final helix. NMR, CD and fluorescence spectroscopy show that the pilotin binds tightly to 18 residues close to the C-terminus of the secretin. These residues, unstructured before binding to the pilotin, become helical on binding. Data collected from crystals of the complex suggests how the secretin peptide binds to the pilotin and further experiments confirm the importance of these C-terminal residues in vivo.
Published in the journal: . PLoS Pathog 8(2): e32767. doi:10.1371/journal.ppat.1002531
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1002531Summary
Gram-negative bacteria secrete virulence factors and assemble fibre structures on their cell surface using specialized secretion systems. Three of these, T2SS, T3SS and T4PS, are characterized by large outer membrane channels formed by proteins called secretins. Usually, a cognate lipoprotein pilot is essential for the assembly of the secretin in the outer membrane. The structures of the pilotins of the T3SS and T4PS have been described. However in the T2SS, the molecular mechanism of this process is poorly understood and its structural basis is unknown. Here we report the crystal structure of the pilotin of the T2SS that comprises an arrangement of four α-helices profoundly different from previously solved pilotins from the T3SS and T4P and known four α-helix bundles. The architecture can be described as the insertion of one α-helical hairpin into a second open α-helical hairpin with bent final helix. NMR, CD and fluorescence spectroscopy show that the pilotin binds tightly to 18 residues close to the C-terminus of the secretin. These residues, unstructured before binding to the pilotin, become helical on binding. Data collected from crystals of the complex suggests how the secretin peptide binds to the pilotin and further experiments confirm the importance of these C-terminal residues in vivo.
Introduction
The secretins are an important group of bacterial membrane proteins whose function is to facilitate the transport of secreted proteins and macromolecular complexes across the outer membrane [1].They are essential components of the type II and type III secretion systems (T2SS and T3SS respectively) and play a key role in the assembly of type IV pili (T4P) and release of filamentous bacteriophages. Determination of the structure of secretins has been confined to low-resolution transmission electron microscopy and cryo EM studies [2]–[5] which show membrane penetrating ring structures with 12–14 fold rotational symmetry. A specialized class of small lipoprotein pilotins bind their cognate secretins and facilitate oligomerization, insertion and proper assembly in the outer bacterial membrane. In this paper we explore the structure and function of the pilotin from D. dadantii (OutS), several other pilot proteins have been described [6]–[12]. Pilotins whose structures have been determined are MxiM (PDB code: 1Y9L) of the T3SS of Shigella flexneri and PilW/PilF (PDB codes: 2VQ2)/2FI7 and 2HO1) of the T4P of Neisseria meningitidis or Pseudomonas aeruginosa [13], [14]. The cracked β-barrel structure of MxiM has been solved in complex with an 18 residue peptide from the cognate secretin MxiD (PDB code: 2JW1) and the authors propose a model for the way MxiM assists MxiD assembly [10], [15]. The other known pilot structure, PilW/PilF, appears to perform a broadly similar function to MxiM, ensuring multimerization of the secretin PilQ into the outer membrane, but has a different architecture comprising six serial α-helical tetratricopeptide repeats [9], [12]. A third auxiliary secretin-binding protein has been characterised structurally, PilP, which is also involved in the assembly or stability of the secretin PilQ of Pseudomonas aeruginosa [16], the structure comprises a sandwich of two sheets each with three anti-parallel β-strands.
The type II secretion system spans both the inner and outer bacterial membranes [17], [18]. It consists of an inner membrane subcomplex, periplasmic pseudopilins and the outer membrane secretin [19]. There have been considerable recent advances in our understanding of the T2SS secretin. First the structure of the N-terminal periplasmic domains N0, N1, and N2 in complex with a nanobody [20] was elucidated (PDB code 3EZJ) and secondly a cryo EM reconstruction of the secretin itself has been described [3]. In the absence of pilotin the D. dadantii secretin (OutD) mislocates to the inner membrane [21]. The pilotin possesses at its N-terminus the characteristic lipoprotein signal sequence (LAAC), with the signal peptidase LspA cleaving site just before the cysteine to which the lipid is covalently attached. The pilotin (OutS) binds to the C-terminal 62 residues of the secretin (OutD) [11], [19]. Here we elucidate the structure of the T2SS pilotin and show that it binds tightly to 18 residues close to the C-terminus of the secretin subunit causing this unstructured region to become helical on forming the complex.
Results/Discussion
Structure of the T2ss Pilotin
To ensure authentic folding and production of soluble Dickeya dadantii pilotin (OutS) in the E. coli periplasm, the PelB secretion sequence was substituted in place of the N-terminal lipidation sequence thereby preventing lipidation of the pilotin. This substitution facilitated protein production and crystallization without compromising secretin-binding [11]. Cleavage of the secretion signal accompanies transport in to the periplasm. The crystal structure of the pilotin was determined using the anomalous scattering from a potassium tetrachloroplatinate derivative and the structure refined at 1.65 Å resolution (Table 1). The two copies of the pilotin subunit in the asymmetric unit of the crystal are virtually identical in structure (root mean square deviation of 94 α-carbon atoms, residues 38 to 132, is 0.243 Å). The structure is clearly defined in the electron density map except for the N-terminal residues preceding Val 38 which presumably form a flexible linker to the lipidation site. The architecture of the pilotin is the remarkable insertion of one α-helical hairpin into a second open α-helical hairpin with bent final helix (Figure 1); this is unlike the two other pilotin structures solved and is profoundly different from any previously described four helix bundle. The first helix of five turns (residues 40–60), is connected to the second of four turns (residues 69–82) by an 8 residue loop. The second loop of 10 residues connects to the third helix of four turns (residues 93–106) which packs against helix one. A short three residue loop which connects helices three and four and the disulfide bridge, between Cys 115 in the second turn of helix four and Cys 61 the first residue of the helix one to helix two loop (Figure 1B), sets the scene for the packing of helix four (residues 111–130). The pronounced bend of 65° in helix four is important for the architecture; the helix has three large hydrophobic residues which are at least partially buried by interactions with hydrophobic residues on the other three helices: Phe 118, Met 122, and Phe 125. The requirement to pack conserved Phe 125 appears to dictate the severe bend of this helical element. In the crystal the pilotin subunits form a dimer, with Arg 63 and Asn 119 (Figure 1B) involved in an electrostatic interface, between subunits, however there is currently no evidence that dimerization occurs in solution or in vivo.
Fig. 1. Structure of the pilotin. 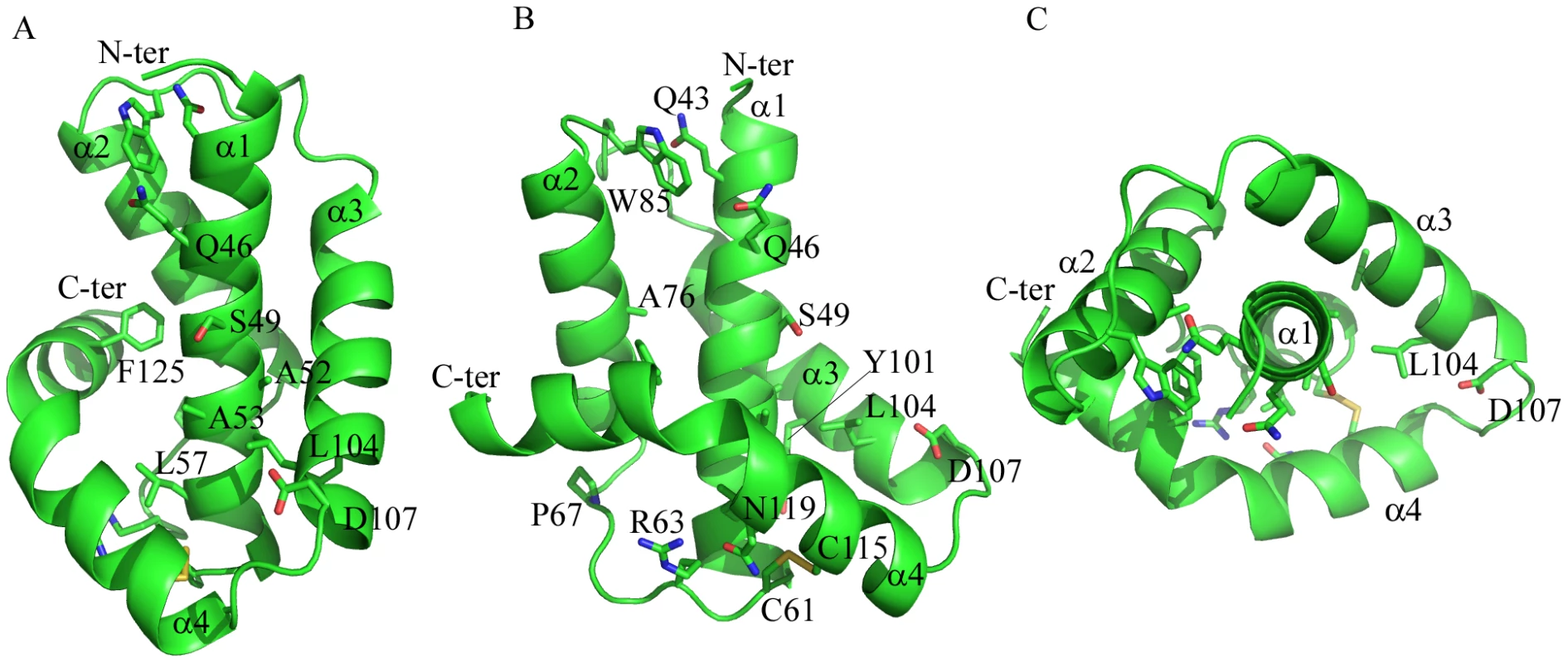
The crystal structure of Dickeya dadantii OutS consists of four α-helices, the last of which is bent. (A) α3 and bent α4 wrap around the anti-parallel hairpin formed by α1 and α2. Conserved residues (see Figure S1 for sequence alignment) are represented as sticks. (B) Rotated about the y-axis by 90°, this view reveals the concave surface formed between helix α1 and helices α3and α4. The disulfide can be seen linking α1 and α4. (C) Rotated around the x-axis by 90°, α1 is surrounded by the other three helices. A52, A53, S49 and D107 are not strictly conserved but the first three are always small and 107 is either D or E suggesting some functional constraint on this region. Q46 is absolutely conserved and may map the other extremity of the binding site. Figure 1 and panels D and E of Figure 3 were produced using PYMOL. Tab. 1. Crystallographic data and refinement statistics for pilotin. 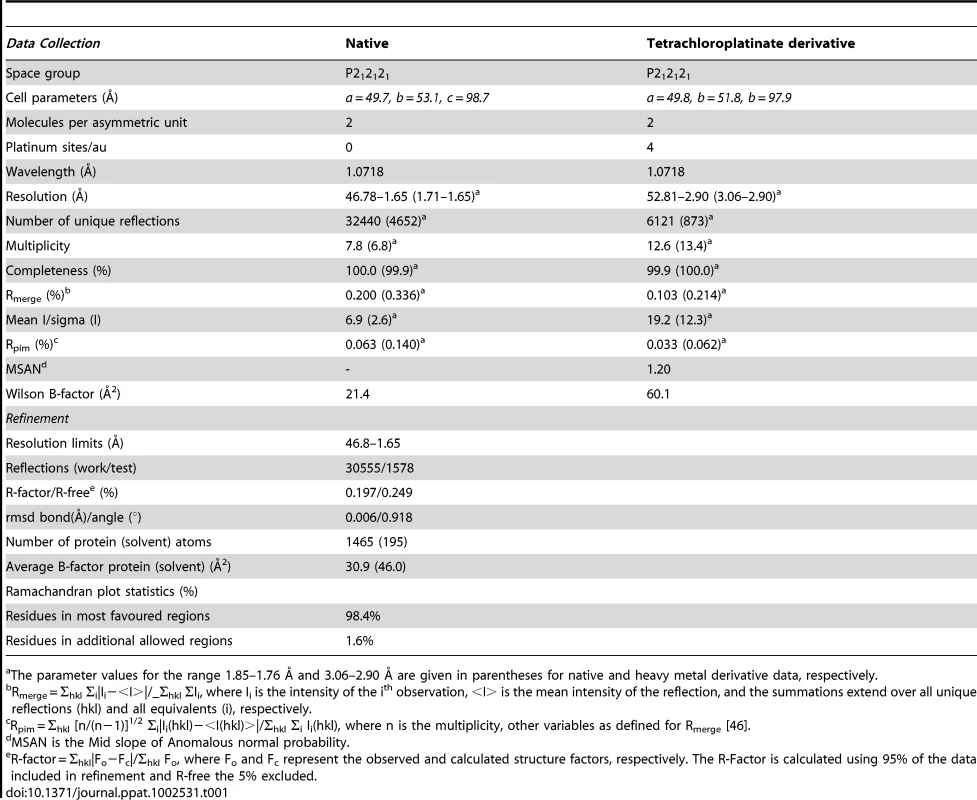
The parameter values for the range 1.85–1.76 Å and 3.06–2.90 Å are given in parentheses for native and heavy metal derivative data, respectively. Sequence Similarity and Structural Similarity
The majority of the 13 absolutely conserved residues in the sequence alignment (Figure S1A) appear to be of structural rather than of direct functional significance. The two highly conserved cysteine residues, 61 and 115, form the disulfide bridge between helices α1 and α4 that stabilizes the correct nested α-helical protein fold of the pilotin is functionally relevant. When a reducing agent was used in pull-down assays or in bacterial two-hybrid tests, the pilotin was unable to bind the cognate secretin (Figure S6). Presumably this is because the disulfide is reduced and the pilotin did not fold correctly in the cytoplasm. Interestingly, the previous mutagenesis analysis revealed several structurally or functionally relevant residues of pilotin (OutS), notably conserved Leu 57, Arg 63 and Ser 97 [11]. Substitution each of these prevented secretin (OutD) targeting to the outer membrane. Conserved Cys 21 covalently attaches to the lipid and this residue is essential for targeting the pilotin to the outer membrane. Conserved Gln 46, on the solvent exposed surface of helix 1 (Figure 1), must also be of functional rather than structural significance and maps to the extremity of the concave surface of the pilotin formed by helices α1, α3 and α4; it is plausible that this concave surface is the binding site for the secretin (Figure 1B). Conserved residues including: Gln 46 and Leu 104 and semi-conserved Leu/Val/Ile 50, Phe/Leu 118 are also in this region. Residues 49, 52 and 53 on the solvent exposed surface of helix α1 are Ser/Ala for the former two and Ala/Gly for the latter, respectively (Figure S1A). This conservation of small residues at these positions is consistent with this region being important in binding as there is no structural reason why larger residues could not be accommodated at these sites unless the secretin binds tight up against the first helix (Figure 1). A DaliLite database search revealed that P40 nucleoprotein has a similar arrangement of α-helices to that of the pilotin. The Dali score was 6.0 and sequence identity 5%. The P40 nucleoprotein domain architecture is however substantially more complex with seven helices instead of the pilotin's four. The concave surface of the corresponding helices of P40 nucleoprotein is occupied by a helix supporting the view that this is the binding site for an α-helix. Mutations of the pilotin binding surface confirm the importance for binding of some of the residues decorating the concave surface (Table S1). Mutating Ser 49 to Arg has a profound effect on binding and the mutants Leu 96 Ala, Leu 100 Ala and Gln 114 Ala have a substantial effect on binding as expected if this is the binding surface (Figure 1B).
Secretin-Binding to the Pilotin in Solution
It had previously been shown that the C-terminal 62 residues of the secretin bound to the pilotin [11]. This secretin peptide was produced with 15N-label as a fusion to GST and then released with PreScision protease. The backbone amide protons were poorly dispersed in the 1H-15N HSQC spectra revealing the C-terminal 62 residues are unstructured in solution (Figure S4). NMR cross-titration studies revealed that only peaks corresponding to residues 691–708 of the secretin peptide were shifted on addition of unlabelled pilotin (Figure S2; with assignment of secretin peptide shown in Figure S3). When the pilotin was 15N-labelled, good dispersion of the backbone amides protons was observed as expected given its folded structure (Figure 2). Titration of the unlabelled 62 residue secretin peptide into the 15N-labelled pilotin produced a large number of peak shifts (Figure 2A). Shift perturbations are extremely sensitive indicators of structural changes and the extent of the changes observed is compatible with the secretin peptide decorating the surface of the pilotin and causing subtle structural rearrangements, perhaps in packing interactions in the hydrophobic core, reflected in chemical shift changes across much of the structure. The secretin peptide binds to the pilotin in a 1∶1 stoichiometric ratio as determined from the NMR titration where the intensity of the shifted peaks of the complex saturate at an equimolar ratio of OutD to OutS. Since only residues 691–708 of the secretin were affected by interaction with pilotin (Figure S2), a synthetic 18 residue peptide corresponding in sequence to these residues was assessed. The pattern of shifts in 1H-15N HSQC pilotin spectrum observed using this synthetic 18 residue peptide was identical to that using the 62 residue secretin peptide (Figure 2B, C) confirming that it is these 18 residues that are those principally involved in binding to the pilotin. Circular dichroism measurements also showed the unstructured nature of the secretin peptide and provided evidence that the peptide becomes helical on binding (Figure 3A). The signal saturates at a stoichiometric ratio of secretin peptide to pilotin. The CD spectra of the pilotin and secretin peptide together correspond to more helical structure than the spectra of the pilotin and secretin peptide summed. The additional helical content can be quantified as 12 residues assuming all secretin and pilotin molecules are in complex, a reasonable assumption given the high affinity of complex formation (see below). The most plausible explanation for this is that 12 residues of the secretin peptide become helical on binding to the pilotin. The helical propensity of the 18 residue secretin peptide was apparent from secondary structure predictions (Jpred [22] and shown on Figure S1B). To estimate the binding affinity of pilotin for the secretin peptide, fluorescence spectroscopy was used. Since the 18 residue secretin peptide has no tryptophan residues, quenching of the fluorescence signal from the single tryptophan residue in the pilotin on addition of the secretin peptide, was used to determine the affinity of binding. The 1∶1 stoichiometric binding ratio can be seen from the saturation of the fluorescence quenching of OutS by an equimolar quantity of the secretin peptide (Figure 3B). The binding of the 18 residue peptide is tight with Kd of 55±20 nM (Figure 3B) and is comparable to that of the T3SS pilotin-secretin complex [10], [15]. 3JHNHA spectra of the complex showed peaks coupled by less than 5 Hz (Figure S5) confirming that at least four residues of the secretin peptide become helical on binding to the pilotin, there may be more, but they are hidden by overlapping peaks.
Fig. 2. Elucidation of secretin-pilotin interactions. 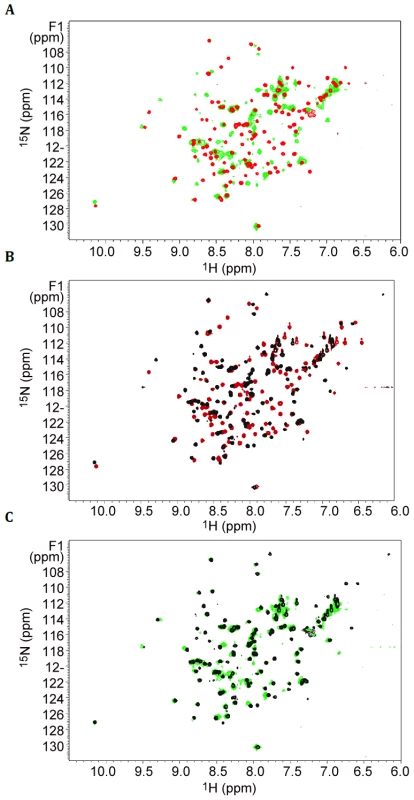
Titration of secretin peptides into 15N labelled pilotin. 1H-15N-HSQC spectra of the pilotin in the absence of secretin (red spectra), in the presence of 62 residue secretin peptide (green) and in the presence of 18 residue peptide (black). Protein concentration was 100 µM. (A) Pilotin in the absence and presence of 62 residue secretin peptide. (B) Pilotin in the absence and presence of the 18 residue secretin peptide. (C) Overlay of the two complexes with secretin peptides showing the 18 residue peptide is behaving in a closely similar way to the 62 residue peptide. Fig. 3. The secretin-pilotin complex. 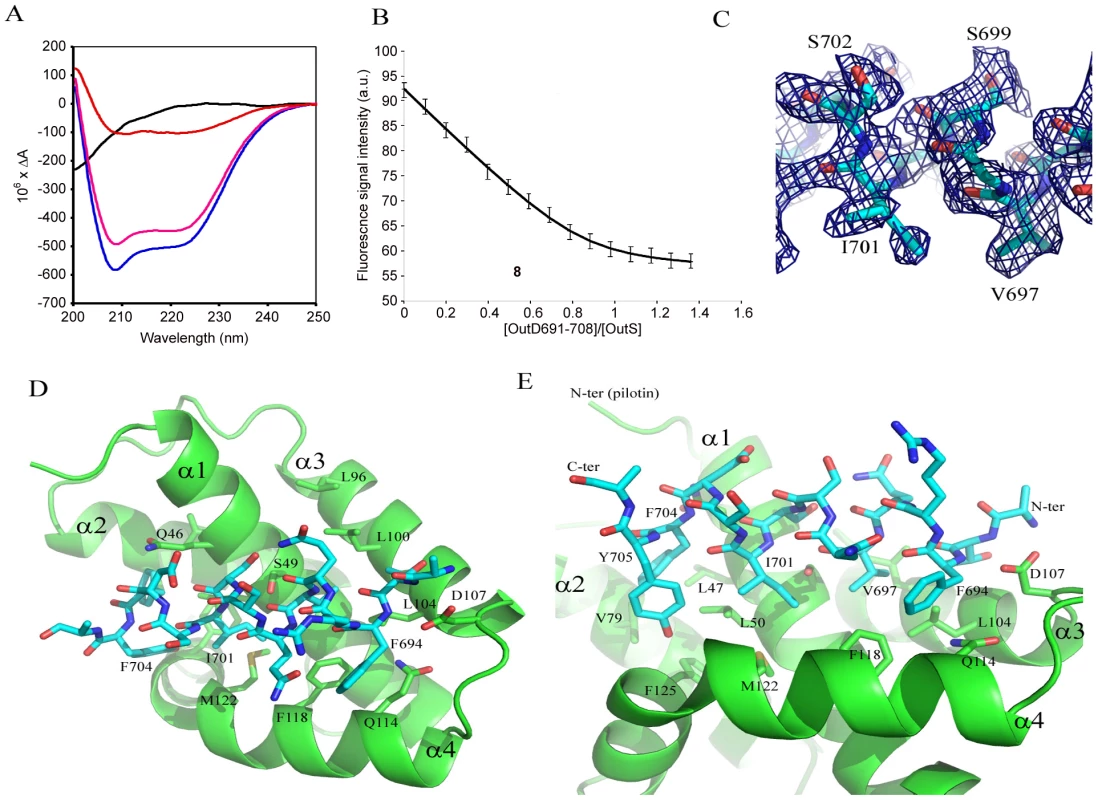
(A) The far uv circular dichroism spectra of the 18 residue secretin peptide alone (black), pilotin alone (pink) and a stoichiometric ratio of both pilotin (OutS) and secretin peptide (OutD691–708) together (blue). The difference between the secretin/pilotin complex and the pilotin only is shown in red. As forming the stoichiometric ratio diluted both proteins by half, these data were multiplied by two to compensate for the dilution factor. The concentration of both pilotin and secretin peptide were 0.55 mM. 3JHNHA evidence of helical conformation is presented in Figure S5. (B) Measurement of the binding affinity of the pilotin OutS for the 18 residue secretin peptide determined using fluorescence spectroscopy. 1 µM pilotin was titrated with 50 µM secretin peptide in to 1 µM pilotin so that there was no dilution of pilotin. The stoichiometry can be seen to be 1∶1 within experimental error. The Kd is 55±20 nM. Details of the equation fitted can be found in Table S1. (C) Part of the simulated annealed omit map showing the quality of the electron density map used to derive the models shown in (D) and (E). (D) Model of the secretin peptide bound to the pilotin, D107 acts as an N-terminal helical cap. (E) Close up showing the hydrophobic nature of the complementary side chains involved in forming the complex. Model of the Pilotin/Secretin Peptide Complex
Crystals of the pilotin/secretin complex were grown, they belong to space group P65 but are twinned (Table S2) and have solvent channels with disordered density within. Nevertheless four pilotin subunits can be found by molecular replacement and density for peptide can be seen occupying the concave surface of the pilotin. The evidence that suggests residues 694 to 704 of the secretin adopt a helical conformation in the electron density map, a part of the simulated annealed omit map is shown in Figure 3C. Ten residues are in helical conformation and the hydrophobic surface of the amphipatic helix interacts with the hydrophobic surface of the pilotin (Figure 3D, E). In this model the methyl groups of T692 and V697 interact with L100 and L104, I701 interacts with L50 and M122; F704 (with L47, V79 and F125) and F694 (with F118, L104 and Q114) interactions occur either side of these central interactions. D107 is the N-terminal helix capping residue, stabilizing the helix dipole of the bound secretin peptide. The peptide binds tight up against Q46, S49, A52 and A53, providing an explanation for their conservation or presence only as small residues in the case of the latter three. Interestingly, the same dimer as described for the non-complexed secretin is seen in these crystals too suggesting that the interaction may have some biological relevance. The quality of the refinement is relatively poor because of the disordered protein and for that reason the structure is being referred to as a model of the secretin peptide/pilotin interaction.
Intrinsically disordered regions of proteins such as these C-terminal residues of the secretin subunit facilitate binding by increasing their capture radius for cognate partner, the so-called fly-casting mechanism [23]. Initial weak binding may draw the secretin and pilotin together and as the secretin peptide folds on the pilotin surface the binding becomes tighter, locking the two together.
Assessment of the Pilotin/Secretin Interaction In Vivo
A series of in vivo experiments were used to test the proposed model of secretin binding to the pilotin. The 62 residue C-terminus of secretin possesses three putative α-helices (Figure 4A) [22]. If the region consisting of the first two C-terminal helices were deleted, the truncated OutDΔC1 behaves like wild type secretin. Firstly, OutDΔC1 was barely detectable in the absence but was well produced in the presence of pilotin OutS (Figure 4B). Secondly, with pilotin, the mutant secretin was mainly recovered in the outer membrane fractions (Figure 4C). Consistent with the outer membrane location, expression of OutDΔC1 in the presence of OutS results in a rather low level of pspA induction. Phage shock protein (psp) response helps to maintain proton motive force in cells under pmf-dissipating stress and is indicative of mislocalization of the secretins in the inner membrane [8], [11]. These results are consistent with the NMR experiments demonstrating that it is the 18 residue C-terminus of OutD which binds tightly to the pilotin. Despite its outer membrane location, OutDΔC1 was unable to restore pectinase secretion in D. dadantii ΔoutD A3559 strain (data not shown) indicating an important functional relevance of the deleted region. Deletion of the extreme C-terminus of secretin resulted in partial stabilization of the truncated OutDΔC2 secretin as judged from the quantity produced in the absence of the pilotin, but prevented its correct targeting to the outer membrane (Figure 4B,C). In the presence of pilotin, the amount of OutDΔC2 was increased indicating that the pilotin can still stabilize and hence interact with the truncated secretin but is not able to target it to the outer membrane. In agreement with this, expression of OutDΔC2 strongly induced pspA even in the presence of pilotin. Deletion of the full 62 residue C-terminus of OutD resulted in neither stabilization nor the correct targeting of the truncated secretin OutDΔC3 as judged by low protein content but high pspA level (Figure 4B).
Fig. 4. In vivo determination of secretin interactions with pilotin. 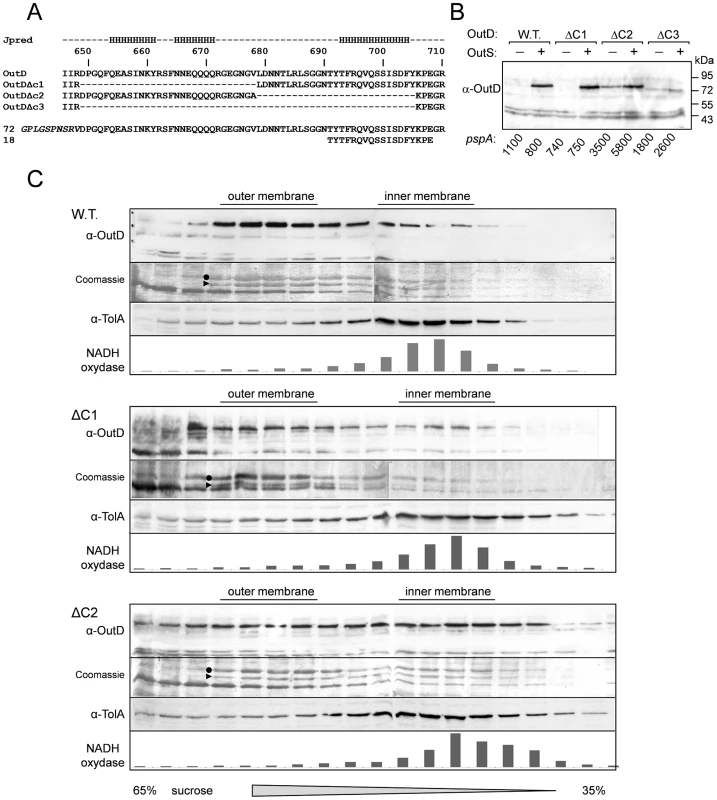
Removal of the extreme C-terminal region of the secretin results in mislocalization of the secretin to the inner membrane (A) Sequence of the C-terminus of Dickeya dadantii secretin (OutD) along with the secondary structure prediction for this region; the predicted three helices are marked “H”. The sequences of the three secretin deletion mutants (OutDΔC1 to OutDΔC3) and the C-terminal secretin peptides used are shown. (B) Stabilization of truncated derivatives of the secretin (OutD) by pilotin (OutS) in vivo. E. coli MC3 cells expressing an OutD derivative (indicated above) and either OutS (+) or empty pACT3 vector (−) were grown for 12 h at 30°C in LB medium and then analyzed by immunoblotting with OutD antibodies. In the same cultures, β-galactosidase activity was assessed to estimate expression of pspA-lacZ. An elevated level of pspA reflects mislocalization of the corresponding secretin derivative to the inner membrane. Equivalent amounts of cell extracts were loaded into each well and used for activity measurement. (C) Pilotin promotes the outer membrane location of the full-length secretin OutD and truncated secretin OutDΔC1 but not OutDΔC2. The whole membrane fraction from E. coli NM522 cells coexpressing the indicated secretin derivatives and pilotin was separated by flotation sucrose gradient centrifugation and analyzed by immunoblotting with OutD- antibodies or stained with Coomassie G-250 to detect the major porins, which reflect the position of the outer membrane. Immunoblotting with TolA-antibodies and NADH-oxydase activity indicate the position of the inner membrane fractions. OmpA is indicated by a triangle and OmpC/F by a dot. Conclusion
The type II secretin/pilotin complex from Klebsiella oxytoca has been imaged by cryo electron microscopy at modest resolution [24]. The secretin subunits form a dodecameric ring with relatively weak radial arms that the authors tentatively assign as the pilotin or pilotin bound to the secretin C-terminus [4], [24]. Comparing the envelope of the complex with that determined by Reichow et al. (2010) of the secretin only [3], the radial arms are located either in the periplasm or in the inner leaflet of the outer membrane. This is the position anticipated from the observation in this work that the pilotin interacts tightly with 18 residues close to the C-terminus of the secretin subunit, so it is entirely plausible that the radial arms seen in the cryo-EM map correspond to the unusual four helix bundle of the type II secretion pilotin bound to the induced C-terminal helix of the secretin subunit.
Recent structural studies have revealed striking structural similarities between components from distant secretion systems and other bacterial cell machines. Besides the expected structural homologies between several conserved components of ancestrally related T2SS and T4P [25]–[27] several other remarkable structural similarities should be mentioned. Notably, the extreme N-terminal N0 domain is shared by secretins from the T2SS, T3SS and T4P but is also structurally related to a domain of lipoprotein DotD from the T4SS, a domain of VgrG from the T6SS and from a TonB dependent receptor FpvA [20], [28]–[30]. Similarly the N1/N2 domains of secretins show a significant structural homology with several ring-forming proteins from the T3SS [20], [31], [32]. It is therefore becoming common to attribute similar function to the proteins or domains of a related structure. Given this background it is remarkable then that the T2SS pilotin described here is profoundly different in architecture to the T3SS pilotin but has similar function. Both the T2SS and the T3SS pilotin bind the extreme C-terminal region of their cognate secretins and this previously unstructured part of the secretin becomes an ordered α-helix on binding. It is therefore remarkable that the corresponding pilotins are different in architecture, one an open β-barrel (T3SS; MxiM) the other an unusual helical bundle (T2SS; OutS). These striking structural differences show that in these systems the pilotins have been evolved independently to play similar roles.
Materials and Methods
Plasmid Construction, Expression, Purification and Protein Analysis
The pET-20b(+) plasmid expressing non-lipidated OutS (residues 26 to 132) fused to N-terminal PelB signal peptide has been constructed previously [11] at a manner as the sequence of mature non-lipidated OutS after cleavage by the signal peptidase LepB is: MDP26VKNT etc. To fuse a C-terminal 6His tag to non-lipidated OutS, SalI site was introduced at the end of outS sequence by using the primer (5′-ctt gac gcc atg cgc acc gtc gac tga ggg gga agc aac tgc) and the reverse complementary one (mutated bases are underlined). Then, by SalI/XhoI digestion the sequence coding for non-lipidated OutS was fused with that coding for 6His. Mutants of OutS were made using Strategene QuikChange and confirmed by sequencing.
To generate OutD truncated derivatives, an Eco47III site and V678A substitution were introduced using the primer (5′-gcgcggcgaaggcaacggagcgctggataacaacaccctgc) and the reverse complementary one. This site and naturally existing NruI and PsiI sites were used to generate OutDΔC1 (Δ650–678), OutDΔC2 (Δ679–705) and OutDΔC3 (Δ650–705) derivatives. To fuse the C-terminal segments of OutD to GST, the corresponding gene fragments were subcloned from pTdB-OD plasmids expressing either OutD, or OutDΔC1, or OutDΔC2, or OutDΔC3 into pGEX-6P-3 or pGEX-3X vectors in frame with the GST coding sequence.
Protein Expression, Purification and Analysis
E. coli BL21(DE3) strain (Stratagene) was used to produce non-lipidated pilotin (OutS) and GST-secretin (OutD) derivatives. Non-lipidated OutS was released from the periplasm by osmotic shock as described previously [33] and purified by size-exclusion chromatography Superdex S75 10/300 GL (GE Healthcare). The OutD peptide was purified and then released from GST-OutD fusion as described previously [34]. For NMR spectroscopy, uniformly 15N - and 13C-labeled pilotin and secretin peptides were produced by growing cell cultures in M9 minimal medium that contained 15N-ammonium chloride and 13C-D-glucose (Cambridge Isotope Laboratories Inc.) as the sources of nitrogen and carbon, respectively. The 18 residue synthetic secretin peptide (residues 691 to 708 inclusive) was purchased from Generon. Cell membrane fractionation by sucrose density gradient centrifugation was performed as described previously [11] with steady-state cultures of E. coli NM522 (Stratagene) expressing OutD derivatives from pTdB-OD and OutS from pACT-S plasmid. The location of outer membrane porins was determined by staining with Coomassie G-250. The position of inner membrane fractions was estimated by immunoblotting with TolA antibodies and NADH oxidase activity. E. coli MC3 strain carrying a pspA-lacZ fusion [35] was used to estimate miss location of OutD derivatives. To assess functionality of OutD derivatives, complementation assays with D. dadantii ΔoutD A3559 strain were used as previously described [36]. SDS-PAGE and immunoblotting were performed as previously [19]. Anti-OutD rabbit serum was raised against entire OutD purified from a recombinant E. coli strain. Anti-TolA serum was kindly provided by J.C. Lazzaroni.
Crystallisation and Structure Determination
Hampton Research sparse matrix screen was used to search for crystallization conditions. Crystals were grown using hanging drop vapour equilibration using 10 mg/ml OutS and a reservoir of 2 M ammonium sulphate, 2% PEG 400 and 0.1 M HEPES pH 7.5. Vitrification of crystals in liquid nitrogen was achieved using the reservoir solution with 2.1 M ammonium sulphate and augmented with 15–25% glycerol. Data were collected at ESRF ID23-1 and processed using MOSFLM [37] and scaled using SCALA [38]. SAD data were collected from a crystal soaked in 25 mM potassium tetrachloroplatinate (K2PtCl4) for 4 days. The structure was solved using PHENIX [39] and COOT [40] and refined using the native data at 1.65 Å resolution and non-crystallographic symmetry restraints. The final model comprises 188 amino acid residues and 208 water molecules. DALILITE [41] was used to search for similar structures, CLUSTALW [42] for sequence alignment and JPRED for secondary structure predictions [22]. Crystals of the complex were grown using a 1.0∶1.1 molar ratio of pilotin: secretin peptide and crystallized using a reservoir of 2 M ammonium sulphate, 0.1 M Tris, pH 8.5. Around 100 complex crystals were screened before a well-diffracting reasonable ordered crystal was found. The pilotin/secretin complex was solved by molecular replacement using data collected at DLS I02 and CCP4/PHENIX/COOT. These crystals appear to be P6522 but are most likely twinned P65 with four pilotin molecules in the asymmetric unit. The packing of the molecules is such that there are large solvent channels running through the crystal lattice. These solvent channels appear to have disordered protein present the modelling of which hampers refinement. The disordered regions do not gain clarity if the lower symmetry space group P32 is used (for more details see Table S2).
Preparation of Proteins for Nmr Spectroscopy and Acquisition of Nmr Spectra
Samples of 0.05 to 0.5 mM labelled proteins in 90% H2O, 10% 2H2O containing 20 mM Tris (pH 7.0) and 150 mM NaCl. All NMR spectra were acquired at 15°C using Bruker Avance 700 - and 600-MHz spectrometers. Assignment of 1H, 15N, and 13C resonances of the backbone was achieved by analysis of HNCACB, CBCA (CO)NH triple resonance experiments [43].
Circular Dichroism
Far-UV CD measurements were made using a Jasco J-715 spectropolarimeter equipped with a PTC-348WI temperature controller. Spectra were recorded in 20 mM Tris, 150 mM NaCl (pH 7.0) at 15°C using 1 mm path length fused silica cuvettes. The spectra are presented as differential absorbance after baseline subtraction. Calculations employed CONTIN, SELCON, and CDSSTR [44].
Fluorescence Spectroscopy
Fluorescence data were collected using a Jasco FP-6300 Spectrofluorometer. To avoid exciting tyrosyl side chains, an excitation wavelength of 290 nm was used. Emission spectra were recorded at 15°C in steps of 2 nm from 310 to 400 nm. The fluorescence signal at 340 nm was plotted to calculate Kd. Pilotin spectra was measured at 1 µM. 50 µM of secretin peptide prepared in 1 µM pilotin was titrated into pilotin solution (for more information see also Table S1 and reference [45]).
Accession codes
Coordinates and structure factor amplitudes have been deposited in the protein databank with the accession codes 3UTK and 3UYM. The sequences of OutS and OutD are available in the UniProt database with accession codes Q01567 and Q01565, respectively.
Supporting Information
Zdroje
1. BayanNGuilvoutIPugsleyAP 2006 Secretins take shape. Mol Micro 60 1 4
2. CollinsRFFryeSAKitmittoAFordRCTonjumT 2004 Structure of the Neisseria meningitidis outer membrane PilQ secretin complex at 12 angstrom resolution. J Biol Chem 279 39750 39756
3. ReichowSLKorotkovKVHolWGJGonenT 2010 Structure of the cholera toxin secretion channel in its closed state. Nature Struct Mol Biol 17 1226 1233
4. ChamiMGuilvoutIGregoriniMRemigyHWMullerSA 2005 Structural insights into the secretin PulD and its trypsin-resistant core. J Biol Chem 280 37732 37741
5. HodgkinsonJLHorsleyAStabatDSimonMJohnsonS 2009 Three-dimensional reconstruction of the Shigella T3SS transmembrane regions reveals 12-fold symmetry and novel features throughout. Nat Struct Mol Biol 16 477 485
6. BurghoutPBeckersFde WitEvan BoxtelRCornelisGR 2004 Role of the pilot protein YscW in the biogenesis of the YscC secretin in Yersinia enterocolitica. J Bacteriol 186 5366 5375
7. CragoAMKoronakisV 1998 Salmonella InvG forms a ring-like multimer that requires the InvH lipoprotein for outer membrane localization. Mol Micro 30 47 56
8. HardieKRSeydelAGuilvoutIPugsleyAP 1996 The secretin-specific, chaperone-like protein of the general secretory pathway: Separation of proteolytic protection and piloting functions. Mol Micro 22 967 976
9. KooJTammamSKuSYSampaleanuLMBurrowsLL 2008 PilF Is an Outer Membrane Lipoprotein Required for Multimerization and Localization of the Pseudomonas aeruginosa Type IV Pilus Secretin. J Bacteriol 190 6961 6969
10. LarioPIPfuetznerRAFreyEACreaghLHaynesC 2005 Structure and biochemical analysis of a secretin pilot protein. EMBO J 24 1111 1121
11. ShevchikVECondemineC 1998 Functional characterization of the Erwinia chrysanthemi OutS protein, an element of a type II secretion system. Microbiology 144 1443219 3228
12. TrindadeMBJobVContreras-MartelCPelicicVDessenA 2008 Structure of a widely conserved type IV pilus biogenesis factor that affects the stability of secretin multimers. J Mol Biol 378 1031 1039
13. DerrickJ 2008 A Pilot Sheds Light on Secretin Assembly. Structure 16 1441 1442
14. KorotkovKVGonenTHolWGJ 2011 Secretins: dynamic channels for protein transport across membranes. Trends Biochem Sci 36 433 443
15. OkonMMoraesTFLarioPICreaghALHaynesCA 2008 Structural Characterization of the Type-III Pilot-Secretin Complex from Shigella flexneri. Structure 16 1544 1554
16. GolovanovAPBalasinghamSTzitzilonisCGoultBTLianLY 2006 The solution structure of a domain from the Neisseria meningitidis lipoprotein PiIP reveals a new beta-sandwich fold. J Mol Biol 364 186 195
17. SandkvistM 2001 Biology of type II secretion. Mol Microbiol 40 271 283
18. JohnsonTLAbendrothJHolWGJSandkvistM 2006 Type II secretion: from structure to function. FEMS Micro Letts 255 175 186
19. ShevchikVERobertBaudouyJCondemineG 1997 Specific interaction between OutD, an Erwinia chrysanthemi outer membrane protein of the general secretory pathway, and secreted proteins. EMBO J 16 3007 3016
20. KorotkovKVPardonESteyaertJHolWGJ 2009 Crystal Structure of the N-Terminal Domain of the Secretin GspD from ETEC Determined with the Assistance of a Nanobody. Structure 17 255 265
21. GuilvoutIChamiMEngelAPugsleyAPBayanN 2006 Bacterial outer membrane secretin PulD assembles and inserts into the inner membrane in the absence of its pilotin. EMBO J 25 5241 5249
22. ColeCBarberJDBartonGJ 2008 The Jpred 3 secondary structure prediction server. Nucl Acid Res 36 W197 W201
23. ShoemakerBAPortmanJJWolynesPG 2000 Speeding molecular recognition by using the folding funnel: The fly-casting mechanism. Proc Natl Acad Sci U S A 97 8868 8873
24. NouwenNRansonNSaibilHWolpensingerBEngelA 1999 Secretin PulD: Association with pilot PulS, structure, and ion-conducting channel formation. Proc Natl Acad Sci U S A 96 8173 8177
25. SampaleanuLMBonannoJBAyersMKooJTammamS 2009 Periplasmic Domains of Pseudomonas aeruginosa PilN and PilO Form a Stable Heterodimeric Complex. J Mol Biol 394 143 159
26. AbendrothJRiceAEMcLuskeyKBagdasarianMHolWGJ 2004 The crystal structure of the periplasmic domain of the type II secretion system protein EpsM from Vibrio cholerae: The simplest version of the ferredoxin fold. J Mol Biol 338 585 596
27. AbendrothJKregerACHolWGJ 2009 The dimer formed by the periplasmic domain of EpsL from the Type 2 Secretion System of Vibrio parahaemolyticus. J Str Biol 168 313 322
28. NakanoNKuboriTKinoshitaMImadaKNagaiH 2010 Crystal Structure of Legionella DotD: Insights into the Relationship between Type IVB and Type II/III Secretion Systems. PLOS Pathog 6 e1001129
29. LeimanPGBaslerMRamagopalUABonannoJBSauderJM 2009 Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc Natl Acad Sci U S A 106 4154 4159
30. Garcia-HerreroAVogelHJ 2005 Nuclear magnetic resonance solution structure of the periplasmic signalling domain of the TonB-dependent outer membrane transporter FecA from Escherichia coli. Mol Micro 58 1226 1237
31. YipCKKimbroughTGFeliseHBVuckovicMThomasNA 2005 Structural characterization of the molecular platform for type III secretion system assembly. Nature 435 702 707
32. WorrallLJVuckovicMStrynadkaNCJ 2010 Crystal structure of the C-terminal domain of the Salmonella type III secretion system export apparatus protein InvA. Protein Sci 19 1091 1096
33. FriesMIhrigJBrocklehurstKShevchikVEPickersgillRW 2007 Molecular basis of the activity of the phytopathogen pectin methylesterase. EMBO J 26 3879 3887
34. LoginFHShevchikVE 2006 The single transmembrane segment drives self-assembly of OutC and the formation of a functional type II secretion system in Erwinia chrysanthemi. J Biol Chem 281 33152 33162
35. BerglerHAbrahamDAschauerHTurnowskyF 1994 Inhibition of lipid biosynthesis induces the expression of the pspa gene. Microbiology 140 1937 1944
36. BouleyJCondemineGShevchikVE 2001 The PDZ domain of OutC and the N-terminal region of OutD determine the secretion specificity of the type II out pathway of Erwinia chrysanthemi. J Mol Biol 308 205 219
37. LeslieAGW 2006 The integration of macromolecular diffraction data. Acta Cryst D62 48 57
38. EvansP 2006 Scaling and assessment of data quality. Acta Cryst D62 72 82
39. AdamsPDAfoninePVBunkocziGChenVBDavisIW 2010 PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Cryst D66 213 221
40. EmsleyPLohkampBScottWGCowtanK 2010 Features and development of Coot. Acta Cryst D66 486 501
41. HolmLKaariainenSRosenstromPSchenkelA 2008 Searching protein structure databases with DaliLite v.3. Bioinformatics 24 2780 2781
42. LarkinMABlackshieldsGBrownNPChennaRMcGettiganPA 2007 Clustal W and clustal X version 2.0. Bioinformatics 23 2947 2948
43. BaxAGrishaevA 2005 Weak alignment NMR: a hawk-eyed view of biomolecular structure. Curr Opin Struct Biol 15 563 570
44. SreeramaNVenyaminovSYWoodyRW 2000 Estimation of protein secondary structure from circular dichroism spectra: Inclusion of denatured proteins with native proteins in the analysis. Anal Biochem 287 243 251
45. MartinSRSchilstraMJ 2008 Circular dichroism and its application to the study of biomolecules. Biophysical Tools for Biologists: Vol 1 in Vitro Techniques San Diego Elsevier Academic Press Inc 263 293
46. WeissMS 2001 Global indicators of X-ray data quality. J Appl Cryst 34 130 135
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2012 Číslo 2- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Mechanisms of Pathogenesis, Infective Dose and Virulence in Human Parasites
- Structural and Functional Insights into the Pilotin-Secretin Complex of the Type II Secretion System
- Characterising the Mucosal and Systemic Immune Responses to Experimental Human Hookworm Infection
- Population Genetic Analyses Reveal the African Origin and Strain Variation of var.
- How Do Microbial Pathogens Make s?
- Substance P Causes Seizures in Neurocysticercosis
- Phagosomal Rupture by Results in Toxicity and Host Cell Death
- Absence of HIV-1 Evolution in the Gut-Associated Lymphoid Tissue from Patients on Combination Antiviral Therapy Initiated during Primary Infection
- Five Questions about Viral Trafficking in Neurons
- Selecting an Invertebrate Model Host for the Study of Fungal Pathogenesis
- A Co-Opted DEAD-Box RNA Helicase Enhances Tombusvirus Plus-Strand Synthesis
- Biochemical Properties of Highly Neuroinvasive Prion Strains
- ClpP1 and ClpP2 Function Together in Protein Degradation and Are Required for Viability and During Infection
- Discrete Cyclic di-GMP-Dependent Control of Bacterial Predation versus Axenic Growth in
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Discrete Cyclic di-GMP-Dependent Control of Bacterial Predation versus Axenic Growth in
- Characterising the Mucosal and Systemic Immune Responses to Experimental Human Hookworm Infection
- How Do Microbial Pathogens Make s?
- Substance P Causes Seizures in Neurocysticercosis
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



