-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaNews from the Fungal Front: Wall Proteome Dynamics and Host–Pathogen Interplay
article has not abstract
Published in the journal: . PLoS Pathog 8(12): e32767. doi:10.1371/journal.ppat.1003050
Category: Pearls
doi: https://doi.org/10.1371/journal.ppat.1003050Summary
article has not abstract
Introduction
In Candida albicans, like in Saccharomyces cerevisiae, the basal layer of the mature cell wall consists of a network of β-1,3 - and β-1,6-glucans and chitin and functions as a skeletal layer. This basal layer is covered by an external layer of highly glycosylated, covalently anchored wall proteins radiating from the cell surface, which are directly involved in the first contacts between the fungal pathogen and host cells. The majority of the covalently bound wall proteins are modular glycosylphosphatidylinositol (GPI)-proteins. In their final form, wall-bound GPI-proteins usually consist of a C-terminal, truncated GPI-anchor that attaches them to the β-glucan layer, followed by a heavily glycosylated serine/threonine-rich spacer domain that often includes repeats, and an N-terminally located functional domain protruding from the cell surface [1]. At any given time-point >20 different covalently bound wall proteins can be identified [2], [3] that are involved in processes such as adhesion, biofilm formation, wall remodeling, iron acquisition, and coping with immune responses. Importantly, the wall proteome is highly dynamic and continuously adapts to the specific conditions that C. albicans encounters in the host environment. In this review we examine the role of wall proteins in infection-related processes and assess their potential as targets for antifungal and vaccine development.
Why Do Most Wall Proteins Form Families?
C. albicans is able to thrive in many host niches, including the skin, mucosal surfaces, the bloodstream, and internal organs. Wall proteins are subject to the surrounding conditions and come into contact with highly diverse, niche-associated, extracellular matrix proteins from the host as well as with bacterial surface proteins. This probably explains the evolution of many wall protein families with individual members showing optimal functionality dependent on environmental conditions and infection sites [1]. For example, the environmental pH strongly affects the wall proteome, revealing the preferred usage of specific family members at acidic and neutral pH [4]. Interestingly, invasive growth is generally associated with hyphal growth, and comparison of the wall proteomes of yeast and hyphal cells revealed a core set of hypha-associated wall proteins under various hyphal growth-inducing conditions (Als3, Hwp1, Hwp2, Hyr1, Plb5, and Sod5) [2], [5]. The two largest wall protein families are the Als family [6] and the Hyr/Iff family [7]. The family of agglutinin-like sequence (ALS) proteins consists of eight GPI-modified, elongated, broad-specificity adhesins with an immunoglobulin-like N-terminal domain that can interact with a wide variety of host proteins [8]. Some Als proteins possess amyloid-forming sequences, which could play a role in forming biofilms [9]. Fascinatingly, Als3 has multiple functions, including ferritin binding [10] as well as binding to E-cadherin, thereby facilitating iron uptake and active internalization of C. albicans by host cells, respectively [11]. This supports that proteins of a family may share a particular function, but might also have additional functions that are not conserved throughout the family. Intriguingly, Hyr1, one of the 12 GPI-proteins belonging to the Iff/Hyr family, is strongly hypha-associated and confers resistance to neutrophil killing [12] through its N-terminal domain. Although the domain structure within the family is variable, the N-terminal domain is strongly conserved in all family members (Figure 1) [7]. This hints at a more general, niche-specific role of the family in evading immune cells under different growth conditions.
Fig. 1. Key concepts of the C. albicans wall proteome. 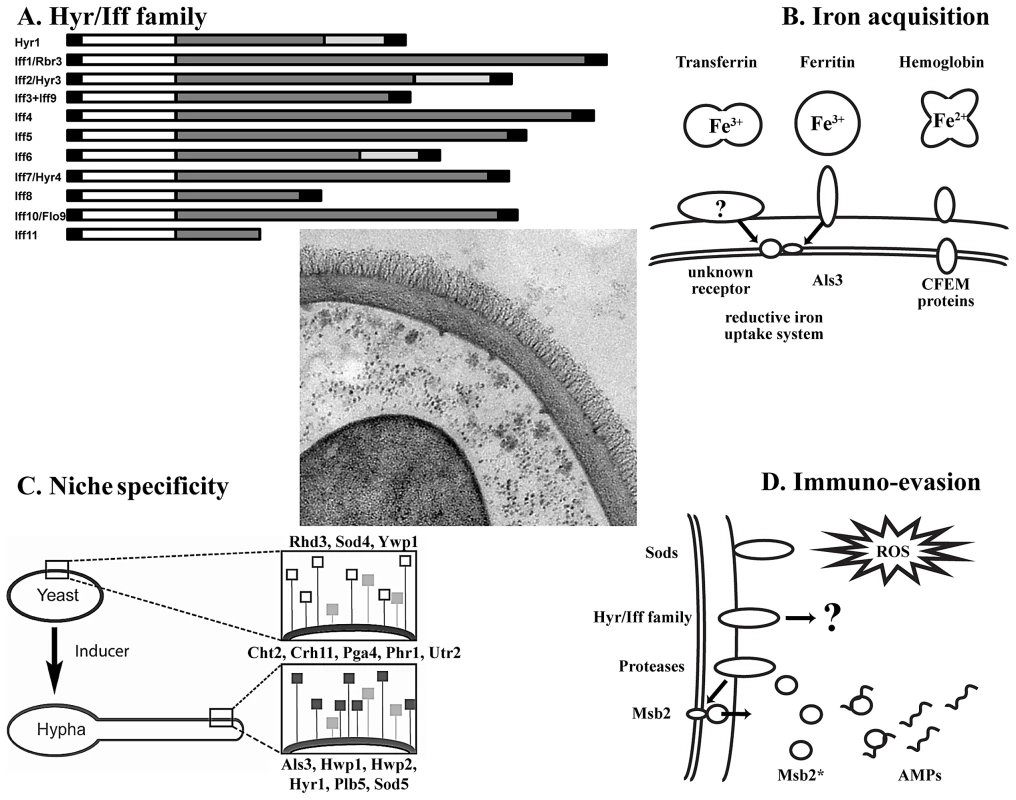
Center: TEM picture of the cell wall and its proteins (courtesy of Iuliana V. Ene and Alistair J.P. Brown, Aberdeen). (A) Domain structure of the Hyr/Iff family (adapted from [7]). From left to right: N-terminal signal peptide; white box, conserved domain; dark grey box, Ser/Thr-rich region; light grey box, Asp/Gly-rich region; black box, GPI-anchor addition signal. (B) Wall proteins implicated in iron acquisition from host proteins. Membrane and wall-bound CFEM proteins are able to bind hemoglobin, while Als3 is the receptor for ferritin. It is unknown if there exists a receptor for transferrin. Bound hemoglobin is taken up by endocytosis, while iron from ferritin and transferrin is sequestered via the reductive iron uptake system. (C) Effect of yeast-to-hypha transition on the wall proteome with yeast-associated (top; open squares), morphotype-independent (middle; grey squares) and hypha-associated (bottom; black squares) proteins [2]. (D) Interaction of wall proteins with the immune system. Wall-resident superoxide dismutases (Sods) detoxify reactive oxygen species (ROS) to H2O2, which is subsequently converted into H2O and O2 by catalase activity [20]. Proteins of the Hyr/Iff family confer resistance to neutrophil and phagocyte killing through an unknown mechanism [12]. Possibly, like in S. cerevisiae, proteases situated on the cell wall process the trans-membrane signaling protein Msb2 and liberate the extracellular domain Msb2*. Msb2* is able to bind to antimicrobial peptides (AMPs) in a dose-dependent manner and confers resistance [21]. What Is the Role of Wall Proteins in Iron Acquisition?
One of the most restricted nutrients in the human body is iron. Because of its reactive nature, but also in order to restrict growth of invading microorganisms, free iron is highly limited in the host and mainly found in association with proteins, either as a prosthetic group like in hemoglobin and myoglobin, stored inside ferritin, transported by transferrin, or liganded by lactoferrin. C. albicans has evolved a number of strategies to scavenge iron from these complexes. Of the five Rbt5 family proteins, which belong to the CFEM superfamily and are characterized by an internal domain containing eight invariantly spaced cysteines [13], Csa1, Pga7, Pga10, and Rbt5 are found attached both to the plasma membrane and the wall, while Csa2 is secreted [3], [14]–[16]. It has been shown that Csa1, Pga10, and Rbt5 are involved in heme binding [17]. As the expression of CSA1, CSA2, PGA7, PGA10, and RBT5 is co-regulated under various conditions, including iron restriction, the question arises whether the Rbt5 family proteins might act as a relay system, similar to bacterial iron uptake systems [18]. As mentioned above, Als3 is also important for iron acquisition as a receptor for ferritin, an iron-storage host molecule that contains about 30% of the total human iron pool. Without Als3, C. albicans is unable to grow with ferritin as its sole iron source [10].
Which Wall Proteins Allow C. albicans to Cope with the Host Immune Response?
C. albicans has evolved various mechanisms to avoid or counteract the immune response. The cell wall is the first line of defense, but also a target for the immune system due to its immunogenic epitopes. For example, the receptor dectin-1, which is mainly expressed on dendritic cells and macrophages, recognizes the β-glucan of the wall and leads to the activation of pro-inflammatory cytokines [19]. However, the mannoprotein coat largely prevents the detection of the underlying β-glucan layer. Additionally, the wall protein Hyr1 effectively reduces immune cell killing of C. albicans [12]. In support of its protective role, heterologous expression of Hyr1 in Candida glabrata also mitigates immune cell killing, suggesting a direct function of the protein. C. albicans also has two wall-bound, morphotype-associated superoxide dismutases (Sod4, Sod5) [14]. These cell wall–resident superoxide dismutases (Sods) are essential for dealing with extracellular ROS (reactive oxygen species), resulting from the oxidative burst, a general mechanism of immune cells to kill invading pathogens. As expected, SOD4 and SOD5 knockout mutants are more susceptible to oxidative stress [20]. Sod6, another GPI-anchored member of the Sod family, has not been detected in proteomic screens, and gene deletion did not reveal a clear phenotype [20]. Antimicrobial peptides, like histatins, defensins, and cathelicidins, belong to the arsenal of host defense mechanisms as well. Recently, the shedding of the extracellular part of the plasma membrane-bound signaling mucin Msb2, which is involved in maintaining cell wall integrity, has been shown to convey resistance to histatin-5 and the cathelicidin LL-37 in a dose-dependent manner [21]. This processing and shedding is likely mediated by secretory aspartyl proteases (Saps) [22], but there is no evidence that the GPI-modified, wall-resident proteins Sap9 and Sap10 are involved [21], [22].
How Do the Wall and its Proteins Cope with Surface Stress?
Cell shape is mainly determined by the skeletal polysaccharides of the wall, which are important for resisting the internal turgor pressure and shielding the cell from external mechanical forces. Nonetheless, remodeling of the wall is required, for example, during isotropic growth and cell separation, and for coping with surface stress. Remodeling of the wall is mediated by wall - and plasma membrane-resident, carbohydrate-active enzymes that detach, re-arrange, and re-attach carbohydrates. The main wall-bound proteins involved are a chitinase (Cht2), transglucosylases (Phr1, Phr2, Pga4), and chitin transglycosylases (Crh11, Utr2). The secretory aspartyl proteases Sap9 and 10, and Pir1, a predicted β-glucan cross-linking protein, also seem to be involved [3], [23]. In contrast to Sap1 to 8, Sap9 and 10 are GPI-modified, yapsin-like proteases that are retained at the cell surface [23]. Interestingly, Sap9 has been implicated in the processing and shedding of other wall proteins, most notably, the chitinase Cht2 and Pir1 [24]. The levels of wall-bound Sap9 seem largely morphotype-independent, but its levels increase in conjunction with surface stress conditions as observed in response to fluconazole [3].
Strikingly, when C. albicans is grown on a poor carbon source such as lactate (found in the vaginal fluid and together with acetate maintaining its acidic pH [4]), or on a mixture of lactate and glucose, the cell wall gets significantly thinner and more flexible. Importantly, these alterations are accompanied by substantial changes in the wall proteome [25]. This and other studies have identified a core set (Crh11, Phr1, Phr2, Pga4, Sap9, Utr2) of wall-remodeling proteins that is conserved in the response to several surface-stress conditions ([3], [25] and (Heilmann et al., unpublished data). Conceivably, this protein set could also be important to survive other surface stresses, including membrane-perturbing antimicrobial peptides found in body fluids, epithelial layers, and immune cells. The functional domains of these proteins are conserved in the Ascomycotina, suggesting similar importance for other fungi as well.
Which Wall Proteins Are Promising Targets for Vaccine Development?
A vaccine that could be administered to high-risk groups, e.g., pre-surgery, or to women suffering from recurrent vaginitis, would be an important asset. As stated earlier, the functional domain of wall proteins is almost exclusively situated in the N-terminal region, while the C-terminal part is mainly of structural importance. This is reflected in the various vaccines that are currently being developed (reviewed in [1], [26]). For example, mice immunized with the recombinantly expressed N-terminal domain of Als3 become resistant to infections by C. albicans as well as Staphylococcus aureus [27]. The N-terminal domains of Als1 and Hyr1, and a short immunogenic peptide from the N-terminal domain of Hwp1 conjugated to a β-1,2-linked mannotrioside, have been used similarly as C. albicans vaccines [1], [12]. Notably, these four vaccine targets are strongly associated with hyphae, suggesting that hyphal epitopes might be more easily recognized by the immune system as a threat, since they are associated with the breaching of host tissue. Invasive growth in vivo is not only associated with hyphal growth, but probably also with iron restriction and thus with increased levels of the iron acquisition proteins in the wall [15]. Relevantly, all five members of the Rbt5 family contain an identical sequence (with Csa1 containing four copies) that could represent a prime target. Developing this approach further, it is conceivable to combine immunogenic epitopes from the N-terminal functional region of a selection of wall proteins in a single recombinant protein for use as a multi-component vaccine. In summary, the evolution of wall protein families in the human fungal pathogen C. albicans allows survival in diverse host niches and has resulted in an impressive plasticity of the wall proteome. The exposure of wall proteins on the surface together with their critical functions, and the use of single - or multi-component vaccines, makes them promising targets for combating fungal infections.
Zdroje
1. KlisFM, de KosterCG, BrulS (2011) A mass spectrometric view of the fungal wall proteome. Future Microbiol 6 : 941–951.
2. HeilmannCJ, SorgoAG, SiliakusAR, DekkerHL, BrulS, et al. (2011) Hyphal induction in the human fungal pathogen Candida albicans reveals a characteristic wall protein profile. Microbiology 157 : 2297–2307.
3. SorgoAG, HeilmannCJ, DekkerHL, BekkerM, BrulS, et al. (2011) Effects of fluconazole on the secretome, the wall proteome, and wall integrity of the clinical fungus Candida albicans. Eukaryot Cell 10 : 1071–1081.
4. SosinskaGJ, de KoningLJ, de GrootPW, MandersEM, DekkerHL, et al. (2011) Mass spectrometric quantification of the adaptations in the wall proteome of Candida albicans in response to ambient pH. Microbiology 157 : 136–146.
5. StaabJF, BradwaySD, FidelPL, SundstromP (1999) Adhesive and mammalian transglutaminase substrate properties of Candida albicans Hwp1. Science 283 : 1535–1538.
6. HoyerLL, GreenCB, OhSH, ZhaoX (2008) Discovering the secrets of the Candida albicans agglutinin-like sequence (ALS) gene family–a sticky pursuit. Med Mycol 46 : 1–15.
7. BoisrameA, CornuA, Da CostaG, RichardML (2011) Unexpected role for a serine/threonine-rich domain in the Candida albicans Iff protein family. Eukaryot Cell 10 : 1317–1330.
8. SalgadoPS, YanR, TaylorJD, BurchellL, JonesR, et al. (2011) Structural basis for the broad specificity to host-cell ligands by the pathogenic fungus Candida albicans. Proc Natl Acad Sci U S A 108 : 15775–15779.
9. LipkePN, GarciaMC, AlsteensD, RamsookCB, KlotzSA, et al. (2012) Strengthening relationships: amyloids create adhesion nanodomains in yeasts. Trends Microbiol 20 : 59–65.
10. AlmeidaRS, BrunkeS, AlbrechtA, ThewesS, LaueM, et al. (2008) the hyphal-associated adhesin and invasin Als3 of Candida albicans mediates iron acquisition from host ferritin. PLoS Pathog 4: e1000217.
11. LiuY, FillerSG (2011) Candida albicans Als3, a multifunctional adhesin and invasin. Eukaryot Cell 10 : 168–173.
12. LuoG, IbrahimAS, SpellbergB, NobileCJ, MitchellAP, et al. (2010) Candida albicans Hyr1p confers resistance to neutrophil killing and is a potential vaccine target. J Infect Dis 201 : 1718–1728.
13. KulkarniRD, KelkarHS, DeanRA (2003) An eight-cysteine-containing CFEM domain unique to a group of fungal membrane proteins. Trends Biochem Sci 28 : 118–121.
14. SorgoAG, HeilmannCJ, DekkerHL, BrulS, de KosterCG, et al. (2010) Mass spectrometric analysis of the secretome of Candida albicans. Yeast 27 : 661–672.
15. SosinskaGJ, de GrootPW, Teixeira de MattosMJ, DekkerHL, de KosterCG, et al. (2008) Hypoxic conditions and iron restriction affect the cell-wall proteome of Candida albicans grown under vagina-simulative conditions. Microbiology 154 : 510–520.
16. WeissmanZ, ShemerR, ConibearE, KornitzerD (2008) An endocytic mechanism for haemoglobin-iron acquisition in Candida albicans. Mol Microbiol 69 : 201–217.
17. WeissmanZ, KornitzerD (2004) A family of Candida cell surface haem-binding proteins involved in haemin and haemoglobin-iron utilization. Mol Microbiol 53 : 1209–1220.
18. BraunV, HantkeK (2011) Recent insights into iron import by bacteria. Curr Opin Chem Biol 15 : 328–334.
19. BrownGD, NeteaMG (2012) Exciting developments in the immunology of fungal infections. Cell Host Microbe 11 : 422–424.
20. FrohnerIE, BourgeoisC, YatsykK, MajerO, KuchlerK (2009) Candida albicans cell surface superoxide dismutases degrade host-derived reactive oxygen species to escape innate immune surveillance. Mol Microbiol 71 : 240–252.
21. Szafranski-SchneiderE, SwidergallM, CottierF, TielkerD, RomanE, et al. (2012) Msb2 shedding protects Candida albicans against antimicrobial peptides. PLoS Pathog 8: e1002501.
22. PuriS, KumarR, ChadhaS, TatiS, ContiHR, et al. (2012) Secreted aspartic protease cleavage of Candida albicans Msb2 activates Cek1 MAPK signaling affecting biofilm formation and oropharyngeal candidiasis. PLoS One 7: e46020.
23. AlbrechtA, FelkA, PichovaI, NaglikJR, SchallerM, et al. (2006) Glycosylphosphatidylinositol-anchored proteases of Candida albicans target proteins necessary for both cellular processes and host-pathogen interactions. J Biol Chem 281 : 688–694.
24. SchildL, HeykenA, de GrootPW, HillerE, MockM, et al. (2010) Proteolytic cleavage of covalently linked cell wall proteins by Candida albicans Sap9 and Sap10. Eukaryot Cell 10 : 98–109.
25. EneIV, HeilmannCJ, SorgoAG, WalkerLA, de KosterCG, et al. (2012) Carbon source-induced reprogramming of the cell wall proteome and secretome modulates the adherence and drug resistance of the fungal pathogen Candida albicans. Proteomics doi: 10.1002/pmic.201200228.
26. VecchiarelliA, PericoliniE, GabrielliE, PietrellaD (2012) New approaches in the development of a vaccine for mucosal candidiasis: progress and challenges. Front Microbiol 3 : 294.
27. SpellbergB, IbrahimAS, YeamanMR, LinL, FuY, et al. (2008) The antifungal vaccine derived from the recombinant N terminus of Als3p protects mice against the bacterium Staphylococcus aureus. Infect Immun 76 : 4574–4580.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2012 Číslo 12- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Virus-Encoded microRNAs: An Overview and a Look to the Future
- Reactive Oxygen Species Production and Survivorship in with Artificial Infection Types
- Zinc Exploitation by Pathogenic Fungi
- Attenuated Typhimurium Lacking the Pathogenicity Island-2 Type 3 Secretion System Grow to High Bacterial Numbers inside Phagocytes in Mice
- The Polyfunctionality of Human Memory CD8+ T Cells Elicited by Acute and Chronic Virus Infections Is Not Influenced by Age
- How the Fly Balances Its Ability to Combat Different Pathogens
- MiniCD4 Microbicide Prevents HIV Infection of Human Mucosal Explants and Vaginal Transmission of SHIV in Cynomolgus Macaques
- Bidirectional Transfer of RNAi between Honey Bee and : Gene Silencing Reduces Population
- Global Gene Transcriptome Analysis in Vaccinated Cattle Revealed a Dominant Role of IL-22 for Protection against Bovine Tuberculosis
- Morphogenesis in Fungal Pathogenicity: Shape, Size, and Surface
- Inflammatory Responses Associated with the Induction of Cerebral Malaria: Lessons from Experimental Murine Models
- News from the Fungal Front: Wall Proteome Dynamics and Host–Pathogen Interplay
- Blood Flukes Exploit Peyer's Patch Lymphoid Tissue to Facilitate Transmission from the Mammalian Host
- Parallels in Intercellular Communication in Oomycete and Fungal Pathogens of Plants and Humans
- Influenza Human Monoclonal Antibody 1F1 Interacts with Three Major Antigenic Sites and Residues Mediating Human Receptor Specificity in H1N1 Viruses
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Influenza Human Monoclonal Antibody 1F1 Interacts with Three Major Antigenic Sites and Residues Mediating Human Receptor Specificity in H1N1 Viruses
- Parallels in Intercellular Communication in Oomycete and Fungal Pathogens of Plants and Humans
- Virus-Encoded microRNAs: An Overview and a Look to the Future
- Reactive Oxygen Species Production and Survivorship in with Artificial Infection Types
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



