-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaMouse Y-Linked and Are Expressed during the Male-Specific Interphase between Meiosis I and Meiosis II and Promote the 2 Meiotic Division
The mouse Y chromosome genes Zfy1 and Zfy2 were first identified in the late 1980s during the search for the gene on the Y that triggers male development; they encode proteins that regulate the expression of other genes to which they bind via a ‘zinc finger’ domain. We have now discovered that these genes play important roles during spermatogenesis. Zfy2 proved to be essential for the efficient operation of a ‘checkpoint’ during the first meiotic division that identifies and kills cells that would otherwise produce sperm with an unbalanced chromosome set. Female meiosis, which does not have an equivalent checkpoint, generates a significant proportion of eggs with an unbalanced chromosome set. In the present study we show that Zfy2 also has a major role in ensuring that the second meiotic division occurs, with Zfy1 and a related gene, Zfx, on the X chromosome providing some support. In order to fulfil this function all three genes are expressed in the ‘interphase’ stage between the two divisions. In female meiosis there is no interphase stage between the two meiotic divisions but in this case essential functions during the divisions are supported by stored RNAs, so an interphase is not needed.
Published in the journal: . PLoS Genet 10(6): e32767. doi:10.1371/journal.pgen.1004444
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004444Summary
The mouse Y chromosome genes Zfy1 and Zfy2 were first identified in the late 1980s during the search for the gene on the Y that triggers male development; they encode proteins that regulate the expression of other genes to which they bind via a ‘zinc finger’ domain. We have now discovered that these genes play important roles during spermatogenesis. Zfy2 proved to be essential for the efficient operation of a ‘checkpoint’ during the first meiotic division that identifies and kills cells that would otherwise produce sperm with an unbalanced chromosome set. Female meiosis, which does not have an equivalent checkpoint, generates a significant proportion of eggs with an unbalanced chromosome set. In the present study we show that Zfy2 also has a major role in ensuring that the second meiotic division occurs, with Zfy1 and a related gene, Zfx, on the X chromosome providing some support. In order to fulfil this function all three genes are expressed in the ‘interphase’ stage between the two divisions. In female meiosis there is no interphase stage between the two meiotic divisions but in this case essential functions during the divisions are supported by stored RNAs, so an interphase is not needed.
Introduction
Historically the realisation that there were spermatogenic factors on the human and mouse Y chromosomes distinct from the testis determinant came from the study of Y deletion variants [1], [2]. However, it was not until the search for the testis determinant that Y-encoded genes began to be identified; amongst these were the human and mouse Y genes encoding zinc finger transcription factors cloned in the late 1980s [3]–[5]. Subsequent progress in assigning spermatogenic gene functions to mouse Y-encoded genes was thwarted by a failure to disrupt Y gene functions using the emerging gene targeting techniques that had proved successful in disrupting X and autosomal gene functions, compounded by the paucity of genomic sequence data for the mouse Y chromosome. To circumvent these problems the Mitchell and Burgoyne labs established a collaboration with the aim of identifying mouse Y gene functions using a Y ‘transgene rescue’ strategy whereby Y genes were added to Y deletion variants with defined spermatogenic failure. In the context of Y genes mapping to the short arm (Yp), three XO male mouse models with diminishing Yp gene complements were utilised (Figure 1): XSxraO in which the X carries the Yp-derived sex-reversal factor Tp(Y)1CtSxr-a that provides an almost complete Yp gene complement [6], XSxrbO males where the X carries an Sxra derivative Tp(Y)1CtSxr-b in which a 1.3Mb deletion (ΔSxr-b) has removed the majority of the Yp gene complement [6], [7], and XOSry males in which the only Yp gene present is an autosomally located Sry transgene [8].
Fig. 1. The XO and XY*X mouse models. 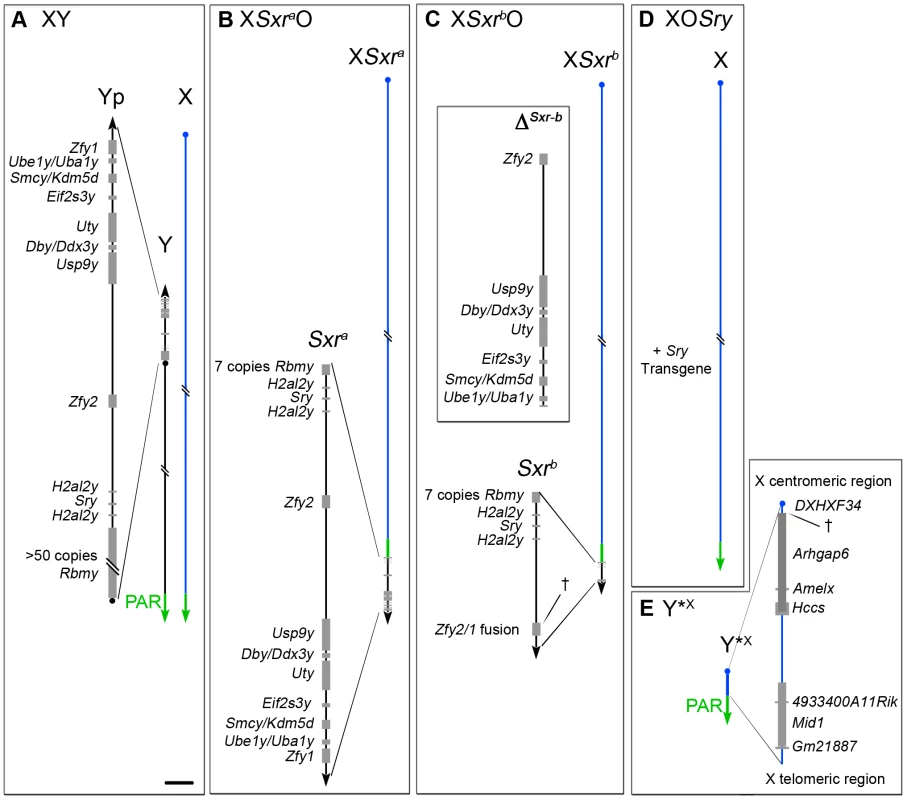
A. XY. The Y short arm (Yp) gene complement of an XY male (represented to scale in the magnified view) comprises seven single copy genes, two duplicated genes and one multi copy gene. The pseudoautosomal region (PAR) located distally on the Y long arm mediates pairing and crossing over with the X PAR during meiosis to generate the XY sex bivalent. B–D. The diminishing Yp gene complements for the three XO male mouse models that lack the Y long arm. B. XSxraO. The Yp-derived Sxra sex-reversal factor, attached distal to the X PAR provides an almost complete Yp gene complement. C. XSxrbO. The Sxra-derived deletion variant Sxrb has a 1.3 Mb deletion (ΔSxr-b) removing 6 single copy genes and creating a Zfy2/1 fusion gene spanning the deletion breakpoint (†). D. XOSry. This model has only one Y chromosome gene, namely the testis determinant Sry provided as an autosomally located transgene. E. Y*X. This mini sex-chromosome is an X chromosome with a deletion from just proximal to Amelx to within the DXHXF34 repeat adjacent to the X centromere. † represents the deletion breakpoint. This X chromosome derivative has a complete PAR that can pair with the PAR of XSxra, XSxrb or X to form a ‘minimal sex bivalent’. Scale bar for magnified views is 150 kb. The latter two Yp-deficient models have a marked block in spermatogonial proliferation, and in 2001 we reported that this block could be circumvented by the addition of Eif2s3y; this Y-linked gene encodes a protein almost identical to that encoded by the X-linked gene Eif2s3x - a subunit of the essential translation initiation factor EIF2 [8]. Paradoxically, in both Eif2s3y rescue models the majority of spermatocytes complete meiosis I, whereas in the XSxraO ‘control’ there is a very efficient apoptotic elimination of spermatocytes at the first meiotic metaphase (MI) [9]–[11]; this apoptosis is assumed to be triggered by an MI spindle assembly checkpoint (SAC) response to the univalent X at MI [12]. This suggested that a Yp gene that was deleted or inactivated in Sxrb was necessary for an efficient apoptotic response to the univalent X, although a markedly reduced apoptotic response remained.
To identify the Yp gene that promoted the MI spermatocyte apoptosis, transgenes were tested by adding them to XOSry males that carried an X-linked Eif2s3y transgene (here denoted as XEOSry), but none of Yp genes completely removed by ΔSxr-b (Figure 1C) reinstated the apoptotic response. Focus then shifted onto Zfy1 and Zfy2 because the ΔSxr-b deletion breakpoints lie within these two genes, creating a transcribed Zfy2/Zfy1 fusion gene with the encoded protein almost identical to that encoded by Zfy1 [7], [13]. Introducing an X-linked Zfy2 transgene into XEOSry males reinstated the apoptotic response but addition of Zfy1 had no discernible effect [11].
Further studies of the Eif2s3y rescue models XEOSry and XESxrbO revealed that although most primary spermatocytes evaded the apoptotic response and completed meiosis I to form diploid secondary spermatocytes that entered interphase (“interphasic secondary spermatocytes”), very few secondary spermatocytes recondensed their chromosomes and underwent meiosis II [14]. We can envisage three factors that individually, or in combination, could be responsible for the meiosis II impairment: (1) The triggering of the MI SAC by the univalent X, (2) the reduced apoptotic response, and (3) the lack of a Yp gene or genes that promotes meiosis II. It is assumed that the apoptotic elimination is in some way a consequence of the prior triggering of the MI SAC [12], but with as yet no information on the molecular link between Zfy2 expression and the apoptotic response, factors (1) and (2) are confounded. We therefore sought to check for a Yp gene requirement in a situation where the MI SAC and apoptotic response are circumvented. We have previously shown that the apoptotic elimination of MI spermatocytes in XSxraO males can largely be circumvented by adding a minute X chromosome derivative (denoted Y*X for historical reasons) comprising a complete PAR, an X PAR boundary, a very short X-specific region and an X centromere [15]–[19] (Figure 1E). In the majority of MI spermatocytes this Y*X mini-chromosome and XSxra had formed a sex bivalent (indicative of prior PAR synapsis and crossing over) and thus evaded the MI SAC/apoptotic elimination [19]. In the present study we therefore added this chromosome to the XEOSry, XESxrbO and XSxraO models in order to assess if the near complete Yp gene complement of Sxra promotes meiosis II more effectively than the two depleted Yp gene complements. This proved to be the case so we then proceeded to use Yp transgene addition to identify the Yp genes responsible for meiosis II completion.
Results
Yp-encoded genetic information promotes meiosis II
We will abbreviate the three models with Y*X used in this study as XY*XSxra, XEY*XSxrb and XEY*XSry. Their Yp gene complements are as shown in Figure 1B–D, except for the addition of the Eif2s3y transgene to the X of the latter two models (denoted XE).
For comparison with the published data on ploidy frequency of post-meiotic cells in the XESxrbO and XEOSry models [14] we have processed the Y*X complemented males at 6 weeks of age. We used a combination of centromere (CREST) and chromosome axial element (SYCP3) immunostaining, which allows PAR-PAR synapsis to be distinguished from associations between the X-derived Y*X centromere and the X centromere (Figure 2). This revealed an average of 74.9% PAR-PAR synapsis with no significant difference between the three Y*X complemented models and the remainder of cells either having the X and Y*X PARs unpaired, or lacking an identifiable Y*X (Table 1). In the latter case it is likely that the tiny Y*X chromosome was lost during cell spreading.
Fig. 2. Efficiency of XY synapsis in the XY*X males with varying Yp complements. 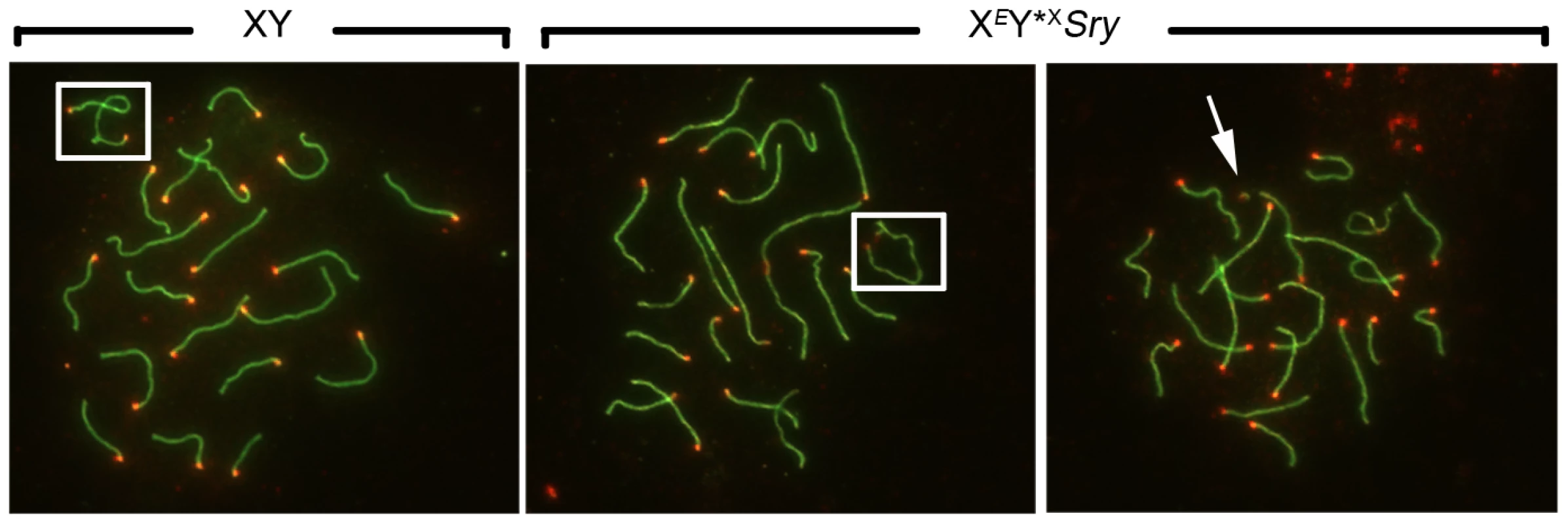
Spread pachytene spermatocytes from 6 week old XY and XEY*XSry testes stained with antibodies against SYCP3 (green) and CREST (red). Frames show PAR-PAR sex chromosome synapsis in XY and XEY*XSry males; the arrow points to an unsynapsed Y*X chromosome in an XEY*XSry male. Tab. 1. X-Y pairing efficiency in pachytene spermatocytes. 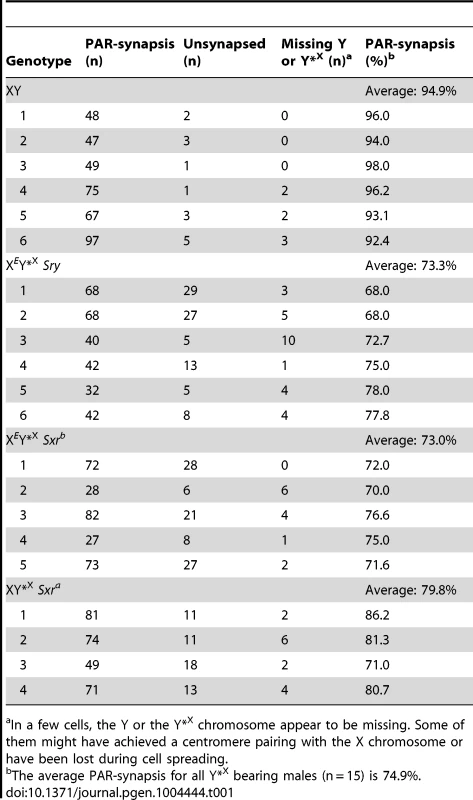
In a few cells, the Y or the Y*X chromosome appear to be missing. Some of them might have achieved a centromere pairing with the X chromosome or have been lost during cell spreading. To assess the efficiency of the meiotic divisions, we analyzed the ploidy of spermatids in SYCP3 and DAPI-stained spermatogenic cell spreads. The DAPI nuclear morphology of diploid spermatids is indistinguishable from that of interphasic secondary spermatocytes, but the latter have a characteristic SYCP3 staining pattern [11] and were excluded. It is important to bear in mind when assessing the consequences of the Y*X additions that in an average of 25.1% of MI cells from all three models the X fails to achieve PAR synapsis with the Y*X (Table 1); these cells will be subject to efficient apoptotic elimination when Zfy2 is present, but not when Zfy2 is absent. Our strategy was therefore to adjust the haploid frequencies of all the Zfy2-negative males carrying Y*X by ‘removing’ the products of the 25.1% of MI cells that did not achieve PAR-PAR synapsis (see Table S1). The adjusted frequencies are presented in Figure 3; the unadjusted frequencies are available in Table S2.
Fig. 3. Zfy1 and Zfy2 promote meiosis-II in the presence of the sex chromosome pairing partner Y*X. 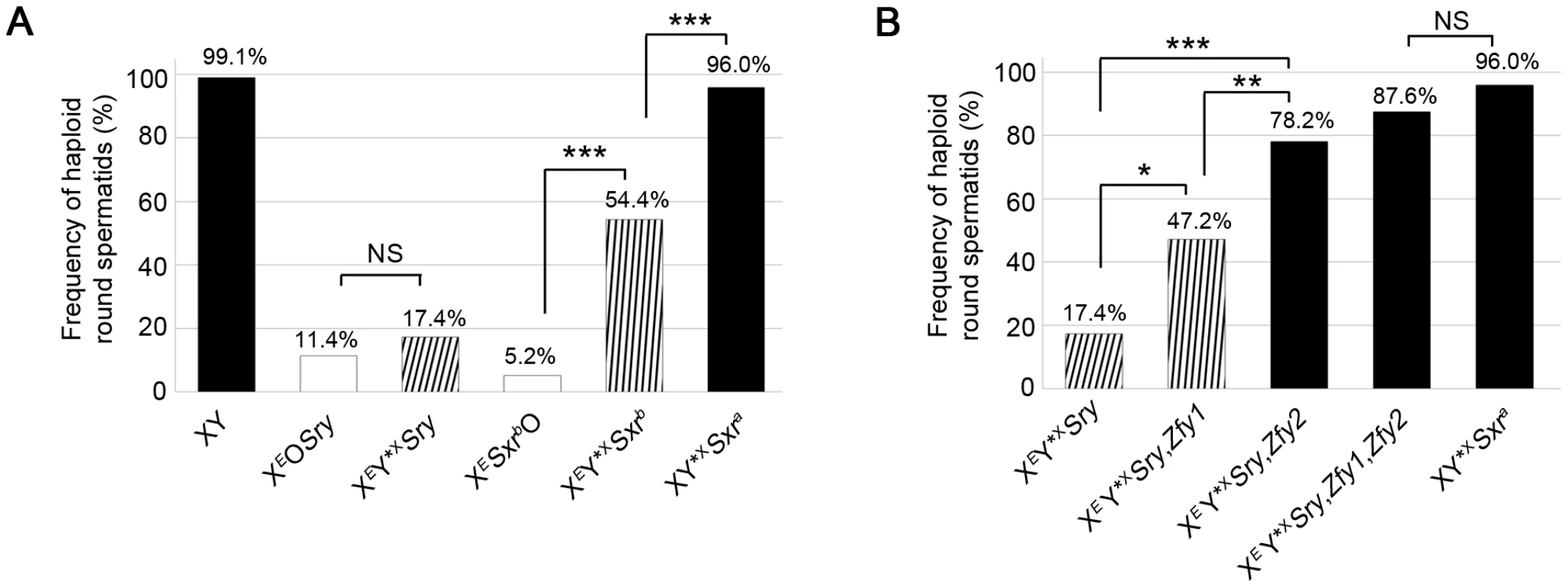
Data collected after DNA quantitation of spermatids using DAPI fluorescence intensity measurement on SYCP3-labelled testis cell spreads. Pooled data expressed as percentages are shown for each genotype (n = 4). Key: in black, models with robust apoptotic elimination of MI spermatocytes with X univalents; in white, XO models with markedly reduced apoptotic response; striped, XY*X models with markedly reduced apoptotic response in which the frequency was adjusted to remove spermatids derived from MI spermatocytes that had not formed an X-Y*X bivalent by PAR-PAR synapsis (see Table S1). A. Percentage of haploid round spermatids found in testis of 6 week old XO and XY*X males with various Yp chromosome gene contents. The data for the two XO male genotypes derive from Vernet et al., 2012 [14]. The Yp-derived Sxrb (which includes a Zfy2/1 fusion gene encoding a ZFY1-like protein) and Sxra (which includes Zfy1 and Zfy2) promote meiosis II in the presence of Y*X; Sxra is substantially more effective than Sxrb. B. Percentage of haploid round spermatids found in testis of 6 week old XEY*XSry males with X-linked Zfy transgene additions. Zfy1, and to a greater extent Zfy2, promote meiosis II. NS, Non significant; *p≤0.05; **p≤0.01; ***p≤0.001. Strikingly, there was no significant increase in haploid frequency in XEY*XSry (17.4%) relative to XEOSry (11.4%); in marked contrast the haploid frequency had significantly increased (P = 0.00059) in XEY*XSxrb (54.4%) relative to XESxrbO (5.2%) (Figure 3A). This was an unexpected result because there was no indication from the two XO models that Sxrb potentiated meiosis II; indeed XESxrbO had a lower haploid spermatid frequency than XEOSry. We conclude that meiosis II is not potentiated by the formation of a sex bivalent per se, but there is genetic information in Sxrb that in the context of a sex bivalent promotes the completion of meiosis II.
Sxrb has a very depleted Yp gene complement so we next wanted to assess the consequences of the Y*X addition in the context of Sxra, which provides a near complete Yp gene complement. This proved to have a much more potent effect than in the Sxrb context with the haploid frequency increasing to 96% (P = 0.00082) (Figure 3A). We conclude that there is genetic information on mouse Yp that promotes meiosis II when a sex bivalent is formed, and this is provided more effectively by Sxra than Sxrb.
Addition of Zfy1 and/or Zfy2 to XEY*XSry promotes meiosis II
The protein-coding gene content of Sxrb is thought to be limited to a few copies of Rbmy, two copies of H2al2y, Sry and a Zfy2/1 fusion gene spanning the Sxrb deletion breakpoint [7], [20], [21]. Because interphasic secondary spermatocytes are a very transient cell type in normal testes, there is no published information on expression of these genes at this stage. However, by RNA in situ analysis Rbmy transcripts are not detected beyond early pachytene [22] and H2al2y does not appear until step 6 round spermatids (Figure S1), so they are unlikely to be transcribed in interphasic secondary spermatocytes. The Sry transcripts present in the adult mouse testis are circular transcripts that are thought to be untranslated [23], [24]. Our initial focus was therefore on the Zfy2/1 fusion gene, which is known to be transcribed during early prophase, is presumed to be silenced by meiotic sex chromosome inactivation (MSCI, [25]) at the beginning of pachytene, as are Zfy1 and Zfy2 in normal males, but is transcribed post-meiotically [7], [26]. The Sxrb deletion breakpoint is located within a 95 bp region of sequence identity between intron 5 of Zfy2 and Zfy1, and the protein encoded by the fusion gene is predicted to be identical to that encoded by Zfy1 except for the 16th amino acid where a leucine replaces a phenylalanine [7], [11].
Because the Zfy2/1 fusion gene encodes a protein nearly identical to that of Zfy1 we first added a Zfy1 transgene to XEY*XSry to see if this mimicked the effect of Sxrb in promoting the second meiotic division in the presence of Y*X. This proved to be the case in that the proportion of haploid spermatids increased significantly (P = 0.01219) from 17.4% to 47.2% (Figure 3B).
Based on their DNA sequences, Zfy1 and Zfy2 are expected to produce transcription factors that will bind to the same target genes. We therefore also generated XEY*XSry males that were transgenic for Zfy2, and this addition increased the haploid frequency from 17.4% to 78.2% (P = 0.00011), which is significantly higher (P = 0.00665) than that achieved with the Zfy1 transgene (47.2%). Thus the Zfy2 transgene promotes meiosis II more effectively than the Zfy1 transgene. Both transgenes are single copy and inserted on the X chromosome, but we cannot assess relative transcript levels in interphasic secondary spermatocytes because of our inability to adequately purify this rare cell type. However we have previously established by qRT-PCR that the transcript level for the Zfy1 transgene is higher than that for the Zfy2 transgene in testes from 17.5 day-old XEOSry carriers [11], so we would expect a similar excess of Zfy1 transcripts in interphasic secondary spermatocytes. We were therefore surprised that the Zfy2 transgene had a markedly greater effect. With the addition of both transgenes the frequency of haploid spermatids increased to 87.6% (Figure 3B; Table S2C). These results point to the combined activity of Zfy1 and Zfy2 as important for promoting meiosis II.
Zfy1, Zfy2 and Zfx are transcribed during the interphase prior to meiosis II
Transcription of Zfy1 and Zfy2 is reportedly testis specific, at least post-natally [27]–[29]. Recently we have shown that in the adult testis this transcription is limited to germ cells, starting in leptotene spermatocytes, with more robust transcription in zygotene spermatocytes, followed by silencing in pachytene spermatocytes as a consequence MSCI; there was no resumption of transcription prior to MI, but transcription was shown to have resumed in (Y-bearing) round spermatids [7]. However, no data are available for interphasic secondary spermatocytes [14]. To assess transcription of Zfy1 and Zfy2 in these cells we used Zfy1 and Zfy2 RNA-FISH on DAPI - and SYCP3-stained testis cell spreads from XY males and confirmed the presence of the Y chromosome using Zfy1 and Zfy2 DNA-FISH. Zfy1 and Zfy2 transcription was detected in 45% and 27%, respectively, of the Y-bearing secondary spermatocytes (Figure 4; Table 2). As expected, Zfy1 and Zfy2 DNA-FISH signals were not observed in half of the secondary spermatocytes (X-bearing) and these also lacked Zfy1 and Zfy2 RNA-FISH signals. However, 92% of these X-bearing secondary spermatocytes were transcribing the related X-linked gene Zfx (Figure 4; Table 2).
Fig. 4. The mouse Zfy and Zfx genes are transcribed in interphasic secondary spermatocytes. 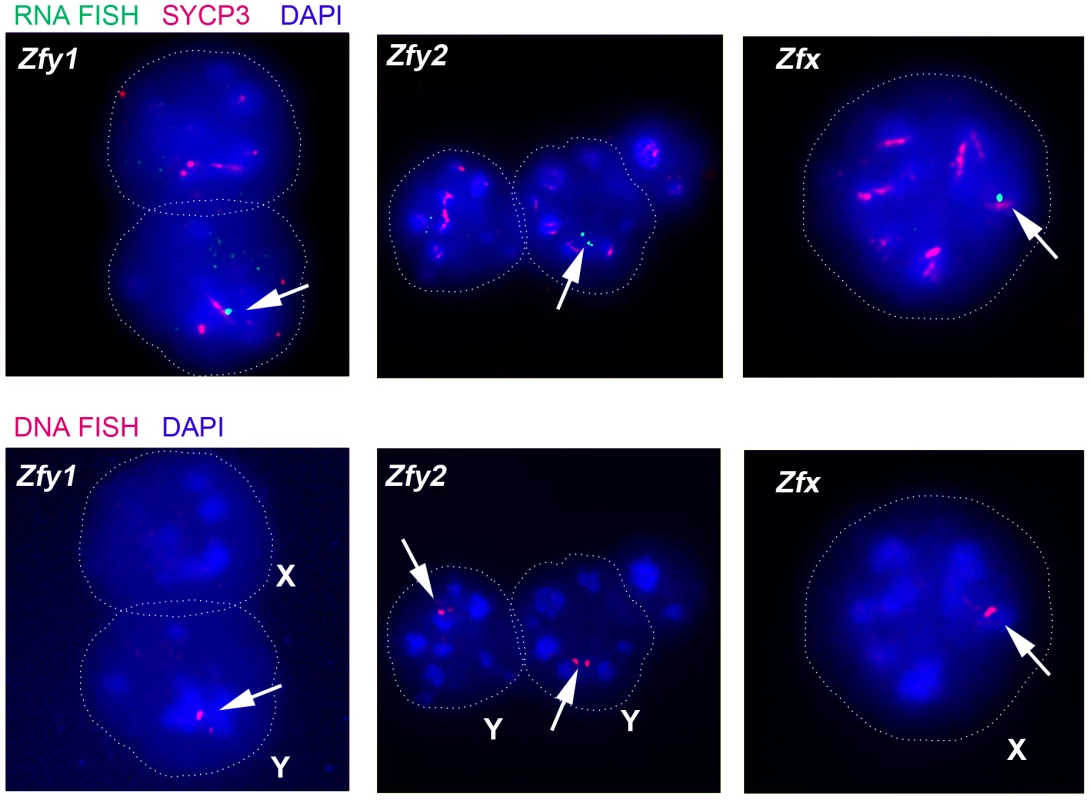
Representative images of interphasic secondary spermatocyte nuclei are shown hybridized with RNA FISH probes specific for Zfy1, Zfy2 or Zfx (arrows, top panels). Interphasic secondary spermatocytes were distinguished from diploid spermatids by staining spread spermatogenic cells from 6-week old XY males with an antibody against SYCP3 (red, top panels). The appropriate localization of the RNA FISH probe to the encoding genes was confirmed by DNA FISH (arrows, bottom panels). Nuclei are stained with DAPI (blue). X- or Y-bearing secondary spermatocytes are respectively represented by an X or a Y next to the cell. Tab. 2. X and Y-linked ‘<i>Zf</i>’ gene expression by RNA-FISH in spread interphasic secondary spermatocytes from 6 week old XY male. 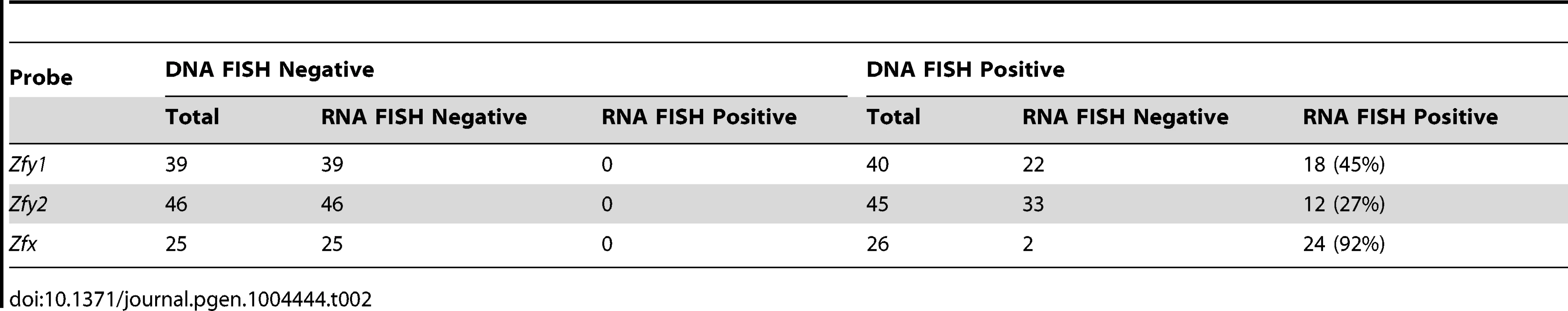
Our finding that Zfx is also expressed in interphasic secondary spermatocytes raised the question as to whether Zfx also promotes the second meiotic division. We had available a Zfx transgenic line with 7 copies of a Zfx genomic BAC inserted on an autosome. As expected for an autosomally located X-chromosome-derived transgene it was exempt from MSCI, and was expressed in pachytene cells (Figure S2A,B); like the endogenous Zfx gene it was expressed in interphasic secondary spermatocytes (Figure S2C). We added this transgene to XEY*XSry males and the proportion of haploid spermatids increased from 15.0% (in non-transgenic siblings) to 79.9%, showing that Zfx can promote meiosis II (Figure 5A,B; Table S2D); we therefore consider it likely that the endogenous Zfx also has a minor role in promoting meiosis II.
Fig. 5. Zfx over-expression promotes meiosis II. 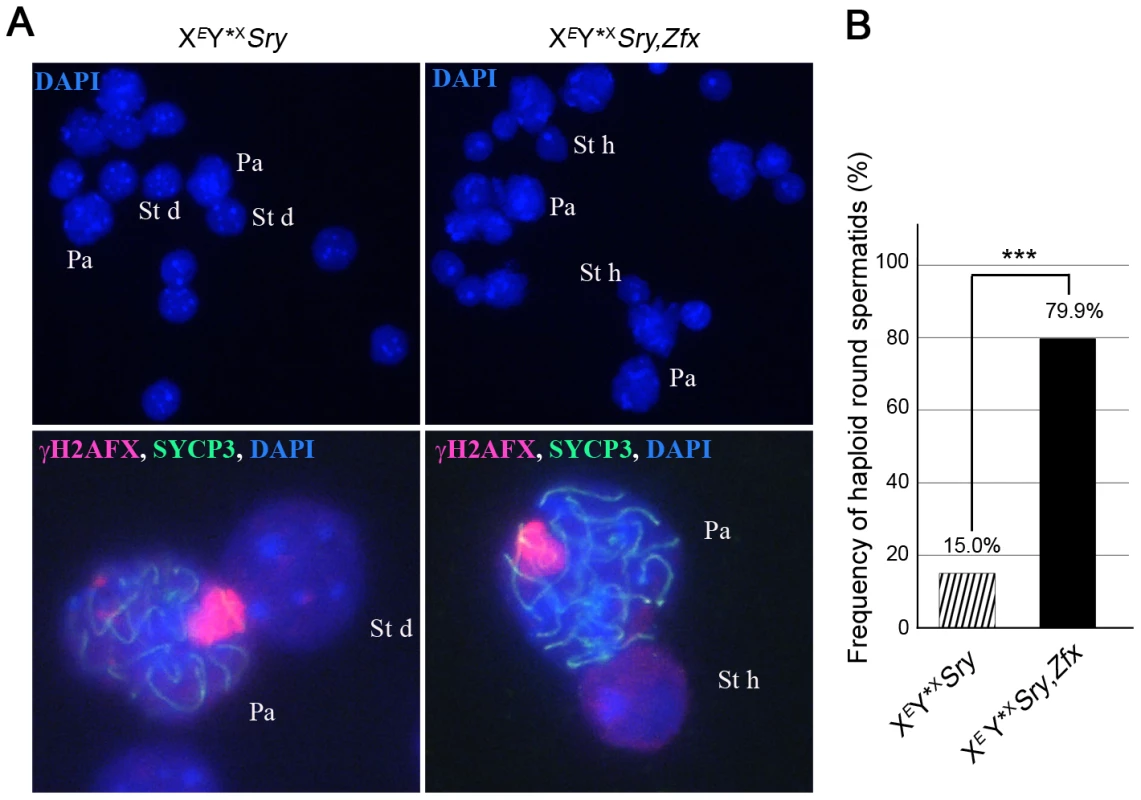
A. Spread cells found in testis of 6-week old XEY*XSry males without or with Zfx transgene. Pachytene (Pa), diploid spermatid (St d) and haploid spermatids (St h) nuclei are stained with DAPI (top panel) and higher magnifications are shown additionally labelled with γH2AFX, and SYCP3 antibodies (bottom panel). B. Percentage of haploid round spermatids found in mice from panel A (see also Table S2D). Key: in black, XEY*XSry,Zfx transgenic males have a robust apoptotic elimination of the ∼25% of MI spermatocytes that have an X univalent (see Figure S5); striped, XEY*XSry males have a markedly reduced apoptotic response so the frequency was adjusted as detailed in Table S1. The addition of the Zfx transgene significantly increases (***p≤0.00001) the proportion of haploid round spermatids. The Zfy2 transactivation domain is much more potent than that of Zfy1
We were struck by the much more potent effect of Zfy2 as compared to Zfy1 in promoting meiosis II. Based on an in vitro assay it was previously reported that Zfy2 encodes a protein with a much more potent transactivation (TA) domain than that of Zfx, but Zfy1 was not assayed at that time [30]. We have therefore used a similar in vitro assay to compare the transactivation domains of mouse Zfx, Zfy1, Zfy2 and the autosomal Zfa (originating from a retroposed X transcript [31]–[35]), and have also compared these with the transactivation domains of human ZFX and ZFY (Figure 6). We confirmed the expression of all the ZF-Gal4 fusion proteins by western blot analysis (Figure S3). The assay revealed that the TA domain of mouse Zfy1 has a similar activity to human ZFX and ZFY. Strikingly, the mouse Zfy2 TA domain is 5.5-fold more active than that of mouse Zfy1, and is ∼10-fold more active than that of mouse Zfx. The TA domain of the putative ZFA protein proved to have a very weak TA activity. A single nucleotide deletion near the beginning of the ZFY/ZFX open reading frame of Zfa actually makes it very unlikely to translate a protein that includes the zinc finger DNA binding domain, which would preclude binding to target genes. Zfa is now flagged as a pseudogene in Genbank (accession no. NR_037920).
Fig. 6. Zfy2 acidic domain is a much more potent transactivator than other ‘Zf’ acidic domains. 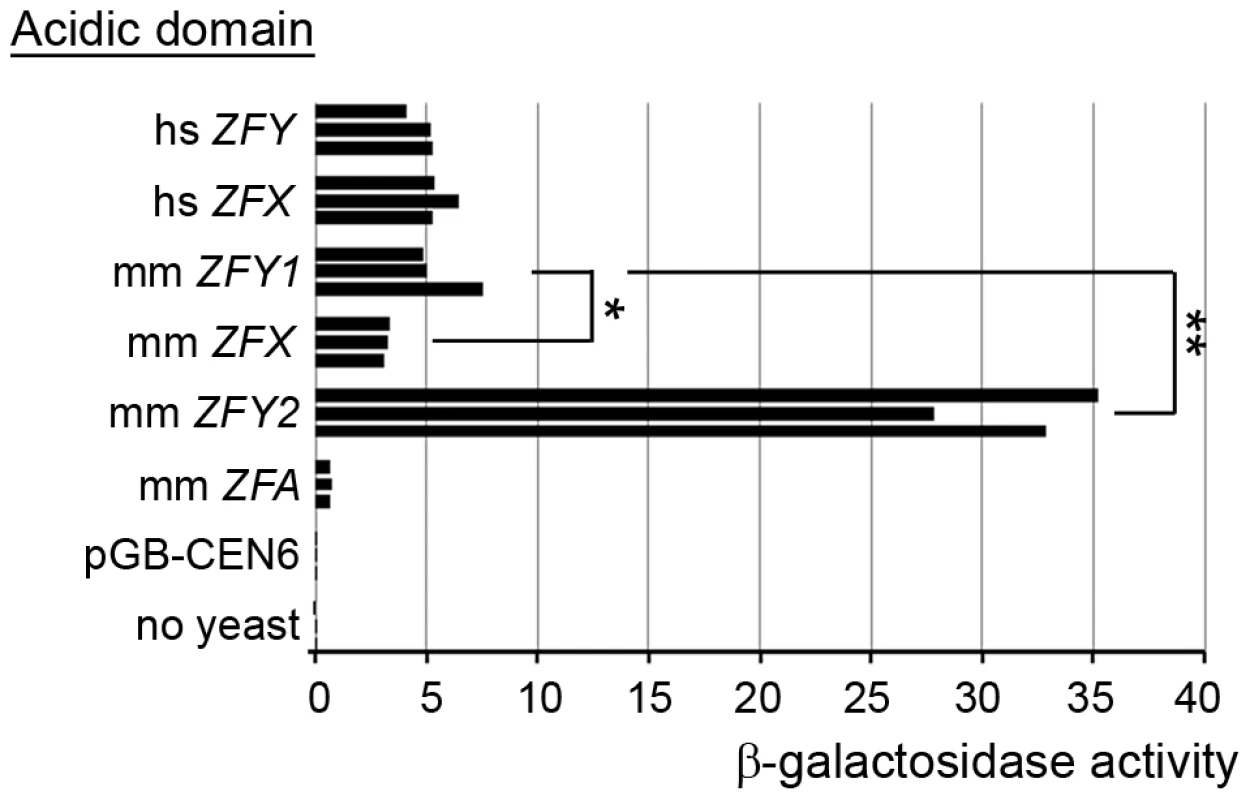
Levels of β-galactosidase induced by the Gal4-DNA-binding domain on its own (pGB-CEN6; negative control) or fused to an acidic domain from one of six different ZF isoforms from human (hs) or mouse (mm). Among the mouse sex-linked genes, Zfy2 has a substantially more potent activation domain than Zfy1, and Zfx is significantly less potent than Zfy1. mm ZFA derives from the autosomal Zfa gene that originated from a retroposed X transcript. *p≤0.05; **p≤0.01. Discussion
Our previous study of meiotic progression in the three XO male models with varying Yp gene complements revealed that the majority of spermatocytes in the Yp gene deficient models XEOSry and XESxrbO reached the interphase that precedes meiosis II; however, they failed to recondense their chromosomes to enable completion of meiosis II and instead formed diploid round spermatids [14]. Formally, the failure to undergo meiosis II could be a consequence of the prior triggering of the MI SAC by the univalent X, the reduced apoptotic response due to the absence of Zfy2, or the lack of a Yp gene or genes that promotes meiosis II (Figure 7A). The aim of the present study was to check specifically for a Yp gene requirement by circumventing the MI SAC and apoptotic responses; for this we added a minute PAR-bearing X chromosome derivative (Y*X) to all three XO models to enable formation of a sex bivalent without altering the Yp gene complement. This established that there is genetic information present in Sxra and Sxrb that promotes meiosis II, but Sxra was more effective (Figure 7B). Yp transgene additions to XEY*XSry males then identified Zfy1 and Zfy2 as the Yp genes responsible with Zfy2 having the more potent effect (Figure 7C).
Fig. 7. A summary of the meiotic outcome in XO and XY*X males with varying Yp gene content. 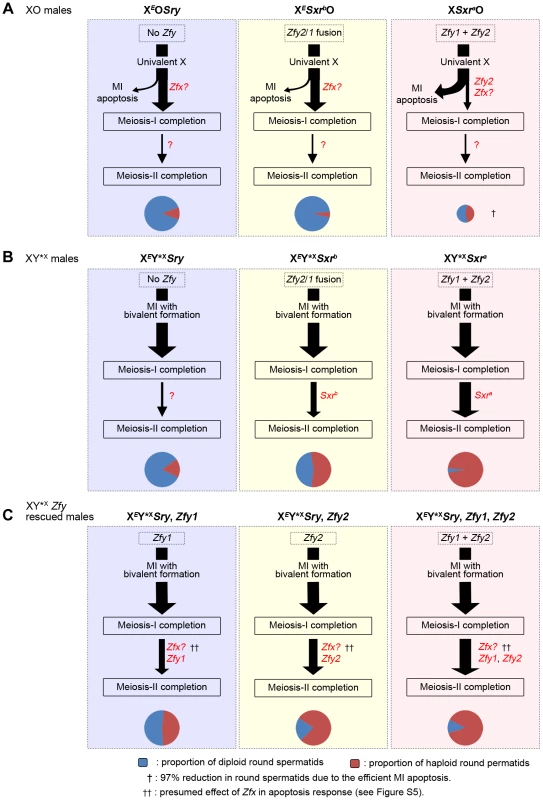
Throughout this figure the thickness of the arrows indicates the proportion of cells progressing from one step to the next and the cheeses at the bottom represent the proportion of haploid and diploid spermatids. Size of the cheese indicates the relative success of the different models in meiosis completion. A. XO models. In XEOSry and XESxrbO males the majority of spermatocytes complete meiosis I because of the reduced apoptotic response at MI due to the absence of Zfy2 [11]. Zfx expression is likely responsible for the residual apoptotic response (see Figure S5). The majority of spermatocytes then arrest at the interphase between meiosis I and meiosis II. This could be a consequence of the prior triggering of the MI SAC by the univalent X, the reduced apoptotic response due to the absence of Zfy2, or the lack of a Yp gene or genes that promotes meiosis II. In XSxraO males there is a very efficient apoptotic elimination of spermatocytes at MI so that very few complete meiosis I and this results in a 97% reduction in the number of spermatids. This precludes any firm conclusion as to a role for Yp genes for completion of meiosis II because the apoptotic elimination may have had a bias towards removing MI cells that were otherwise destined to arrest at the following interphase. B. XY*X models. In these models the spermatocytes that form a sex bivalent circumvent the MI SAC/apoptotic response and complete meiosis I. This reveals that Sxra strongly promotes meiosis II, thus confirming that a gene or genes on Yp promotes meiosis II. Surprisingly Sxrb, which did not promote meiosis II in XESxrbO males, does so now that the apoptotic response is circumvented by formation of an X-Y*X sex bivalent. C. The XY*XSry ‘Zf’ transgene addition models. These transgene additions revealed that Zfy1 and Zfy2 are the genes on Yp that promote meiosis II with Zfy2 the more effective. Sxrb includes the Zfy2/1 fusion gene that encodes a ZFY1-like protein, whereas Sxra includes Zfy1 and Zfy2, thus explaining the more potent effect of Sxra in promoting meiosis II. Zfx over-expression also promotes meiosis II (Figure 5) making it likely that the endogenous Zfx also does so to some degree. We attribute the difference in potency between Sxra and Sxrb to the presence of Zfy1 and Zfy2 in Sxra whereas Sxrb only has the Zfy2/Zfy1 fusion gene that encodes a protein almost identical to Zfy1 [7], [11]). However, we were struck by the fact that Sxrb did not promote meiosis II in the absence of Y*X; this implies that the triggering of the MI SAC, and/or the reduced apoptotic response, impairs progression through meiosis II. In order to distinguish between these possibilities it is informative to consider what happens in XO females where there is an MI SAC response but no apoptotic response. XO female mice are fertile and produce XO (and XX) daughters, so that some XO oocytes must complete meiosis I and meiosis II. Furthermore, although some X univalents achieve bipolar attachment to the spindle (which is expected to satisfy the MI SAC), this is not a prerequisite for the completion of meiosis I [36]. This is in agreement with accumulating data for female mice showing that the MI SAC does not maintain arrest until all kinetochores have achieved appropriate attachments to the spindle - anaphase can proceed in the presence of one (or a few) univalents [37]–[41]. Thus XO oocytes can complete meiosis I to generate MII oocytes with either an XX sex chromosome complement (i.e. two X chromatids) or lacking an X chromosome, both of which should be able to complete meiosis II without triggering an MII SAC response. This suggests that in the three XO male models the triggering of the MI SAC per se would not impair meiosis II. We therefore favour the view that the addition of the Y*X to the XESxrbO model increases the haploid spermatid frequency from 5.2% to 54.4% because the formation of an XSxrb/Y*X bivalent avoids the reduced apoptotic response. We envisage that the reduced DNA damage at MI as a consequence of the reduced apoptotic response is usually insufficient to trigger elimination at MI, but is sufficient to trigger a G2/M DNA damage checkpoint (reviewed in [42]) at the post meiosis I interphase and block progression to MII. The arrested interphase cells then enter spermiogenesis as diploid spermatids. Unfortunately we cannot use these models to assess the ultimate fate of the diploid spermatids, because in the absence of the Y long arm there is marked over-expression of X and Y genes due to the absence of the repressive effects of Sly, which is present in >50 copies on the Y long arm [43], [44], and this (together with Yp gene deficiency) results in severely perturbed spermiogenesis [14]. Figure 8 summarises how we see these MI and G2/MII checkpoint responses operating in males with a normal ‘Zf’ gene complement (XSxraO, XY*XSxra and XY).
Fig. 8. A combined MI SAC and G2/MII checkpoint model to explain the consequences of the male-specific apoptotic response to spermatocytes with univalent chromosomes at MI. 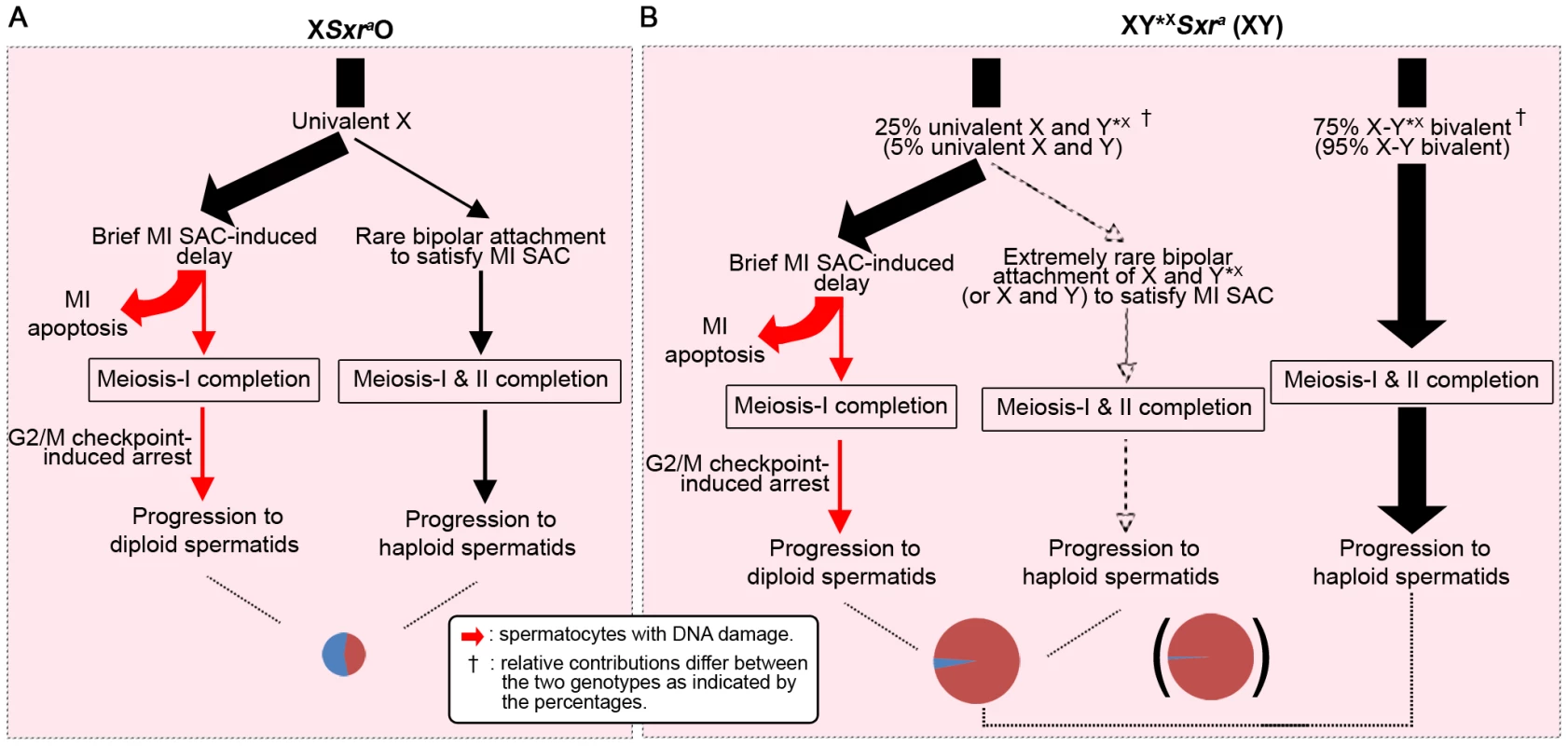
To illustrate the model we consider the consequences of these two checkpoint responses in XSxraO, XY*XSxra and XY males, all of which have a complete ‘Zf’ gene complement. Red arrows denote DNA damage. The cheeses at the bottom represent the proportion of haploid and diploid spermatids. A. In XSxraO males each MI spermatocyte will have a univalent X that is expected to trigger the MI SAC and cause a brief delay in MI progression. We propose that this delay is detected by the surrounding Sertoli cell, which initiates a robust Zfy2+Zfx-dependent apoptotic response. In order to explain the mix of diploid and haploid spermatids originating from the very few surviving MI spermatocytes we propose: 1) Rare MI spermatocytes complete meiosis I with apoptotic DNA damage that triggers a G2/MII checkpoint in the subsequent interphase and blocks progression to MII – these interphasic secondary spermatocytes then enter spermiogenesis to form diploid spermatids. [The number of such cells was elevated in the XO models lacking Zfy2 in Figure 7]. 2) In rare cases some MI spermatocytes evade the MI SAC and apoptosis by achieving bipolar attachment to the spindle – these complete meiosis I and meiosis II to form haploid spermatids. B. In XY*XSxra males (25% of MI spermatocytes with univalent X and Y*X) and XY males (5% of MI spermatocytes with univalent X and Y) the MI cells with univalents will follow the pathways 1) and 2) above, although the likelihood of both univalents achieving bipolar attachment to the spindle will be much lower. The MI cells that have formed a sex bivalent will progress through both divisions to form haploid spermatids (unless they have a pair of autosomal univalents in which case pathways 1 and 2 apply). The finding that both Zfy2 and Zfy1 promoted meiosis II was surprising because at MI only Zfy2 promotes the robust apoptotic elimination of spermatocytes with a univalent X chromosome [11]. What is the basis for the resurrection of Zfy1 function in the short interval between MI and meiosis II? Our previous RNA FISH analyses of nascent nuclear transcripts on spread spermatogenic cells from normal XY males [7] have established that Zfy1 and Zfy2 are transcribed in all mid-late zygotene nuclei, but this ceased in pachytene nuclei – an expected consequence of meiotic sex chromosome inactivation (MSCI – reviewed by [25]), and remained undetectable right through MI. The role of Zfy2 in the apoptotic response to univalence at MI must therefore be a consequence of transcriptional changes mediated by ZFY2 translated from these zygotene transcripts. Zfy1 and Zfy2 have the same predicted DNA target sequences, so if both are robustly expressed during zygotene, why is the apoptotic role limited to Zfy2? A plausible explanation is provided by our finding that during the pre-pachytene phase of transcription, alternative splicing of Zfy transcripts leads to ∼81% of Zfy1 transcripts lacking exon 6 with the encoded protein lacking transactivation (TA) activity, whereas ∼96% of Zfy2 transcripts have exon 6 and thus a functional TA domain [7]. Our current TA domain analysis further demonstrates that the few full length Zfy1 transcripts that are produced during zygotene generate a protein with a much less potent TA domain than that of Zfy2. In view of this it is reasonable to conclude that Zfy1 function in meiosis II is based on the de novo transcription in interphasic secondary spermatocytes (and that this also applies to Zfy2). It also implies that at this stage there is a greater preponderance of Zfy1 transcripts with exon 6 that encodes the TA domain; this is supported by our previous finding of a 3.7-fold increase in such transcripts in pubertal testes between 20dpp and 27dpp, which covers the period when transcripts from interphasic secondary spermatocytes and round spermatids should progressively increase as a proportion of the total testis RNA [7].
Given that Zfx, Zfy1 and Zfy2-encoded transcription factors are predicted to bind the same target sequences, it is to be expected that in tissues where they are all expressed they will regulate the transcription of the same genes. The extent to which they transactivate target genes will be dependent on the relative potency of their TA domains (Zfx<Zfy1<Zfy2); the protein encoded by Zfy1 lacking exon 6 would be expected to bind but not transactivate, and could thus function as a competitive inhibitor of the three full length ZF proteins [7]. In XY males, all three sex-linked ‘Zf’ genes are transcribed in zygotene spermatocytes with a predominance of Zfy1 transcripts lacking exon 6 (Figure S4 and [7]), and in interphasic secondary spermatocytes (Figure 4 and Table 2) in which full length Zfy1 transcripts are thought to be more prevalent. Does the Zfx transgene addition support the expectation that Zfx will contribute to ‘Zf’-mediated functions at MI (indirect) and during meiosis II (potentially direct)? We have already presented the data showing a marked promotion of meiosis II by the Zfx transgene (Figure 5) and there is also a marked promotion of the apoptotic response to X univalence at MI (Figure S5), which clearly demonstrates that Zfx is able to contribute to these functions. [The marked promotion of these functions in both cases is unsurprising given that the transgene is present in 7 copies and that its autosomal location is associated with extension of transcription through to just prior to MI, together with exemption from the MSCI-dependent repression that affects the X and Y chromatin of interphasic secondary spermatocytes (see below).] Thus it is reasonable to conclude that the endogenous Zfx also contributes to these functions; indeed, in the XEOSry model there is some MI apoptosis [11] and in the XEY*XSry model there is some progression through meiosis II (17.4% haploid spermatids, Figure 3A). A role for the endogenous Zfx in spermatogenesis has also been suggested based on the reduced sperm count in Zfx knockout males, although this effect is confounded with severe growth deficiency [45].
Although Monesi reported transcription in interphasic secondary spermatocytes in the 1960s [46], [47], other than our finding that there is de novo transcription of the multi-copy mouse Y gene Sly in interphasic secondary spermatocytes [48], we are not aware of any published data giving information on which genes are actively transcribed at this stage. It has previously been concluded based on Cot1 RNA FISH assessments of global transcription in interphasic secondary spermatocytes that the autosomal chromatin is actively transcribed, whereas the X and Y chromatin remains substantially repressed; the repression of the sex chromosomes has been shown to be dependent on the prior MSCI, and is carried through into round spermatids (‘post-meiotic sex chromosome repression’) [49]–[51]. This raises the possibility that the de novo transcription of the ‘Zf’ gene family represents a selective reactivation. As a first look at this issue we assessed de novo transcription of Mtm1 (X-linked) and Uty (Y-linked) in interphasic secondary spermatocytes and round spermatids, with Zfx serving as a positive control. This revealed that these two genes are also transcribed in interphasic secondary spermatocytes, but it is noteworthy that for all three genes the frequency of RNA FISH positive cells was higher in interphasic secondary spermatocytes (Zfx 90%; Mtm1 67%; Uty 88%) than in round spermatids (Zfx 32%; Mtm1 36%; Uty 60%) (Table S3). These preliminary data are consistent with: (1) there being a partial relaxation of sex chromosome silencing during the late diplotene–MI period, counterbalanced by the global transcriptional repression associated with the condensation of the metaphase chromosomes, (2) the decondensation of the chromosomes in interphasic secondary spermatocytes allowing strong transcription from the autosomes, but weaker transcription from the sex chromosomes because of the MSCI carry-over effect; and (3) further repression of the sex chromosomes in round spermatids due to the repressive chromatin changes driven by the multi-copy Y gene Sly [44].
The marked increase in transactivation activity of Zfy2 relative to Zfx and Zfy1 (Figure 6 and [30]) raises some interesting questions in an evolutionary context. The autosomal ‘Zf’ precursor of Zfy and Zfx is thought to have been added to the PAR after the separation of the eutherian and marsupial lineages 193–186 million years ago, and that with further PAR additions and rearrangements it became located in the non-recombining regions of the X-Y pair [52], [53]. In eutherian mammals the X-linked genes with retained Y-linked homologues are typically exempt from X dosage compensation, suggesting a constraining dosage requirement in somatic tissues, and in most eutherian mammals this is known or is presumed to be true for Zfx and Zfy [54], [55]. However, around 40–70 million years ago in the myomorph rodent lineage, Zfx became subject to X-dosage compensation and the Zfy-encoded proteins diverged [30], [55]–[59]. Furthermore, the divergence in Zfy protein sequence is more marked in the highly acidic amino terminal TA domain that activates target genes, than in the carboxy terminal zinc finger domain that mediates binding to DNA. Here we have shown that in Mus musculus this divergence is associated with increased TA activity and that this is much more marked in Zfy2 than in Zfy1. Given that in mature male mice expression of the Zfy genes has only been detected in testes [27]–[29], specifically in the germ-line [7], this implies that there was a strong selective force in spermatogenic cells for improved TA activity. This male germ-line specific selective force is likely to have been MSCI, which will have affected Zfx as well as Zfy1 and Zfy2. For a zinc finger transcription factor needed for meiosis II that is dependent on transcription during the brief interphase between meiosis I and meiosis increasing the transactivation activity would be a major advantage. The TA domain of Zfx is likely precluded from responding to the selection because of a dosage sensitive role in somatic cells. On the other hand, the spermatogenic cell specific expression of the Y-encoded genes in the post natal testis allowed their TA domains to increase in activity, but the TA domain of Zfy2 has responded much more than that of Zfy1.
Given the importance of ‘Zf’ gene transcription during the interphase between meiosis I and meiosis II in male meiosis, it is intriguing that female mice (and female mammals generally) have no interphase between the two meiotic divisions. We previously hypothesized that the presence of an interphase between the two meiotic divisions in male mammals would be essential if there are meiosis II critical genes on the sex chromosomes, because they would have been transcriptionally silenced (MSCI) during the preceding ∼8 days [14]. However, the two meiotic divisions in females are dependent on RNAs produced and stored during oocyte growth [60], and it may be this dependence on stored RNAs that has enabled female meiosis to dispense with the interphase.
In conclusion, our present findings provide evidence for a specific requirement for Zfx and Zfy expression in the interphase between meiosis I and meiosis II, for meiosis II to be efficiently completed. We have also provided additional evidence for a marked divergence in the functionality of the three ‘Zf’-encoded transcription factors, with Zfx providing a dosage constrained somatic role with only a minor contribution to sex-linked ‘Zf’ gene function in spermatogenesis, Zfy1 developing a dual role in spermatogenesis via alternative splicing to produce activatory and repressive proteins (see also [7]), and Zfy2 becoming a super-active transcription factor (see also [30]) to enable it to function in the face of the repressive effects of MSCI and the linked post-meiotic sex chromosome repression. There are undoubtedly further sex-linked ‘Zf’ gene functions to be discovered so the identification of the direct targets of the ‘Zf’-encoded transcription factors is a high priority. This has been thwarted by a failure to obtain specific antibodies for chromatin immunoprecipitation analyses, but transgenes encoding tagged versions of the proteins should provide a way forward. We have also presented a case for their being a G2/M DNA damage checkpoint operating in the interphase between meiosis I and meiosis II that prevents progression to MII if there is unrepaired DNA damage present; our XO mouse models together with the recent first report of a successful method for the targeted disruption of a single copy Y gene [61], will be invaluable for investigating this further. Some intriguing recent data obtained with the XESxrbO model suggested that following the injection of diploid spermatids (almost certainly together with interphasic secondary spermatocytes) into eggs (“ROSI”), a proportion of the cells completed the second meiotic division in the egg, thus avoiding triploidy which is lethal in early pregnancy [62]. The egg provides the cellular machinery for DNA damage repair by non-homologous end joining (NHEJ) [63], which would be expected to release the proposed G2/M DNA damage checkpoint arrest in the XESxrbO model. However, this pathway of repair is inherently mutagenic [64], which may have important ramifications for the use of ROSI with cells harboring such DNA damage if they were unintentionally used when treating human male infertility.
Materials and Methods
Ethics statement
All animal procedures were in accordance with the United Kingdom Animal Scientific Procedures Act 1986 and were subject to local ethical review.
Mice
Aside from the mice with Sxra or Sxrb attached to the Y*X chromosome (see section (1) below), the mice in this study have an outbred MF1 (NIMR colony) background. The XY*X males with varying Yp gene complements (Figure 1) were produced by either 1 or 2 below, and the Zfy and Zfx transgene additions to XEY*XSry males are described in 3.
-
(i) Mating XX females to ‘XSxraY*’ males, or (ii) Mating XX females homozygous for the X-linked Eif2s3y transgene to ‘XSxrbY*’ males. The fathers used in these crosses are unique genotypes generated specifically for this study to enable a more efficient production of males carrying the X chromosome derivative (Y*X, see below) and the Yp-derived sex reversal (Sxr) factors. These unique genotypes are males [15], [16], [18] with either the Tp(Y)1CtSxr-a sex-reversal factor [65], or the Tp(Y)1CtSxr-b sex-reversal factor [6], [13], attached distal to the X PAR (denoted XSxraY* and XSxrbY* respectively). XSxrY* males have a Y chromosome ‘hijacked’ by an X centromere attached distal to a rearranged PAR. One of the recombinant sex chromosomes generated is the minute ‘Y*X’ chromosome comprising a complete PAR with an X PAR boundary, a very limited amount of X-specific DNA, and an X centromere (Figure 1E). In these XSxrY* males the recombination event generating the Y*X adds the Sxra or Sxrb factor distal to the Y*X PAR. In producing the XSxrY* fathers the Sxr factors were passed through females heterozygous for the X-autosome translocation T(X;16)16H as previously described [66], [67], and this introduced some ‘non-MF1’ genetic background. The resulting XEY*XSxrb and XY*XSxra offspring were 87.5% MF1. A detailed description of the production and characteristics of these XSxraY* and XSxrbY* males is in preparation.
-
Mating XY*X females [15], [16], [18] carrying an X-linked GFP transgene marker [68] to: (i) ‘XEYΔSrySry’ males that have the X carrying an Eif2s3y Y-genomic BAC transgene [8], a Y-chromosome with an 11 kb deletion removing Sry (dl1Rlb) [69], [70], and an autosomally located Sry transgene [Tg(Sry)2Ei] [71]; this cross produces XEOSry and XEY*XSry males that do not exhibit GFP florescence when examined using GFP goggles (FHS/EF-2G2: Biological Equipment Maintenance and Service Ltd, Budapest, Hungary). (ii) ‘XEYSxrb’ males that have the X-linked Eif2s3y transgene and a Y-chromosome with Sxrb attached distal to the PAR; this cross produces GFP-negative XESxrbO and XEY*XSxrb males with Sxrb attached to the X PAR. (iii) ‘XYSxra’ males that have a Y-chromosome with Sxra attached distal to the PAR; this cross produces GFP-negative XSxraO and XY*XSxra males with Sxra attached to the X PAR.
MF1 XYRIII males were used as normal controls; YRIII is the strain of Y chromosome from which Sxrb and Sxra derive.
-
Zfy1 or Zfy2 transgenes inserted on autosomes are expressed during pachytene, which results in pachytene stage IV apoptosis and consequent sterility [26], so we have used X-located single copy Zfy1 and Zfy2 transgenes that are silenced along with the endogenous X and Y genes at the beginning of pachytene. The Zfy and Zfx transgene additions involved cross 2(i) except that the XEYΔSrySry male also carried either (i) 1 copy of an X-located Zfy1 - Uba1y BAC (RP24-327G6) transgene [26], (ii) 1 copy of a Zfy2 BAC inserted by cassette mediated exchange (CME) into the Hprt locus on the X chromosome [11], [26], or (iii) 7 copies of an autosomally located Zfx BAC (RP23-269L6) transgene.
Genotyping and copy number estimation
Crosses 1(i)-2(iii) above generate XO males as well as the XY*X males with varying Yp complements. As a guide to the presence of Y*X we utilised PCRs for X-linked Prdx4 (absent in Y*X), Amelx (present in Y*X) and Myog (on chromosome 1) for normalisation. Two PCR reactions were used to detect the presence of Y*X and the number of X-chromosomes. An 82-bp Prdx4 and a 162-bp Amelx fragment were amplified using primers Prdx4-F and Prdx4-R together with primers Amelx-F and Amelx-R. The 162-bp Amelx fragment and a 246-bp Myog fragment were amplified using Amelx primers together with Myog primers Om1a and Om1b [72]. Primer sequences are described in Table S4. The following conditions were used: 95°C for 5 min, followed by 28 cycles of 95°C for 30 sec, 60°C for 20 sec and 72°C for 30 sec, with a final extension at 72°C for 5 min. Products were separated on a 3.5% (w/v) agarose gel and the genotype inferred from the relative intensities of the PCR products: XO 1 Prdx4 + 1 Amelx + 2 Myog, XY*X 1 Prdx4 + 2 Amelx + 2 Myog, XX 2 Prdx4 + 2 Amelx + 2 Myog and XXY*X 2 Prdx4 + 3 Amelx + 2 Myog.
For mice typed as Y*X positive that provided material for the present study the presence of the Y*X was confirmed either by examination of Giemsa-stained bone marrow chromosome spreads to check for the presence of the very small Y*X chromosome, by SYCP3 and CENT immunostaining of testis cell spreads (see below), or by quantitative PCR using Prdx4-F, Prdx4-R, Amelx-F and Amelx-R primers with the following genotypes as controls: XX, XO and XY*X. Om1a and Om1b primers were used for normalisation.
Copy number estimation was done by quantitative PCR as previously described in Royo et al 2010 with slight modification. A SacBII amplicon obtained using SacBII-F and SacBII-R primers (Table S4), match the backbone of the Zfx-bearing vector. A XZfy1/Uba1yY sample was used as a reference, because it bears a known transgene copy number of one (Zfy1-7; [26]) and the backbone of the Zfy1/Uba1y-bearing vector contains SacBII ORF. Reactions were normalised against amplification of the Atr gene. The difference in PCR cycles with respect to Atr (ΔCt) for a given experimental sample was subtracted from the mean ΔCt of the reference samples (XZfy1/Uba1yY) (ΔΔCt). The transgene copy number was calculated as the mean of the power 2 (ΔΔCt).
Pairing efficiency
Pairing efficiency between X and Y*X was assessed on surface-spread spermatogenic cells preparation from 6-week-old testes. Briefly, a portion of frozen testicular tissue (approximately 10 mg) was defrosted and macerated in 0.2 ml RPMI 1640 solution (Invitrogen Corporation, Gibco) to produce a thin cell suspension. One drop of cell suspension was applied on a pre-boiled microscope slide, mixed with five drops of 4.5% sucrose solution and allowed to stand for one hour in a humid chamber at room temperature. The cells were permeabilized by adding three drops of 0.05% Triton X-100 solution for 10 min, after which ten drops of 2% formaldehyde solution (TAAB) containing 0.02% SDS pH 8.4 were added for 30 min. The slides were then dipped briefly in distilled water and air-dried. After rehydration in PBS the slides were soaked in PBST-BSA (PBS containing 0.1% Tween 20 and 0.15% BSA) for 1 hour and incubated overnight at 37°C with rabbit polyclonal anti-SYCP3 (1∶300; Abcam) and an anti-centromere (CREST) antibody (1∶500; Antibodies Inc.) diluted in PBST-BSA. Slides were washed in PBST, incubated with chicken anti-rabbit Alexa 488 (1∶500; Molecular Probes) and goat anti-human Alexa 594 (1∶500; Molecular Probes) diluted in PBS for 1 h at 37°C and washed in PBST. Pairing efficiency was evaluated on a Leica microscope after staining the cell nuclei with 4′,6-diamidino-2-phenylindole (DAPI) diluted in the mounting medium (Vectashield with DAPI; Vector).
At least four mice per genotype were used and pairing efficiency was assessed for ∼50 pachytene spermatocytes that were identified based on their DAPI nuclear morphology and their full autosomal synapsis identified by the synaptonemal complex (SYCP3) staining pattern. We classified the pairing of the Y*X in three categories: (i) clear PAR-PAR pairing of the X-chromosome with the Y*X chromosome, (ii) the Y*X chromosome clearly identifiable as a univalent chromosome, and (iii) no Y*X chromosome could be identified (most likely lost during cell spreading).
Ploidy analysis on testis cell spreads
Nuclear DNA content was measured on surface-spread spermatogenic cells from 6 week old testes as described previously [11], [14] using SYCP3 staining and DAPI fluorescence intensity measurements. Antibody against γH2AFX (1∶500; Upstate) was used to identify the sex body of pachytene spermatocytes [73].
Fluorescence in situ hybridization (FISH)
RNA-FISH for nascent nuclear transcripts from Zfy1, Zfy2 and Zfx was performed as previously described [7], [74] using spread testis cells from adult MF1 male mice. Zfx RNA FISH was also carried out on spread testis cells from XY,Zfx transgenics. The Zfy2-specific probe was BAC CITB-288D7 (Research Genetics), the Zfy1-specific probe was a modified version of BAC RP24-498K8 (CHORI) from which we had removed the entire Uba1y gene by recombineering, the Zfx-specific probe was BAC BMQ-372M23 (CHORI), the Mtm-specific probe was BAC RP24-287E17 (CHORI) and the Uty-specific probe was BAC CITB-246A22 (Research Genetics). Zfy1, Zfy2 and Zfx RNA FISH signals were confirmed with DNA FISH as described previously [74]. Antibody against SYCP3 (1∶100; Abcam) was used to identify secondary spermatocytes as previously describe [11].
In vitro transactivation assay
ZF TA domain-Gal4 fusion-protein constructs were made by inserting cDNA segments encoding the different acidic domains into the NcoI and SalI, or NdeI and SalI, restriction sites of the vector pGBK-CEN6, a single-copy version of pGBKT7 (Clontech), downstream of the Gal4 DNA-binding domain and the c-myc epitope tag of the vector. pGBK-CEN6 without an insert was included as a negative control. We used a low-copy origin because we had previously noticed that the expression of an acidic domain that strongly transactivates (Zfy2 and Gal4) inhibits yeast growth [7], and this has been described for the overexpression of Gal4 [75]. Validating our strategy, yeast transformed with the different acidic domain constructs, including Zfy2, all showed similar growth rates (as did the Gal4 acidic domain – data not shown). To create pGBK-CEN6, we replaced the 2 µ high-copy origin of pGBKT7 with the ARS4/CEN6 low-copy origin from pDEST22 (Invitrogen), by recombineering in the E.coli strain DY380 [76].
Acidic domains from human ZFY and mouse Zfy1 and Zfy2 were transferred to pGBK-CEN6 from pGBKT7 constructs as described previously [7]. The acidic domains from human ZFX and mouse Zfx were amplified from testis cDNAs and mouse Zfa was amplified from genomic DNA. PCR-amplified inserts were shown to be without error by sequencing recombinants. Primers used were Zfx: o4472/o4109, with respectively NcoI and SalI adaptors, and ZFX: o4473/o4109 and Zfa: o4471/o4109, with respectively NdeI and SalI adaptors (Table S4). One recombinant was selected for each construct and transformed into the S. cerevisiae strain Y187, in which the β-galactosidase gene is under the control of the Gal4-responsive Gal1 promoter. Three single transformed colonies were picked from SD/-trp agar plates and grown separately in liquid culture to an OD600 of 0.9–1.26 in SD/-trp liquid minimal medium. The β-galactosidase assay was performed on 1 OD600 unit of the culture using the permeabilized cell assay [77].
Statistical analysis
For ploidy frequency the differences between genotypes were assessed by one tail student t-test assuming unequal variances after angular transformation of percentages, using Excel (Microsoft) software. For the transactivation assay one tail student t-test assuming unequal variances was performed on the β-Galactosidase activity.
Supporting Information
Zdroje
1. BurgoynePS, LevyER, McLarenA (1986) Spermatogenic failure in male mice lacking H-Y antigen. Nature 320 : 170–172.
2. TiepoloL, ZuffardiO (1976) Localization of factors controlling spermatogenesis in the nonfluorescent portion of the human Y chromosome long arm. Human Genetics 34 : 119–124.
3. PageDC, MosherR, SimpsonEM, FisherEMC, MardonG, et al. (1987) The sex-determining region of the human Y chromosome encodes a finger protein. Cell 51 : 1091–1104.
4. MardonG, MosherR, DistecheCM, NishiokaY, McLarenA, et al. (1989) Duplication, deletion, and polymorphism in the sex-determining region of the mouse Y chromosome. Science 243 : 78–80.
5. MardonG, PageDC (1989) The sex-determining region of the mouse Y chromosome encodes a protein with a highly acidic domain and 13 zinc fingers. Cell 56 : 765–770.
6. MazeyratS, SautN, SargentCA, GrimmondS, LongepiedG, et al. (1998) The mouse Y chromosome interval necessary for spermatogonial proliferation is gene dense with syntenic homology to the human AZFa region. Hum Mol Genet 7 : 1713–1724.
7. DecarpentrieF, VernetN, MahadevaiahSK, LongepiedG, StreichembergerE, et al. (2012) Human and mouse ZFY genes produce a conserved testis-specific transcript encoding a zinc finger protein with a short acidic domain and modified transactivation potential. Hum Mol Genet 21 : 2631–2645.
8. MazeyratS, SautN, GrigorievV, MahadevaiahSK, OjarikreOA, et al. (2001) A Y-encoded subunit of the translation initiation factor Eif2 is essential for mouse spermatogenesis. Nat Genet 29 : 49–53.
9. KotMC, HandelMA (1990) Spermatogenesis in XO,Sxr mice: role of the Y chromosome. J Exp Zool 256 : 92–105.
10. SutcliffeMJ, DarlingSM, BurgoynePS (1991) Spermatogenesis in XY, XYSxra and XOSxra mice: a quantitative analysis of spermatogenesis throughout puberty. Mol Reprod Dev 30 : 81–89.
11. VernetN, MahadevaiahSK, OjarikreOA, LongepiedG, ProsserHM, et al. (2011) The Y-encoded gene Zfy2 acts to remove cells with unpaired chromosomes at the first meiotic metaphase in male mice. Curr Biol 21 : 787–793.
12. BurgoynePS, MahadevaiahSK, TurnerJM (2009) The consequences of asynapsis for mammalian meiosis. Nat Rev Genet 10 : 207–216.
13. SimpsonEM, PageDC (1991) An interstitial deletion in mouse Y chromosomal DNA created a transcribed Zfy fusion gene. Genomics 11 : 601–608.
14. VernetN, MahadevaiahSK, EllisPJ, de RooijDG, BurgoynePS (2012) Spermatid development in XO male mice with varying Y chromosome short arm gene content: evidence for a Y gene controlling the initiation of sperm morphogenesis. Reproduction 144 : 433–445.
15. EicherEM, HaleDW, HuntPA, LeeBK, TuckerPK, et al. (1991) The mouse Y* chromosome involves a complex rearrangement, including interstitial positioning of the pseudoautosomal region. Cytogenet Cell Genet 57 : 221–230.
16. HaleDW, HuntPA, TuckerPK, EicherEM (1991) Synapsis and obligate recombination between the sex chromosomes of male laboratory mice carrying the Y* rearrangement. Cytogenetics and Cell Genetics 57 : 231–239.
17. BurgoynePS, EvansEP (2000) A high frequency of XO offspring from X(Paf)Y* male mice: evidence that the Paf mutation involves an inversion spanning the X PAR boundary. Cytogenet Cell Genet 91 : 57–61.
18. BurgoynePS, MahadevaiahSK, PerryJ, PalmerSJ, AshworthA (1998) The Y* rearrangement in mice: new insights into a perplexing PAR. Cytogenet Cell Genet 80 : 37–40.
19. BurgoynePS, MahadevaiahSK, SutcliffeMJ, PalmerSJ (1992) Fertility in mice requires X-Y pairing and a Y-chromosomal “spermiogenesis” gene mapping to the long arm. Cell 71 : 391–398.
20. Alfoldi JE (2008) Sequence of the mouse Y chromosome [Thesis (Ph. D.)]. Massachusetts Institute of Technology
21. FergusonL, EllisPJ, AffaraNA (2009) Two novel mouse genes mapped to chromosome Yp are expressed specifically in spermatids. Mamm Genome 20 : 193–206 doi: 10.1007/s00335-009-9175-8
22. LeeJ, HongJ, KimE, KimK, KimSW, et al. (2004) Developmental stage-specific expression of Rbm suggests its involvement in early phases of spermatogenesis. Mol Hum Reprod 10 : 259–264.
23. CapelB, SwainA, NicolisS, HackerA, WalterM, et al. (1993) Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell 73 : 1019–1030.
24. DolciS, GrimaldiP, GeremiaR, PesceM, RossiP (1997) Identification of a promoter region generating Sry circular transcripts both in germ cells from male adult mice and in male mouse embryonal gonads. Biol Reprod 57 : 1128–1135.
25. TurnerJM (2007) Meiotic sex chromosome inactivation. Development 134 : 1823–1831.
26. RoyoH, PolikiewiczG, MahadevaiahSK, ProsserH, MitchellM, et al. (2010) Evidence that meiotic sex chromosome inactivation is essential for male fertility. Curr Biol 20 : 2117–2123.
27. HansenMA, NielsenJE, TanakaM, AlmstrupK, SkakkebaekNE, et al. (2006) Identification and expression profiling of 10 novel spermatid expressed CYPT genes. Mol Reprod Dev 73 : 568–579.
28. NagamineCM, ChanK, HakeLE, LauYF (1990) The two candidate testis-determining Y genes (Zfy-1 and Zfy-2) are differentially expressed in fetal and adult mouse tissues. Genes Dev 4 : 63–74.
29. NagamineCM, ChanKM, KozakCA, LauYF (1989) Chromosome mapping and expression of a putative testis-determining gene in mouse. Science 243 : 80–83.
30. MardonG, LuohSW, SimpsonEM, GillG, BrownLG, et al. (1990) Mouse Zfx protein is similar to Zfy-2: each contains an acidic activating domain and 13 zinc fingers. Mol Cell Biol 10 : 681–688.
31. AshworthA, SkeneB, SwiftS, Lovell-BadgeR (1990) Zfa is an expressed retroposon derived from an alternative transcript of the Zfx gene. Embo J 9 : 1529–1534.
32. McCarreyJR, DilworthDD, SharpRM (1992) Semiquantitative analysis of X-linked gene expression during spermatogenesis in the mouse: ethidium bromide staining of RT-PCR products. Genetic Analysis - Techniques and Applications 9 : 117–123.
33. LuohSW, PageDC (1994) The structure of the Zfx gene on the mouse X chromosome. Genomics 19 : 310–319.
34. MitchellM, SimonD, AffaraN, Ferguson-SmithM, AvnerP, et al. (1989) Localization of murine X and autosomal sequences homologous to the human Y located testis-determining region. Genetics 121 : 803–809.
35. EricksonRP, ZwingmanT, AoA (1993) Gene expression, X-inactivation, and methylation during spermatogenesis: the case of Zfa, Zfx, and Zfy in mice. Molecular Reproduction and Development 35 : 114–120.
36. HuntP, LeMaireR, EmburyP, SheeanL, MrozK (1995) Analysis of chromosome behavior in intact mammalian oocytes: monitoring the segregation of a univalent chromosome during female meiosis. Hum Mol Genet 4 : 2007–2012.
37. NagaokaSI, HodgesCA, AlbertiniDF, HuntPA (2011) Oocyte-Specific Differences in Cell-Cycle Control Create an Innate Susceptibility to Meiotic Errors. Curr Biol 21 : 651–7 doi: 10.1016/j.cub.2011.03.003
38. GuiL, HomerH (2012) Spindle assembly checkpoint signalling is uncoupled from chromosomal position in mouse oocytes. Development 139 : 1941–1946.
39. KolanoA, BrunetS, SilkAD, ClevelandDW, VerlhacMH (2012) Error-prone mammalian female meiosis from silencing the spindle assembly checkpoint without normal interkinetochore tension. Proc Natl Acad Sci U S A 109: E1858–1867.
40. LaneSI, YunY, JonesKT (2012) Timing of anaphase-promoting complex activation in mouse oocytes is predicted by microtubule-kinetochore attachment but not by bivalent alignment or tension. Development 139 : 1947–1955.
41. SebestovaJ, DanylevskaA, NovakovaL, KubelkaM, AngerM (2012) Lack of response to unaligned chromosomes in mammalian female gametes. Cell Cycle 11 : 3011–3018.
42. BurgoynePS, MahadevaiahSK, TurnerJM (2007) The management of DNA double-strand breaks in mitotic G(2), and in mammalian meiosis viewed from a mitotic G(2) perspective. Bioessays 29 : 974–986.
43. CocquetJ, EllisPJ, MahadevaiahSK, AffaraNA, VaimanD, et al. (2012) A genetic basis for a postmeiotic x versus y chromosome intragenomic conflict in the mouse. PLoS Genet 8: e1002900.
44. CocquetJ, EllisPJ, YamauchiY, MahadevaiahSK, AffaraNA, et al. (2009) The multicopy gene Sly represses the sex chromosomes in the male mouse germline after meiosis. PLoS Biol 7: e1000244.
45. LuohSW, BainPA, PolakiewiczRD, GoodheartML, GardnerH, et al. (1997) Zfx mutation results in small animal size and reduced germ cell number in male and female mice. Development 124 : 2275–2284.
46. MonesiV (1964) Ribonucleic Acid Synthesis during Mitosis and Meiosis in the Mouse Testis. J Cell Biol 22 : 521–532.
47. MonesiV (1965) Synthetic activities during spermatogenesis in the mouse. Experimental Cell Research 39 : 197–224.
48. ReynardLN, CocquetJ, BurgoynePS (2009) The Multi-Copy Mouse Gene Sycp3-Like Y-Linked (Sly) Encodes an Abundant Spermatid Protein That Interacts with a Histone Acetyltransferase and an Acrosomal Protein. Biol Reprod 81 : 250–7 doi: 10.1095/biolreprod.108.075382
49. MuellerJL, MahadevaiahSK, ParkPJ, WarburtonPE, PageDC, et al. (2008) The mouse X chromosome is enriched for multicopy testis genes showing postmeiotic expression. Nat Genet 40 : 794–799.
50. NamekawaSH, ParkPJ, ZhangLF, ShimaJE, McCarreyJR, et al. (2006) Postmeiotic sex chromatin in the male germline of mice. Curr Biol 16 : 660–667.
51. TurnerJM, MahadevaiahSK, EllisPJ, MitchellMJ, BurgoynePS (2006) Pachytene asynapsis drives meiotic sex chromosome inactivation and leads to substantial postmeiotic repression in spermatids. Dev Cell 10 : 521–529.
52. van RheedeT, BastiaansT, BooneDN, HedgesSB, de JongWW, et al. (2006) The platypus is in its place: nuclear genes and indels confirm the sister group relation of monotremes and Therians. Mol Biol Evol 23 : 587–597.
53. GravesJA (1995) The origin and function of the mammalian Y chromosome and Y-borne genes–an evolving understanding. Bioessays 17 : 311–320.
54. Schneider-GädickeA, Beer-RomeroP, BrownLG, NussbaumR, PageDC (1989) The ZFX gene on the human X chromosome escapes X-inactivation and is closely related to ZFY, the putative sex determinant on the Y chromosome. Cell 57 : 1247–1258.
55. JegalianK, PageDC (1998) A proposed path by which genes common to mammalian X and Y chromosomes evolve to become X inactivated. Nature 394 : 776–780.
56. PamiloP, BianchiNO (1993) Evolution of the Zfx and Zfy genes: rates and interdependence between the genes. Molecular Biology and Evolution 10 : 271–281.
57. TuckerPK, AdkinsRM, RestJS (2003) Differential rates of evolution for the ZFY-related zinc finger genes, Zfy, Zfx, and Zfa in the mouse genus Mus. Mol Biol Evol 20 : 999–1005.
58. AshworthA, RastanS, Lovell-BadgeR, KayG (1991) X-chromosome inactivation may explain the difference in viability of XO humans and mice. Nature 351 : 406–408.
59. ShimminLC, ChangBH, LiWH (1994) Contrasting rates of nucleotide substitution in the X-linked and Y-linked zinc finger genes. J Mol Evol 39 : 569–578.
60. LiL, ZhengP, DeanJ (2010) Maternal control of early mouse development. Development 137 : 859–870.
61. WangH, HuYC, MarkoulakiS, WelsteadGG, ChengAW, et al. (2013) TALEN-mediated editing of the mouse Y chromosome. Nat Biotechnol 31 : 530–532.
62. YamauchiY, RielJM, StoytchevaZ, WardMA (2014) Two Y genes can replace the entire Y chromosome for assisted reproduction in the mouse. Science 343 : 69–72.
63. DerijckA, van der HeijdenG, GieleM, PhilippensM, de BoerP (2008) DNA double-strand break repair in parental chromatin of mouse zygotes, the first cell cycle as an origin of de novo mutation. Hum Mol Genet 17 : 1922–1937.
64. LieberMR (2008) The mechanism of human nonhomologous DNA end joining. J Biol Chem 283 : 1–5.
65. CattanachBM, PollardCE, HawkesSG (1971) Sex reversed mice : XX and XO males. Cytogenetics 10 : 318–337.
66. CattanachBM, EvansEP, BurtenshawMD, BarlowJ (1982) Male, female and intersex development in mice of identical chromosome constitution. Nature 300 : 445–446.
67. McLarenA, MonkM (1982) Fertile females produced by inactivation of X chromosome of ‘sex-reversed’ mice. Nature 300 : 446–448.
68. HadjantonakisAK, GertsensteinM, IkawaM, OkabeM, NagyA (1998) Non-invasive sexing of preimplantation stage mammalian embryos. Nat Genet 19 : 220–222.
69. GubbayJ, CollignonJ, KoopmanP, CapelB, EconomouA, et al. (1990) A gene mapping to the sex-determining region of the mouse Y chromosome is a member of a novel family of embryonically expressed genes. Nature 346 : 245–250.
70. GubbayJ, VivianN, EconomouA, JacksonD, GoodfellowP, et al. (1992) Inverted repeat structure of the Sry locus in mice. Proc Natl Acad Sci U S A 89 : 7953–7957.
71. MahadevaiahSK, OdorisioT, ElliottDJ, RattiganA, SzotM, et al. (1998) Mouse homologues of the human AZF candidate gene RBM are expressed in spermatogonia and spermatids, and map to a Y chromosome deletion interval associated with a high incidence of sperm abnormalities. Hum Mol Genet 7 : 715–727.
72. WrightWE, SassoonDA, LinVK (1989) Myogenin, a factor regulating myogenesis, has a domain homologous to MyoD. Cell 56 : 607–617.
73. MahadevaiahSK, TurnerJMA, BaudatF, RogakouEP, de BoerP, et al. (2001) Recombinational DNA double-strand breaks in mice precede synapsis. Nature Genetics 27 : 271–276.
74. MahadevaiahSK, CostaY, TurnerJM (2009) Using RNA FISH to study gene expression during mammalian meiosis. Methods Mol Biol 558 : 433–444.
75. GillG, PtashneM (1988) Negative effect of the transcriptional activator GAL4. Nature 334 : 721–724.
76. LeeEC, YuD, Martinez de VelascoJ, TessarolloL, SwingDA, et al. (2001) A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics 73 : 56–65.
77. GuarenteL (1983) Yeast promoters and lacZ fusions designed to study expression of cloned genes in yeast. Methods Enzymol 101 : 181–191.
Štítky
Genetika Reprodukční medicína
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 6
-
Všechny články tohoto čísla
- Inflammation: Gone with Translation
- Recombination Accelerates Adaptation on a Large-Scale Empirical Fitness Landscape in HIV-1
- Caspase Inhibition in Select Olfactory Neurons Restores Innate Attraction Behavior in Aged
- Accurate, Model-Based Tuning of Synthetic Gene Expression Using Introns in
- A Novel Peptidoglycan Binding Protein Crucial for PBP1A-Mediated Cell Wall Biogenesis in
- Ancient DNA Analysis of 8000 B.C. Near Eastern Farmers Supports an Early Neolithic Pioneer Maritime Colonization of Mainland Europe through Cyprus and the Aegean Islands
- The Epidermal Growth Factor Receptor Critically Regulates Endometrial Function during Early Pregnancy
- Introgression from Domestic Goat Generated Variation at the Major Histocompatibility Complex of Alpine Ibex
- Netrins and Wnts Function Redundantly to Regulate Antero-Posterior and Dorso-Ventral Guidance in
- Coordination of Wing and Whole-Body Development at Developmental Milestones Ensures Robustness against Environmental and Physiological Perturbations
- Phenotypic Dissection of Bone Mineral Density Reveals Skeletal Site Specificity and Facilitates the Identification of Novel Loci in the Genetic Regulation of Bone Mass Attainment
- Deep Evolutionary Comparison of Gene Expression Identifies Parallel Recruitment of -Factors in Two Independent Origins of C Photosynthesis
- Loss of UCP2 Attenuates Mitochondrial Dysfunction without Altering ROS Production and Uncoupling Activity
- Translational Regulation of Specific mRNAs Controls Feedback Inhibition and Survival during Macrophage Activation
- Rosa26-GFP Direct Repeat (RaDR-GFP) Mice Reveal Tissue- and Age-Dependence of Homologous Recombination in Mammals
- Abnormal Type I Collagen Post-translational Modification and Crosslinking in a Cyclophilin B KO Mouse Model of Recessive Osteogenesis Imperfecta
- : Clonal Reinforcement Drives Evolution of a Simple Microbial Community
- Reviving the Dead: History and Reactivation of an Extinct L1
- Defective iA37 Modification of Mitochondrial and Cytosolic tRNAs Results from Pathogenic Mutations in TRIT1 and Its Substrate tRNA
- Early Back-to-Africa Migration into the Horn of Africa
- Aberrant Autolysosomal Regulation Is Linked to The Induction of Embryonic Senescence: Differential Roles of Beclin 1 and p53 in Vertebrate Spns1 Deficiency
- Microbial Succession in the Gut: Directional Trends of Taxonomic and Functional Change in a Birth Cohort of Spanish Infants
- Integrated Pathway-Based Approach Identifies Association between Genomic Regions at CTCF and CACNB2 and Schizophrenia
- Genetic Determinants of Long-Term Changes in Blood Lipid Concentrations: 10-Year Follow-Up of the GLACIER Study
- Palaeosymbiosis Revealed by Genomic Fossils of in a Strongyloidean Nematode
- Early Embryogenesis-Specific Expression of the Rice Transposon Enhances Amplification of the MITE
- PINK1-Mediated Phosphorylation of Parkin Boosts Parkin Activity in
- OsHUS1 Facilitates Accurate Meiotic Recombination in Rice
- Genetic Background Drives Transcriptional Variation in Human Induced Pluripotent Stem Cells
- Pervasive Divergence of Transcriptional Gene Regulation in Caenorhabditis Nematodes
- N-WASP Is Required for Structural Integrity of the Blood-Testis Barrier
- The Transcription Factor TFII-I Promotes DNA Translesion Synthesis and Genomic Stability
- An Operon of Three Transcriptional Regulators Controls Horizontal Gene Transfer of the Integrative and Conjugative Element ICE in B13
- Digital Genotyping of Macrosatellites and Multicopy Genes Reveals Novel Biological Functions Associated with Copy Number Variation of Large Tandem Repeats
- ATRA-Induced Cellular Differentiation and CD38 Expression Inhibits Acquisition of BCR-ABL Mutations for CML Acquired Resistance
- The EJC Binding and Dissociating Activity of PYM Is Regulated in
- JNK Controls the Onset of Mitosis in Planarian Stem Cells and Triggers Apoptotic Cell Death Required for Regeneration and Remodeling
- Mouse Y-Linked and Are Expressed during the Male-Specific Interphase between Meiosis I and Meiosis II and Promote the 2 Meiotic Division
- Rasa3 Controls Megakaryocyte Rap1 Activation, Integrin Signaling and Differentiation into Proplatelet
- Transcriptional Control of Steroid Biosynthesis Genes in the Prothoracic Gland by Ventral Veins Lacking and Knirps
- Souffle/Spastizin Controls Secretory Vesicle Maturation during Zebrafish Oogenesis
- The POU Factor Ventral Veins Lacking/Drifter Directs the Timing of Metamorphosis through Ecdysteroid and Juvenile Hormone Signaling
- The First Endogenous Herpesvirus, Identified in the Tarsier Genome, and Novel Sequences from Primate Rhadinoviruses and Lymphocryptoviruses
- Sequence of a Complete Chicken BG Haplotype Shows Dynamic Expansion and Contraction of Two Gene Lineages with Particular Expression Patterns
- Background Selection as Baseline for Nucleotide Variation across the Genome
- CPF-Associated Phosphatase Activity Opposes Condensin-Mediated Chromosome Condensation
- The Effects of Codon Context on Translation Speed
- Glycogen Synthase Kinase (GSK) 3β Phosphorylates and Protects Nuclear Myosin 1c from Proteasome-Mediated Degradation to Activate rDNA Transcription in Early G1 Cells
- Regulation of Gene Expression in Autoimmune Disease Loci and the Genetic Basis of Proliferation in CD4 Effector Memory T Cells
- Muscle Structure Influences Utrophin Expression in Mice
- BLMP-1/Blimp-1 Regulates the Spatiotemporal Cell Migration Pattern in
- Identification of Late Larval Stage Developmental Checkpoints in Regulated by Insulin/IGF and Steroid Hormone Signaling Pathways
- Transport of Magnesium by a Bacterial Nramp-Related Gene
- Sgo1 Regulates Both Condensin and Ipl1/Aurora B to Promote Chromosome Biorientation
- The HY5-PIF Regulatory Module Coordinates Light and Temperature Control of Photosynthetic Gene Transcription
- The Rim15-Endosulfine-PP2A Signalling Module Regulates Entry into Gametogenesis and Quiescence Distinct Mechanisms in Budding Yeast
- Regulation of Hfq by the RNA CrcZ in Carbon Catabolite Repression
- Loss of a Neural AMP-Activated Kinase Mimics the Effects of Elevated Serotonin on Fat, Movement, and Hormonal Secretions
- Positive Feedback of Expression Ensures Irreversible Meiotic Commitment in Budding Yeast
- Hecate/Grip2a Acts to Reorganize the Cytoskeleton in the Symmetry-Breaking Event of Embryonic Axis Induction
- Regulatory Mechanisms That Prevent Re-initiation of DNA Replication Can Be Locally Modulated at Origins by Nearby Sequence Elements
- Speciation and Introgression between and
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Early Back-to-Africa Migration into the Horn of Africa
- PINK1-Mediated Phosphorylation of Parkin Boosts Parkin Activity in
- OsHUS1 Facilitates Accurate Meiotic Recombination in Rice
- An Operon of Three Transcriptional Regulators Controls Horizontal Gene Transfer of the Integrative and Conjugative Element ICE in B13
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



