-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaTwo Distinct Repressive Mechanisms for Histone 3 Lysine 4 Methylation through Promoting 3′-End Antisense Transcription
Histone H3 di - and trimethylation on lysine 4 are major chromatin marks that correlate with active transcription. The influence of these modifications on transcription itself is, however, poorly understood. We have investigated the roles of H3K4 methylation in Saccharomyces cerevisiae by determining genome-wide expression-profiles of mutants in the Set1 complex, COMPASS, that lays down these marks. Loss of H3K4 trimethylation has virtually no effect on steady-state or dynamically-changing mRNA levels. Combined loss of H3K4 tri - and dimethylation results in steady-state mRNA upregulation and delays in the repression kinetics of specific groups of genes. COMPASS-repressed genes have distinct H3K4 methylation patterns, with enrichment of H3K4me3 at the 3′-end, indicating that repression is coupled to 3′-end antisense transcription. Further analyses reveal that repression is mediated by H3K4me3-dependent 3′-end antisense transcription in two ways. For a small group of genes including PHO84, repression is mediated by a previously reported trans-effect that requires the antisense transcript itself. For the majority of COMPASS-repressed genes, however, it is the process of 3′-end antisense transcription itself that is the important factor for repression. Strand-specific qPCR analyses of various mutants indicate that this more prevalent mechanism of COMPASS-mediated repression requires H3K4me3-dependent 3′-end antisense transcription to lay down H3K4me2, which seems to serve as the actual repressive mark. Removal of the 3′-end antisense promoter also results in derepression of sense transcription and renders sense transcription insensitive to the additional loss of SET1. The derepression observed in COMPASS mutants is mimicked by reduction of global histone H3 and H4 levels, suggesting that the H3K4me2 repressive effect is linked to establishment of a repressive chromatin structure. These results indicate that in S. cerevisiae, the non-redundant role of H3K4 methylation by Set1 is repression, achieved through promotion of 3′-end antisense transcription to achieve specific rather than global effects through two distinct mechanisms.
Published in the journal: . PLoS Genet 8(9): e32767. doi:10.1371/journal.pgen.1002952
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002952Summary
Histone H3 di - and trimethylation on lysine 4 are major chromatin marks that correlate with active transcription. The influence of these modifications on transcription itself is, however, poorly understood. We have investigated the roles of H3K4 methylation in Saccharomyces cerevisiae by determining genome-wide expression-profiles of mutants in the Set1 complex, COMPASS, that lays down these marks. Loss of H3K4 trimethylation has virtually no effect on steady-state or dynamically-changing mRNA levels. Combined loss of H3K4 tri - and dimethylation results in steady-state mRNA upregulation and delays in the repression kinetics of specific groups of genes. COMPASS-repressed genes have distinct H3K4 methylation patterns, with enrichment of H3K4me3 at the 3′-end, indicating that repression is coupled to 3′-end antisense transcription. Further analyses reveal that repression is mediated by H3K4me3-dependent 3′-end antisense transcription in two ways. For a small group of genes including PHO84, repression is mediated by a previously reported trans-effect that requires the antisense transcript itself. For the majority of COMPASS-repressed genes, however, it is the process of 3′-end antisense transcription itself that is the important factor for repression. Strand-specific qPCR analyses of various mutants indicate that this more prevalent mechanism of COMPASS-mediated repression requires H3K4me3-dependent 3′-end antisense transcription to lay down H3K4me2, which seems to serve as the actual repressive mark. Removal of the 3′-end antisense promoter also results in derepression of sense transcription and renders sense transcription insensitive to the additional loss of SET1. The derepression observed in COMPASS mutants is mimicked by reduction of global histone H3 and H4 levels, suggesting that the H3K4me2 repressive effect is linked to establishment of a repressive chromatin structure. These results indicate that in S. cerevisiae, the non-redundant role of H3K4 methylation by Set1 is repression, achieved through promotion of 3′-end antisense transcription to achieve specific rather than global effects through two distinct mechanisms.
Introduction
Packaging of eukaryotic DNA with histones has a generally repressive effect on transcription [1]. Histones themselves are subject to a variety of post-translational modifications, such as acetylation, methylation and ubiquitinylation. These modifications correlate with specific states of transcription, as well as with the activity of other DNA-linked processes, such as chromosome segregation and DNA repair [2], [3]. Among the epigenetic marks, histone methylation has been extensively associated with both activation and repression of genes in euchromatic and heterochromatic regions respectively [4]. Methylation of histone H3 on lysine 4 (H3K4) for example, has been linked to transcriptional activation in many eukaryotic species. Vertebrates possess several H3K4 methyltransferases related to the SET domain of yeast Set1 and Drosophila Trx (MLL family) [5]. These methyltransferases are responsible for mono - (H3K4me1), di - (H3K4me2) and trimethylation (H3K4me3) of H3K4 [6]. Di - and trimethylation of H3K4 is generally restricted to euchromatin and genome-wide studies in metazoan cells have revealed high levels of histone acetylation and H3K4 methylation in promoter regions of active genes [7], [8], [9], [10], [11]. H3K4me2 and H3K4me3 are thought to facilitate transcription through the recruitment of general transcription factors [12] and cofactors [13] or by preventing repressors from binding to chromatin [14]. The precise mechanism through which the various H3K4 methylation states contribute to control of gene expression are not fully understood.
In Saccharomyces cerevisiae, H3K4 methylation is carried out by the Set1 complex, COMPASS [15], which is composed of the catalytic subunit Set1 and at least six other components (Swd1, Swd2, Swd3, Bre2, Sdc1 and Spp1) [16], [17], [18], [19]. Loss or inactivation of individual subunits differentially affects the methylation state of H3K4. Swd1, Swd2 and Swd3 are required for COMPASS stability and their disruption affects all three H3K4 methylation states. Bre2 and Sdc1 promote the efficient di - and trimethylation of H3K4, while inactivation of Spp1 only affects H3K4 trimethylation [20], [21], [22]. In addition, monoubiquitylation of Swd2 has recently been shown to mediate the trans-tail process between H2B ubiquitylation and H3K4 trimethylation, by controlling the recruitment of the Spp1 subunit [23]. Set1 has been found to be predominantly associated with the coding regions of highly transcribed RNA polymerase II genes and the presence of trimethylated H3K4 correlates with Set1 occupancy [24] and transcription rate [25]. Genome-wide studies in yeast indicate that active transcription is characteristically accompanied by histone H3K4 trimethylation at the 5′-end of genes and by H3K4 dimethylation and monomethylation at nucleosomes positioned further downstream in the transcription unit [25].
Although H3K4 trimethylation has been linked to transcription initiation and elongation in yeast [6], [21], [26], its precise role in transcription as well as the role of H3K4 mono - and dimethylation remain poorly understood. This is in part because previous genome-wide analyses of the effects of H3K4 methylation loss have yielded conflicting results [6], [27], [28], [29] [30]. While two studies suggested a global reduction in transcription when H3K4 methylation is abolished [27], [28], a third study reported and focused on only 480 very marginally down-regulated genes, even though twice as many genes were observably upregulated upon applying the same selection criteria [6]. The most recent study also reported roughly 300 genes up-regulated and 100 down-regulated [30]. A more statistically stringent study that included adequate replicate experiments showed that 200 genes become up-regulated upon loss of SET1, with virtually no down-regulation observed [29], suggesting that H3K4 methylation may actually play a more prominent role in repression than in activation of protein-coding genes.
Recently, a form of RNA-mediated transcriptional repression has been reported in S. cerevisiae, that is independent of the RNAi machinery which is absent from budding yeast. Ty1, PHO84 and GAL1/10 expression have been shown to be regulated by antisense RNA transcription [31], [32], [33]. For PHO84, it was found that expression of PHO84 antisense RNA from an ectopic PHO84 gene copy was able to trigger silencing of the endogenous PHO84 gene [34]. Production of the PHO84 antisense RNA was found to be positively regulated by Set1 [34] potentially linking H3K4 methylation to non-coding RNA (ncRNA) regulation. Genome-wide analysis has recently revealed the existence of hundreds of previously uncharacterized ncRNAs in mammals [35], [36], [37], [38] and in yeast [39], [40], that either stably exist or are rapidly degraded by the RNA surveillance pathway. Strikingly, most of these newly identified transcripts initiate from nucleosome-free regions associated with bidirectional promoters of protein-coding genes or regions in the body or close to the 3′-ends of protein-coding genes [40]. Regulation of ncRNAs is far from understood.
Here we present an extensive genome-wide analysis that discriminates between the roles of the different H3K4 methylation states. While preventing H3K4 trimethylation on its own has no effect on mRNA expression of coding genes, 1% of coding genes are derepressed upon combined loss of di - and trimethylation. Further analyses indicate distinct roles for these two marks in repression of coding genes through mechanisms that are mediated through 3′-end antisense transcription.
Results
Loss of H3K4 dimethylation correlates with increased expression of a subset of genes
Previous genome-wide analyses of the effects of losing H3K4 methylation [6], [27], [28], [29] [30] focused on loss of all three H3K4 methylation states simultaneously, either through deletion of the gene that codes for the H3K4 methyltransferase, SET1 or through substitution of H3K4 with alanine or arginine. To investigate whether there are separate roles for H3K4 mono-, di - and trimethylation, we made use of the fact that mutating different components of the Set1 complex, COMPASS, results in different methylation states. First, the methylation status of H3K4 was assessed in strains with deletions of the non-essential members of the complex, in the single genetic background used for this study (BY4741). An additional strain was included that carries a mutation that prevents monoubiquitylation of the essential subunit Swd2 (swd2K68,69R), resulting in a severe reduction of H3K4me3 [23]. Histones were purified from each strain and their H3K4 methylation status was checked with antibodies specific for each methylated state (Figure 1A). As expected from previous results (see the introduction), deletion of SET1, SWD1 or SWD3 abolishes mono-, di - and trimethylation of H3K4. Deletion of BRE2 or SDC1 results in a complete loss of H3K4me3, a significant decrease of H3K4me2 but no change in H3K4me1, while inactivation of SPP1 or mutating SWD2 (swd2K68,69R) results in a severe and specific decrease of H3K4me3 (Figure 1A).
Fig. 1. Loss of H3K4 di- and trimethylation results in upregulation of a subset of genes. 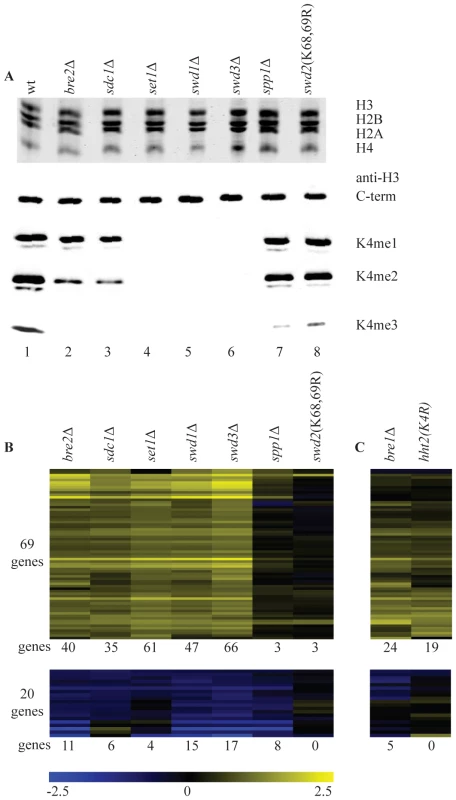
(A) Commassie-stained gel of purified histones from the indicated strains (top) and western blots with antibodies directed against H3 carboxy-terminus and the different H3K4 methylation states (bottom). (B) Hierarchical clustering of all genes with significantly changed mRNA expression (p-value less than 0.01 and fold-change versus wild-type more than 1.7) in at least two COMPASS mutants. Fold-change of mRNA expression in mutant versus wild-type is indicated by the colour bar as log2 values. Number of genes below each heatmap correspond to the genes called significant in each mutant. (C) Genes depicted in the same order as in B for the H3K4R point mutant and bre1Δ. The same strains were analyzed in parallel by long oligo DNA microarray expression-profiling, targeting the coding strand of virtually all yeast genes. Throughout this study all microarray analyses were performed with four replicates (two independent cultures, each measured in duplicate, Materials and Methods). In addition, controls were included that allow detection of global changes in the entire mRNA population [41]. Such global changes were not detected. In agreement with the most recent studies of SET1 deletion on its own [29] [30], expression of only a minority of genes is affected in the different COMPASS mutants. Within the entire set of deletion mutants, 89 genes changed significantly in at least two mutants (p-value lower than 0.01 and fold-change versus wild-type more than 1.7), with 69 genes showing increased expression and only 20 exhibiting decrease (Figure 1B). Deletion of any of the five core subunits Set1, Swd1, Swd3, Bre2 and Sdc1 leads essentially to the same expression profile (Figure 1B and Figure S1).
It is interesting to compare the changes in gene expression to the H3K4 methylation states observed in the different mutants. Virtually no significant changes in gene expression are observed in spp1Δ or in the swd2K68,69R mutant (Figure 1B) that both show a specific and severe decrease of H3K4 trimethylation (Figure 1A). Changes in gene expression are observed in bre2Δ and sdc1Δ, where H3K4 dimethylation is significantly diminished on top of the loss of trimethylation, (Figure 1A, 1B). The additional loss of H3K4 monomethylation, as observed in set1Δ, swd1Δ or swd3Δ (Figure 1A), does not lead to additional changes in gene expression (Figure 1B). Because of the correlation between their location and transcription rates [25], H3K4 methylation marks in yeast have generally been associated with transcription activation. The main effect of mutating COMPASS components in S. cerevisiae is nevertheless derepression (Figure 1B). Furthermore, the effect is only strong upon loss of dimethylation on top of trimethylation loss, which on its own has little effect.
To distinguish whether the repressive effect of COMPASS is related to H3K4 methylation or is due to an unidentified methylation target of Set1, a H3K4 point mutant was analyzed. The predominant effect is up-regulation (Figure 1C) and the overlap with the COMPASS-repressed genes is highly significant (p-value 3.1*10−27, hypergeometric test). An apparently lower number of genes is derepressed in the H3K4 point mutant. As analyzed later, this is likely related to the H3/H4 histone dosage effect of the strain used to generate the point mutant. To nevertheless investigate the possibility that Set1 repression is mediated by a target other than H3K4, SET1 was deleted in the H3K4 point mutant strain. DNA microarray analysis of the double mutant shows a completely epistatic relationship with no additional effect of deleting SET1 in the H3K4 point mutant strain (Figure S2). This confirms that the repressive effect of COMPASS observed here is mediated through H3K4.
It has been previously shown that H3K4 di - and tri-, but not monomethylation states are controlled by the Rad6/Bre1-mediated monoubiquitylation of histone H2BK123 via a trans-tail pathway involving ubiquitylation of Swd2 [23], [42], [43], [44], [45]. To investigate whether the repressive effects of H3K4 methylation are mediated by this pathway, a bre1Δ strain was analyzed. Changes in gene expression in bre1Δ matches the COMPASS mutants profiles with a highly significant overlap (p-value of 1.0*10−37, hypergeometric test) (Figure 1C). The repressive effects observed here therefore correspond to the action of the entire pathway starting from ubiquitylation of histone H2B and leading to di - and trimethylation of H3K4.
Repression dynamics are subtly affected by loss of H3K4 methylation
Since the experiments described above deal with steady-state changes in mRNA levels, we next asked whether the absence of H3K4 methylation would affect the kinetics of gene expression changes. This is based on the proposal that H3K4me3 may have a memory function, bookmarking genes that require rapid induction under specific growth conditions, both in mammals [46] and yeast [24]. For this purpose, wild-type (wt), set1Δ (absence of all three H3K4 methylation states) and spp1Δ (lack of H3K4 trimethylation only) were expression-profiled at multiple time-points during the transition from post-diauxic shift to early log phase, a transition during which a large number of genes change expression levels [47]. During this transition, expression of approximately 3400 genes change significantly in wt cells, covering a broad range of gene expression dynamics (Figure 2A). No major differences in the transcription kinetics between wt and the two mutant strains are observed. This indicates that disruption of H3K4 methylation or H3K4 trimethylation on its own does not have a global effect on the dynamics of transcription (Figure 2A), even though most active genes exhibit H3K4me3 marks [6], [11], [27]. These results also agree with the lack of a global effect after removing the H3K4me3 mark under steady-state conditions (Figure 1B).
Fig. 2. Loss of H3K4 di- and trimethylation leads to a delay in repression kinetics for a subset of genes. 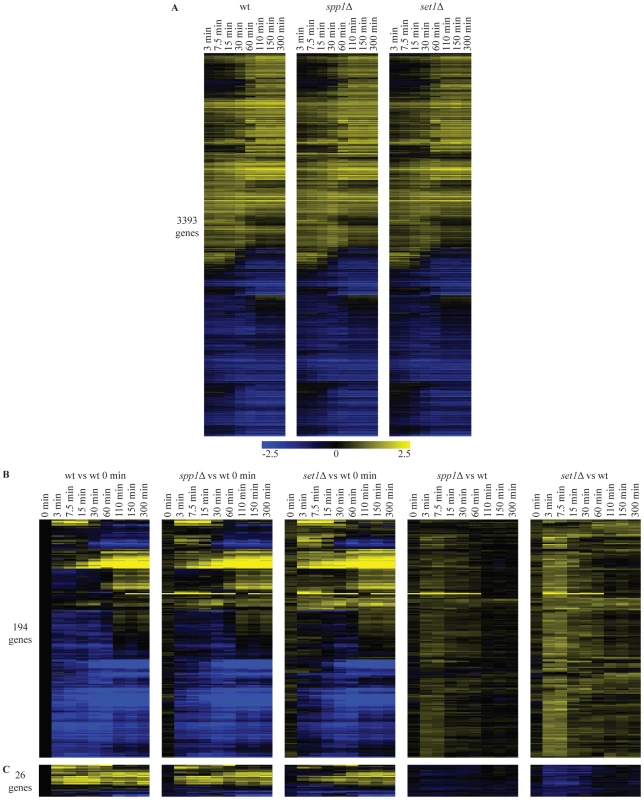
(A) Hierarchical clustering of all genes with significant changes in mRNA expression during the shift from low to high glucose in any of the wt, spp1Δ and set1Δ time-courses. The log2 values correspond to the difference with the zero time point of each time-course. (B) Hierarchical clustering of genes with delayed repression compared to wt. These genes were identified based on statistically significant differences between the mutant and wt time-courses (Materials and Methods). The first three panels show the differences in expression versus the wt zero time point. The last two panels (spp1Δ vs wt) and (set1Δ vs wt) depict the differences between mutant and wt for each different time point, by subtracting the log base 2 gene expression ratios of the wt time-course from the mutant time-course. (C) Hierarchical clustering of genes that show delay in activation. A detailed statistical analysis for genes showing differences in their induction or repression kinetics in the mutants was also performed among the 3400 genes that change significantly during the time-course experiment. In the set1Δ (loss of all three H3K4 methylation states) time-course, 220 genes show statistically significant differences in their expression kinetics compared to the corresponding time points in wt (compare Figure 2B, 2C first and third panel). The vast majority of these (194 genes - Figure 2B) exhibit defective repression, observed as delayed repression or faster activation. Only a minority of genes exhibit an activation defect (26 genes - Figure 2C). To facilitate visualization of these mostly quite subtle changes, the wt time-course was subtracted from each mutant time-course. This results in the right-hand panels of Figure 2B and 2C, showing for each time-point, the difference in expression levels for each mutant relative to the wt at the same time point. For spp1Δ (loss of H3K4me3), only 15 genes exhibit any differences in their expression kinetics (Figure 2B, 2C, second panel). These all belong to the 220 genes with slightly altered kinetics in the set1Δ time-course. In agreement with the steady-state analysis, the effects detected in the time-course experiments are thus virtually all attributable to the complete loss of methylation observed in set1Δ, rather than to the specific loss of H3K4me3 observed in spp1Δ. The results concur with a repressive role for COMPASS on mRNA expression of a subset of genes, as observed in the steady-state experiments too (Figure 1) with an extremely significant overlap between the affected genes (p-value 6*10−35), as expected.
Unconventional methylation patterns at the 3′-end of COMPASS-repressed genes
We next investigated whether there are any particular characteristics shared by the set of genes upregulated upon mutation of COMPASS components (Figure 1B). In agreement with a recent analysis of set1Δ [29], statistically significant enrichment for location close to telomeres is observed (Figure S3). Among the 69 COMPASS-repressed genes, 10 are telomere-proximal (within 15 kb) Figure S3 and Table S1). Although this enrichment is significant, in most cases the expression of adjacent genes was not found to be affected by the deletion of COMPASS subunits. For instance, PHO11, SNO4, MCH2, SOR2, YGL258W-A and PHO12, that are located between 4 and 10 kb from the telomeric DNA on different chromosomes (Table S1) are all flanked by genes that are not affected by the absence of Set1. This, as well as the small number of all telomere - proximal genes being derepressed in the COMPASS mutants makes it unlikely that the observed derepression of telomere-proximal genes is only caused by loss of the Sir-dependent telomeric position effect [19], [48], [49], [50], [51], [52].
As the effect of COMPASS deletions is attributable to H3K4 methylation (Figure 1C), the H3K4 methylation patterns of COMPASS-repressed genes were examined using chromatin immunoprecipitation data from a wt strain from the same genetic background, grown under similar conditions [53]. Intriguingly, the di - and trimethylation patterns of the 69 COMPASS-repressed genes (Figure 3A) deviate from the average gene which has enrichment of H3K4me3 around the transcription start site (Figure S4) [25], [53]. Instead, the majority of COMPASS-repressed genes show enrichment of H3K4me3 at the 3′-end or in the body of the gene. In the minority of cases where 5′-end enrichment is observed, this is accompanied by a second trimethylation peak at the 3′ end. To exclude that the deviating localization of peaks is not due to measurement noise or signal originating from neighbouring genes, the methylation profiles are averaged in Figure 3B only for those genes that have a greater than 2-fold enrichment of H3K4 methylation on any portion of the gene. This average pattern for COMPASS-repressed genes shows a clear enrichment of H3K4me3 at the 3′ end, followed by H3K4me2 enrichment in the gene body, which is in turn followed by H3K4me1 further towards the 5′-end. Genes repressed by COMPASS therefore show abberant H3K4 methylation patterns that are characterized by a reversed orientation of the normal H3K4 methylation pattern observed for active genes [3].
Fig. 3. COMPASS-repressed genes have aberrant H3K4 methylation patterns, indicative of 3′-end antisense transcription. 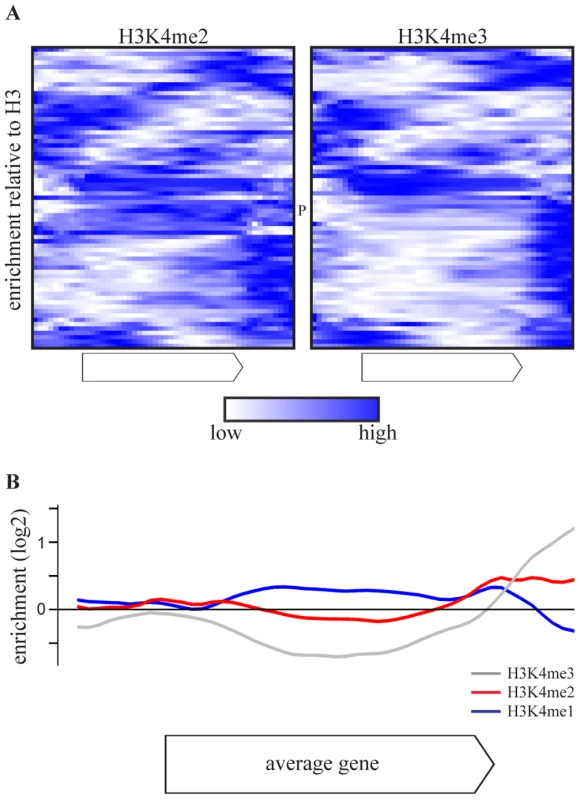
(A) Heatmaps of enrichment of H3K4me2(left) and H3K4me3(right) over H3 in the gene body and flanking regions of the 69 COMPASS repressed genes, based on [53]. The enrichments are rescaled for each individual gene with blue and white corresponding to the highest and lowest enrichment, respectively. PHO84 is marked by P. (B) The average enrichment of H3K4me1 (blue), H3K4me2 (red) and H3K4me3 (grey) over H3, for the set of 47 COMPASS-repressed genes that show at least a two-fold enrichment of H3K4me2 or H3K4me3 somewhere across the gene or flanking region [53]. Promotion of 3′-end antisense transcription by Set1 contributes to repression on coding genes through two distinct mechanisms
A plausible explanation for the H3K4 di - and trimethylation peaks at the 3′-ends of COMPASS-repressed genes is the presence of antisense transcription initiation at the 3′-end of the coding region, leading to non-coding transcription over the same genomic location but in the opposite direction of the sense transcription. The DNA microarrays used in the previous experiments are coding strand-specific and do not detect anti-sense transcripts. However, two recent genome-wide surveys of non-coding transcripts [39], [40], do detect antisense RNAs for more than 85% of the COMPASS-repressed genes (Table S2). Interestingly, PHO84 belongs to the group of COMPASS-repressed genes identified here (Figure 3A, marked with P) and has been shown to be regulated by antisense RNA transcripts originating from its 3′-end both in cis and in trans [32], [34]. We therefore investigated the manner in which 3′-end antisense transcription may be involved in Set1-mediated repression.
One hallmark of the mechanism of repression of PHO84 is the contribution of the antisense transcript itself rather than only the process of antisense transcription. Stabilization of the antisense transcript by deletion of the exosome component RRP6 [32] is sufficient to repress sense PHO84 transcription. To test whether COMPASS repression is mediated by 3′-end antisense transcripts, an rrp6Δ profile was generated and compared to set1Δ. Deletion of RRP6 affects expression of 117 coding genes in total (p<0.01, fold-change>1.7) and does not have a general effect on all COMPASS-repressed genes (Figure 4A). In agreement with previous studies however, a significant down-regulation of PHO84 is observed (marked P in Figure 4A). Lack of down-regulation of the other COMPASS-repressed genes in rrp6Δ may be simply due to an already repressed state in wt. Since these genes are derepressed in set1Δ, the possible involvement of antisense transcripts in repressing all COMPASS-affected genes was further tested by analysis of an rrp6Δ set1Δ double mutant (Figure 4A). The double mutant expression-profile reveals two classes of COMPASS-repressed genes. On the smaller group of genes (Figure 4A, marked with a black bar), that includes PHO84 as well as several other phosphate-related genes, an epistatic effect is observed in rrp6Δ set1Δ, whereby the upregulation in set1Δ is lost in the double mutant. This implies that the antisense transcript mediated repression of sense genes is not unique for PHO84, but is shared with functionally related genes. Such genes are the exception however. The largest group of Set1-repressed genes behaves in a different manner, still showing derepression in the double mutant, similar to their behaviour upon deletion of SET1 on its own. This therefore likely represents a distinct mechanism of COMPASS repression.
Fig. 4. COMPASS repression is mediated through 3′-end antisense transcriptional gene silencing. 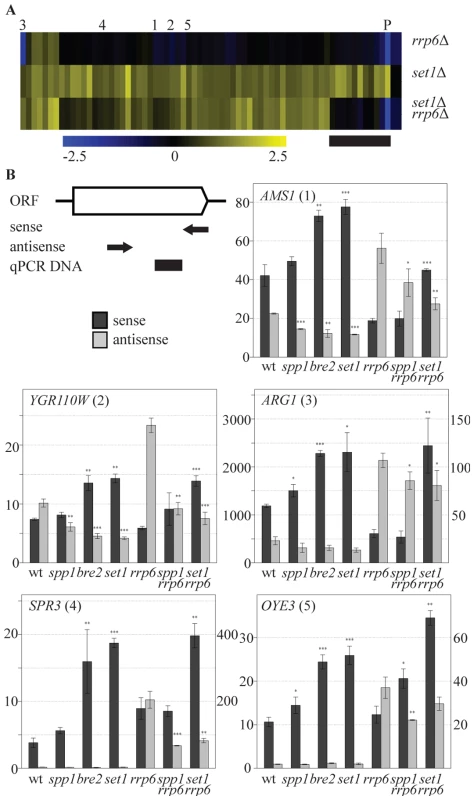
(A) Hierarchical clustering of the 69 COMPASS-repressed genes in the rrp6Δ, set1Δ and the set1Δ rrp6Δ strains. PHO84 is marked by P and AMS1, YGR110W, ARG1, SPR3 and OYE3 are marked by 1 to 5, respectively. The black bar marks the subset of genes where the two mutations are epistatic. Three quarters of these genes are related to phosphate metabolism. (B) Sense and antisense RNA levels analyzed by qPCR in indicated backgrounds. The schematic representation of the follow-up genes shows the relative positions of the primers used for strand-specific reverse transcription reactions in arrows, while the black box indicates the location of the DNA fragment produced during the qPCR. Error bars reflect standard deviations of an average signal obtained from at least two independent experiments. The significance of the difference in expression changes observed between the mutant cells and the corresponding background strain (rrp6Δ in the case of spp1Δrrp6Δ and set1Δrrp6Δ, wt for the others), was evaluated using Student's t-test (*P 0.01–0.05; **P 0.001–0.01; ***P<0.001). In order to understand the mechanism by which COMPASS represses coding transcription in an exosome-independent manner, five representative genes from this group, AMS1, YGR110W, ARG1, SPR3 and OYE3 (indicated by 1 to 5 in Figure 4A), were analyzed in greater detail. These genes represent different functional categories, different telomeric proximities and different types of antisense transcripts, as suggested by the genome-wide datasets. The first three genes contain antisense stable unannotated transcripts (SUTs), while the other two have antisense cryptic unstable transcripts (CUTs) [40]. The location of H3K4 methylation patterns [53] corresponds to the location of the transcription initiation sites of these antisense transcription units (Figure S5). The effects of different COMPASS mutants on both sense and antisense transcription of these genes were analyzed by quantitative RT-PCR using strand-specific primers (Figure 4B).
Sense transcript upregulation of the five genes is observed in set1Δ (Figure 4B), that exhibits loss of all H3K4 methylation marks (Figure 1A), in bre2Δ (Figure 4B), that exhibits loss of all H3K4me3 and most H3K4me2 (Figure 1A) and in set1Δ combined with rrp6Δ (Figure 4B), all in agreement with the sense-specific microarray results (Figure 1B, Figure 4A). In bre2Δ and set1Δ, upregulation of sense transcription is accompanied by a decrease in antisense transcription (Figure 4B, panels 1–3). As expected, changes in antisense CUT transcription are not evident without prior stabilization by the exosome deletion (Figure 4B, panels 4,5). For all five genes, stabilisation of antisense transcripts does occur in rrp6Δ (Figure 4B), but these increased antisense levels do not necessarily result in more repression of sense transcription (Figure 4B) as is clearly the case for PHO84 ([32] and Figure 4A). This confirms that an increase in antisense transcript levels through rrp6Δ-dependent stabilisation is not the mechanism of COMPASS repression for these genes. Rather, the data suggest that it is the process of antisense transcription itself that represses the sense transcription.
The Set1 repressive effect is mediated through 3′-end antisense transcription
Because sense and 3′end antisense transcription seem coupled [54], it is difficult to distinguish whether the increased sense transcription in COMPASS mutants is caused, or is followed, by a decrease in 3′-end antisense transcription. One way of addressing this directly is to eliminate 3′-end antisense transcription by other means than through disruption of COMPASS. For this purpose strong terminator sequences were introduced downstream of the five model genes analyzed in Figure 4B, either as insertions between antisense promoters and the end of the ORF, or as replacement of complete intergenic sequences. Neither approach resulted in loss of 3′-end antisense transcription, which agrees with the recent finding that terminators can function as promoters [55] . Disruption of 3′-end antisense transcription was then attempted by removal of all, or a significant part of the intergenic region. Complete loss of all antisense transcription was only observed for the YGR110W intergenic deletion mutant, which we further analyzed in depth (YGR110W-ingdel, Figure 5).
Fig. 5. Set1 represses sense transcription through promotion of antisense transcription. 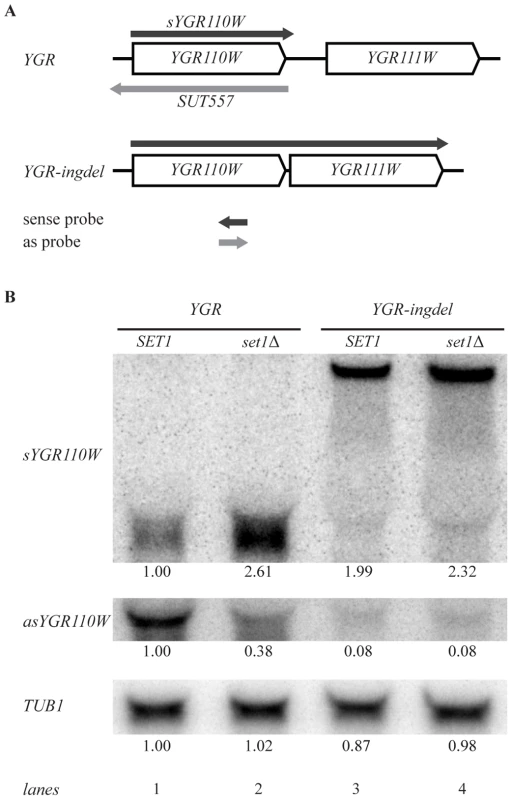
(A) Scheme showing YGR110W and YGR111W genes before (YGR) and after (YGR-ingdel) deletion of their intergenic region. The sense YGR110W transcript (sYGR110W), as well as the longer sense transcript in the YGR-ingdel strain are shown in dark grey, while the antisense transcript (SUT557) is shown in light grey. The position and 5′-3′ direction of the strand-specific probes used to detect the transcripts are also shown. (B) Autoradiographs of Northern blots hybridized with the strand-specific DNA probes designed to detect the sense (sYGR110W) or antisense (asYGR110W) transcripts of YGR110W in the YGR and YGR-ingdel strains with wild-type (SET1) or deleted SET1 (set1Δ). An autoradiograph of the same blot hybridized with a tubulin probe (TUB1) was used as loading control. Quantitation of the bands are shown below each panel relative to the wt (SET1 YGR) strain for TUB1 and relative to the wt (SET1 YGR) strain and the loading control for the sYGR110W and asYGR110W panels. Strand-specific Northern blot analysis of YGR110W-ingdel shows that loss of antisense transcription (Figure 5B, asYGR110W, lane 1 versus lane 3), is accompanied by derepression of sense transcription (Figure 5B, sYGR110W). This demonstrates that 3′-end antisense transcription results in repression of sense transcription. Furthermore, introduction of SET1 deletion into the YGR110W-ingdel strain, does not result in significant further derepression as is observed in the presence of 3′-end antisense transcription (Figure 5B, lanes 1 and 2 versus lanes 3 and 4). This agrees with the proposal that the repressive effect of COMPASS on coding genes is a result of promoting 3′-end antisense transcription.
H3K4me3 promotes 3′-end antisense transcription and H3K4me2 contributes to coding gene repression
The results presented in Figure 4B and Figure 5B imply a positive role for Set1 on antisense transcription. SET1 deletion results in loss of H3K4me1, me2 and me3 (Figure 1A). SPP1 deletion (loss of H3K4me3 only), has little effect on sense transcript levels (Figure 1, Figure 2, Figure 4B). SPP1 deletion does result in decreased antisense transcripts as observed either in the presence or absence of RRP6 (Figure 4B). Our results indicate that H3K4 trimethylation, which is found at the 3′-end of these genes, has a role in promoting 3′-end antisense transcription. This effect is not absolute however. Antisense transcripts are reduced in the SET1 RRP6 double deletion compared to rrp6Δ, but are not completely absent. This indicates that antisense transcription is promoted by, but not fully dependent on, H3K4me3. Since spp1Δ still exhibits wt levels of H3K4me2 (Figure 1A) and virtually no derepression of sense transcription (Figure 1B and Figure 4B), this indicates that it is the H3K4me2 mark which is most important for repression of sense transcription on these genes. Together, the results of these experiments are consistent with a model, whereby the majority of COMPASS-repressed genes are maintained in an inactive state through 3′-end antisense transcription that is in part promoted through H3K4me3 at the 3′-end, and in turn deposits a repressive H3K4me2 mark further into the body of the gene.
COMPASS repression is mimicked by reducing nucleosome levels
We next asked what determines the specificity of the effects observed upon mutation of COMPASS. H3K4 methylation marks all active genes and approximately one third of all genes exhibit antisense transcripts [40], yet only a subset are affected by deleting COMPASS subunits (Figure 1). It has recently been proposed that the transcription factor Reb1 may drive non-coding transcription, either from neighbouring genes [33] or from the promoter of the antisense transcript itself [56]. Reb1 binding sites are found downstream of only three of the 69 COMPASS-repressed genes. There is also no statistically significant enrichment for Reb1 binding sites in the ORFs or flanking regions of genes up-regulated in the COMPASS mutants. Both observations suggest that the specificity of Set1 repressive effects is not generally linked to Reb1. In addition, no other putative regulatory motifs could be detected in these regions using different search algorithms [57].
An alternative explanation for the specificity of COMPASS repressive effects is that specificity is dictated by increased sensitivity of specific genes to a particular chromatin structure which is influenced by H3K4 methylation. While profiling strains with altered histone expression levels we noted an interesting correlation with the collection of COMPASS mutants. To investigate this, a strain bearing single copies of the histone H3 and H4 genes under control of their native promoters [58] was analyzed. The two-fold reduction in mRNA levels of H3 and H4 in this strain (Figure 6B, marked HHT2 and HHF2) is accompanied by slightly decreased H3 and H4 protein levels (Figure 6A). Interestingly, this results in upregulation of a specific subset of genes that strongly correspond to the genes upregulated upon SET1 deletion (p-value 1.4*10−23, Figure 6B). Although the overlap is highly statistically significant, it is not complete and does not extend to PHO84 for example, in agreement with the proposal for a distinct repressive mechanism for such genes (Figure 4A). SET1 deletion does not globally affect nucleosome levels (data not shown), and antisense transcript levels are not reduced in the single copy H3 H4 strain (Figure 6C). Besides antisense transcription, a second common property of the genes affected by loss of COMPASS function is therefore sensitivity to histone abundance. Since histone abundance affects nucleosome density [59], this suggests that Set1 may repress genes by effecting nucleosome density. As is discussed below, one manner in which this may be achieved is through the repressive H3K4me2 mark that is laid down through 3′-end antisense transcription.
Fig. 6. COMPASS-repressed genes are derepressed upon decrease in nucleosome content. 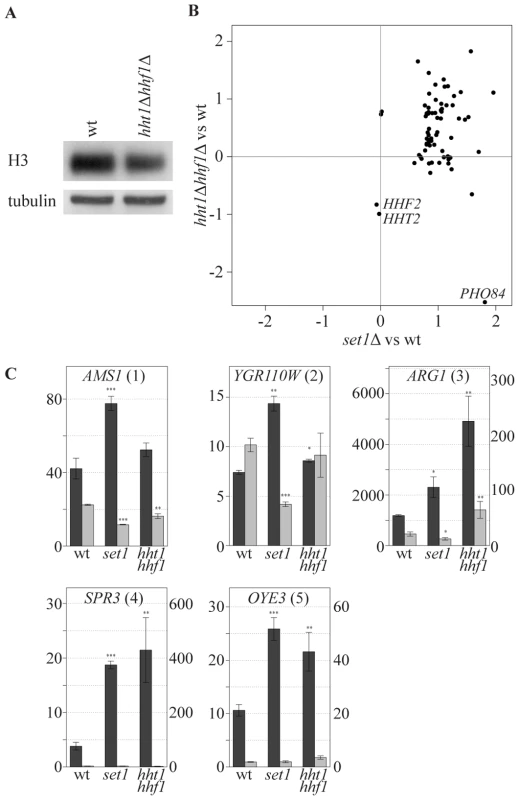
(A) The cellular expression level of H3 was analyzed by Western blotting in wt and hht1Δ hhf1Δ cells. (B) Correlation of the effects, as measured by log base 2 ratios, of low nucleosomes levels (hht1Δ hhf1Δ) and SET1 deletion (set1Δ) on the COMPASS-repressed genes versus wt cells. LYS2 is excluded as it is used as an auxotrophic marker for the hht1Δ hhf1Δ strain. PHO84 is marked, as it is significantly repressed in the low nucleosome strain. The two histone genes are also depicted verifying the expected 2-fold reduction in their mRNA levels. (C) Strand-specific qPCR analysis, as in Figure 4B, for the indicated backgrounds. Discussion
Repressive role of COMPASS in S. cerevisiae
The results presented here add to a number of reports that indicate that the major non-redundant role of COMPASS in S. cerevisiae is repression of coding genes [34], [56]. Early genome-wide analyses of set1Δ yielded conflicting results, in two cases pointing to global positive effects [27], [28] and in one case ignoring the prevalence of specific repressive effects [6]. Some of the differences between these studies and the current one can in retrospect be attributed to use of double-stranded cDNA arrays, less convenient for discriminating between sense and ant-sense effects, as well as to normalisation issues. The analyses presented here, using strand-specific techniques, with replicate experiments for a variety of different mutants under both steady-state and dynamic conditions, indicates that removal of H3K4me3, a global mark of active transcription, has no global effect. The repressive effects observed on a specific subset of genes agree with the most recent other genome-wide analyses of set1Δ [29] [30], as well as with the fact that deletion of SET1 is not lethal. Gene Ontology analysis of the affected genes reveals an overrepresentation of vitamin metabolism (essentially thiamin biosynthesis) and spore wall assembly (Table S3) in agreement with the cell wall and stationary phase defects previously observed in set1Δ cells [51].
COMPASS and antisense transcription
What is the mechanism of the observed repression? Despite the fact that COMPASS-repressed genes show a significant enrichment for telomeric-proximal localization (Figure S3 and [29]), Set1-dependent repression of these genes due to a telomere position effect can probably be ruled out since the derepression observed in set1Δ only affects a small percentage of individual genes within these regions. Only a few of the affected genes are close to telomeres and only few telomere-proximal genes are affected. Analysis of methylation patterns (Figure 3 and [53]), non-coding RNA maps (Table S2 and [39], [40]) and the comparison of mutants with different methylation states support a model whereby COMPASS mediates repression of coding genes by promoting the expression of 3′-end antisense transcripts through deposition of H3K4me3 at their 3′-end.
An involvement of Set1 in promotion of 3′-end antisense transcription, resulting in a repressive effect on sense transcription has been reported for PHO84, which is repressed through the presence of antisense transcripts [34]. Our results are consistent with a repressive role for Set1 on PHO84. The genome-wide nature of our experiments indicates however that the majority of Set1 affected genes are repressed through a different mechanism, independent of the level of antisense RNA transcripts. Rather, for the majority of Set1-regulated genes, repression is caused by the process of antisense transcription itself. This mechanism is therefore related to the recently reported attenuation in GAL10-GAL1 activation which is also facilitated through cryptic transcription [56]. One major difference is that for the mechanism reported here, COMPASS is required to maintain antisense transcription whereas this does not seem to be the case for the cryptic transcription observed at the GAL10-GAL1 locus [56].
Distinct roles for H3K4me2 and H3K4me3
The comparison of different COMPASS mutations carried out here, facilitates distinguishing between the roles of the different H3K4 methylation states. Mutants with grossly lowered or completely absent H3K4me3 exhibit decreased antisense transcription. However this only results in derepression of the coding gene if H3K4me2 is also abolished. The positive role of H3K4me3 on antisense transcription fits with the correlation observed between the presence of this mark and promoter activity of coding genes [7], [8], [9], [10], [11], [25]. Most non-coding RNAs originate from nucleosome-free regions (NFRs) shared with protein-coding transcripts [40]. This is also the case for three of the five COMPASS-repressed genes analyzed here in detail (AMS1, ARG1 and OYE3). Interestingly, despite sharing a NFR with the downstream protein-coding gene, loss of H3K4 trimethylation causes reduction of antisense transcription without affecting transcription of the flanking protein-coding gene in each case. This fits with the observation that bidirectional transcription from a single NFR, originates from two distinct preinitiation complex recruitment sites [60]. This may indicate the presence of redundant mechanisms for maintaining protein-coding gene transcription in the absence of H3K4me3, which are lacking for the antisense non-coding transcription originating from the same NFR. Another, non mutually exclusive mechanism, can be that lack of H3K4 trimethylation in the antisense transcription start site increases the recruitment of corepressor complexes, such as Rpd3S [61], that repress the expression of the non-coding transcript, but not that of the coding gene [62].
The results also indicate a role for the H3K4me2 mark in facilitating repression. This agrees with several recent studies suggesting mechanisms through which H3K4me2 may play a repressive role. For example, it has recently been reported that H3K4me2 in the body of active genes is recognized by the Set3 complex, leading to histone deacetylation, a repressive chromatin state [63]. A different histone deacetylase, Rpd3, has been implicated in the repressive role involving Set1 on the GAL10-GAL1 locus [56]. Furthermore, methylation of H3K4 protects against an H3 tail endopeptidase recently described in S. cerevisiae and humans that facilitates transcription initiation and precedes histone eviction [64], [65]. All these possible mechanisms fit with the observation made here that globally reducing H3 and H4 levels mimics the derepression of COMPASS mutants. The degree of overlap between the COMPASS mutants' profiles and the histone depletion profile also give an explanation for the specificity of the COMPASS repression. Genes repressed by COMPASS have antisense transcription, but are also sensitive to nucleosome density.
Sensitivity to histone depletion may not be the only reason for the lack of genome-wide effects upon COMPASS mutation. Functional redundancy may also contribute. One of the prevalent ideas for a general role of H3K4 methylation in S. cerevisiae is that transcription-associated H3K4 methylation, as well as deposition of the histone variant H2A.Z, antagonizes the local spread of Sir-dependent silent chromatin into adjacent euchromatic regions [52], [66], [67]. It has recently been shown that H2A.Z deposition and Set1 cooperate to prevent Sir-dependent repression of a large number of genes located across the genome [29]. This functional redundancy between H3K4 methylation and H2AZ deposition may thus buffer transcription from changes in euchromatin, thereby minimizing the observed effects of H3K4 methylation loss.
This work offers a plausible explanation for how a transcription factor, previously thought to positively contribute to transcription, can nevertheless exert a negative effect, through promoting antisense transcription. The opposite has previously been shown for regulation of IME4. Here a repressor complex binds to the promoter of an antisense transcription unit in the 5′-end of IME4 and by repressing the antisense transcription, facilitates the in cis sense transcription activation [68]. Although further work is required to pinpoint the mechanisms further downstream of H3K4 di - and trimethylation, COMPASS exemplifies the growing insight that the roles of histone modifications in gene expression are non-linear [69] and context-dependent [70].
Materials and Methods
Microarray data is accessible through the public microarray database ArrayExpress (http://www.ebi.ac.uk/arrayexpress/) under accession number E-TABM-486. The accession numbers below refer to detailed protocols in ArrayExpress.
Strains and primers
Strains and primers used in this study are described in Tables S4 and S5 respectively. The YGR110W-ingdel strain was created by first inserting a cassette containing the Sp HIS5 gene in reverse orientation flanked by the Ag TEF promoter, terminator sequences and loxP sites from plasmid pUG27 [71] to replace the intergenic region between YGR110W and YGR111W using YGR110W_HIS5_F and YGR110W_HIS5_R primers. Subsequently the cassette was floxed out by transforming the strain with the plasmid pSH47 and expressing Cre recombinase as previously described [72].
Histone purification and Western blotting
Histones were purified as described [42], subjected to 16% SDS-polyacrylamide gel electrophoresis, and either Coomassie Blue stained or transferred to 0.2 µm ProtranR nitrocellulose. Antibodies used to detect mono-, di - and trimethylated H3K4 and histone H3 were from Abcam.
Cultures
Two independent colonies of each strain were first inoculated and grown overnight in synthetic complete medium with 2% glucose. For the mid-log/steady-state experiment, larger cultures were inoculated the next day at an OD600 of 0.15 in fresh medium, allowed to grow at 30°C and harvested at OD600 0.6, (P-UMCU-36). For the time-course experiment, overnight cultures were used to inoculate 50 ml cultures at an OD600 of 0.15. These were allowed to deplete glucose by growing for 24 hours and were used the next day to start 500 ml cultures at an OD600 of 0.15 in fresh medium for the time-course sampling (P-UMCU-47).
RNA isolation and amplification
Total RNA isolation was by hot acid phenol (P-UMCU-37) and cleaned up using RNeasy (Qiagen). Before amplification, external RNA controls were added to total RNA to check for global shifts in mRNA levels [41]. cRNA amplification and labelling using amino-allyl UTP was performed on a Caliper robot system (P-UMCU-38).
Microarrays and hybridizations
Each sample was generated twice, as independent biological replicates. These were hybridized in dye-swap against a common wt reference RNA (P-UMCU-39) on oligo-arrays that represented each gene twice (P-UMCU-34). After scanning (P-UMCU-40), raw data were extracted with Imagene (Biodiscovery) (P-UMCU-42).
Data analysis
Since spike-in of external RNA controls revealed no global changes in the mRNA population [41] for the mid-log experiment, non-background corrected data were normalized with print-tip LOESS [73] on gene probes with a span of 0.4 (P-UMCU-41). For the time-course, all features, including negative and external controls (EC) except EC 4, 6 and 8, were used for the estimation of the LOESS curve (P-UMCU-46). Probes flagged as absent, or with a nearly saturated signal were not used to estimate the LOESS curve. For differential expression analysis, the LIMMA package [74] was used. Mitochondrial-encoded genes and Ty elements were excluded due to their high biological variation. Genes with an FDR-adjusted p-value less than 0.01 and a fold-change of more than 1.7 were considered significant. These thresholds are based on systematic analyses of the variation observed in a collection of more than 100 wt expression profiles [75]. For the time-course experiment, changes were considered significant if they fulfilled these criteria for two consecutive time-points. Hierarchical clustering was by MeV [76], using standard correlation and average linkage. Analysis of overlap between genelists of significantly changing genes of two expression profiles was by hypergeometric test. For the GO and transcription factor enrichment analysis, a right-sided Fisher's exact test was used and the p-values were corrected for multiple testing using Bonferroni. The GO annotations were obtained from SGD.
For the H3K4 methylation ChIP-chip analysis, the data are from [53]. For each gene, a region corresponding to the ORF plus 500 bps in both directions was used. The ORF was divided into 30 bins of equal length and the flanking regions in 10 bins each. A loess algorithm [77] with a span of 0.2 was used to estimate the enrichment of the methylation marks for every bin.
Reverse Transcription and qPCR
cDNAs of sense RNA or antisense RNA were generated by SuperScript III Reverse transcriptase (Invitrogen) from total RNAs using gene and strand-specific primers. For each gene, cDNAs obtained from the reverse transcription of sense or antisense RNA were quantified by a real-time qPCR with gene-specific primers corresponding to a 150 bp fragment (Figure 4B). The same primers were used to quantify sense and antisense cDNA of each gene. The position and the sequence of each primer are indicated in Figure 4B and Table S5.
Northern blotting
The strand-specific DNA probes used to detect the presence of sense and antisense transcripts of YGR110W are shown schematically in Figure 5A. First a cold PCR product template was obtained using primers 3′qYGR110W and 5′qYGR110W. Subsequently the hot ssDNA probes for detection of the sense and antisense transcripts were generated from the template using the first or the second primer, respectively, in linear PCR reactions. Quantitation of the radioactive signal was performed using ImageQuant (Molecular Dynamics).
Supporting Information
Zdroje
1. KornbergRD, LorchY (1999) Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell 98 : 285–294.
2. JenuweinT, AllisCD (2001) Translating the histone code. Science 293 : 1074–1080.
3. LiB, CareyM, WorkmanJL (2007) The role of chromatin during transcription. Cell 128 : 707–719.
4. LachnerM, JenuweinT (2002) The many faces of histone lysine methylation. Curr Opin Cell Biol 14 : 286–298.
5. RuthenburgAJ, AllisCD, WysockaJ (2007) Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell 25 : 15–30.
6. Santos-RosaH, SchneiderR, BannisterAJ, SherriffJ, BernsteinBE, et al. (2002) Active genes are tri-methylated at K4 of histone H3. Nature 419 : 407–411.
7. BarskiA, CuddapahS, CuiK, RohTY, SchonesDE, et al. (2007) High-resolution profiling of histone methylations in the human genome. Cell 129 : 823–837.
8. BernsteinBE, KamalM, Lindblad-TohK, BekiranovS, BaileyDK, et al. (2005) Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 120 : 169–181.
9. KimJ, HakeSB, RoederRG (2005) The human homolog of yeast BRE1 functions as a transcriptional coactivator through direct activator interactions. Mol Cell 20 : 759–770.
10. RohTY, CuddapahS, CuiK, ZhaoK (2006) The genomic landscape of histone modifications in human T cells. Proc Natl Acad Sci U S A 103 : 15782–15787.
11. SchneiderR, BannisterAJ, MyersFA, ThorneAW, Crane-RobinsonC, et al. (2004) Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat Cell Biol 6 : 73–77.
12. VermeulenM, MulderKW, DenissovS, PijnappelWW, van SchaikFM, et al. (2007) Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell 131 : 58–69.
13. TavernaSD, LiH, RuthenburgAJ, AllisCD, PatelDJ (2007) How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol 14 : 1025–1040.
14. BergerSL (2007) The complex language of chromatin regulation during transcription. Nature 447 : 407–412.
15. DehePM, GeliV (2006) The multiple faces of Set1. Biochem Cell Biol 84 : 536–548.
16. BriggsSD, BrykM, StrahlBD, CheungWL, DavieJK, et al. (2001) Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev 15 : 3286–3295.
17. MillerT, KroganNJ, DoverJ, Erdjument-BromageH, TempstP, et al. (2001) COMPASS: a complex of proteins associated with a trithorax-related SET domain protein. Proc Natl Acad Sci U S A 98 : 12902–12907.
18. NagyPL, GriesenbeckJ, KornbergRD, ClearyML (2002) A trithorax-group complex purified from Saccharomyces cerevisiae is required for methylation of histone H3. Proc Natl Acad Sci U S A 99 : 90–94.
19. RoguevA, SchaftD, ShevchenkoA, PijnappelWW, WilmM, et al. (2001) The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. Embo J 20 : 7137–7148.
20. DehePM, DichtlB, SchaftD, RoguevA, PamblancoM, et al. (2006) Protein interactions within the Set1 complex and their roles in the regulation of histone 3 lysine 4 methylation. J Biol Chem 281 : 35404–35412.
21. MorillonA, KarabetsouN, NairA, MellorJ (2005) Dynamic lysine methylation on histone H3 defines the regulatory phase of gene transcription. Mol Cell 18 : 723–734.
22. SchneiderJ, WoodA, LeeJS, SchusterR, DuekerJ, et al. (2005) Molecular regulation of histone H3 trimethylation by COMPASS and the regulation of gene expression. Mol Cell 19 : 849–856.
23. Vitaliano-PrunierA, MenantA, HobeikaM, GeliV, GwizdekC, et al. (2008) Ubiquitylation of the COMPASS component Swd2 links H2B ubiquitylation to H3K4 trimethylation. Nat Cell Biol 10 : 1365–1371.
24. NgHH, RobertF, YoungRA, StruhlK (2003) Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell 11 : 709–719.
25. PokholokDK, HarbisonCT, LevineS, ColeM, HannettNM, et al. (2005) Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122 : 517–527.
26. TavernaSD, IlinS, RogersRS, TannyJC, LavenderH, et al. (2006) Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription at a subset of targeted ORFs. Mol Cell 24 : 785–796.
27. BernsteinBE, HumphreyEL, ErlichRL, SchneiderR, BoumanP, et al. (2002) Methylation of histone H3 Lys 4 in coding regions of active genes. Proc Natl Acad Sci U S A 99 : 8695–8700.
28. BoaS, CoertC, PattertonHG (2003) Saccharomyces cerevisiae Set1p is a methyltransferase specific for lysine 4 of histone H3 and is required for efficient gene expression. Yeast 20 : 827–835.
29. VenkatasubrahmanyamS, HwangWW, MeneghiniMD, TongAH, MadhaniHD (2007) Genome-wide, as opposed to local, antisilencing is mediated redundantly by the euchromatic factors Set1 and H2A.Z. Proc Natl Acad Sci U S A 104 : 16609–16614.
30. GuillemetteB, DrogarisP, LinHH, ArmstrongH, Hiragami-HamadaK, et al. (2011) H3 lysine 4 is acetylated at active gene promoters and is regulated by H3 lysine 4 methylation. PLoS Genet 7: e1001354 doi:10.1371/journal.pgen.1001354.
31. BerrettaJ, PinskayaM, MorillonA (2008) A cryptic unstable transcript mediates transcriptional trans-silencing of the Ty1 retrotransposon in S. cerevisiae. Genes Dev 22 : 615–626.
32. CamblongJ, IglesiasN, FickentscherC, DieppoisG, StutzF (2007) Antisense RNA stabilization induces transcriptional gene silencing via histone deacetylation in S. cerevisiae. Cell 131 : 706–717.
33. HouseleyJ, RubbiL, GrunsteinM, TollerveyD, VogelauerM (2008) A ncRNA modulates histone modification and mRNA induction in the yeast GAL gene cluster. Mol Cell 32 : 685–695.
34. CamblongJ, BeyrouthyN, GuffantiE, SchlaepferG, SteinmetzLM, et al. (2009) Trans-acting antisense RNAs mediate transcriptional gene cosuppression in S. cerevisiae. Genes Dev 23 : 1534–1545.
35. CoreLJ, WaterfallJJ, LisJT (2008) Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322 : 1845–1848.
36. HeY, VogelsteinB, VelculescuVE, PapadopoulosN, KinzlerKW (2008) The antisense transcriptomes of human cells. Science 322 : 1855–1857.
37. PrekerP, NielsenJ, KammlerS, Lykke-AndersenS, ChristensenMS, et al. (2008) RNA exosome depletion reveals transcription upstream of active human promoters. Science 322 : 1851–1854.
38. SeilaAC, CalabreseJM, LevineSS, YeoGW, RahlPB, et al. (2008) Divergent transcription from active promoters. Science 322 : 1849–1851.
39. NeilH, MalabatC, d'Aubenton-CarafaY, XuZ, SteinmetzLM, et al. (2009) Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature 457 : 1038–1042.
40. XuZ, WeiW, GagneurJ, PerocchiF, Clauder-MunsterS, et al. (2009) Bidirectional promoters generate pervasive transcription in yeast. Nature 457 : 1033–1037.
41. van de PeppelJ, KemmerenP, van BakelH, RadonjicM, van LeenenD, et al. (2003) Monitoring global messenger RNA changes in externally controlled microarray experiments. EMBO Rep 4 : 387–393.
42. DehePM, PamblancoM, LucianoP, LebrunR, MoinierD, et al. (2005) Histone H3 lysine 4 mono-methylation does not require ubiquitination of histone H2B. J Mol Biol 353 : 477–484.
43. DoverJ, SchneiderJ, Tawiah-BoatengMA, WoodA, DeanK, et al. (2002) Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J Biol Chem 277 : 28368–28371.
44. LeeJS, ShuklaA, SchneiderJ, SwansonSK, WashburnMP, et al. (2007) Histone crosstalk between H2B monoubiquitination and H3 methylation mediated by COMPASS. Cell 131 : 1084–1096.
45. SunZW, AllisCD (2002) Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418 : 104–108.
46. KouskoutiA, TalianidisI (2005) Histone modifications defining active genes persist after transcriptional and mitotic inactivation. Embo J 24 : 347–357.
47. RadonjicM, AndrauJC, LijnzaadP, KemmerenP, KockelkornTT, et al. (2005) Genome-wide analyses reveal RNA polymerase II located upstream of genes poised for rapid response upon S. cerevisiae stationary phase exit. Mol Cell 18 : 171–183.
48. CordaY, SchramkeV, LongheseMP, SmokvinaT, PaciottiV, et al. (1999) Interaction between Set1p and checkpoint protein Mec3p in DNA repair and telomere functions. Nat Genet 21 : 204–208.
49. HechtA, LarocheT, Strahl-BolsingerS, GasserSM, GrunsteinM (1995) Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell 80 : 583–592.
50. KroganNJ, DoverJ, KhorramiS, GreenblattJF, SchneiderJ, et al. (2002) COMPASS, a histone H3 (Lysine 4) methyltransferase required for telomeric silencing of gene expression. J Biol Chem 277 : 10753–10755.
51. NislowC, RayE, PillusL (1997) SET1, a yeast member of the trithorax family, functions in transcriptional silencing and diverse cellular processes. Mol Biol Cell 8 : 2421–2436.
52. Santos-RosaH, BannisterAJ, DehePM, GeliV, KouzaridesT (2004) Methylation of H3 lysine 4 at euchromatin promotes Sir3p association with heterochromatin. J Biol Chem 279 : 47506–47512.
53. KirmizisA, Santos-RosaH, PenkettCJ, SingerMA, VermeulenM, et al. (2007) Arginine methylation at histone H3R2 controls deposition of H3K4 trimethylation. Nature 449 : 928–932.
54. XuZ, WeiW, GagneurJ, Clauder-MunsterS, SmolikM, et al. (2011) Antisense expression increases gene expression variability and locus interdependency. Mol Syst Biol 7 : 468.
55. MurraySC, Serra BarrosA, BrownDA, DudekP, AylingJ, et al. (2012) A pre-initiation complex at the 3′-end of genes drives antisense transcription independent of divergent sense transcription. Nucleic Acids Res 40 : 2432–2444.
56. PinskayaM, GourvennecS, MorillonA (2009) H3 lysine 4 di - and tri-methylation deposited by cryptic transcription attenuates promoter activation. Embo J 28 : 1697–1707.
57. WijayaE, YiuSM, SonNT, KanagasabaiR, SungWK (2008) MotifVoter: a novel ensemble method for fine-grained integration of generic motif finders. Bioinformatics 24 : 2288–2295.
58. DaiJ, HylandEM, YuanDS, HuangH, BaderJS, et al. (2008) Probing nucleosome function: a highly versatile library of synthetic histone H3 and H4 mutants. Cell 134 : 1066–1078.
59. HanM, GrunsteinM (1988) Nucleosome loss activates yeast downstream promoters in vivo. Cell 55 : 1137–1145.
60. RheeHS, PughBF (2012) Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature 483 : 295–301.
61. QuanTK, HartzogGA (2010) Histone H3K4 and K36 methylation, Chd1 and Rpd3S oppose the functions of Saccharomyces cerevisiae Spt4-Spt5 in transcription. Genetics 184 : 321–334.
62. ChurchmanLS, WeissmanJS (2011) Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature 469 : 368–373.
63. KimT, BuratowskiS (2009) Dimethylation of H3K4 by Set1 recruits the Set3 histone deacetylase complex to 5′ transcribed regions. Cell 137 : 259–272.
64. DuncanEM, Muratore-SchroederTL, CookRG, GarciaBA, ShabanowitzJ, et al. (2008) Cathepsin L proteolytically processes histone H3 during mouse embryonic stem cell differentiation. Cell 135 : 284–294.
65. Santos-RosaH, KirmizisA, NelsonC, BartkeT, SaksoukN, et al. (2009) Histone H3 tail clipping regulates gene expression. Nat Struct Mol Biol 16 : 17–22.
66. MeneghiniMD, WuM, MadhaniHD (2003) Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell 112 : 725–736.
67. ZhangH, RobertsDN, CairnsBR (2005) Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell 123 : 219–231.
68. HongayCF, GrisafiPL, GalitskiT, FinkGR (2006) Antisense transcription controls cell fate in Saccharomyces cerevisiae. Cell 127 : 735–745.
69. LenstraTL, BenschopJJ, KimT, SchulzeJM, BrabersNA, et al. (2011) The specificity and topology of chromatin interaction pathways in yeast. Mol Cell 42 : 536–549.
70. SimsRJ3rd, ReinbergD (2008) Is there a code embedded in proteins that is based on post-translational modifications? Nat Rev Mol Cell Biol 9 : 815–820.
71. GueldenerU, HeinischJ, KoehlerGJ, VossD, HegemannJH (2002) A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res 30: e23.
72. GuldenerU, HeckS, FielderT, BeinhauerJ, HegemannJH (1996) A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res 24 : 2519–2524.
73. YangYH, DudoitS, LuuP, LinDM, PengV, et al. (2002) Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res 30: e15.
74. Smyth GK, Gentleman R, Carey V, Dudoit S, Irizarry R, et al.. (2005) Limma: linear models for microarray data. Bioinformatics and Computational Biology Solutions using R and Bioconductor: Springer, New York. pp. 397–420.
75. van WageningenS, KemmerenP, LijnzaadP, MargaritisT, BenschopJJ, et al. (2010) Functional overlap and regulatory links shape genetic interactions between signaling pathways. Cell 143 : 991–1004.
76. SaeedAI, SharovV, WhiteJ, LiJ, LiangW, et al. (2003) TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34 : 374–378.
77. Team RDC (2007) R: A Language and Environment for Statistical Computing; Computing RFfS, editor. Vienna, Austria.
Štítky
Genetika Reprodukční medicína
Článek Genome-Wide Association Study for Serum Complement C3 and C4 Levels in Healthy Chinese SubjectsČlánek Role of Transposon-Derived Small RNAs in the Interplay between Genomes and Parasitic DNA in RiceČlánek Tetraspanin Is Required for Generation of Reactive Oxygen Species by the Dual Oxidase System inČlánek An Essential Role of Variant Histone H3.3 for Ectomesenchyme Potential of the Cranial Neural CrestČlánek A Mimicking-of-DNA-Methylation-Patterns Pipeline for Overcoming the Restriction Barrier of BacteriaČlánek A Genome-Wide Association Study Identifies Five Loci Influencing Facial Morphology in Europeans
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 9
-
Všechny články tohoto čísla
- Heterozygous Mutations in DNA Repair Genes and Hereditary Breast Cancer: A Question of Power
- GWAS of Diabetic Nephropathy: Is the GENIE out of the Bottle?
- The Conflict within and the Escalating War between the Sex Chromosomes
- Proteome-Wide Analysis of Disease-Associated SNPs That Show Allele-Specific Transcription Factor Binding
- Exome Sequencing Identifies Rare Deleterious Mutations in DNA Repair Genes and as Potential Breast Cancer Susceptibility Alleles
- A Gene Family Derived from Transposable Elements during Early Angiosperm Evolution Has Reproductive Fitness Benefits in
- Genome-Wide Association Study for Serum Complement C3 and C4 Levels in Healthy Chinese Subjects
- Role of Transposon-Derived Small RNAs in the Interplay between Genomes and Parasitic DNA in Rice
- Co-Evolution of Mitochondrial tRNA Import and Codon Usage Determines Translational Efficiency in the Green Alga
- SIRT6/7 Homolog SIR-2.4 Promotes DAF-16 Relocalization and Function during Stress
- CNV Formation in Mouse Embryonic Stem Cells Occurs in the Absence of Xrcc4-Dependent Nonhomologous End Joining
- Tetraspanin Is Required for Generation of Reactive Oxygen Species by the Dual Oxidase System in
- Citrullination of Histone H3 Interferes with HP1-Mediated Transcriptional Repression
- Variation in Genes Related to Cochlear Biology Is Strongly Associated with Adult-Onset Deafness in Border Collies
- The Long Non-Coding RNA Affects Chromatin Conformation and Expression of , but Does Not Regulate Its Imprinting in the Developing Heart
- Rif2 Promotes a Telomere Fold-Back Structure through Rpd3L Recruitment in Budding Yeast
- Is a Metastasis Susceptibility Gene That Suppresses Metastasis by Modifying Tumor Interaction with the Cell-Mediated Immunity
- The p38/MK2-Driven Exchange between Tristetraprolin and HuR Regulates AU–Rich Element–Dependent Translation
- Rare Copy Number Variants Contribute to Congenital Left-Sided Heart Disease
- A Genetic Basis for a Postmeiotic X Versus Y Chromosome Intragenomic Conflict in the Mouse
- An Essential Role of Variant Histone H3.3 for Ectomesenchyme Potential of the Cranial Neural Crest
- Characterization of Inducible Models of Tay-Sachs and Related Disease
- Hominoid-Specific Protein-Coding Genes Originating from Long Non-Coding RNAs
- Transcriptional Repression of Hox Genes by HP1/HPL and H1/HIS-24
- Integrative Genomic Analysis Identifies Isoleucine and CodY as Regulators of Virulence
- Convergence of the Transcriptional Responses to Heat Shock and Singlet Oxygen Stresses
- Genomics of Adaptation during Experimental Evolution of the Opportunistic Pathogen
- Enrichment of HP1a on Drosophila Chromosome 4 Genes Creates an Alternate Chromatin Structure Critical for Regulation in this Heterochromatic Domain
- Vsx2 Controls Eye Organogenesis and Retinal Progenitor Identity Via Homeodomain and Non-Homeodomain Residues Required for High Affinity DNA Binding
- The Long Path from QTL to Gene
- TCF7L2 Modulates Glucose Homeostasis by Regulating CREB- and FoxO1-Dependent Transcriptional Pathway in the Liver
- The Non-Flagellar Type III Secretion System Evolved from the Bacterial Flagellum and Diversified into Host-Cell Adapted Systems
- Complex Chromosomal Rearrangements Mediated by Break-Induced Replication Involve Structure-Selective Endonucleases
- Factors That Promote H3 Chromatin Integrity during Transcription Prevent Promiscuous Deposition of CENP-A in Fission Yeast
- A Mimicking-of-DNA-Methylation-Patterns Pipeline for Overcoming the Restriction Barrier of Bacteria
- Determinants of Human Adipose Tissue Gene Expression: Impact of Diet, Sex, Metabolic Status, and Genetic Regulation
- Genome-Wide Association Studies Identify Heavy Metal ATPase3 as the Primary Determinant of Natural Variation in Leaf Cadmium in
- Tethering of the Conserved piggyBac Transposase Fusion Protein CSB-PGBD3 to Chromosomal AP-1 Proteins Regulates Expression of Nearby Genes in Humans
- A Genome-Wide Association Study Identifies Five Loci Influencing Facial Morphology in Europeans
- Normal DNA Methylation Dynamics in DICER1-Deficient Mouse Embryonic Stem Cells
- H4K20me1 Contributes to Downregulation of X-Linked Genes for Dosage Compensation
- The NDR Kinase Scaffold HYM1/MO25 Is Essential for MAK2 MAP Kinase Signaling in
- Coevolution within and between Regulatory Loci Can Preserve Promoter Function Despite Evolutionary Rate Acceleration
- New Susceptibility Loci Associated with Kidney Disease in Type 1 Diabetes
- SWI/SNF-Like Chromatin Remodeling Factor Fun30 Supports Point Centromere Function in
- A Response Regulator Interfaces between the Frz Chemosensory System and the MglA/MglB GTPase/GAP Module to Regulate Polarity in
- Functional Variants in and Involved in Activation of the NF-κB Pathway Are Associated with Rheumatoid Arthritis in Japanese
- Two Distinct Repressive Mechanisms for Histone 3 Lysine 4 Methylation through Promoting 3′-End Antisense Transcription
- Genetic Modifiers of Chromatin Acetylation Antagonize the Reprogramming of Epi-Polymorphisms
- UTX and UTY Demonstrate Histone Demethylase-Independent Function in Mouse Embryonic Development
- A Comparison of Brain Gene Expression Levels in Domesticated and Wild Animals
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Enrichment of HP1a on Drosophila Chromosome 4 Genes Creates an Alternate Chromatin Structure Critical for Regulation in this Heterochromatic Domain
- Normal DNA Methylation Dynamics in DICER1-Deficient Mouse Embryonic Stem Cells
- The NDR Kinase Scaffold HYM1/MO25 Is Essential for MAK2 MAP Kinase Signaling in
- Functional Variants in and Involved in Activation of the NF-κB Pathway Are Associated with Rheumatoid Arthritis in Japanese
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



