-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaA Gene Family Derived from Transposable Elements during Early Angiosperm Evolution Has Reproductive Fitness Benefits in
The benefits of ever-growing numbers of sequenced eukaryotic genomes will not be fully realized until we learn to decipher vast stretches of noncoding DNA, largely composed of transposable elements. Transposable elements persist through self-replication, but some genes once encoded by transposable elements have, through a process called molecular domestication, evolved new functions that increase fitness. Although they have conferred numerous adaptations, the number of such domesticated transposable element genes remains unknown, so their evolutionary and functional impact cannot be fully assessed. Systematic searches that exploit genomic signatures of natural selection have been employed to identify potential domesticated genes, but their predictions have yet to be experimentally verified. To this end, we investigated a family of domesticated genes called MUSTANG (MUG), identified in a previous bioinformatic search of plant genomes. We show that MUG genes are functional. Mutants of Arabidopsis thaliana MUG genes yield phenotypes with severely reduced plant fitness through decreased plant size, delayed flowering, abnormal development of floral organs, and markedly reduced fertility. MUG genes are present in all flowering plants, but not in any non-flowering plant lineages, such as gymnosperms, suggesting that the molecular domestication of MUG may have been an integral part of early angiosperm evolution. This study shows that systematic searches can be successful at identifying functional genetic elements in noncoding regions and demonstrates how to combine systematic searches with reverse genetics in a fruitful way to decipher eukaryotic genomes.
Published in the journal: . PLoS Genet 8(9): e32767. doi:10.1371/journal.pgen.1002931
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002931Summary
The benefits of ever-growing numbers of sequenced eukaryotic genomes will not be fully realized until we learn to decipher vast stretches of noncoding DNA, largely composed of transposable elements. Transposable elements persist through self-replication, but some genes once encoded by transposable elements have, through a process called molecular domestication, evolved new functions that increase fitness. Although they have conferred numerous adaptations, the number of such domesticated transposable element genes remains unknown, so their evolutionary and functional impact cannot be fully assessed. Systematic searches that exploit genomic signatures of natural selection have been employed to identify potential domesticated genes, but their predictions have yet to be experimentally verified. To this end, we investigated a family of domesticated genes called MUSTANG (MUG), identified in a previous bioinformatic search of plant genomes. We show that MUG genes are functional. Mutants of Arabidopsis thaliana MUG genes yield phenotypes with severely reduced plant fitness through decreased plant size, delayed flowering, abnormal development of floral organs, and markedly reduced fertility. MUG genes are present in all flowering plants, but not in any non-flowering plant lineages, such as gymnosperms, suggesting that the molecular domestication of MUG may have been an integral part of early angiosperm evolution. This study shows that systematic searches can be successful at identifying functional genetic elements in noncoding regions and demonstrates how to combine systematic searches with reverse genetics in a fruitful way to decipher eukaryotic genomes.
Introduction
Recent technological advances have enabled the sequencing of hundreds of eukaryotic genomes, emphasizing that protein-coding genes constitute only a small fraction of the DNA [1], [2]. Since then, attention has increasingly shifted to deciphering other parts of the genome, the so-called non-coding regions that are largely composed of transposable elements (TEs). Unlike canonical genes, TEs can persist without benefiting fitness by replicating through transposition within the genome [3], [4]. There are two major classes of TEs, which transpose by fundamentally different mechanisms: retrotransposons, by reverse transcription of an RNA intermediate; and DNA transposons, by cut and paste transposition. TEs in each active family encode the proteins they need for transposition, which differ by replication strategy and superfamily [5].
Although traditionally viewed as selfish [3], [4], evidence continues to mount that TEs affect how genomes evolve in a variety of beneficial ways. One example is molecular domestication, where TEs are repurposed into new genes or other sequences with novel functions [6]–[10]. TE genes are adapted for transposition-related functions, yet they have molecular properties, such as DNA binding and protein-protein interaction, that can be used as raw genetic material and co-opted to perform new functions. Domesticated TEs (DTEs) are different from TEs and similar to canonical genes both in terms of their structure and activity, and in that they are subject to phenotypic selection. Most known DTEs have co-linear orthologs and have lost transposition-related features such as flanking terminal repeats and transposase catalytic activity [8], [10]. However, other features of the original TE that contribute to the beneficial function are maintained, such as conserved domains. DTEs perform various beneficial functions; for example, many DTEs derived from DNA transposons are transcription factors [7], while others are involved in centromere binding, chromosome segregation, meiotic recombination, heterochromatin formation, TE silencing, programmed genome rearrangement, V(D)J recombination, genome stability, and translational regulation [8], [11]. Molecular domestication has helped to spur remarkable evolutionary innovations, including the mammalian placenta and the vertebrate adaptive immune system [8].
To date, most DTEs have been discovered fortuitously by forward genetics. In plants for example, the FHY3 (FAR-RED ELONGATED HYPOCOTYL 3) family and DAYSLEEPER are the only well-characterized DTEs. FHY3 and FAR1 (FAR-RED IMPAIRED RESPONSE 1), two members of the FHY3 family, were identified in screens for far-red light mutants [12]–[14]. DAYSLEEPER, which is essential to plant development, was identified in a yeast one-hybrid screen [15]. Even if only a small fraction of the tens of thousands of TE-like genes in plant genome are DTEs rather than TEs, the total number of DTEs may be much higher than currently reported, suggesting that many more may await discovery and that traditional genetic methods may be insufficient to find them, for example due to functional redundancy.
These limitations can be overcome by direct bioinformatic searches of genomic data in which DTE genes are discriminated from TEs using genomic signatures that result from differences between how TEs and DTEs function and evolve, such as differences in expression, microsynteny, evolutionary rate, phylogeny, repetitiveness, and TE termini [2], [16]–[21]. But while theoretically sound, bioinformatics-based searches for DTE genes have not yet been confirmed experimentally. To assess the validity of this approach, we investigated a family of DTEs, MUSTANG (MUG), identified in a previous bioinformatic search of plant genomes [17], using a reverse genetic approach. MUG sequences are similar to ancestral TEs called Mutator-like elements (MULEs), but unlike MULEs, MUG genes lack signature terminal sequences, are collinear in monocots and eudicots, are functionally constrained, and are differentially expressed [17], [22], [23]. Here, we show that MUG genes have been conserved throughout angiosperm evolution and experimentally validate that they are functional in Arabidopsis thaliana by showing that they are essential to flower development and plant fitness.
Results
MUG subfamilies are conserved among angiosperms
To investigate the size and distribution of the MUG gene family across a wide range of taxa, we searched for complete sets of MUG paralogs in nine angiosperm species with high quality full-genome sequencing projects at an advanced stage or completed (Figure S1). Phylogenetic analysis revealed that MUG consists of two major subfamilies, MUGA and MUGB, both present in all nine species, but with different patterns of lineage-specific diversification (Figure 1, Figure S2). We found a minimum of four and a maximum of eight MUG genes per species, including the eight previously identified in A. thaliana [17], which we call At-MUG1 to At-MUG8. There may be additional MUG genes in unassembled parts of the genomes that we could not identify. These results are consistent with previous analyses of sugarcane (Saccarum officinarum and Saccarum spontaneum) [19], [24], which found 5 MUGA and 10 MUGB genes, referred to as “Class III” and “Class IV”, respectively, and of grapevine (Vitis vinifera) [21], which found 5 MUGA (we found only 4) and 3 MUGB genes, referred to as MUGvine1–5 and MUGvine6–8, respectively.
Fig. 1. MUSTANG phylogeny and gene structure in A. thaliana. 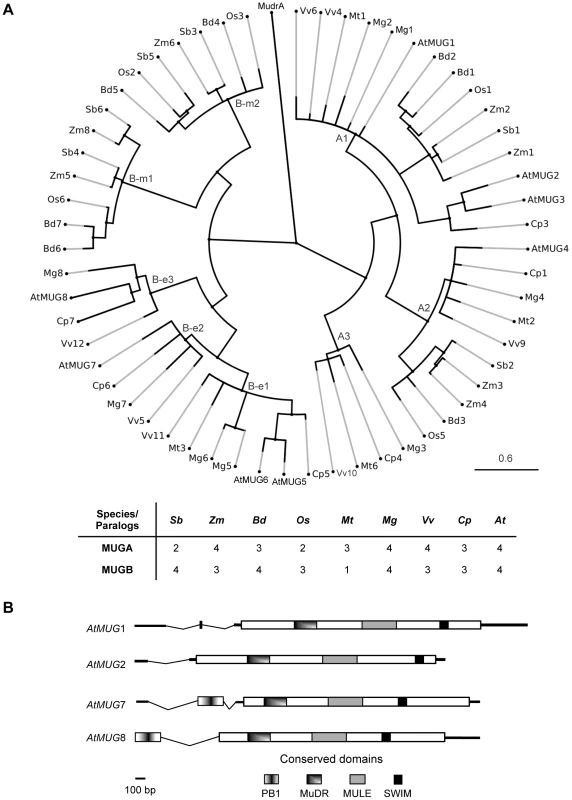
(A) MUG phylogeny in nine angiosperm species. At, Arabidopsis thaliana; Bd, Brachypodium distachyon; Cp, Carica papaya; Mg, Mimulus guttatus; Mt, Medicago truncatula; Os, Oryza sativa; Sb, Sorghum bicolor; Vv, Vitis vinifera; Zm, Zea mays. See Figure S1 for sequences and Table S5 for locus IDs. All bootstrap values are >70% (not shown). Cp7 is truncated; its position is approximate. (B) Graphical representation of At-MUG1, At-MUG2, At-MUG7, and At-MUG8 gene transcripts. Bold horizontal lines represent transcripts, dips introns, and rectangles coding sequences. Consistent with previous reports showing that MUG genes were present prior to the monocot-eudicot split [17], [19], [24], we found that the MUGA subfamily diversified prior to the divergence of monocots and eudicots into three extant clades, A1 to A3. Clade A1 has at least one member in each examined species and includes At-MUG1, At-MUG2, and At-MUG3, the latter two belonging to a Brassicales-specific subclade. Clade A2, which includes At-MUG4, has exactly one copy in every species, except Zea mays, in which it appears to have a recent duplication. Clade A3 differs from A1 and A2. Although it was the first of these three clades to diverge, apparently prior to the monocot-eudicot split, it has no member in the examined monocot species, nor is there a clade A3 member in A. thaliana.
Conversely, the diversification of the MUGB subfamily did not occur until after the monocot-eudicot split, into two monocot-specific clades (B-m1 and B-m2) and three eudicot-specific clades (B-e1 to B-e3). Each examined monocot species has one or two members of each monocot-specific clade. The branches leading to these clades are long, suggesting that B-m1 and B-m2 may have diverged early in monocot evolution. Similarly, all examined eudicots have one or two members in the eudicot-specific clade B-e1, which includes At-MUG5 and At-MUG6. Clades B-e2, which includes At-MUG7, and B-e3, which includes At-MUG8, each consist of exactly one member from each examined eudicot species, except Medicago truncatula, which has no member in either B-e2 or B-e3. Clade B-e3 is the most divergent MUG clade. Together, these results suggest that most MUG clades are conserved within lineages, with MUGA clades encompassing both monocots and eudicots and MUGB clades specific to monocots or eudicots.
To further investigate the origin of the MUG family and its distribution among plant taxa, we searched expressed sequence tag (EST) libraries deposited at the Ancestral Angiosperm Genome Project (AAGP; http://ancangio.uga.edu) and the National Center for Biotechnology Information (NCBI) dbEST [25]. We identified multiple ESTs similar to A. thaliana MUG genes in all five ancestral angiosperm species represented in the AAGP (Table S1), as well as in every order of angiosperm in dbEST with a sufficiently large library (5,000 ESTs or more), except Acorales, Alismatales, and Liliales (Table S2). No putative MUG ESTs were found in any gymnosperm in the AAGP or dbEST, nor in any other taxa outside angiosperms, although putative MULEs were found in all taxa. Consistent with these EST results, the sequences most similar to MUG in the genomes of Selaginella moellendorffii (a primitive vascular plant) and Physcomitrella patens (a moss) are repetitive and contain premature stop codons, characteristics indicative of TEs. Phylogenetic analyses of aligned selections of ESTs and genomic sequences confirmed these results (data not shown).
All MUG genes contain three conserved domains, an N-terminal MuDR DNA-binding domain (Pfam PF03108) [26], a core MULE transposase domain (PF10551), and a C-terminal SWIM zinc-finger domain (PF04434) (Figure 1B, Figure S2). The same domain architecture is found in diverse transposases of the MULE superfamily [27]. MUGA and MUGB members encode all three domains in a single exon. In addition, MUGB genes have an additional short 5′ exon encoding the Phox and Bem1p (PB1) domain (PF00564).
MUGA and MUGB subfamilies have synonymous nucleotide substitution rates (dN/dS) of 0.12 and 0.10, respectively, which are significantly less than one (p = 0.0045 and p = 0.0085, respectively), showing that they are under purifying selection.
Creation of MUG double mutants for reverse genetics
To test for MUG functionality in A. thaliana, we used a reverse genetics approach. Two independently derived mutant alleles with T-DNA insertions into the coding regions were obtained from the Arabidopsis Biological Research Center, Ohio State University. When grown under standard conditions, homozygous single mutants show no obvious differences from wild-type (data not shown). This suggests that MUG genes may be functionally redundant, as is often observed in plant gene families. To test this, we crossed single mutants within each subfamily to obtain homozygous double mutants. The mug1 mug2 (MUGA subfamily) and mug7 mug8 (MUGB subfamily) double mutants exhibit strong phenotypes, consistent with the hypothesis that MUG1 has some degree of functional redundancy with MUG2, and MUG7 with MUG8.
MUG double mutants have severe developmental and reproductive defects
To characterize the double mutant phenotypes, we measured a number of traits that reflect plant fitness at various life stages (Table S3). We compared two different allelic combinations each of mug1 mug2 and mug7 mug8 mutants to wild-type A. thaliana plants, ecotype Col-0 (Figure S3). When grown under standard laboratory conditions, the double mutants differ dramatically from wild-type at all developmental stages from germination to senescence. They exhibit phenotypes that include reduced plant size, an increased incidence of aborted seeds, reduced seed amount, and delays in developmental timing and flowering (Figure 2A). Although the two double mutants have defects in several similar traits, the degree of severity for some traits is stronger in mug7 mug8 than in mug1 mug2. mug1 mug2 yields 36% of the wild-type seed set, whereas mug7 mug8 yields only 7%. mug1 mug2 stems are more than 2 times shorter than wild-type, whereas those of mug7 mug8 are more than 27 times shorter. Interestingly, mug1 mug2 has a ratio of stem height to rosette diameter (r = 3.44) similar to wild-type (r = 3.89), whereas mug7 mug8 has a much-reduced ratio (r = 0.37) (Figure 2B). Application of exogenous gibberellic acid (GA) failed to rescue reduced stem height or delayed flowering phenotypes (data not shown). The growth defect seems to be restricted to light conditions, since both double mutants show hypocotyls that etiolate normally (data not shown).
Fig. 2. Phenotypic analysis of mug1 mug2 and mug7 mug8 in A. thaliana. 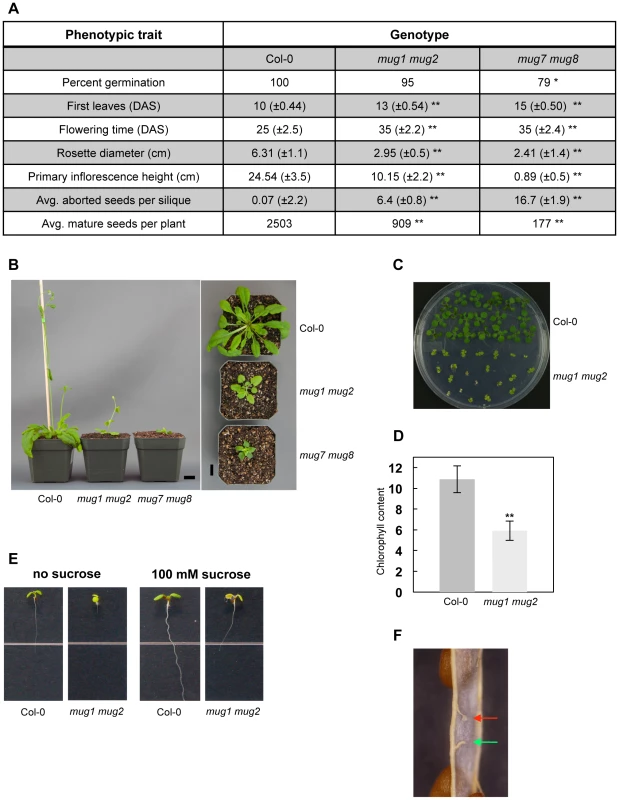
(A) Phenotypes of wild-type (Col-0), mug1 mug2, and mug7 mug8 based on the following traits: 1) Germination (%); 2) First leaves recorded as number of days after sterilization (DAS); 3) Flowering time (DAS); 4) Rosette leaf coloration; 5) Rosette diameter (cm) at 42 DAS; 6) Height (cm) of primary inflorescence at 42 DAS; 7) Average number of aborted seeds. Measurements based on 60 plants per genotype. Statistical significance based on two-sample student t-test; α = 0.05; * p<0.01; ** p<0.001. (B) Growth phenotype of Col-0, mug1 mug2, and mug7 mug8 at 40 DAS. Scale = 1 cm. (C) Image of an MS plate (0.8% agar w/v; 1% sucrose w/v) containing 17-day-old Col-0 and mug1 mug2 seedlings. (D) Chlorophyll accumulation in Col-0 and mug1 mug2 seedlings. Bars represents standard deviations of quadruple experiments. 200 mg or approximately 10 Col-0 or 30 mug1 mug2 seedlings per experiment. (E) Phenotypes of mug1 mug2 under sucrose conditions. Twelve-day-old seedlings of Col-0 and mug1 mug2 were grown on standard MS medium without sucrose or with 100 mM sucrose. (F) Close up of a dissected mug1 mug2 silique showing a normal funiculus from which the attached mature seed was released (green arrow) and a funiculus attached to undeveloped ovule tissues (red arrow). Some traits are unique to each double mutant. mug7 mug8 leaves are slightly curly, whereas mug1 mug2 leaves have a pale yellow-green coloration, which is most evident in seedlings but remains visible in mature plants (Figure 2B, 2C). mug1 mug2 seedlings contain only about half the chlorophyll per unit mass of wild-type seedlings (Figure 2D). They also fail to develop beyond the cotyledon stage in the absence of exogenous sucrose, and even with exogenous sucrose their growth is restricted (Figure 2E).
Each double mutant has defects in floral development and organ morphology that result in reduced fertility (Figure 3). The flowers are smaller than wild-type. Floral abnormalities are most dramatic at anthesis, especially in mug7 mug8 (Figure 3). The gynoecium becomes highly elongated relative to the anther filaments (Figure 3C). Scanning electron microscopy (SEM) showed that the surface cell layers of certain flower organs, including the stigmatic hair-bearing region and the anthers, are deflated and have abnormal shapes, giving the flowers a shriveled appearance (Figure 3F). These observations suggest that mug7 mug8 flowers may undergo premature senescence. Anthers are bilocular structures that normally produce and hold pollen grains, and upon maturity dehisce longitudinally to release the pollen. Most mug7 mug8 anthers are flat and contain no pollen, and even when they do contain pollen, the locules usually fail to furrow and dehisce, preventing the pollen from being released (Figure 3L). The female organs of mug7 mug8 are also defective, since applying wild-type pollen onto the mutant stigma did not rescue the fertility. mug1 mug2 has less severe defects (Figure 3). The flowers are not shriveled and both the female and male organs are functional and capable of participating in self-fertilization. However, the anthers do exhibit restricted dehiscence, with only one of the two furrows opening successfully (Figure 3K). The siliques produced by both double mutants have a smaller maximum size than wild-type, restricting the number of seeds they bear, and they have a high incidence of undeveloped ovules, attenuating the seed yield (Figure 2A, 2F).
Fig. 3. Flower structure of wild-type, mug1 mug2, and mug7 mug8 in A. thaliana. 
(A–C) Bright-field micrographs of dissected flowers shown of (A) wild-type, (B) mug1 mug2, and (C) mug7 mug8. Scale = 0.5 mm. (D–F) SEM micrographs of dissected flowers of (D) wild-type, (E) mug1 mug2, and (F) mug7 mug8. Scale = 0.5 mm. (G–I) SEM micrographs of dissected flowers at higher magnification showing the stigma papillae of (G) wild-type, (H) mug1 mug2, and (I) mug7 mug8. Scale = 100 µm. (J–L) SEM micrographs of dissected flowers at higher magnification showing one anther and pollen of (J) wild-type, (K) mug1 mug2, and (L) mug7 mug8. Scale = 50 µm. Discussion
In silico screens like that in which the MUG gene family was identified [17] have the potential to detect many novel DTEs, but to be convincing their predictions must be validated experimentally. To do so, we exploited a fundamental difference between TE genes and canonical genes. Although TEs can produce phenotypes, for instance by inserting into and disabling a canonical gene, they are not known to themselves encode beneficial functions [11]. Therefore, knocking out a TE gene should have either no effect on fitness or should increase it, whereas knocking out a canonical gene, such as a DTE, may reduce the fitness of the organism. We utilized this difference by examining traits closely tied to fitness in mug T-DNA insertion mutants.
Fitness is a measure of how much a genotype contributes to the next generation in a given environment and includes components of survival and reproduction [28]. Both of these aspects are compromised in mug mutants, and certain phenotypes are especially striking. First, the survival of mug mutants is compromised by defects in their physiology (Figure 2, Figure 3). mug7 mug8 plants are severe dwarfs, which may disadvantage them in various environments, such as where space or light is limited. The size of mug1 mug2 plants is less reduced, but they have other serious defects. They contain only approximately half of the wild-type amount of chlorophyll per unit mass and their roots barely elongate in the absence of exogenous sucrose (Figure 2C–2E). Sucrose, the major product of photosynthesis, plays a key role in sugar signaling pathways and is required to supply metabolic energy to the roots [29]. Reduced chlorophyll concentrations could impair chloroplast activity and sucrose production, so may be one explanation of why mug1 mug2 plants require exogenous sucrose, without which they would be unlikely to survive to maturity in wild environments.
Second, the reproduction of mug mutants is compromised by defects in the floral organ development, fecundity, and reproductive timing (Figure 2A, Figure 3). These defects are particularly severe in mug7 mug8 mutants. They produce little or no pollen, their stamens and pistil do not elongate normally, preventing contact and limiting self-pollination, and their gynoecium is defective, limiting fertilization even if pollen does adhere to the stigmatic papillae (Figure 3). This combination of defects renders mug7 mug8 mutants nearly sterile, with an average seed yield of only 7% of wild-type, even under optimal laboratory conditions.
The observed phenotypes may result from defects in a variety of physiological processes, so the function of MUG is not yet clear; however, patterns of conservation and expression do provide a few clues. Multiple lines of evidence, in addition to the mutant phenotypes, show that it is highly unlikely that MUG genes function in transposition. They lack the TE termini required for mobilization and are collinear in multiple genomes [17]; Most lack intact DDE motifs, which are required to catalyze transposition (Figure S2, Table S4) [27], [30]; Searches of publicly available data show that, unlike TEs [31], they are not targeted by small RNAs or silenced by DNA methylation [32]–[34], but are instead expressed in diverse tissues in A. thaliana, sugarcane [19], [24], [35], rice [36], and other angiosperms (Table S1, Table S2). The only other known MULE-like DTEs, the FHY3 family, are transcription factors [30], [37], [38], a common function among domesticated DNA transposons [8], [11]. Like FHY3, the WRKY-GCM1, MULE, and SWIM domains of MUG genes are highly conserved, including key active site residues (Figure S2), suggesting that they may also have a function involving DNA binding, such as transcription regulation.
The severe defects and reduced fitness associated with mutations to MUG genes may explain why they are well conserved. They appear to be ubiquitous among all angiosperms, including basal angiosperms, but are absent from non-angiosperms (Table S1, S2), suggesting that MUG was domesticated during early angiosperm evolution to perform what evolved to become a key function. Consistent with previous studies [17], [19], [24], it is clear from their phylogeny that the two MUG subfamilies diverged early in angiosperm evolution, prior to the monocot-eudicot split (Figure 1). However, phylogenetic and phenotypic differences between MUGA and MUGB suggest that they may have originated from more than one domestication event, similar to the FHY3 family [30]. If all MUG genes did descend from a single domestication event, then while MUGA must have diversified prior to the monocot-eudicot split, MUGB must have acquired a PB1 exon and evolved rapidly prior to the monocot-eudicot split, yet not diversified until after the split. Alternately, if the MUGB subfamily descended from a different domestication event than MUGA, it may have acquired the PB1 domain either through transduplication prior to domestication [39], gene fusion during domestication [40], or exon shuffling subsequent to domestication. There may even have been more than two domestication events; in particular, Clade A3 has an unusual phylogenetic pattern, possibly reflecting a separate origin. Although multiple domestication events may help explain differences between the evolution and phenotypes of the two subfamilies, additional evidence, such as from phylogenetic studies of closely related MULEs or the functional characterization of additional MUG genes, will be required to resolve this question.
In summary, our results suggest the MUG family originated from TE genes adopting an adaptive function early in flowering plant evolution and are now conserved among angiosperms. Serious defects in mug mutants show that these genes make important contributions to fitness through roles in plant growth, flower development, and reproduction. The approach we used, of evaluating the fitness consequences of mutations to predicted DTEs, effectively couples in silico searches of genomic data with experimental validation. In the future, we expect that similar studies will enable a more complete characterization of TE-derived sequences that have been co-opted to provide fitness benefits.
Materials and Methods
MUG homologs
To confirm the protein sequences of the eight previously identified A. thaliana MUG genes [17], the sequences of At-MUG1 and At-MUG3 through At-MUG7 were determined from gene models supported by publicly available full-length cDNAs at The Arabidopsis Information Resource (TAIR) database, release 10 [41]: At-MUG1, TAIR AT3G04605, GenBank AY074390; At-MUG3, AT1G06740, AK221278; At-MUG4, AT5G16505, AY059842; At-MUG5, AT3G06940, AF462806; At-MUG6, AT5G48965, AY136382; At-MUG7, AT3G05850, BT008628. At-MUG2 (AT2G30640) and At-MUG8 (AT5G34853) have no publicly available full-length cDNA sequence, so we predicted their structure from genomic DNA using FGENESH (dicot setting) [42]. The predicted sequence of At-MUG2 was confirmed by RT-PCR (data not shown) and both At-MUG2 and At-MUG8 are consistent with available EST and mRNA-seq data [32], [41].
To identify additional MUG homologs, we performed comprehensive genomic searches in nine angiosperm genomes: A. thaliana, Carica papaya, V. vinifera, M. truncatula, Mimulus guttatus, Sorghum bicolor, Z. mays, Oryza sativa var. japonica, and Brachypodium distachyon. Because the identity between MUGA and MUGB sequences is low, a representative amino acid query was chosen for each subfamily: At-MUG1 for MUGA, At-MUG7 for MUGB.
In A. thaliana, O. sativa, and S. bicolor, we used BLASTP to search the protein databases of TAIR10 [41], the Rice Annotation Project (RAP-DB) [43], and Sbi 1.4 [44], respectively. To ensure that we found all MUG genes present in the datasets, we calibrated E-value thresholds to well below those needed to find all known MUG genes [17] as well as a few non-MUG sequences, and we validated the results using phylogenetic analyses. At each resulting locus, the highest-ranked annotated gene model was chosen. Conserved domains were identified using the NCBI Conserved Domain Database [45]. Gene models terminating in truncated domains were extended using FGENESH [42] where possible. Because TE-like genes (including DTEs) are commonly filtered from protein databases, we confirmed the results in A. thaliana using a TBLASTN [46] search of the TAIR10 Genes database, which includes loci annotated as “transposable element genes”. In S. bicolor, we confirmed the results using a TBLASTN [46] search of the unmasked genome assembly [44].
The remaining six genomes had limited gene models, so instead of searching protein databases, we searched whole genome assemblies: C. papaya, Hawaii Papaya Genome Project, 2007 release [47]; V. vinifera, Genoscope, March 2010, 12X assembly [48]; M. truncatula, Medicago Genome Sequencing Consortium, release Mt3.5 [49]; M. guttatus, Joint Genome Institute (JGI) assembly v1.0 and gene annotation v1.1 [50]; Z. mays, Maizesequence.org, release 4a.53 [51]; B. distachyon, JGI 8x assembly release v1.0 of strain Bd21 [50], [52]. We used TBLASTN [46] to identify potential MUG homologs and FGENESH [42] to predict corresponding gene models. Conserved domains were identified as above.
Phylogenetic analyses
We conducted phylogenetic analyses on the amino acid sequences identified in the genome searches, including all putative MUG homologs as well as a sample of putative MURA sequences. Putative MURAs typically formed larger, low-identity clusters and the sequences often contained premature stop codons or frameshifts, even within conserved domains, which is characteristic of TEs. Clusters of putative MURA sequences with greater than 95% identity were represented by a single sequence. Alignments were generated using MUSCLE [53], curated with Gblocks [54], and phylogenetic analysis performed using PhyML (500 bootstraps) [55], [56]. All putative MURA sequences were non-monophyletic to the MUG tree. Z. mays MURA (GenBank AAA81535.1) was used as the outgroup. Synonymous substitution rates were calculated using PAML CODEML [57].
To determine the taxonomic distribution of MUG homologs, we conducted TBLASTN searches of NCBI dbEST libraries of each major seed plant subgroup not represented in the genome searches, using At-MUG2 and At-MUG7 queries as above (Table S2). We identified putative MUG ESTs by looking in each species for small clusters of top-ranked ESTs with low E-values and high identity, consistent with results of previously identified MUG homologs. We validated MUG and MURA assignments by phylogenetic analyses, using subsets of ESTs that could be aligned.
Plant material
The mutants mug1-1 (GK_514B01), mug1-2 (GK_293B02), mug2-3 (SALK_090878), mug2-4 (SALK_055071), mug7-1 (SALK_012814), mug7-5 (GK_378C04), mug8-1 (GK_244B09), and mug8-2 (GK_155E09) were obtained from GABI-Kat (http://www.gabi-kat.de) [58] and SALK (http://www.arabidopsis.org/abrc) [59] T-DNA insertion populations. The positions of the insertion sites in double mutants used in the phenotypic analyses were confirmed by sequencing of the allele-specific PCR products (data not shown). Wild-type ecotype Col-0 seeds were originally obtained from Lehle Seeds (www.arabidopsis.com).
Seeds were sterilized using a 50% bleach solution for 5 min, then washed once in 95% ethanol and 3 times in sterile water. Sterilized seeds were sown on half-strength Murashige and Skoog (MS) [60] medium plates containing 0.8% agar (w/v) and 1% sucrose (w/v). Plated seeds were stratified in the dark for 3 days at 4°C and kept on plates for 2 weeks in a growth chamber (Conviron model E15) at 22°C under a 16 h light/8 h dark photoperiod, ∼100 µmol quanta/m2/s light intensity, 60% relative humidity. Seedlings were transplanted to soil with a composition of PRO-MIX (Premier Tech Horticulture, Quebec, Canada): vermiculite: perlite of 2∶1∶1 in 2 ½ inch square pots and returned to the growth chamber.
Phenotypic analyses
We selected eight traits for phenotypic analyses previously shown to reflect plant fitness [61]–[65]. A detailed description of each trait is presented in Table S3. Statistical analysis was performed by two-sample t-tests using the wild-type control (α = 0.05).
Scanning electron microscopy was performed using a Hitachi S4700 Field Emission-STEM microscope. Wild-type and homozygous mutant inflorescences were fixed overnight in 2% glutaraldehyde, washed, and dehydrated using a series of graded ethanol solutions (30 to 100%). Dried samples were sputter coated with gold-platinum. Inflorescences were also photographed using an Olympus DP71 camera attached to an Olympus MVX10 stereomicroscope.
For the sucrose assay, Col-0 and mutant seeds were sown on MS medium plates with or without 100 mM sucrose. Seeds were stratified for 3 days in the dark at 4°C and transferred to a growth chamber with settings as above for 12 days with plates vertically-oriented, after which each plate was photographed using a Nikon D3100 camera and scored manually.
To measure total chlorophyll content, 200 mg of 17-day-old wild-type and mutant seedlings were extracted by shaking overnight in the dark in 1 ml of 80% acetone. Chlorophyll levels were measured using a 4050 Ultrospec II UV/Vis spectrophotometer (LKB Biochrom) and the total amount of chlorophyll was determined using MacKinney's coefficients [66] and the equation: chlorophyll a+b = 7.15×OD660 nm+18.71×OD647 nm.
Supporting Information
Zdroje
1. AGI (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 : 796–815.
2. LanderES, LintonLM, BirrenB, NusbaumC, ZodyMC, et al. (2001) Initial sequencing and analysis of the human genome. Nature 409 : 860–921.
3. DoolittleWF, SapienzaC (1980) Selfish genes, the phenotype paradigm and genome evolution. Nature 284 : 601–603.
4. OrgelLE, CrickFH (1980) Selfish DNA: the ultimate parasite. Nature 284 : 604–607.
5. WickerT, SabotF, Hua-VanA, BennetzenJL, CapyP, et al. (2007) A unified classification system for eukaryotic transposable elements. Nat Rev Genet 8 : 973–982.
6. VolffJ-N (2006) Turning junk into gold: domestication of transposable elements and the creation of new genes in eukaryotes. Bioessays 28 : 913–922.
7. FeschotteC (2008) Transposable elements and the evolution of regulatory networks. Nat Rev Genet 397–405.
8. SinzelleL, IzsvakZ, IvicsZ (2009) Molecular domestication of transposable elements: From detrimental parasites to useful host genes. Cell Mol Life Sci 66 : 1073–1093.
9. VolffJN (2009) Cellular genes derived from Gypsy/Ty3 retrotransposons in mammalian genomes. Ann N Y Acad Sci 1178 : 233–243.
10. Hua-VanA, Le RouzicA, BoutinTS, FileeJ, CapyP (2011) The struggle for life of the genome's selfish architects. Biology direct 6 : 19.
11. FeschotteC, PrithamEJ (2007) DNA transposons and the evolution of eukaryotic genomes. Annu Rev Genet 41 : 331–368.
12. WhitelamGC, JohnsonE, PengJ, CarolP, AndersonML, et al. (1993) Phytochrome A null mutants of Arabidopsis display a wild-type phenotype in white light. Plant Cell 5 : 757–768.
13. HudsonM, RingliC, BoylanMT, QuailPH (1999) The FAR1 locus encodes a novel nuclear protein specific to phytochrome A signaling. Genes Dev 13 : 2017–2027.
14. HudsonME, LischDR, QuailPH (2003) The FHY3 and FAR1 genes encode transposase-related proteins involved in regulation of gene expression by the phytochrome A-signaling pathway. Plant J 34 : 453–471.
15. BundockP, HooykaasP (2005) An Arabidopsis hAT-like transposase is essential for plant development. Nature 436 : 282–284.
16. ZdobnovE, CampillosM, HarringtonE, TorrentsD, BorkP (2005) Protein coding potential of retroviruses and other transposable elements in vertebrate genomes. Nucleic Acids Res 33 : 946–954.
17. CowanR, HoenD, SchoenD, BureauT (2005) MUSTANG is a novel family of domesticated transposase genes found in diverse angiosperms. Mol Biol Evol 22 : 2084–2089.
18. MuehlbauerGJ, BhauBS, SyedNH, HeinenS, ChoS, et al. (2006) A hAT superfamily transposase recruited by the cereal grass genome. Mol Genet Genomics 275 : 553–563.
19. SaccaroNL, Van SluysM-A, de Mello VaraniA, RossiM (2007) MudrA-like sequences from rice and sugarcane cluster as two bona fide transposon clades and two domesticated transposases. Gene 392 : 117–125.
20. PiriyapongsaJ, Marino-RamirezL, JordanIK (2007) Origin and evolution of human microRNAs from transposable elements. Genetics 176 : 1323–1337.
21. BenjakA, ForneckA, CasacubertaJM (2008) Genome-wide analysis of the “cut-and-paste” transposons of grapevine. PLoS ONE 3: e3107 doi:10.1371/journal.pone.0003107..
22. LeQH, WrightS, YuZ, BureauT (2000) Transposon diversity in Arabidopsis thaliana. Proc Natl Acad Sci U S A 97 : 7376–7381.
23. YuZ, WrightSI, BureauTE (2000) Mutator-like elements in Arabidopsis thaliana. Structure, diversity and evolution. Genetics 156 : 2019–2031.
24. RossiM, AraujoPG, de JesusEM, VaraniAM, Van SluysMA (2004) Comparative analysis of Mutator -like transposases in sugarcane. Molecular genetics and genomics : MGG 272 : 194–203.
25. BoguskiMS, LoweTM, TolstoshevCM (1993) dbEST–database for “expressed sequence tags”. Nat genet 4 : 332–333.
26. BabuMM, IyerLM, BalajiS, AravindL (2006) The natural history of the WRKY-GCM1 zinc fingers and the relationship between transcription factors and transposons. Nucleic Acids Res 34 : 6505–6520.
27. Hua-VanA, CapyP (2008) Analysis of the DDE Motif in the Mutator Superfamily. J Mol Evol 67 : 670–681.
28. OrrHA (2009) Fitness and its role in evolutionary genetics. Nature reviews Genetics 10 : 531–539.
29. SmithAM, StittM (2007) Coordination of carbon supply and plant growth. Plant, cell & environment 30 : 1126–1149.
30. LinR, DingL, CasolaC, RipollDR, FeschotteC, et al. (2007) Transposase-derived transcription factors regulate light signaling in Arabidopsis. Science 318 : 1302–1305.
31. SimonSA, MeyersBC (2011) Small RNA-mediated epigenetic modifications in plants. Curr Opin Plant Biol 14 : 148–155.
32. ListerR, O'MalleyRC, Tonti-FilippiniJ, GregoryBD, BerryCC, et al. (2008) Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133 : 523–536.
33. ZhangX, YazakiJ, SundaresanA, CokusS, ChanSW, et al. (2006) Genome-wide high-resolution mapping and functional analysis of DNA methylation in arabidopsis. Cell 126 : 1189–1201.
34. GregoryBD, O'MalleyRC, ListerR, UrichMA, Tonti-FilippiniJ, et al. (2008) A link between RNA metabolism and silencing affecting Arabidopsis development. Developmental cell 14 : 854–866.
35. de AraujoP, RossiM, de JesusE, SaccaroN, KajiharaD, et al. (2005) Transcriptionally active transposable elements in recent hybrid sugarcane. Plant J 44 : 707–717.
36. JiaoY, DengXW (2007) A genome-wide transcriptional activity survey of rice transposable element-related genes. Genome Biol 8: R28.
37. OuyangX, LiJ, LiG, LiB, ChenB, et al. (2011) Genome-Wide Binding Site Analysis of FAR-RED ELONGATED HYPOCOTYL3 Reveals Its Novel Function in Arabidopsis Development. Plant Cell 23 : 2514–2535.
38. LinR, TengY, ParkH-J, DingL, BlackC, et al. (2008) Discrete and essential roles of the multiple domains of Arabidopsis FHY3 in mediating phytochrome A signal transduction. Plant Physiol 148 : 981–992.
39. HoenDR, ParkKC, ElroubyN, YuZ, MohabirN, et al. (2006) Transposon-mediated expansion and diversification of a family of ULP-like genes. Mol Biol Evol 23 : 1254–1268.
40. CordauxR, UditS, BatzerMA, FeschotteC (2006) Birth of a chimeric primate gene by capture of the transposase gene from a mobile element. P Natl Acad Sci U S A 103 : 8101–8106.
41. SwarbreckD, WilksC, LameschP, BerardiniTZ, Garcia-HernandezM, et al. (2008) The Arabidopsis Information Resource (TAIR): gene structure and function annotation. Nucleic acids res 36: D1009–1014.
42. SalamovAA, SolovyevVV (2000) Ab initio gene finding in Drosophila genomic DNA. Genome Res 10 : 516–522.
43. TanakaT, AntonioBA, KikuchiS, MatsumotoT, NagamuraY, et al. (2008) The Rice Annotation Project Database (RAP-DB): 2008 update. Nucleic Acids Res 36: D1028–1033.
44. PatersonAH, BowersJE, BruggmannR, DubchakI, GrimwoodJ, et al. (2009) The Sorghum bicolor genome and the diversification of grasses. Nature 457 : 551–556.
45. Marchler-BauerA, LuS, AndersonJB, ChitsazF, DerbyshireMK, et al. (2011) CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res 39: D225–229.
46. AltschulSF, GishW, MillerW, MyersEW, LipmanDJ (1990) Basic local alignment search tool. Journal of molecular biology 215 : 403–410.
47. MingR, HouS, FengY, YuQ, Dionne-LaporteA, et al. (2008) The draft genome of the transgenic tropical fruit tree papaya (Carica papaya Linnaeus). Nature 452 : 991–996.
48. JaillonO, AuryJM, NoelB, PolicritiA, ClepetC, et al. (2007) The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449 : 463–467.
49. CannonSB, SterckL, RombautsS, SatoS, CheungF, et al. (2006) Legume genome evolution viewed through the Medicago truncatula and Lotus japonicus genomes. Proceedings of the National Academy of Sciences of the United States of America 103 : 14959–14964.
50. GoodsteinDM, ShuS, HowsonR, NeupaneR, HayesRD, et al. (2012) Phytozome: a comparative platform for green plant genomics. Nucleic acids research 40: D1178–1186.
51. SchnablePS, WareD, FultonRS, SteinJC, WeiF, et al. (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326 : 1112–1115.
52. TIBI (2010) Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463 : 763–768.
53. EdgarRC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic acids research 32 : 1792–1797.
54. CastresanaJ (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Molecular biology and evolution 17 : 540–552.
55. DereeperA, GuignonV, BlancG, AudicS, BuffetS, et al. (2008) Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36: W465–469.
56. GuindonS, GascuelO (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic biology 52 : 696–704.
57. YangZ (2007) PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24 : 1586–1591.
58. RossoMG, LiY, StrizhovN, ReissB, DekkerK, et al. (2003) An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol Biol 53 : 247–259.
59. AlonsoJM, StepanovaAN, LeisseTJ, KimCJ, ChenH, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 : 653–657.
60. MurashigeT, SkoogF (1962) A Revised Medium for Rapid Growth and Bio Assays with Tobacco Tissue Cultures. Physiologia plantarum 15 : 473–497.
61. Alonso-BlancoC, AartsM, BentsinkL, KeurentjesJ, ReymondM, et al. (2009) What has natural variation taught us about plant development, physiology, and adaptation? Plant Cell Online 21 : 1877.
62. ShindoC, BernasconiG, HardtkeCS (2008) Intraspecific competition reveals conditional fitness effects of single gene polymorphism at the Arabidopsis root growth regulator BRX. New Phytol 180 : 71–80.
63. DonohueK (2002) Germination timing influences natural selection on life-history characters in Arabidopsis thaliana. Ecology 83 : 1006–1016.
64. PigliucciM, SchlichtingCD (1996) Reaction norms of Arabidopsis IV. Relationships between plasticity and fitness. Heredity 76(Pt 5):427–436.
65. ShawRG, ChangS-M (2006) Gene action of new mutations in Arabidopsis thaliana. Genetics 172 : 1855–1865.
66. MackinneyG (1941) Absorption of light by chlorophyll solutions. J Biol Chem 140 : 315–322.
Štítky
Genetika Reprodukční medicína
Článek Genome-Wide Association Study for Serum Complement C3 and C4 Levels in Healthy Chinese SubjectsČlánek Role of Transposon-Derived Small RNAs in the Interplay between Genomes and Parasitic DNA in RiceČlánek Tetraspanin Is Required for Generation of Reactive Oxygen Species by the Dual Oxidase System inČlánek An Essential Role of Variant Histone H3.3 for Ectomesenchyme Potential of the Cranial Neural CrestČlánek A Mimicking-of-DNA-Methylation-Patterns Pipeline for Overcoming the Restriction Barrier of BacteriaČlánek A Genome-Wide Association Study Identifies Five Loci Influencing Facial Morphology in Europeans
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 9
-
Všechny články tohoto čísla
- Heterozygous Mutations in DNA Repair Genes and Hereditary Breast Cancer: A Question of Power
- GWAS of Diabetic Nephropathy: Is the GENIE out of the Bottle?
- The Conflict within and the Escalating War between the Sex Chromosomes
- Proteome-Wide Analysis of Disease-Associated SNPs That Show Allele-Specific Transcription Factor Binding
- Exome Sequencing Identifies Rare Deleterious Mutations in DNA Repair Genes and as Potential Breast Cancer Susceptibility Alleles
- A Gene Family Derived from Transposable Elements during Early Angiosperm Evolution Has Reproductive Fitness Benefits in
- Genome-Wide Association Study for Serum Complement C3 and C4 Levels in Healthy Chinese Subjects
- Role of Transposon-Derived Small RNAs in the Interplay between Genomes and Parasitic DNA in Rice
- Co-Evolution of Mitochondrial tRNA Import and Codon Usage Determines Translational Efficiency in the Green Alga
- SIRT6/7 Homolog SIR-2.4 Promotes DAF-16 Relocalization and Function during Stress
- CNV Formation in Mouse Embryonic Stem Cells Occurs in the Absence of Xrcc4-Dependent Nonhomologous End Joining
- Tetraspanin Is Required for Generation of Reactive Oxygen Species by the Dual Oxidase System in
- Citrullination of Histone H3 Interferes with HP1-Mediated Transcriptional Repression
- Variation in Genes Related to Cochlear Biology Is Strongly Associated with Adult-Onset Deafness in Border Collies
- The Long Non-Coding RNA Affects Chromatin Conformation and Expression of , but Does Not Regulate Its Imprinting in the Developing Heart
- Rif2 Promotes a Telomere Fold-Back Structure through Rpd3L Recruitment in Budding Yeast
- Is a Metastasis Susceptibility Gene That Suppresses Metastasis by Modifying Tumor Interaction with the Cell-Mediated Immunity
- The p38/MK2-Driven Exchange between Tristetraprolin and HuR Regulates AU–Rich Element–Dependent Translation
- Rare Copy Number Variants Contribute to Congenital Left-Sided Heart Disease
- A Genetic Basis for a Postmeiotic X Versus Y Chromosome Intragenomic Conflict in the Mouse
- An Essential Role of Variant Histone H3.3 for Ectomesenchyme Potential of the Cranial Neural Crest
- Characterization of Inducible Models of Tay-Sachs and Related Disease
- Hominoid-Specific Protein-Coding Genes Originating from Long Non-Coding RNAs
- Transcriptional Repression of Hox Genes by HP1/HPL and H1/HIS-24
- Integrative Genomic Analysis Identifies Isoleucine and CodY as Regulators of Virulence
- Convergence of the Transcriptional Responses to Heat Shock and Singlet Oxygen Stresses
- Genomics of Adaptation during Experimental Evolution of the Opportunistic Pathogen
- Enrichment of HP1a on Drosophila Chromosome 4 Genes Creates an Alternate Chromatin Structure Critical for Regulation in this Heterochromatic Domain
- Vsx2 Controls Eye Organogenesis and Retinal Progenitor Identity Via Homeodomain and Non-Homeodomain Residues Required for High Affinity DNA Binding
- The Long Path from QTL to Gene
- TCF7L2 Modulates Glucose Homeostasis by Regulating CREB- and FoxO1-Dependent Transcriptional Pathway in the Liver
- The Non-Flagellar Type III Secretion System Evolved from the Bacterial Flagellum and Diversified into Host-Cell Adapted Systems
- Complex Chromosomal Rearrangements Mediated by Break-Induced Replication Involve Structure-Selective Endonucleases
- Factors That Promote H3 Chromatin Integrity during Transcription Prevent Promiscuous Deposition of CENP-A in Fission Yeast
- A Mimicking-of-DNA-Methylation-Patterns Pipeline for Overcoming the Restriction Barrier of Bacteria
- Determinants of Human Adipose Tissue Gene Expression: Impact of Diet, Sex, Metabolic Status, and Genetic Regulation
- Genome-Wide Association Studies Identify Heavy Metal ATPase3 as the Primary Determinant of Natural Variation in Leaf Cadmium in
- Tethering of the Conserved piggyBac Transposase Fusion Protein CSB-PGBD3 to Chromosomal AP-1 Proteins Regulates Expression of Nearby Genes in Humans
- A Genome-Wide Association Study Identifies Five Loci Influencing Facial Morphology in Europeans
- Normal DNA Methylation Dynamics in DICER1-Deficient Mouse Embryonic Stem Cells
- H4K20me1 Contributes to Downregulation of X-Linked Genes for Dosage Compensation
- The NDR Kinase Scaffold HYM1/MO25 Is Essential for MAK2 MAP Kinase Signaling in
- Coevolution within and between Regulatory Loci Can Preserve Promoter Function Despite Evolutionary Rate Acceleration
- New Susceptibility Loci Associated with Kidney Disease in Type 1 Diabetes
- SWI/SNF-Like Chromatin Remodeling Factor Fun30 Supports Point Centromere Function in
- A Response Regulator Interfaces between the Frz Chemosensory System and the MglA/MglB GTPase/GAP Module to Regulate Polarity in
- Functional Variants in and Involved in Activation of the NF-κB Pathway Are Associated with Rheumatoid Arthritis in Japanese
- Two Distinct Repressive Mechanisms for Histone 3 Lysine 4 Methylation through Promoting 3′-End Antisense Transcription
- Genetic Modifiers of Chromatin Acetylation Antagonize the Reprogramming of Epi-Polymorphisms
- UTX and UTY Demonstrate Histone Demethylase-Independent Function in Mouse Embryonic Development
- A Comparison of Brain Gene Expression Levels in Domesticated and Wild Animals
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Enrichment of HP1a on Drosophila Chromosome 4 Genes Creates an Alternate Chromatin Structure Critical for Regulation in this Heterochromatic Domain
- Normal DNA Methylation Dynamics in DICER1-Deficient Mouse Embryonic Stem Cells
- The NDR Kinase Scaffold HYM1/MO25 Is Essential for MAK2 MAP Kinase Signaling in
- Functional Variants in and Involved in Activation of the NF-κB Pathway Are Associated with Rheumatoid Arthritis in Japanese
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



