-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaChromatin Remodeling in Development and Disease: Focus on CHD7
article has not abstract
Published in the journal: . PLoS Genet 6(7): e32767. doi:10.1371/journal.pgen.1001010
Category: Perspective
doi: https://doi.org/10.1371/journal.pgen.1001010Summary
article has not abstract
A central goal of developmental biology is to understand the mechanisms whereby undifferentiated, pluripotent cells differentiate into mature, organized cells and tissues. Recent advances in mouse genetics, embryonic stem cell manipulation, RNA inhibition, and genomics have now made it possible to explore these mechanisms with an unprecedented level of resolution. One major class of proteins, the chromodomain helicase DNA-binding (CHD) family of ATP-dependent chromatin remodelers, has emerged as important regulators of cellular differentiation. CHD proteins are thought to function in the nucleus via binding to DNA and regulating gene transcription. CHD7, a member of the CHD family, encodes a protein mutated in human CHARGE syndrome, a multiple anomaly disorder that affects hearing, vision, and cardiac, craniofacial, and nervous system development [1], [2]. Previous reports have shown tissue and developmental stage specific expression of CHD7 [3], [4], and there is evidence that CHD7 binds thousands of sites in the genome [5]. It is thus a major challenge to identify specific mechanisms of CHD7 action in pluripotency versus differentiation.
A study in this issue of PLoS Genetics [6] used genomics approaches to explore CHD7 binding sites and interacting partners in embryonic stem (ES) cells. The authors applied chromatin immunoprecipitation followed by next generation sequencing (ChIP-Seq) in ES cells under several different growth conditions. They found 10,483 chromatin sites bound by CHD7, most of which appear to be enhancer regions. CHD7 co-localized at these sites with potentially unique combinations of other DNA binding proteins, including p300, Oct4, Sox2, Nanog, Smad1, and STAT3 (see Table 1 for a list of genes and their official symbols; for the purposes of this Review, genes are referred to as cited in the original papers). In addition, genes directly regulated by CHD7 were subtly downregulated in ES cells, and these genes were far more ES cell–specific than either genes that did not change or those that decreased upon loss of CHD7. Despite the predominance of CHD7 binding sites in ES cell–specific genes, ES cell pluripotency, self-renewal, and reprogramming did not appear sensitive to CHD7 dosage. Thus, CHD7 seems to act as a rheostat of ES cell–specific gene expression without overtly controlling ES cell function.
Tab. 1. List of genes cited in the text and their official symbols. 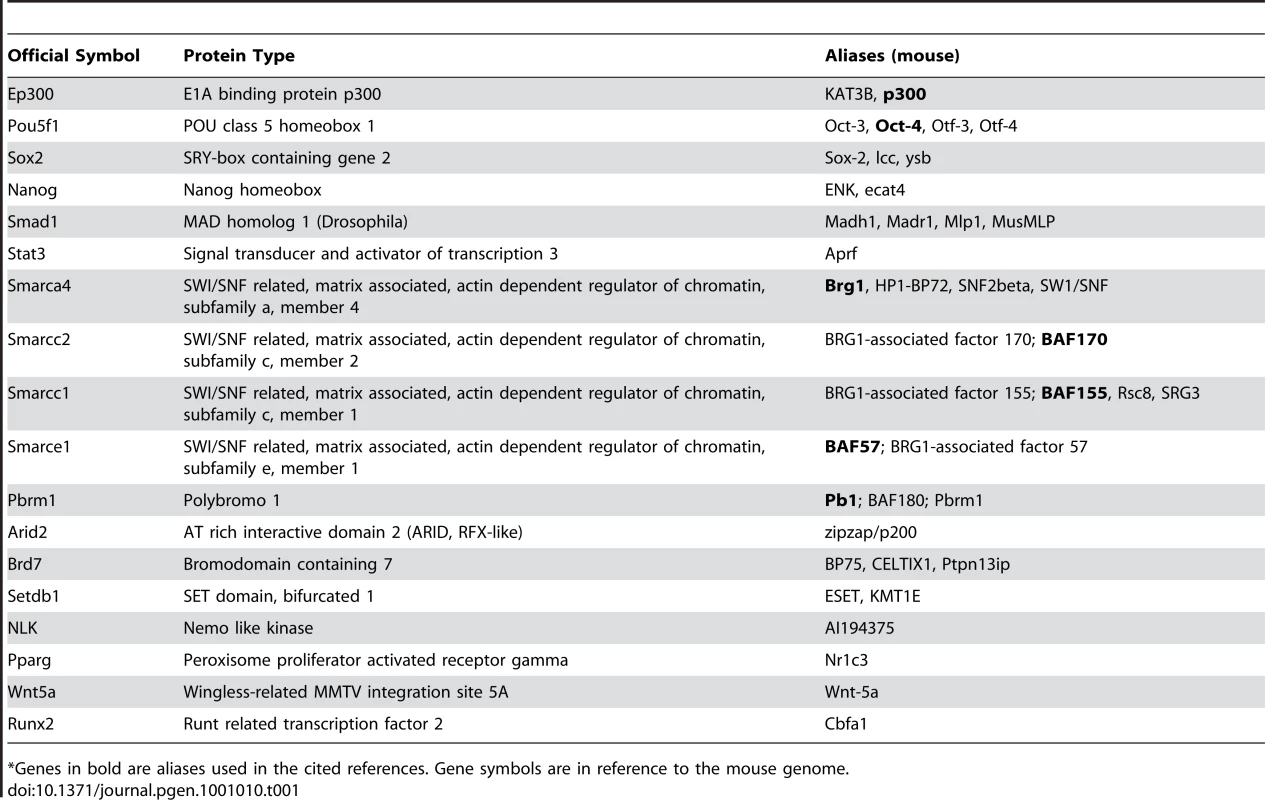
*Genes in bold are aliases used in the cited references. Gene symbols are in reference to the mouse genome. Like ES cells, neural crest–derived cells are pluripotent, since they differentiate into numerous cell types including craniofacial mesenchyme, heart, peripheral nervous system, and melanocytes. It has long been proposed that neural crest abnormalities contribute to the pathophysiology of CHARGE syndrome, but this had not been directly tested. In a related study, Bajpai et al. [7] explored CHD7 function in Xenopus, and found that CHD7 is vital for proper migration of neural crest in developing branchial arches. They also showed that human ES cells form a pluripotent population of cells that express markers of neural crest and can migrate to craniofacial mesenchyme and heart after transplantation into the chick neural tube. Migration of these neural crest–like cells (NCLCs) in vitro was also sensitive to CHD7 dosage. The authors concluded that CHD7 likely regulates cell migration in the developing neural crest.
Because CHD7 binds to thousands of enhancer sites in specific tissues during development, it is possible that each site binds a unique protein complex whose composition changes over developmental time. Identification and characterization of these complexes is thus akin to separating the wheat (binding sites and complexes that have functional significance) from the chaff (binding sites and complexes that have no functional significance). In the paper by Schnetz et al., the “wheat” appears to be a set of about six factors (Smad1, Nanog, Oct4, Sox2, Stat3, and p300) that co-localize with CHD7 (based on ChIP-Seq data) at specific DNA binding sites called multiple transcription factor loci or “MTLs” [6] (Figure 1). Schnetz et al. also identified overlap between sites occupied by BRG1 and CHD7 in ES cells; however, the binding was not as extensive as for the other six factors [6]. Earlier data showed that BRG1 associates with OCT4, SOX2, and NANOG in ES cells [8], providing further support for the idea that CHD7 binding in ES cells may be generally associated with positive and negative regulation. In the study by Bajpai et al., the “wheat” comprises the BAF and PBAF chromatin remodeling complexes that belong to the SWI/SNF family [7]. They found, by co-immunoprecipitation in human NCLCs, that CHD7 binds numerous PBAF specific subunits, including BRG1, BAF170, BAF155, BAF57, PB1, ARID2, and BRD7 (see Table 1 for official symbols). Interestingly, a previous report showed that CHD7 forms a complex with SETDB1, NLK, and PPAR-gamma that binds to DNA in a Wnt-5a responsive manner and promotes Runx2 transcription during osteoblast formation [9].
Fig. 1. Cell type–specific DNA binding complexes containing CHD7. 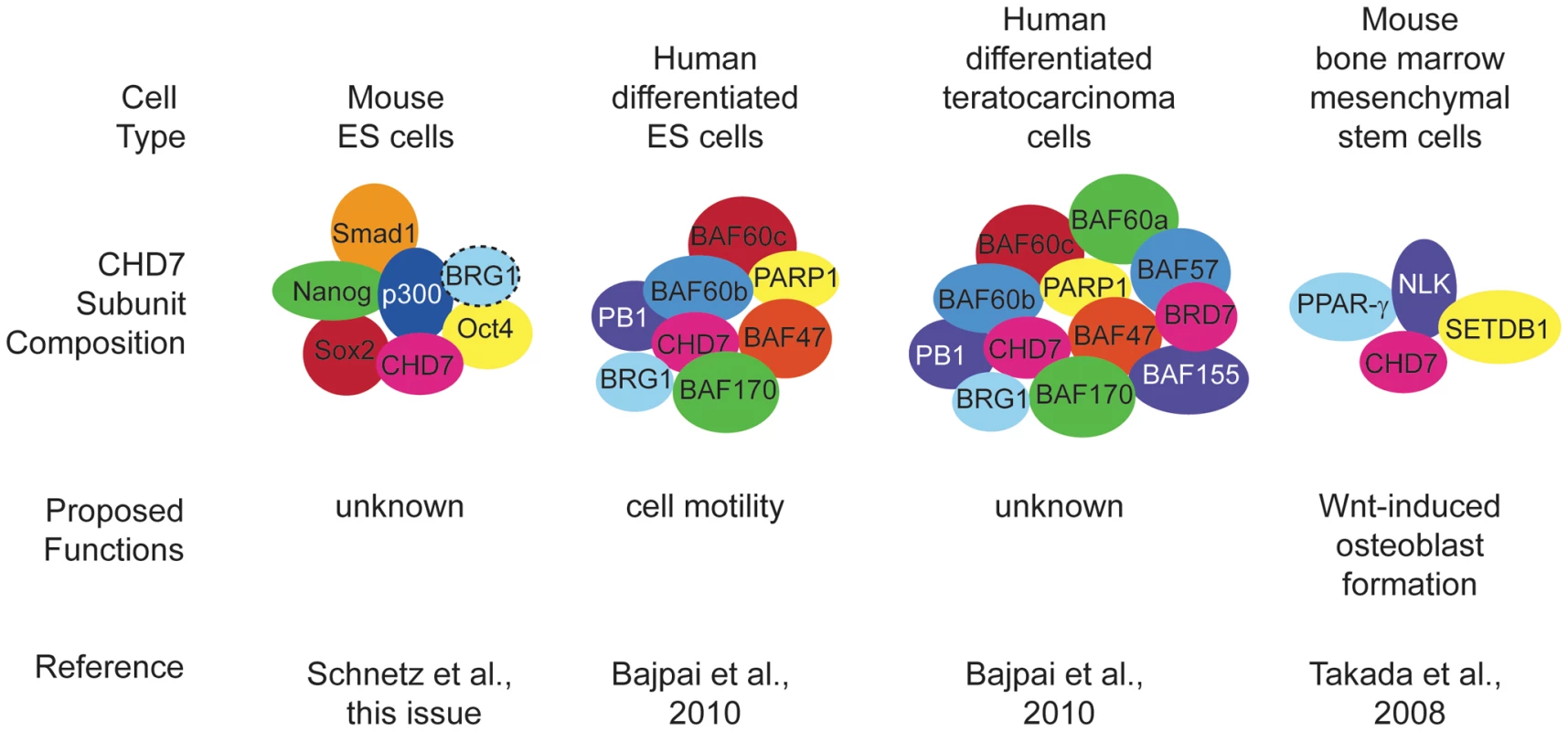
Proposed functions for CHD7 include cell motility and fate decisions. These functions appear to be unique to specific stem cell populations. Hatched lines around BRG1 in ES cells indicate that only some of the complexes have this protein as a component. It is interesting to compare these recent and earlier studies for evidence of CHD7 function in pluripotency versus cellular fate determination and differentiation. CHD7 does not appear to regulate pluripotency in mouse ES cells [6] or human NCLCs [7], but is required for normal proliferation of olfactory neural stem cells [10]. CHD7 also seems to promote one or more aspects of cellular differentiation. Binding of CHD7-containing complexes activates osteoblast fates over chondrocyte or myocyte differentiation of bone marrow mesenchymal stem cells [9] and promotes migration of neural crest cells in the branchial arches [7]. These observations suggest multiple roles for CHD7 in regulation of cellular progenitor proliferation and/or differentiation (Figure 2). Further investigation is necessary to clarify whether CHD7 has similar or different roles in the various cell types in which it is expressed.
Fig. 2. Cartoon schematic of CHD7 roles in cellular proliferation and/or differentiation. 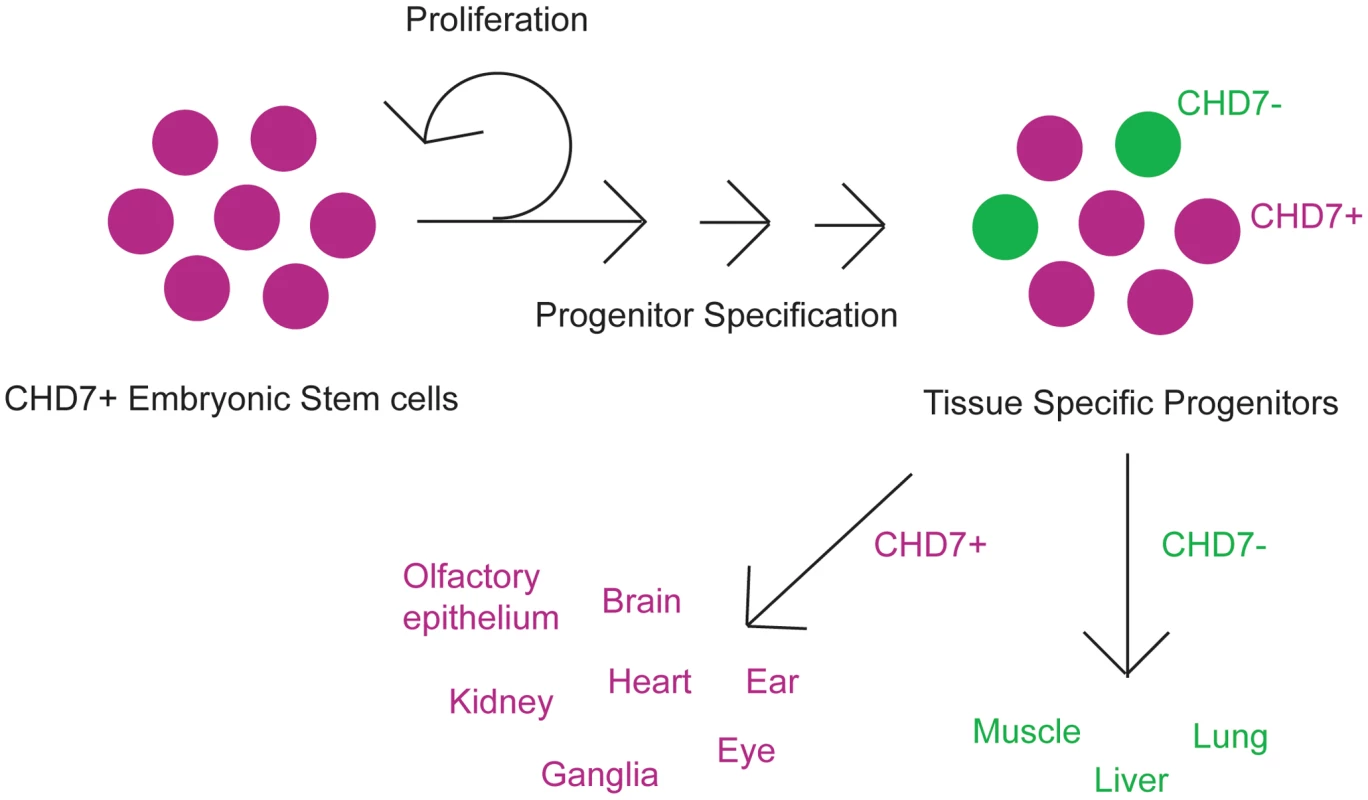
CHD7 is highly expressed in ES cells, and becomes restricted during development to tissue specific progenitor populations. These progenitors give rise to specific cell types in the various organs which are affected in CHARGE syndrome, including neural crest derivatives, central and peripheral nervous systems (ganglia), sensory organs (ear and eye), heart, and kidney. Some organs such as lung, liver, and skeletal muscle express no or very low levels of CHD7. CHD7 and related proteins likely have unique functions in human development. Along with CHD7 and the other eight CHD family members, there are about 30 mammalian genes encoding ATP-dependent chromatin remodeling proteins [11]. ATP-dependent chromatin remodelers differ from other chromatin modifiers (which control DNA methylation and histone acetylation) in that they are genetically non-redundant, haploinsufficient, and increase nucleosome mobility [11]. There are many human developmental disorders known to result from mutations in chromatin modifying proteins, including Rubinstein-Taybi (CBP and EP300), X-linked Alpha Thalassemia (ATRX), and Rett Syndrome (MECP2), among others [12]. Besides CHD7 and CHARGE syndrome, there are few developmental disorders known to result from mutations in ATP dependent remodeling genes. Loss of CHD5 may contribute to the microcephaly and physical features observed in Deletion 1p36 Syndrome [13], although its role is not yet established. Deletion of WSTF (a member of the ISWI class of ATP-dependent chromatin remodelers), along with several other genes in the critical interval on chromosome 7q11, results in Williams-Beuren syndrome, which presents with cardiac defects, craniofacial dysmorphisms, and developmental/cognitive impairments [14].
Accurate identification of CHD7 gene targets, interacting partners, binding sites, and complex subunit composition is going to require cooperative, detailed studies using high-throughput approaches and model organisms. One could argue that each cell type in a given tissue might have unique CHD7 binding sites and protein complexes that change over time. Finding these is a daunting task, and it seems more likely that common factors, targets, and molecular genetic pathways will emerge. It will be of great interest to know, for example, whether CHD7 regulates specific signaling pathways during organogenesis and whether those pathways can be manipulated by molecular intervention. In addition, there is no information yet on CHD7 protein domain-specific functions, over-expression phenotypes, or potential roles for CHD7 in cancer or aging. These topics are sure to occupy researchers for many years to come, and such efforts will likely allow us to harvest some wheat and make a nice, tasty loaf of bread.
Zdroje
1. VissersLE
van RavenswaaijCM
AdmiraalR
HurstJA
de VriesBB
2004 Mutations in a new member of the chromodomain gene family cause CHARGE syndrome. Nat Genet 36 955 957
2. ZentnerGE
LaymanWS
MartinDM
ScacheriPC
2010 Molecular and phenotypic aspects of CHD7 mutation in CHARGE syndrome. Am J Med Genet A 152A 674 686
3. BosmanEA
PennAC
AmbroseJC
KettleboroughR
StempleDL
2005 Multiple mutations in mouse Chd7 provide models for CHARGE syndrome. Hum Mol Genet 14 3463 3476
4. HurdEA
CapersPL
BlauwkampMN
AdamsME
RaphaelY
2007 Loss of Chd7 function in gene-trapped reporter mice is embryonic lethal and associated with severe defects in multiple developing tissues. Mamm Genome 18 94 104
5. SchnetzMP
BartelsCF
ShastriK
BalasubramanianD
ZentnerGE
2009 Genomic distribution of CHD7 on chromatin tracks H3K4 methylation patterns. Genome Res 19 590 601
6. SchnetzMP
HandokoL
Akhtar-ZaidiB
BartelsCF
PereiraCF
2010 CHD7 targets active gene enhancer elements to modulate ES cell-specific gene expression. PLoS Genet 6(7) e1001023 doi:10.1371/journal.pgen.1001023
7. BajpaiR
ChenDA
Rada-IglesiasA
ZhangJ
XiongY
2010 CHD7 cooperates with PBAF to control multipotent neural crest formation. Nature 463 958 962
8. HoL
JothiR
RonanJL
CuiK
ZhaoK
2009 An embryonic stem cell chromatin remodeling complex, esBAF, is an essential component of the core pluripotency transcriptional network. Proc Natl Acad Sci U S A 106 5187 5191
9. TakadaI
MiharaM
SuzawaM
OhtakeF
KobayashiS
2007 A histone lysine methyltransferase activated by non-canonical Wnt signalling suppresses PPAR-gamma transactivation. Nat Cell Biol 9 1273 1285
10. LaymanWS
McEwenDP
BeyerLA
LalaniSR
FernbachSD
2009 Defects in neural stem cell proliferation and olfaction in Chd7 deficient mice indicate a mechanism for hyposmia in human CHARGE syndrome. Hum Mol Genet 18 1909 1923
11. HoL
CrabtreeGR
2010 Chromatin remodelling during development. Nature 463 474 484
12. De SarioA
2009 Clinical and molecular overview of inherited disorders resulting from epigenomic dysregulation. Eur J Med Genet 52 363 372
13. BagchiA
PapazogluC
WuY
CapursoD
BrodtM
2007 CHD5 is a tumor suppressor at human 1p36. Cell 128 459 475
14. PeoplesRJ
CiscoMJ
KaplanP
FranckeU
1998 Identification of the WBSCR9 gene, encoding a novel transcriptional regulator, in the Williams-Beuren syndrome deletion at 7q11.23. Cytogenet Cell Genet 82 238 246
Štítky
Genetika Reprodukční medicína
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2010 Číslo 7
-
Všechny články tohoto čísla
- Question and Answer: An Anniversary Interview with Jane Gitschier
- Multi-Variant Pathway Association Analysis Reveals the Importance of Genetic Determinants of Estrogen Metabolism in Breast and Endometrial Cancer Susceptibility
- Tinkering Evolution of Post-Transcriptional RNA Regulons: Puf3p in Fungi as an Example
- The Importance of Imprinting in the Human Placenta
- Regulator of G Protein Signaling 3 Modulates Wnt5b Calcium Dynamics and Somite Patterning
- Lysosomal Dysfunction Promotes Cleavage and Neurotoxicity of Tau
- Combinatorial Binding Leads to Diverse Regulatory Responses: Lmd Is a Tissue-Specific Modulator of Mef2 Activity
- Variation, Sex, and Social Cooperation: Molecular Population Genetics of the Social Amoeba
- Comparative Analysis of DNA Replication Timing Reveals Conserved Large-Scale Chromosomal Architecture
- The Fitness Landscapes of -Acting Binding Sites in Different Promoter and Environmental Contexts
- Cohesin Is Limiting for the Suppression of DNA Damage–Induced Recombination between Homologous Chromosomes
- Genome-Wide Analysis Reveals Novel Genes Essential for Heme Homeostasis in
- Genome-Wide Meta-Analysis for Serum Calcium Identifies Significantly Associated SNPs near the Calcium-Sensing Receptor () Gene
- Rad3 Decorates Critical Chromosomal Domains with γH2A to Protect Genome Integrity during S-Phase in Fission Yeast
- Quantitative and Molecular Genetic Analyses of Mutations Increasing Life Span
- Association of Variants at with Chronic Kidney Disease and Kidney Stones—Role of Age and Comorbid Diseases
- Breast Cancer DNA Methylation Profiles Are Associated with Tumor Size and Alcohol and Folate Intake
- Calpain 8/nCL-2 and Calpain 9/nCL-4 Constitute an Active Protease Complex, G-Calpain, Involved in Gastric Mucosal Defense
- A Collection of Target Mimics for Comprehensive Analysis of MicroRNA Function in
- A Genome-Wide Analysis Reveals No Nuclear Dobzhansky-Muller Pairs of Determinants of Speciation between and , but Suggests More Complex Incompatibilities
- Microevolution of during Prolonged Infection of Single Hosts and within Families
- Id4, a New Candidate Gene for Senile Osteoporosis, Acts as a Molecular Switch Promoting Osteoblast Differentiation
- CHD7 Targets Active Gene Enhancer Elements to Modulate ES Cell-Specific Gene Expression
- Chromatin Remodeling in Development and Disease: Focus on CHD7
- Extensive DNA End Processing by Exo1 and Sgs1 Inhibits Break-Induced Replication
- Requirement of Male-Specific Dosage Compensation in Females—Implications of Early X Chromosome Gene Expression
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- CHD7 Targets Active Gene Enhancer Elements to Modulate ES Cell-Specific Gene Expression
- Extensive DNA End Processing by Exo1 and Sgs1 Inhibits Break-Induced Replication
- Question and Answer: An Anniversary Interview with Jane Gitschier
- Multi-Variant Pathway Association Analysis Reveals the Importance of Genetic Determinants of Estrogen Metabolism in Breast and Endometrial Cancer Susceptibility
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



