-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaA Collection of Target Mimics for Comprehensive Analysis of MicroRNA Function in
Many targets of plant microRNAs (miRNAs) are thought to play important roles in plant physiology and development. However, because plant miRNAs are typically encoded by medium-size gene families, it has often been difficult to assess their precise function. We report the generation of a large-scale collection of knockdowns for Arabidopsis thaliana miRNA families; this has been achieved using artificial miRNA target mimics, a recently developed technique fashioned on an endogenous mechanism of miRNA regulation. Morphological defects in the aerial part were observed for ∼20% of analyzed families, all of which are deeply conserved in land plants. In addition, we find that non-cleavable mimic sites can confer translational regulation in cis. Phenotypes of plants expressing target mimics directed against miRNAs involved in development were in several cases consistent with previous reports on plants expressing miRNA–resistant forms of individual target genes, indicating that a limited number of targets mediates most effects of these miRNAs. That less conserved miRNAs rarely had obvious effects on plant morphology suggests that most of them do not affect fundamental aspects of development. In addition to insight into modes of miRNA action, this study provides an important resource for the study of miRNA function in plants.
Published in the journal: . PLoS Genet 6(7): e32767. doi:10.1371/journal.pgen.1001031
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1001031Summary
Many targets of plant microRNAs (miRNAs) are thought to play important roles in plant physiology and development. However, because plant miRNAs are typically encoded by medium-size gene families, it has often been difficult to assess their precise function. We report the generation of a large-scale collection of knockdowns for Arabidopsis thaliana miRNA families; this has been achieved using artificial miRNA target mimics, a recently developed technique fashioned on an endogenous mechanism of miRNA regulation. Morphological defects in the aerial part were observed for ∼20% of analyzed families, all of which are deeply conserved in land plants. In addition, we find that non-cleavable mimic sites can confer translational regulation in cis. Phenotypes of plants expressing target mimics directed against miRNAs involved in development were in several cases consistent with previous reports on plants expressing miRNA–resistant forms of individual target genes, indicating that a limited number of targets mediates most effects of these miRNAs. That less conserved miRNAs rarely had obvious effects on plant morphology suggests that most of them do not affect fundamental aspects of development. In addition to insight into modes of miRNA action, this study provides an important resource for the study of miRNA function in plants.
Introduction
MicroRNAs (miRNAs) are a class of small RNA (sRNA) molecules that has recently emerged as a key regulator of gene activity. In plants, miRNAs are released from larger precursors (pri-miRNAs) in the nucleus mainly, by DICER-LIKE1 (DCL1) [1]. The resulting sRNA duplex is methylated and translocated to the cytoplasm where it can be loaded into an RNA-induced silencing complex (RISC) that includes a member of the ARGONAUTE (AGO) family as catalytic component. The RISC can then recognize mRNAs containing sequences complementary to the loaded miRNA [2]. In plants, cleavage of the target mRNA is an important mechanism for plant miRNA action, but there are also direct effects on protein accumulation, as reported for many animal miRNAs [3]–[11].
The spatio-temporal expression pattern of miRNA genes is regulated to a large extent at the transcriptional level, and different members of a miRNA family can have distinct, specialized expression domains [12]–[17]. An additional layer of regulation in miRNA action has been reported by Franco-Zorrilla and colleagues [18]. IPS1 (INDUCED BY PHOSPHATE STARVATION 1) encodes a non-coding RNA with a short motif that is highly complementary to the sequence of miR399, which like IPS1 is involved in the response to phosphate starvation [19]–[23]. In contrast to regular miRNA target sites, the IPS1 sequence contains a three-nucleotide insertion in the center, corresponding to the position where normally miRNA-guided cleavage takes place, and this bulge in the miRNA/target pair prevents endonucleolytic cleavage of IPS1 transcripts. This results in sequestration of RISCmiR399, leading to a reduction of miR399 activity. A similar phenomenon, negative regulation of small RNA activity by a partially complementary mRNA, has been recently described in bacteria as well [24], [25].
MiRNA target mimicry can be exploited to study the effects of reducing the function of entire miRNA families [18]. Simultaneous inactivation of all miRNA family members by constructing multiply mutant lines has so far been achieved for only two relatively small families [16], [26]. Plant target mimics are conceptually similar to miRNA sponges, used to reduce miRNA activity in animals. MiRNA sponges are transcripts containing multiple miRNA binding sites that compete with endogenous target mRNAs, thereby reducing the efficiency of the corresponding miRNA [27]. Although in animals perfect-match miRNA binding sites seems sufficient to sequester miRNAs [28], such optimal sites would be generally cleaved in plants, and they would not succeed in sequestering the miRNA-loaded RISC. Consistent with this, plants overexpressing non-modified versions of miR156 and miR319 target genes show much milder phenotypes than plants expressing the corresponding target mimics [18], [29], [30]. Modifications of the miRNA binding site that prevent cleavage but still allow miRNA binding are therefore required to reduce miRNA activity in plants.
Here, we present a collection of transgenic plants expressing artificial target mimics designed to knockdown the majority of Arabidopsis thaliana miRNA families. One fifth of these lines have obvious morphological defects, which is in the same range as the approximately 10% of miRNA knockouts that caused phenotypic abnormalities or lethality in Caenorhabditis elegans [31]. We found a clear correlation between the evolutionary conservation of plant miRNA families and their effect on aerial plant morphology.
Results/Discussion
Design of target mimics
We generated artificial target mimics for 73 different families or subfamilies of miRNAs and expressed them in Arabidopsis thaliana plants under the control of the constitutive 35S CaMV promoter. As described [18], we modified the 23 nucleotide, miR399-complementary motif in IPS1. The different constructs, and the corresponding transgenic lines, are named “MIM”, followed by the numeric identifier of the targeted miRNA family or subfamily. We targeted all miRNA families reported in miRBase (http://microrna.sanger.ac.uk/sequences/index.shtml) and ASRP (http://asrp.cgrb.oregonstate.edu) [32] at the beginning of 2007, plus some of the miRNAs described subsequently [33]. The majority of the analyzed families have only been described in Arabidopsis thaliana and Arabidopsis lyrata [34], [35]. The remaining families are shared with other angiosperms, and less than a quarter has been detected in non-flowering plants, including gymnosperms, ferns or mosses [32], [33], [36], [37]. A complete list of MIM constructs, and the primer pairs used to generate them, can be found in Table S1. For miRNA target predictions, see [8], [33], unless stated otherwise.
A single artificial target mimic could be designed for most miRNA families. The mature miRNAs produced by members of the miR169 and miR171 families differ slightly, and different target mimics were designed for these subfamilies. Two target mimics were also designed for the miR161 family, which produce two mature miRNAs that have only partially overlapping sequences, and that target similar subsets of the PPR gene family [38]. Conversely, some miRNA families have very similar sequences and overlapping in vivo targets (e.g., miR159/319, miR156/157 and miR170/171a), and artificial target mimics might not be able to unambiguously discriminate between different miRNAs.
In some cases, the sequence of the bulge in the miRNA/target mimic pair had to be modified. For example, maintaining the original central sequence of IPS1 in MIM172 could have reconstituted a cleavage site for miR172. Consistent with such modifications being important, plants expressing the appropriately modified version of MIM172 showed an altered phenotype (see below), whereas plants expressing an initial version of MIM172 in which a putative miR172 cleavage site was present (MIM172cs) did not. Moreover, plants expressing a MIM172 version with only a single-nucleotide mismatch corresponding to position 11 of the mature miRNA (MIM172sn) did not show any abnormal phenotype either, suggesting that the three-nucleotide bulge is required for target mimic activity (Figure 1).
Fig. 1. Requirement of a bulge at the cleavage site for target mimicry. 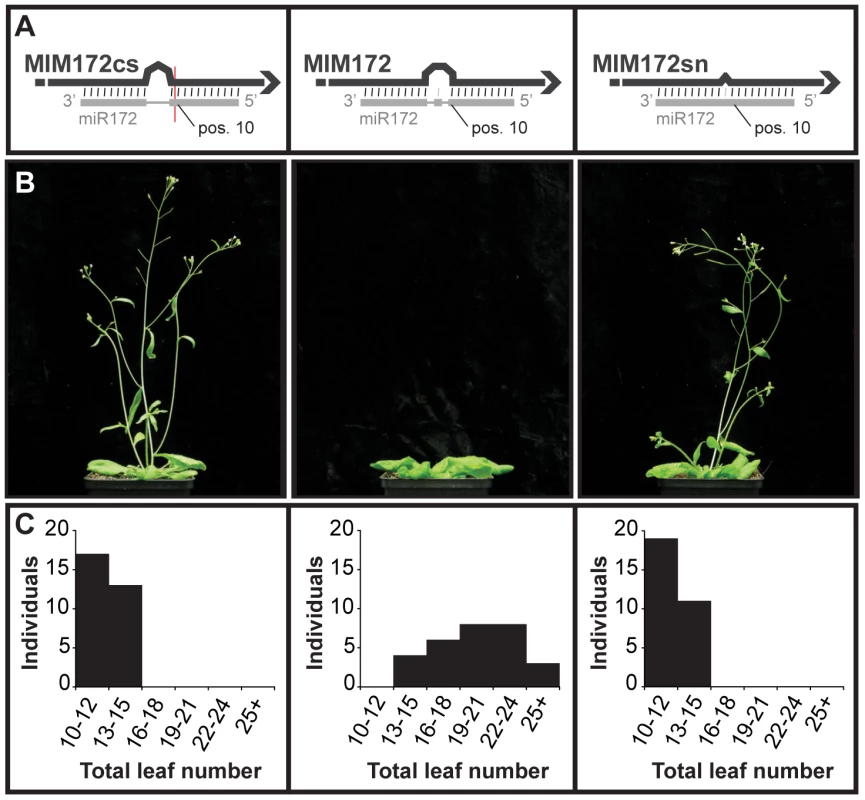
(A) A target mimic with an unmodified central sequence (MIM172cs), which retained complementarity to the central portion of miR172 across the cleavage site (red line) opposite position 10 to 11 of the miRNA, did not change flowering time. Modification of the central sequence (TCTA to GAGT; MIM172) restored a three nucleotide bulge found in IPS1 and generated a functional target mimic, causing a delay in flowering. However, a single nucleotide mismatch introduced into the center of an authentic miR172 target site (MIM172sn), but without a bulge, was not sufficient to reduce miR172 activity. (B) Four-week old plants grown at 23°C in long days. MIM172cs and MIM172sn are phenotypically indistinguishable from wild-type Col-0 plants (see also Figure S1B). (C) Distribution of flowering times of primary transformants grown in the same conditions; compare with Col-0 plants transformed with an empty binary vector in Figure S2. Effects of target mimics on morphology and development
We generated at least 20 independent transformants for each of 75 separate constructs. Of these, 15, targeting 14 different families, caused reproducible phenotypes in the shoot system of the plants, which are described below. Phenotypic alterations were consistent across most, if not all, independent transformants examined for each construct. An example of the phenotypic variation among primary transformants is shown in the histograms in Figure 1. An overview of all lines with morphological defects is given in Table 1, together with the main target genes of the corresponding miRNA family and a list of other taxa in which they can be found. The phenotypes of MIM156 and MIM319 plants have been briefly described before [18], [39]. All miRNA families whose inactivation resulted in visible phenotypical alterations are conserved among the angiosperms, and most of them are also found in non-flowering plants.
Tab. 1. Artificial target mimics causing visible phenotypes. 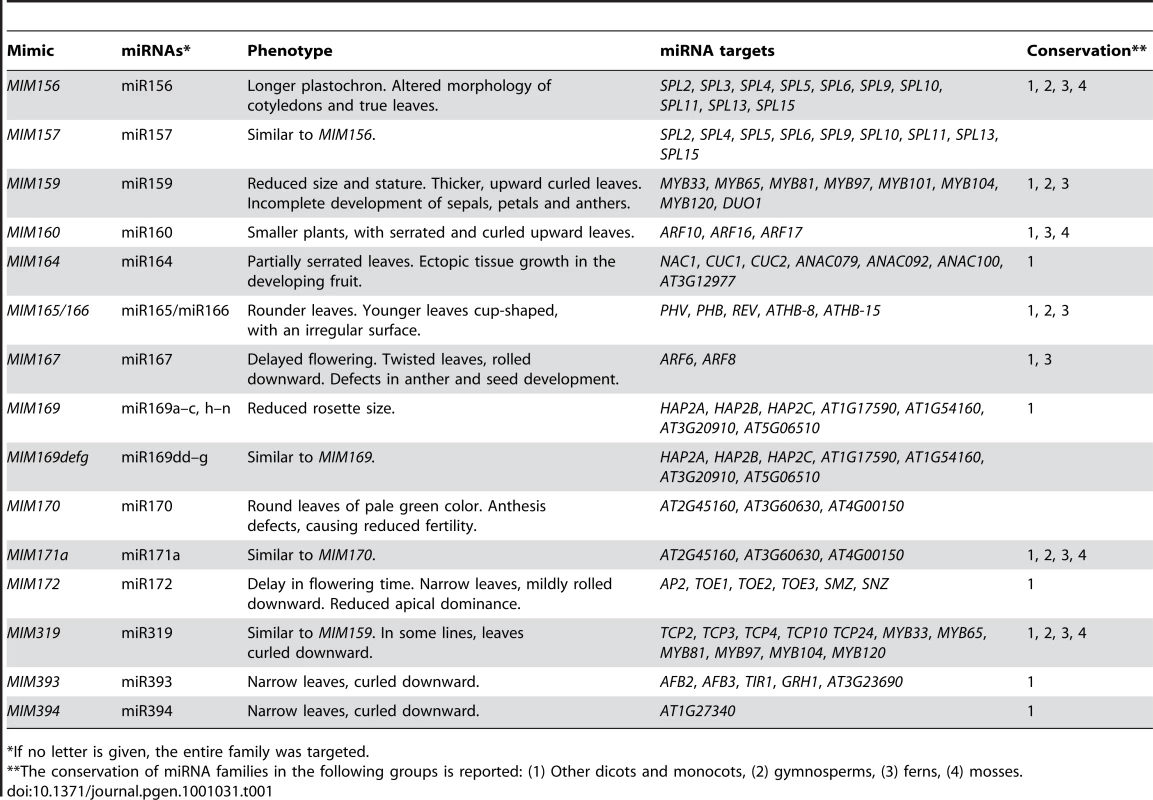
*If no letter is given, the entire family was targeted. MIM156 and MIM157 plants (Figure 2) had reduced leaf initiation rates, such that they flowered at about the same time as wild type, but with only two or three true leaves. This phenotype is similar to what is seen in plants carrying non-targetable versions of SPL9 or SPL10, two of the miR156/157 targets, and opposite of plants overexpressing miR156b or spl9 spl15 double mutants [10], [40]–[42]. In addition, these plants had bent, spoon-shaped cotyledons. The few rosette leaves were characterized by serrated margins, indicating adult leaf identity, consistent with a role of miR156 and its targets in controlling phase change [30].
Fig. 2. Leaf rosettes of target mimic expressing plants. 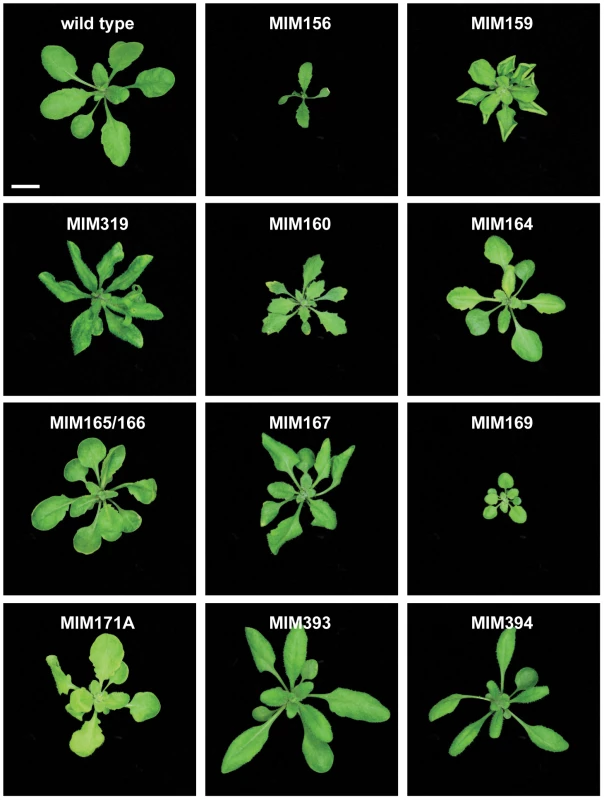
Three-week-old plants. Bar corresponds to 1 cm for all panels. MIM159 plants had extensive pleiotropic defects, and similar phenotypes were observed in most MIM319 lines. These plants had reduced stature, with rounder, upward curled leaves (Figure 2), shorter stem internodes, and smaller flowers with short sepals, reduced petals and anthers that did not develop completely. More severe MIM319 lines were progressively smaller, had warped leaves and lacked well-developed petals (Figure 3A). Stem elongation was often completely suppressed (Figure 3B). Most plants had reduced fertility, and this phenotype was particularly severe in MIM319 plants, for which only a few viable seeds could be recovered after they were grown for prolonged periods at 16°C in long days. Both vegetative and floral phenotypes reminiscent of MIM159 defects have been reported for plants that express non-targetable forms of miR159 target genes [29], and in plants doubly mutant for miR159a and miR159b [26]. In particular, upward curled leaves have been observed in plant expressing non-targetable forms of MYB33, which can be targeted both by miR159 and miR319 [43]. Milder MIM319 lines showed different leaf defects, with leaves curled downward (Figure 2). This is consistent with what has been reported for plants that express non-targetable forms of TCP2 and TCP4, which are both exclusive miR319 targets [29], suggesting that target mimics can at least partially discriminate between these two miRNA families.
Fig. 3. Details of defects observed in target mimic expressing plants. 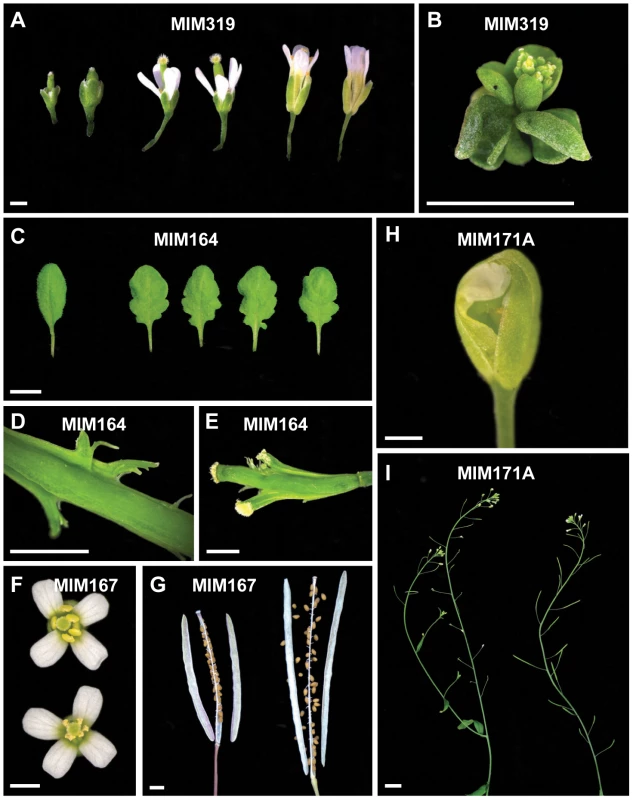
(A) Smaller flowers in severe MIM319 lines. The most strongly affected flowers lacked petals and did not have fully developed anthers (left side); in milder lines, flowers had short sepals, narrow petals, but were fertile (middle). Two flowers from wild type Col-0 are shown on the right side of the panel. (B) Severe MIM159 and MIM319 lines were very small and compact, without any stem elongation. (C) Leaves of MIM164 plants (compared to a leaf from wild type Col-0, on the far left). (D, E) Developing fruits of MIM164 with ectopic growths emanating from valve margins (D), which can develop into pseudo-pistils in severe lines (E). (F) Anthers in MIM167 lines did not mature completely (top), resulting in reduced pollen production (compared to a wild type Col-0 flower, bottom). (G) Seeds of MIM167 plants often do not fill completely, and remained attached to the dried silique (compared to a silique of wild type Col-0, on the right). (H, I) MIM171A lines suffered from defects in the separation of sepals, which prevented emergence of the pistil (H), and caused the plants to be mostly sterile (I, on the left, compared to a wild-type Col-0 plant, on the right). Bars correspond to 1 cm in (A–C) and I, and to 0.1 cm in (D–H). Serrated and hyponastic leaves were seen in MIM160 plants (Figure 2), in agreement with the phenotype of plants that express non-targetable versions of ARF10 or ARF17, two of the three miR160 targets [44], [45]. In addition, MIM160 plants were smaller than wild type. Compared to other constructs, fewer transformants were recovered, consistent with the known requirement of miR160 for seed viability or germination [44].
A different type of leaf serration was caused by MIM164 (Figure 2), similar to what has been reported for plants expressing a non-targetable version of CUC2, one of the miR164 targets, and for plants lacking one of the miR164 isoforms, miR164a [13]. While expression of MIM160 affected the entire leaf, with the serrations being regular and jagged, MIM164 caused mainly serration of the basal part of the leaf, with more irregular and rounded sinuses and teeth (Figure 3C). Although carpel fusion defects have been described for plants lacking miR164c [12], the carpel defects in MIM164 plants seemed to be different, with ectopic growths forming at the valve margins (Figure 3D), resembling those seen in the cuc2-1D mutant, in which a point mutation affects the miR164 complementary motif in CUC2 [46]. In some cases, this tissue could develop into adventitious pistil-like structures (Figure 3E).
Rounder leaves with an irregular surface, which appeared to be hollowed out between the main veins, were caused by MIM165/166. Younger leaves tended also to be cup-shaped (Figure 2). Targets of miR165/166, including the transcription factor-encoding genes PHAVOLUTA and PHABULOSA, control leaf polarity, and dominant mutations that disrupt the miRNA target site in these genes cause severe alterations in leaf morphology [47]–[49].
A substantial delay in flowering was observed in MIM167 plants, which flowered with 20.8±4.2 (mean ± standard deviation; n = 30) leaves in long days, compared to 13.0±0.9 rosette leaves in wild-type plants (Figure S1A and Figure S2). These plants had in addition twisted leaves (Figure 2), as well as defects in the maturation of anthers (Figure 3F) and in the development and shattering of seeds, which often remained attached to the dehiscent siliques (Figure 3G), resulting in reduced seed production and germination (not shown). This is consistent with what has been observed in plants that express a non-targetable form of the miR167 target ARF6 or ARF8. Such plants have smaller leaves and are often sterile due to defects both in ovule and anther development [17]. Effects on flowering time have not been previously associated with miR167 [17], [50], and the late-flowering phenotype of MIM167 plants reveals a new role for this miRNA family.
Two constructs were used to downregulate different subfamilies of miR169 family, whose main targets are HAP transcription factors. MIM169 was designed for miR169a, b, c, h, i, j, k, l, m and n, and MIM169defg for miR169d, e, f and g. Both target mimics reduced the size of transgenic plants (Figure 2).
MiR170 and miR171 target a group of SCARECROW-like transcription factor genes [9], and both MIM170 and MIM171A plants had round, pale leaves (Figure 2), as well as defective flowers, with sepals that did not separate properly, resulting in reduced fertility (Figure 3H and 3I). Expression of target mimics against the b and c members of the miR171 family did not confer any phenotype, suggesting less important roles for these two miRNAs.
MIM172 plants were also late flowering, with 20.0±3.5 (n = 30) rosette leaves in long days (Figure S1B), consistent with the flowering time phenotype of plants that have increased expression of miR172 targets [4], [6], [51]. In addition, leaves of MIM172 plants appeared to be somewhat narrower than those of wild type, and mildly curled downward, and severe MIM172 lines presented reduced apical dominance (not shown). In contrast to plants that express a non-targetable version of AP2 [52], flowers of MIM172 plants were normal. These differential effects could be due to the particularly high levels of miR172 levels during early flower development [6].
MiR393 targets a small group of auxin receptor genes. MIM393 plants had mild defects in leaf morphology, with narrow leaves that were curled downward (Figure 2). Leaf epinasty is often associated with high auxin levels [53], and is consistent with an increase of auxin signaling caused by downregulation of miR393 activity.
Finally, epinastic leaves were observed also in MIM394 plants (Figure 2). MiR394 is predicted to target a gene encoding an F-box protein.
Effects of target mimics on miRNA target genes
Artificial target mimics are thought to sequester their target miRNAs, presumably by stably binding to miRNA-loaded RISCs. To obtain additional evidence for such interactions, we embedded a functional MIM159 site in the 3′-UTR of a triple - Enhanced Yellow Fluorescent Protein (EYFP) reporter; stable recruitment of RISCmiR399 to the mimic site could be expected to interfere with EYFP translation. In 80% of MIM159 expressing T1 plants, as in control plants, the EYFP transgene was completely silenced. In the remaining 20%, we detected EYFP signal that was strongly reduced in the region where MIR159 genes are known to be expressed (Figure 4A) [26]. In addition, these plants presented the typical phenotypic defects of MIM159 plants, confirming that the EYFP:MIM159 construct functions properly as a target mimic.
Fig. 4. Effects of artificial mimics on levels of miRNAs and miRNA targets. 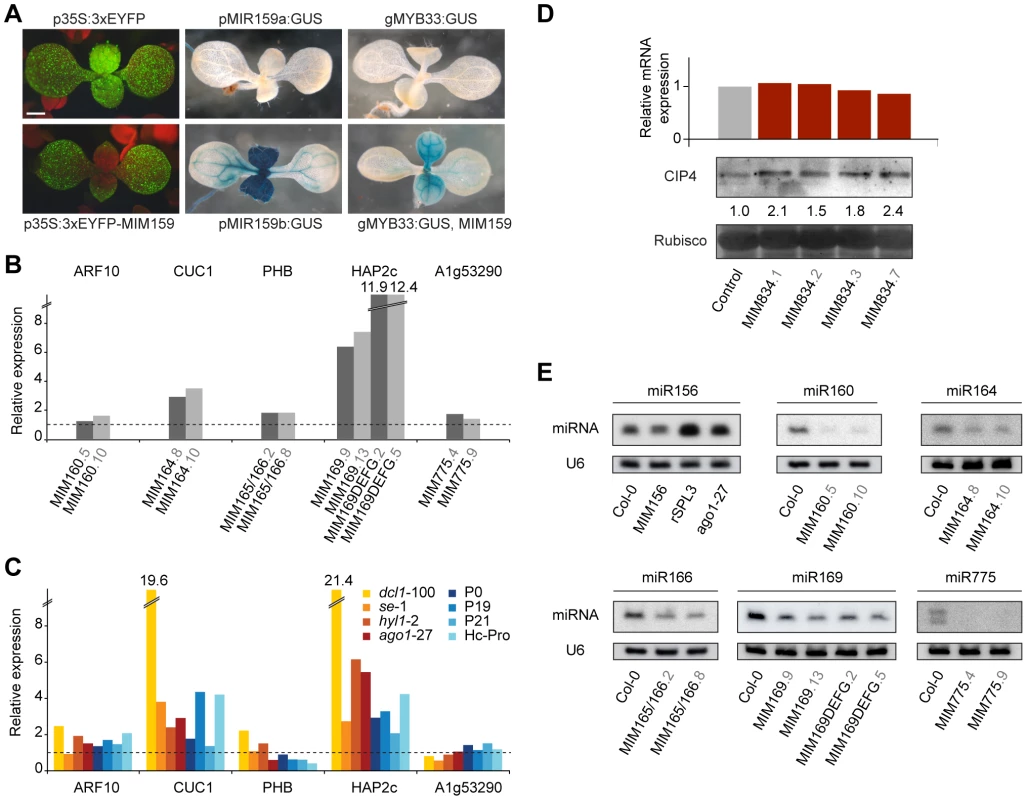
(A) Nine-day-old plants. Introduction of a MIM159 fragment into the 3′ UTR silences a constitutively expressed 3xEYFP in the MIR159 expression domain (compare p35S:3xEYFP and p35S:3xEYFP-MIM159), which is revealed in the pMIR159:GUS lines. MiR159 activity is also indirectly revealed by comparing the effect of expressing MIM159 in a genomic MYB33:GUS line. (B) Transcript levels of select miRNA targets in two independent lines for each MIM construct (represented by bars of different shades of gray). (C) Expression levels of miRNA targets in mutants impaired in miRNA biogenesis or targeting. Expression values are reported as the average of two biological and two technical replicates, and are normalized to the expression levels in wild type Col-0 plants (dotted line). (D) CIP4 mRNA and protein levels in four independent MIM834 lines. Band intensity relative to the wild-type control is reported. (E) Levels of mature miRNAs in several MIM lines. U6 accumulation is shown as control. Increased accumulation of miR156 (lower band in the blot) was observed upon expression of a resistant version of a miR156 target (consistent with what observed for miRNA156a precursor levels in [39]) or inhibition of miRNA activity in the ago1-27 mutants. The decrease in miR156 levels in MIM156 plants is then not an indirect consequence of increased SPL transcript levels. RISCmiRNA sequestration in turn should relieve target genes from miRNA-dependent regulation, resulting in increased levels of the encoded protein. In agreement with such a scenario, activity levels of a genomic MYB33:GUS reporter were markedly increased in MIM159 plants (Figure 4A). In analogy with EYFP:MIM159, reporter activity was increased in the tissues expressing MIR159 genes [26], as expected.
Sequestration of RISCmiR399 by the natural target mimic IPS1 prevents miR399-guided cleavage of PHO2 mRNA, thus increasing PHO2 mRNA levels [18]. To assess the effects of artificial target mimics on the levels of mRNA of miRNA target genes, we tested them by reverse transcription followed by quantitative PCR (qRT-PCR) in a subset of MIM lines. We preferentially analyzed organs in which miRNA abundance was high according to the ASRP database [32], [54], or organs with major phenotypic alterations in MIM lines. Two independent lines were tested for each construct. Among the miRNA targets, we chose ones known to induce phenotypic defects when expressed as non-targetable forms [44], [45], [47] and ones that show altered expression in miRNA biogenesis mutants [32], [54], [55]. PCR products spanned the miRNA target sequence, allowing quantification of the attenuation in slicing activity by the corresponding miRNA. Surprisingly, in most cases there were no major changes in target transcript levels (Figure 4B and Figure S3).
For comparison, we examined the expression of the same miRNA target genes in seedlings of several mutants impaired in small RNA biogenesis and function, including dcl1-100, se-1, hyl1-2 and ago1-27, and in plants overexpressing viral silencing suppressors that are known to counteract the action of the small RNA machinery, including P1/HC-Pro, P0, P19 and p21 [56]–[60]. In most cases, the changes seen in MIM lines correlated with those seen in miRNA biogenesis mutants. Stronger effects were observed only in dcl1-100 plants (Figure 4C). These results are consistent with what has been observed in microarray studies of miRNA biogenesis mutants, including other dcl1 alleles, se and hyl1 [55], .
As in animals, inhibition of translation is an important component of miRNA function in plants [4], [6], [11]. To test whether artificial mimics impact miRNA effects independent of changes in target transcript accumulation, we monitored the protein levels produced by CIP4, a gene that is regulated by miR834 through translational inhibition [5], [62]. In MIM834 lines, CIP4 levels were appreciably increased, while CIP4 mRNA levels were unchanged (Figure 4D). Direct effects on protein translation could explain the absence of a clear correlation between target mRNA levels and plant phenotype in plants expressing artificial target mimics.
Finally, we investigated the levels of mature miRNAs in plants expressing artificial target mimics. In all MIM lines we examined, levels of the targeted miRNA were decreased, suggesting that unproductive interaction of RISCmiRNA with a decoy affects miRNA stability (Figure 4E). Although such an effect has not been observed in case of the endogenous IPS1-miR399 interaction [18], a similar reduction in small RNA levels triggered by a target mimic has been reported in bacteria [24], [25].
Conclusions
We have generated a collection of transgenic plants expressing artificial target mimics designed to reduce activity for most of the known miRNA families in Arabidopsis thaliana. Inhibiting the function of 14 out of 71 miRNA families with target mimics led to morphological abnormalities. All of these families belong to the more abundant and widely conserved miRNA families, which were the first ones to be discovered (Table 1). This agrees with results from experiments in which miRNAs were overexpressed, miRNA target genes were mutated, or miRNA genes were inactivated by conventional knockouts [reviewed in 63]. Together, these findings are consistent with the scenario of frequent birth and death of miRNA genes, with only a few becoming fixed early on during evolution because they acquired a relevant function in plant development [33], [36]. More recently evolved, species-specific miRNAs could instead play a role in adaptation to certain abiotic or biotic challenges, or have no discernable function at all. Some miRNAs are known to regulate physiological traits, and they do not cause morphological abnormalities under standard benign conditions [20], [21], [64]. Such conditional effects would have escaped our screen, as would have defects in the root system of the plant. Moreover, compared to expression of non-targetable forms of miRNA target genes, or miRNA loss-of-function mutants, the defects of MIM plants were often weaker. Examples are the absence of an altered floral phenotype in MIM172 plants, which is seen in plants that express a non-targetable version of AP2 under the control of normal regulatory sequences [52], or the extra-petals phenotype seen in mir164c mutants, but not in MIM164 plants [12]. Another caveat is that some miRNAs might be required for embryonic development, in which case only lines with relatively weak expression of the artificial target mimic might have survived. Such limitations could be overcome by tissue-specific or inducible expression of target mimics. On the other hand, while artificial mimics increase levels of individual miRNA target genes less strongly than what can be achieved by expression of miRNA-resistant forms, mimics have the advantage that they affect all targets simultaneously [18]. Apart from translational regulation [3]–[7], another possibility for the absence of a clear correlation between phenotypic severity and change in mRNA levels of miRNA targets could be that many miRNAs affect their targets only in a small set of cells. In these cases, assaying expression in whole organs would obscure the effects of miRNA downregulation on mRNA levels.
It has recently been suggested that plant miRNAs could also repress the translation of target mRNAs that have only limited sequence complementarity, as often happens in animals [5]. Support for the existence of miRNA binding sites with reduced complementarity in plants comes from an analysis of miR398, which regulates COPPER SUPEROXIDE DISMUTASE (CSD) genes. Certain mutations in the miR398 complementary motif site reduced the effects of miR398 on CSD mRNA, but not on protein levels [3]. We have shown that mimic-like sites, when introduced into the 3′-UTR of a protein-coding gene, not only are active in sequestering the targeted miRNA, but can also reduce protein levels produced by the mRNA linked in cis. This reduction likely occurs at the translational level, since mimic sites are not subject to miRNA-dependent slicing [18]. This observation opens an intriguing scenario in which mRNAs containing mimic-like sites, or possibly other sites with reduced complementarity to miRNAs, are regulated by miRNAs exclusively through translational inhibition. A further level of complexity is added by such sites reducing the effects of an miRNA on other mRNA with a sliceable miRNA targeting motif, similarly to what has been recently proposed in animal systems [65].
Nevertheless, as pointed out before [43], miRNA overexpression and knockout of major target genes normally produce very similar phenotypes, and these are generally the opposite of what is seen in plants with reduced activity of the miRNA. These observations are supported by our finding of extensive similarities between phenotypes caused by target mimics and by expressing resistant forms of individual targets. We conclude that, at least for the instances in which developmental defects could be observed, target genes with extensive complementarity likely account for the majority of miRNA effects, but that in certain cases targets regulated solely through translational inhibition via diverged target sites might be important as well.
Materials and Methods
Plant material
Plants were grown on soil in long days (16 h light/8 hours dark) under a mixture of cool and warm white fluorescent light at 23°C and 65% humidity. The se-1, ago1-27, and hyl1-2 and dcl1-100 mutants have been described [66]–[69]. MIM834 plants were grown on MS media plates supplemented with 1% sucrose for 14 days in long days at 23°C. Plants overexpressing viral proteins Hc-Pro, P0, P19 and P21 were a kind gift from the Carrington lab.
Transgenic lines
Artificial target mimics were generated by modifying the sequence of the IPS1 gene [18]. All target mimics constructs were placed behind the constitutive CaMV 35S promoter in the pGREEN vector conferring resistance to BASTA [70]. For the MYB33:GUS reporter, a MYB33 genomic fragment was PCR amplified, cloned into the TOPO-PCR8 Gateway vector (Invitrogen), and recombined through LR clonase reaction into pGWB433 [71] to generate a GUS translational fusion. The MIM159 construct was introduced into three independent MYB33-GUS T2 lines. A MIM159 site was placed in the 3′-UTR of a triple-EYFP sequence linked to a fragment encoding a nuclear localization signal (NLS) and driven by a CaMV 35S promoter. Constructs were introduced into A. thaliana (accession Col-0) plants by Agrobacterium tumefaciens-mediated transformation [72].
Histochemical assays
Nine-day-old seedlings from three independent T2 lines for all the GUS reporter backgrounds were fixed in acetone 90%. GUS activity was assayed as described [73].
RNA analysis
Total RNA was extracted from 11-day old seedlings and 30-day old inflorescences (47 days for the MIM172 lines), using TRIzol Reagent (Invitrogen). For dcl-100, 13-day old seedlings were collected, to obtain a similar developmental stage compared to the other plants. For real time RT-PCR, two biological replicates with tissue pooled from 8 to 10 plants were assayed from two independent MIM lines per miRNA family or subfamily. Complementary DNA was produced with the RevertAid First Strand cDNA Synthesis Kit (Fermentas), using as starting material 4 µg of total RNA that had been treated with DNase I (Fermentas). PCR was carried out in presence of SYBR Green (Invitrogen) and monitored in real time with the Opticon Continuous Fluorescence Detection System (MJR). Oligonucleotide primers are given in Table S2. Small RNA blots were performed on the same RNA used as template for real time RT-PCR, with DNA oligonucleotides as probes.
Protein analysis
Proteins were extracted from four MIM834 lines using a Tris buffer (50 mM Tris pH 7,5; 150 mM NaCl; 1 mM EDTA; 10% [v/v] Glycerol; 1 mM DTT; 1 mM Pefablock and 1 complete protease inhibitor cocktail [Roche]). Protein concentration was measured using a commercial Bradford assay (BioRad). 50 µg of raw protein extract per sample were resolved on an 8% acrylamide gel. Blotting and antibody incubation were performed as described [5], except that the secondary antibody was incubated for 8 hours at 4°C. Two biological replicates from 4 independent lines were analyzed. Band intensity was measured using the ImageJ software (http://rsbweb.nih.gov/ij/).
Supporting Information
Zdroje
1. KuriharaY
WatanabeY
2004
Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions.
Proc Natl Acad Sci USA
101
12753
12758
2. RamachandranV
ChenX
2008
Small RNA metabolism in Arabidopsis.
Trends Plant Sci
13
368
374
3. DugasDV
BartelB
2008
Sucrose induction of Arabidopsis miR398 represses two Cu/Zn superoxide dismutases.
Plant Mol Biol
67
403
417
4. AukermanMJ
SakaiH
2003
Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes.
Plant Cell
15
2730
2741
5. BrodersenP
Sakvarelidze-AchardL
Bruun-RasmussenM
DunoyerP
YamamotoYY
2008
Widespread translational inhibition by plant miRNAs and siRNAs.
Science
320
1185
1190
6. ChenX
2004
A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development.
Science
303
2022
2025
7. GandikotaM
BirkenbihlRP
HohmannS
CardonGH
SaedlerH
2007
The miRNA156/157 recognition element in the 3′ UTR of the Arabidopsis SBP box gene SPL3 prevents early flowering by translational inhibition in seedlings.
Plant J
49
683
693
8. Jones-RhoadesMW
BartelDP
2004
Computational Identification of plant microRNAs and their targets, including a stress-induced miRNA.
Mol Cell
14
787
799
9. LlaveC
XieZ
KasschauKD
CarringtonJC
2002
Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA.
Science
297
2053
2056
10. SchwabR
PalatnikJF
RiesterM
SchommerC
SchmidM
2005
Specific effects of microRNAs on the plant transcriptome.
Dev Cell
8
517
527
11. VoinnetO
2009
Origin, biogenesis, and activity of plant microRNAs.
Cell
136
669
687
12. BakerCC
SieberP
WellmerF
MeyerowitzEM
2005
The early extra petals1 mutant uncovers a role for microRNA miR164c in regulating petal number in Arabidopsis.
Curr Biol
15
303
315
13. NikovicsK
BleinT
PeaucelleA
IshidaT
MorinH
2006
The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis.
Plant Cell
18
2929
2945
14. NogueiraFT
ChitwoodDH
MadiS
OhtsuK
SchnablePS
2009
Regulation of small RNA accumulation in the maize shoot apex.
PLoS Genet
5
e1000320
doi:10.1371/journal.pgen.1000320
15. RamanS
GrebT
PeaucelleA
BleinT
LaufsP
2008
Interplay of miR164, CUP-SHAPED COTYLEDON genes and LATERAL SUPPRESSOR controls axillary meristem formation in Arabidopsis thaliana.
Plant J
55
65
76
16. SieberP
WellmerF
GheyselinckJ
RiechmannJL
MeyerowitzEM
2007
Redundancy and specialization among plant microRNAs: role of the MIR164 family in developmental robustness.
Development
134
1051
1060
17. WuMF
TianQ
ReedJW
2006
Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction.
Development
133
4211
4218
18. Franco-ZorrillaJM
ValliA
TodescoM
MateosI
PugaMI
2007
Target mimicry provides a new mechanism for regulation of microRNA activity.
Nat Genet
39
1033
1037
19. BurleighSH
HarrisonMJ
1999
The down-regulation of Mt4-like genes by phosphate fertilization occurs systemically and involves phosphate translocation to the shoots.
Plant Physiol
119
241
248
20. ChiouTJ
AungK
LinSI
WuCC
ChiangSF
2006
Regulation of phosphate homeostasis by microRNA in Arabidopsis.
Plant Cell
18
412
421
21. FujiiH
ChiouTJ
LinSI
AungK
ZhuJK
2005
A miRNA involved in phosphate-starvation response in Arabidopsis.
Curr Biol
15
2038
2043
22. MartinAC
del PozoJC
IglesiasJ
RubioV
SolanoR
2000
Influence of cytokinins on the expression of phosphate starvation responsive genes in Arabidopsis.
Plant J
24
559
567
23. ShinH
ShinHS
ChenR
HarrisonMJ
2006
Loss of At4 function impacts phosphate distribution between the roots and the shoots during phosphate starvation.
Plant J
45
712
726
24. Figueroa-BossiN
ValentiniM
MalleretL
FioriniF
BossiL
2009
Caught at its own game: regulatory small RNA inactivated by an inducible transcript mimicking its target.
Genes Dev
23
2004
2015
25. OvergaardM
JohansenJ
Møller-JensenJ
Valentin-HansenP
2009
Switching off small RNA regulation with trap-mRNA.
Mol Microbiol
73
790
800
26. AllenRS
LiJ
StahleMI
DubroueA
GublerF
2007
Genetic analysis reveals functional redundancy and the major target genes of the Arabidopsis miR159 family.
Proc Natl Acad Sci USA
104
16371
16376
27. EbertMS
NeilsonJR
SharpPA
2007
MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells.
Nat Methods
4
721
726
28. GentnerB
SchiraG
GiustacchiniA
AmendolaM
BrownBD
2009
Stable knockdown of microRNA in vivo by lentiviral vectors.
Nat Methods
6
63
66
29. PalatnikJF
AllenE
WuX
SchommerC
SchwabR
2003
Control of leaf morphogenesis by microRNAs.
Nature
425
257
263
30. WuG
PoethigRS
2006
Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3.
Development
133
3539
3547
31. MiskaEA
Alvarez-SaavedraE
AbbottAL
LauNC
HellmanAB
2007
Most Caenorhabditis elegans microRNAs are individually not essential for development or viability.
PLoS Genet
3
e215
doi:10.1371/journal.pgen.0030215
32. GustafsonAM
AllenE
GivanS
SmithD
CarringtonJC
2005
ASRP: the Arabidopsis Small RNA Project Database.
Nucleic Acids Res
33
D637
640
33. RajagopalanR
VaucheretH
TrejoJ
BartelDP
2006
A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana.
Genes Dev
20
3407
3425
34. MaZ
CoruhC
AxtellMJ
2010
Arabidopsis lyrata small RNAs: transient MIRNA and small interfering RNA loci within the Arabidopsis genus.
Plant Cell 22:ePub April 22, 2010
35. FahlgrenN
JogdeoS
KasschauKD
SullivanCM
ChapmanEJ
2010
MicroRNA gene evolution in Arabidopsis lyrata and Arabidopsis thaliana.
Plant Cell 22:ePub April 22, 2010
36. FahlgrenN
HowellMD
KasschauKD
ChapmanEJ
SullivanCM
2007
High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes.
PLoS ONE
2
e219
doi:10.1371/journal.pone.0000219
37. AxtellMJ
BartelDP
2005
Antiquity of microRNAs and their targets in land plants.
Plant Cell
17
1658
1673
38. HowellMD
FahlgrenN
ChapmanEJ
CumbieJS
SullivanCM
2007
Genome-wide analysis of the RNA-DEPENDENT RNA POLYMERASE6/DICER-LIKE4 pathway in Arabidopsis reveals dependency on miRNA - and tasiRNA-directed targeting.
Plant Cell
19
926
942
39. WuG
ParkMY
ConwaySR
WangJW
WeigelD
2009
The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis.
Cell
138
750
759
40. SchwarzS
GrandeAV
BujdosoN
SaedlerH
HuijserP
2008
The microRNA regulated SBP-box genes SPL9 and SPL15 control shoot maturation in Arabidopsis.
Plant Mol Biol
67
183
195
41. WangJW
SchwabR
CzechB
MicaE
WeigelD
2008
Dual effects of miR156-targeted SPL genes and CYP78A5/KLUH on plastochron length and organ size in Arabidopsis thaliana.
Plant Cell
20
1231
1243
42. WangJW
CzechB
WeigelD
2009
miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana.
Cell
138
738
749
43. PalatnikJF
WollmannH
SchommerC
SchwabR
BoisbouvierJ
2007
Sequence and expression differences underlie functional specialization of Arabidopsis microRNAs miR159 and miR319.
Dev Cell
13
115
125
44. LiuPP
MontgomeryTA
FahlgrenN
KasschauKD
NonogakiH
2007
Repression of AUXIN RESPONSE FACTOR10 by microRNA160 is critical for seed germination and post-germination stages.
Plant J
52
133
146
45. MalloryAC
BartelDP
BartelB
2005
MicroRNA-directed regulation of Arabidopsis AUXIN RESPONSE FACTOR17 is essential for proper development and modulates expression of early auxin response genes.
Plant Cell
17
1360
1375
46. LarueCT
WenJ
WalkerJC
2009
A microRNA-transcription factor module regulates lateral organ size and patterning in Arabidopsis.
Plant J
58
450
63
47. MalloryAC
ReinhartBJ
Jones-RhoadesMW
TangG
ZamorePD
2004
MicroRNA control of PHABULOSA in leaf development: importance of pairing to the microRNA 5′ region.
EMBO J
23
3356
3364
48. McConnellJR
EmeryJ
EshedY
BaoN
BowmanJ
2001
Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots.
Nature
411
709
713
49. OchandoI
Jover-GilS
RipollJJ
CandelaH
VeraA
2006
Mutations in the microRNA complementarity site of the INCURVATA4 gene perturb meristem function and adaxialize lateral organs in Arabidopsis.
Plant Physiol
141
607
619
50. RuP
XuL
MaH
HuangH
2006
Plant fertility defects induced by the enhanced expression of microRNA167.
Cell Res
16
457
465
51. SchmidM
UhlenhautNH
GodardF
DemarM
BressanR
2003
Dissection of floral induction pathways using global expression analysis.
Development
130
6001
6012
52. ZhaoL
KimY
DinhTT
ChenX
2007
miR172 regulates stem cell fate and defines the inner boundary of APETALA3 and PISTILLATA expression domain in Arabidopsis floral meristems.
Plant J
51
840
849
53. RomanoCP
CooperML
KleeHJ
1993
Uncoupling auxin and ethylene effects in transgenic tobacco and Arabidopsis plants.
Plant Cell
5
181
189
54. BackmanTW
SullivanCM
CumbieJS
MillerZA
ChapmanEJ
2007
Update of ASRP: the Arabidopsis Small RNA Project database.
Nucleic Acids Res
36
D982
D985
55. AllenE
XieZ
GustafsonAM
CarringtonJC
2005
microRNA-directed phasing during trans-acting siRNA biogenesis in plants.
Cell
121
207
221
56. BaumbergerN
TsaiCH
LieM
HaveckerE
BaulcombeDC
2007
The polerovirus silencing suppressor P0 targets ARGONAUTE proteins for degradation.
Curr Biol
17
1609
1614
57. BortolamiolD
PazhouhandehM
MarroccoK
GenschikP
Ziegler-GraffV
2007
The polerovirus F box protein P0 targets ARGONAUTE1 to suppress RNA silencing.
Curr Biol
17
1615
1621
58. ChapmanEJ
ProkhnevskyAI
GopinathK
DoljaVV
CarringtonJC
2004
Viral RNA silencing suppressors inhibit the microRNA pathway at an intermediate step.
Genes Dev
18
1179
1186
59. DunoyerP
LecellierCH
ParizottoEA
HimberC
VoinnetO
2004
Probing the microRNA and small interfering RNA pathways with virus-encoded suppressors of RNA silencing.
Plant Cell
16
1235
1250
60. PfefferS
DunoyerP
HeimF
RichardsKE
JonardG
2002
P0 of beet Western yellows virus is a suppressor of posttranscriptional gene silencing.
J Virol
76
6815
6824
61. RonemusM
VaughnMW
MartienssenRA
2006
MicroRNA-targeted and small interfering RNA-mediated mRNA degradation is regulated by argonaute, dicer, and RNA-dependent RNA polymerase in Arabidopsis.
Plant Cell
18
1559
1574
62. LanetE
DelannoyE
SormaniR
FlorisM
BrodersenP
2009
Biochemical evidence for translational repression by Arabidopsis microRNAs.
Plant Cell
21
1762
1768
63. WillmannMR
PoethigRS
2007
Conservation and evolution of miRNA regulatory programs in plant development.
Curr Opin Plant Biol
10
503
511
64. SunkarR
KapoorA
ZhuJK
2006
Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance.
Plant Cell
18
2051
2065
65. SeitzH
2009
Redefining microRNA targets.
Curr Biol
19
870
873
66. LaubingerS
SachsenbergT
ZellerG
BuschW
LohmannJU
2008
Dual roles of the nuclear cap-binding complex and SERRATE in pre-mRNA splicing and microRNA processing in Arabidopsis thaliana.
Proc Natl Acad Sci USA
105
8795
8800
67. MorelJB
GodonC
MourrainP
BéclinC
BoutetS
2002
Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance.
Plant Cell
14
629
639
68. RedeíGP
1965
Non-Mendelian megagametogenesis in Arabidopsis.
Genetics
51
857
872
69. VazquezF
GasciolliV
CrétéP
VaucheretH
2004
The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing.
Curr Biol
14
346
351
70. HellensRP
EdwardsEA
LeylandNR
BeanS
MullineauxPM
2000
pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation.
Plant Mol Biol
42
819
832
71. NakagawaT
SuzukiT
MurataS
NakamuraS
HinoT
2007
Improved Gateway binary vectors: high-performance vectors for creation of fusion constructs in transgenic analysis of plants.
Biosci Biotechnol Biochem
71
2095
2100
72. WeigelD
GlazebrookJ
2002
Arabidopsis: A Laboratory Manual.
Cold Spring Harbor, NY
Cold Spring Harbor Laboratory Press.
354
73. BlázquezMA
SoowalL
LeeI
WeigelD
1997
LEAFY expression and flower initiation in Arabidopsis.
Development
124
3835
3844
Štítky
Genetika Reprodukční medicína
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2010 Číslo 7
-
Všechny články tohoto čísla
- Question and Answer: An Anniversary Interview with Jane Gitschier
- Multi-Variant Pathway Association Analysis Reveals the Importance of Genetic Determinants of Estrogen Metabolism in Breast and Endometrial Cancer Susceptibility
- Tinkering Evolution of Post-Transcriptional RNA Regulons: Puf3p in Fungi as an Example
- The Importance of Imprinting in the Human Placenta
- Regulator of G Protein Signaling 3 Modulates Wnt5b Calcium Dynamics and Somite Patterning
- Lysosomal Dysfunction Promotes Cleavage and Neurotoxicity of Tau
- Combinatorial Binding Leads to Diverse Regulatory Responses: Lmd Is a Tissue-Specific Modulator of Mef2 Activity
- Variation, Sex, and Social Cooperation: Molecular Population Genetics of the Social Amoeba
- Comparative Analysis of DNA Replication Timing Reveals Conserved Large-Scale Chromosomal Architecture
- The Fitness Landscapes of -Acting Binding Sites in Different Promoter and Environmental Contexts
- Cohesin Is Limiting for the Suppression of DNA Damage–Induced Recombination between Homologous Chromosomes
- Genome-Wide Analysis Reveals Novel Genes Essential for Heme Homeostasis in
- Genome-Wide Meta-Analysis for Serum Calcium Identifies Significantly Associated SNPs near the Calcium-Sensing Receptor () Gene
- Rad3 Decorates Critical Chromosomal Domains with γH2A to Protect Genome Integrity during S-Phase in Fission Yeast
- Quantitative and Molecular Genetic Analyses of Mutations Increasing Life Span
- Association of Variants at with Chronic Kidney Disease and Kidney Stones—Role of Age and Comorbid Diseases
- Breast Cancer DNA Methylation Profiles Are Associated with Tumor Size and Alcohol and Folate Intake
- Calpain 8/nCL-2 and Calpain 9/nCL-4 Constitute an Active Protease Complex, G-Calpain, Involved in Gastric Mucosal Defense
- A Collection of Target Mimics for Comprehensive Analysis of MicroRNA Function in
- A Genome-Wide Analysis Reveals No Nuclear Dobzhansky-Muller Pairs of Determinants of Speciation between and , but Suggests More Complex Incompatibilities
- Microevolution of during Prolonged Infection of Single Hosts and within Families
- Id4, a New Candidate Gene for Senile Osteoporosis, Acts as a Molecular Switch Promoting Osteoblast Differentiation
- CHD7 Targets Active Gene Enhancer Elements to Modulate ES Cell-Specific Gene Expression
- Chromatin Remodeling in Development and Disease: Focus on CHD7
- Extensive DNA End Processing by Exo1 and Sgs1 Inhibits Break-Induced Replication
- Requirement of Male-Specific Dosage Compensation in Females—Implications of Early X Chromosome Gene Expression
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- CHD7 Targets Active Gene Enhancer Elements to Modulate ES Cell-Specific Gene Expression
- Extensive DNA End Processing by Exo1 and Sgs1 Inhibits Break-Induced Replication
- Question and Answer: An Anniversary Interview with Jane Gitschier
- Multi-Variant Pathway Association Analysis Reveals the Importance of Genetic Determinants of Estrogen Metabolism in Breast and Endometrial Cancer Susceptibility
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



