-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaIs Required for High Efficiency Viral Replication
Viruses often can highly efficiently utilize the host system to make huge amounts of viral proteins for their replication; however, which host factors are needed by the viruses are largely unknown. We analyzed about one hundred Drosophila mutants and found that the pelo mutation in Drosophila inhibited Drosophila C virus (DCV) replication. We found pelo is specifically required for high efficiency synthesis of proteins of a number of viruses, suggesting that inhibition of pelo may mediate a general antiviral activity. We proposed that the function of pelo in quality control of protein synthesis is required for high efficiency viral protein synthesis. Our study presented here identifies a new host factor, pelo, that is specifically required for effective viral replication and it may be a new potential therapeutic target for broad-spectrum antiviral therapy.
Published in the journal: . PLoS Pathog 10(4): e32767. doi:10.1371/journal.ppat.1004034
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1004034Summary
Viruses often can highly efficiently utilize the host system to make huge amounts of viral proteins for their replication; however, which host factors are needed by the viruses are largely unknown. We analyzed about one hundred Drosophila mutants and found that the pelo mutation in Drosophila inhibited Drosophila C virus (DCV) replication. We found pelo is specifically required for high efficiency synthesis of proteins of a number of viruses, suggesting that inhibition of pelo may mediate a general antiviral activity. We proposed that the function of pelo in quality control of protein synthesis is required for high efficiency viral protein synthesis. Our study presented here identifies a new host factor, pelo, that is specifically required for effective viral replication and it may be a new potential therapeutic target for broad-spectrum antiviral therapy.
Introduction
Viruses are the most abundant intracellular pathogens on the earth. They can infect all living organisms and hijack their host factors for replication [1]. In order to withstand virus infections, their hosts have evolved multiple antiviral defense mechanisms [2], [3], [4]. For many years, scientists have been studying host-virus interactions in order to develop new and more effective strategies for the prevention and treatment of viral infection.
Drosophila melanogaster has been shown to be a powerful model system in studying host-pathogen interactions [5]. In addition to the widely appreciated achievement in studying antibacterial and antifungal immunity by using Drosophila [6], there is also growing understanding of Drosophila-virus interactions [7], [8]. Several antiviral innate immunity pathways and their corresponding molecular mechanisms have been deciphered in Drosophila. RNA interference is the major antiviral pathway in Drosophila [9]. Similar to plants, Drosophila can generate both a local and a systemic antiviral RNAi response [10]. However, viruses can counteract host RNAi defense by expressing viral suppressors of RNAi (VSRs), such as FHV-B2, DCV-1A, and Crpv-A [11], [12], [13], [14]. Inducible gene expressions in response to viral infection also contribute to antiviral immunity, including the JAK-STAT pathway and the DExD/H-box helicase Dicer-2-mediated antiviral gene induction [15], [16]. The Toll and IMD pathways are involved in restricting some specific viruses by mechanisms yet to be clarified [17], [18]. Autophagy plays an important antiviral role against the vesicular stomatitis virus (VSV) in Drosophila, which is initiated by Toll-7 after its recognition of the VSV glycoprotein VSV-G [19], [20]. It is apparent that some antiviral mechanisms are conserved between Drosophila and mammals but others are uniquely present in Drosophila.
Hijacking host cellular machineries for viral replication is another major part of host-virus interaction [1], [21]. Studies in Drosophila have revealed that the clathrin-mediated endocytotic pathway is required for viral entry [22], some ribosomal proteins are involved in viral IRES-dependent translation [23], and the coat protein complex I (COPI) coatamer and fatty acid biosynthesis are required to form the intracellular vesicular compartment for viral replication [24]. All of these appear to be commonly involved in host-virus interaction of many different viruses. Targeting these events might be able to interfere with the replication of a broad panel of viruses; however, achieving the goal of virus inhibition without affecting normal cell function is a challenging task. Without exception, viruses have to use cellular protein translation machinery to synthesize their proteins. In many cases, viruses take over the hosts' protein synthesis machinery to make huge amounts of viral proteins for their replication [25]. How can viruses so highly efficiently utilize the cellular system to synthesize their proteins is largely unknown. Information on the cellular factors that are required for high speed synthesis of viral proteins is very limited.
Protein translation is a tightly controlled cellular process, and also is a part of the checking mechanism that eliminates aberrant transcripts and proteins [26]. The pelo-Hbs1 complex (also known as Dom34-Hbs1) recognizes stalled ribosomes caused by defective mRNAs as well as rRNAs and promotes ribosomal subunit dissociation and the release of peptidyl-tRNA [27], [28]. Therefore, the complex participates in quality control for non-go decay (NGD) and non-stop decay (NSD) [29], [30], [31]. ABCE1, a conserved member of the ATP-binding cassette (ABC) family of proteins, is involved in pelo-Hbs1 mediated disassembly of the ribosome in mammalian cells [32]. Recent work has shown that Dom34-Hbs1 is also required for nonstop protein clearance from translocators for normal organelle protein influx [33]. pelo is a highly conserved gene from yeast to human. It contains three eRF1 (eukaryotic translation termination factor 1) domains. X-ray structural analysis of yeast Dom34 and the archaea homolog Pelota reveals that the structure of pelo is similar to eRF1 except for its N-terminal domain [34], [35]. Deletion of Dom34 in yeast led to delayed progression through the G1 phase of the cell cycle, aberrant meiosis, and an altered polyribosome profile [36], [37]. In Drosophila, pelo is required for meiotic cell division and controls germ-line stem cell self-renewal [38], [39]. The mammalian homolog of pelo may be required for progression of the mitotic cell cycle and pelo deficient mice are embryonic lethal [40].
Drosophila C virus (DCV) is the best studied and a relatively simple Drosophila virus. It belongs to the Dicistroviridae family. It is a non-enveloped RNA virus and its capsid is composed of the three major proteins VP1 (33 kDa), VP2 (29 kDa), VP3 (28 kDa), and two minor proteins, VP0 (37.7 kDa) and VP4 (8.5 kDa), with VP0 as a precursor of VP3 and VP4 [41]. DCV contains only one single positive-strand RNA genome which is polyadenylated and with a genome-linked protein at the 5′end [42], [43]. DCV replicates rapidly after injection into adult flies and causes host death in as few as 3 days making it an ideal pathogen for performing a death screen [44]. In an effort to search for host factors that are required for the viral replication, we carried out a forward genetic screen for Drosophila mutants that are more resistant or sensitive to DCV infection-caused death. As a result, we found that pelo deficiency can mediate DCV resistance. Further characterization revealed that pelo is not involved in the known antiviral pathways in Drosophila. Our mechanistic studies showed that pelo is required for high efficiency synthesis of DCV capsid proteins and thus DCV replication; the function of pelo in the dissociation of stalled 80S ribosomes is at least part of the underlying mechanism that allows for high speed synthesis of DCV capsid proteins.
Results
pelo deficiency in Drosophila mediates resistance to DCV infection-caused death
To identify genes that are involved in host-virus interactions, we screened about 100 P-element insertion fly lines, generated by mobilizing a P {Mae-UAS.6.11} transposon to random autosomal sites from the X chromosome, for their sensitivity to DCV-induced death. The mutants were infected by septic injury with DCV to identify lines that are more resistant or sensitive to DCV infection than wild-type. The survival curve of a group of mutant lines is shown as an example of the screen (Figure S1). Two virus-resistant lines were screened out and we were able to identify the P-element insertion site in one of the lines numbered R32. The resistance to DCV induced death of R32 is shown in Figure 1A. The P-element insertion site of R32 is in the 5′UTR of the pelo gene and thus may disrupt pelo expression (Figure 1B). Since the genes of Pka-C1, hoip, and CG31710 are also located near the P-element insertion site, we used qRT-PCR to determine the mRNA expression levels of these and the pelo genes. We found that the expression of pelo is reduced while the other three genes are not changed in the R32 line (Figure 1C). We then generated a specific polyclonal antibody to pelo and used it to confirm that the protein level of pelo was really reduced in R32 (Figure 1C).
Fig. 1. pelo deficiency mediates DCV resistance in adult flies. 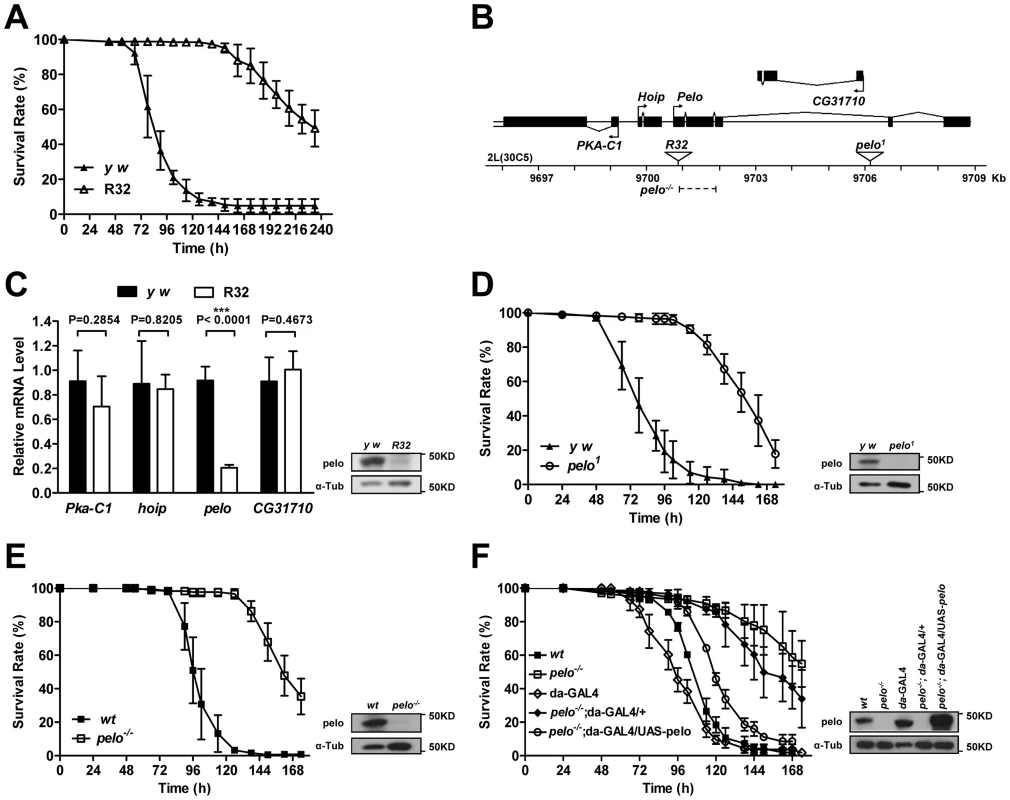
(A) R32 is resistant to DCV infection. 2–4 days old flies were injected in the thorax with DCV and then monitored every 8 hours for mortality. y w was used as a genetic background control. Results are the mean ± SD of three independent experiments with 60 flies in each group. (B) Schematic diagram of the pelo deficient flies. The triangles represent the P-element insertion site in R32 and pelo1 flies respectively. The dashed line shows the deletion region in pelo−/−. (C) Relative mRNA levels of Pka-C1, hoip, pelo, and CG31710 were measured by qRT-PCR. Expression levels of each gene were normalized to Rp49 and shown as the relative values compared to y w. Data are the mean ± SD of three independent experiments. The protein expression levels of pelo were measured by immunoblotting with anti-pelo antibody and α-Tub (α-tubulin) was used as a loading control. (D–F) Survival curve of flies of indicated genotype after DCV infection. Experiment was performed as described in A. pelo1 was backcrossed into the y w genetic background. pelo−/− was generated by imprecise excision of the P-element from the line R32 and a precise P-element excision line was used as the wild-type (wt) control. R32 fly is healthy but male-sterile, which is consistent with a previously reported pelo deficient line called pelo1 [38], [39]. pelo1 has a P-element inserted in the third intron of pelo (Figure 1B), which disrupts pelo protein expression (Figure 1D). To collect more evidence for or against the role of pelo in DCV sensitivity, we compared the sensitivity of pelo1 and its control line to DCV infection. pelo1 is resistant to DCV infection (Figure 1D), supporting the conclusion obtained from the R32 line that pelo deficiency causes DCV resistance. In order to better study the role of pelo in DCV sensitivity, we generated a pelo knockout line by deleting exons 1 and 2 (Figure 1B), which completely eliminated pelo protein expression (Figure 1E). Consistently, the pelo−/− line had DCV resistant phenotype (Figure 1E). To unambiguously demonstrate that pelo deficiency causes DCV resistance in Drosophila, we rescued the expression of pelo in pelo−/− flies by using a UAS-pelo transgene and a ubiquitous da-GAL4 driver (Figure 1F). The rescued line became more sensitive to DCV infection and thus had a phenotype similar to wild-type flies (Figure 1F). It is reported that bacterial symbiont Wolbachia increases resistance of Drosophila to RNA viral infections [45]. To examine whether Wolbachia infection influences our experiments, we measured Wolbachia infection status in our fly lines. All the fly lines we used except pelo1 are infected with Wolbachia (Figure S2A) and the infection levels are almost the same (Figure S2B), suggesting that the DCV resistant phenotype in pelo deficient flies is not caused by Wolbachia infection. Altogether, these data demonstrated that pelo deficiency results in DCV resistance in Drosophila.
pelo deficiency inhibits DCV replication in adult flies and Drosophila S2 cells
To characterize the pelo deficiency-mediated antiviral effect, we measured the viral titer at different time points in the virus-infected wild-type and pelo−/− flies before their death and found that the titer in the mutant flies were dramatically reduced compared to wild-type flies (Figure 2A). Consistent with the lower viral titer in pelo−/− flies (Figure 2A), the amounts of viral RNA and capsid proteins were also reduced in pelo−/− flies (Figures 2B and 2C). The antibody of DCV was produced by inoculation of rabbit with the purified virus, so that it can recognize all the viral capsid proteins. Based on molecular weight and mass spectrum analysis described later, we knew that the upper band is VP0 and the lower band is the mixture of VP1, VP2, and VP3. We also could detect a weak band of a molecular mass of 20 KD when in long exposure but not the 8.5 KD band corresponding to VP4 (data not shown). This decreased viral load correlated with the increased survival rate of the pelo mutant at later stages of DCV infection (Figure 1E). Thus, pelo deficiency-caused resistance to DCV infection-induced death is due to the fact that pelo deficiency limits DCV replication.
Fig. 2. pelo deficiency inhibits viral replication. 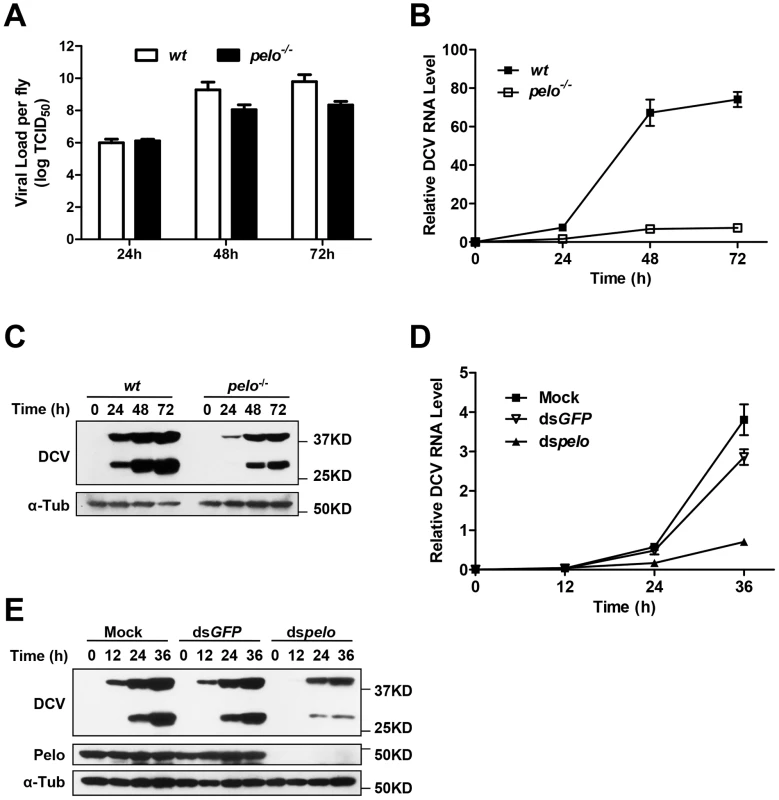
(A–C) Flies were challenged with DCV and then three pools of ten flies were collected at the indicated time points post-infection. The viral titer was determined by end-point dilution (A). The accumulation of DCV RNA was measured by qRT-PCR. Data represent the mean ± SD of triplicates (B). The accumulation of DCV capsid proteins was measured by immunoblotting with anti-DCV antibody (C). (D and E) S2 cells were untreated (Mock) or treated with dsRNAs against GFP (dsGFP) or pelo (dspelo) for 6 days and then infected with DCV (MOI = 0.1). The accumulation of DCV RNA (D) or capsid proteins (E) was measured as described in B and C, respectively. In order to study pelo deficiency-mediated resistance to DCV infection at the cellular level, we evaluated whether Drosophila S2 cells can be used in this study. pelo expression can be effectively knocked-down by RNAi in S2 cells (Figure 2E). pelo knockdown S2 cells have apparently normal morphology and do not have any noticeable changes in cell proliferation when compared with mock treated or control RNAi (dsGFP) treated S2 cells (Figure S3). We infected the control and pelo knockdown S2 cells with DCV and measured the viral RNA by qRT-PCR and the viral proteins by antibodies against viral capsid proteins. As shown in Figures 2D and 2E, both viral RNA and capsid proteins in pelo knockdown cells were much less than that in control cells. Thus the S2 cell line is a suitable culture cell system for studying the mechanism of pelo deficiency-mediated inhibition of DCV replication.
pelo is not involved in the known Drosophila antiviral mechanisms
There are several antiviral mechanisms in Drosophila, including RNAi, autophagy, antiviral gene expression regulated by Dicer-2, JAK-STAT, and NF-KB [8]. Since reported studies showed that the NF-KB pathway and autophagy can restrict certain viruses but not DCV in Drosophila [17], [18], [19], we explored whether pelo can work as a negative regulator of the other antiviral mechanisms. Because RNAi is the major defense mechanism against viral infection in Drosophila, we first tested whether loss of pelo affects siRNA-mediated gene silencing. S2 cells were co-transfected with Firefly and Renilla luciferase reporters and then treated with dsRNA targeting the Firefly luciferase. RNAi efficiency was measured by the decrease in Firefly luciferase activity relative to that of the control Renilla luciferase. We found that pelo deficiency had no effect on dsRNA-mediated inhibition of Firefly luciferase expression (Figure 3A). It is known that the induction of Vago by DCV infection is dependent on the Dicer-2 pathway [16] and we found that pelo deficiency did not further up-regulate Vago in DCV infected flies (Figures 3B), indicating that pelo deficiency did not affect Dicer-2 pathway. DCV-induced JAK-STAT activation can be measured by the expression of its target gene vir-1 [15]. The induction of vir-1 by DCV in pelo−/− flies was not enhanced but even reduced when compared with wild-type flies (Figure 3C). However, when pelo knockdown S2 cells were used, we did not detect any effect of pelo deficiency on DCV induced expression of vir-1 (Figure S4). While we do not know why there is difference between fly and S2 cells in JAK-STAT activation, these data still excluded the possibility that DCV resistance in pelo deficient flies or cells is due to an enhancement of JAK-STAT activation.
Fig. 3. pelo deficiency has no effect on siRNA-mediated gene silencing, Dicer-2 or JAK-STAT mediated antiviral gene expression, and the entry of DCV into S2 cells. 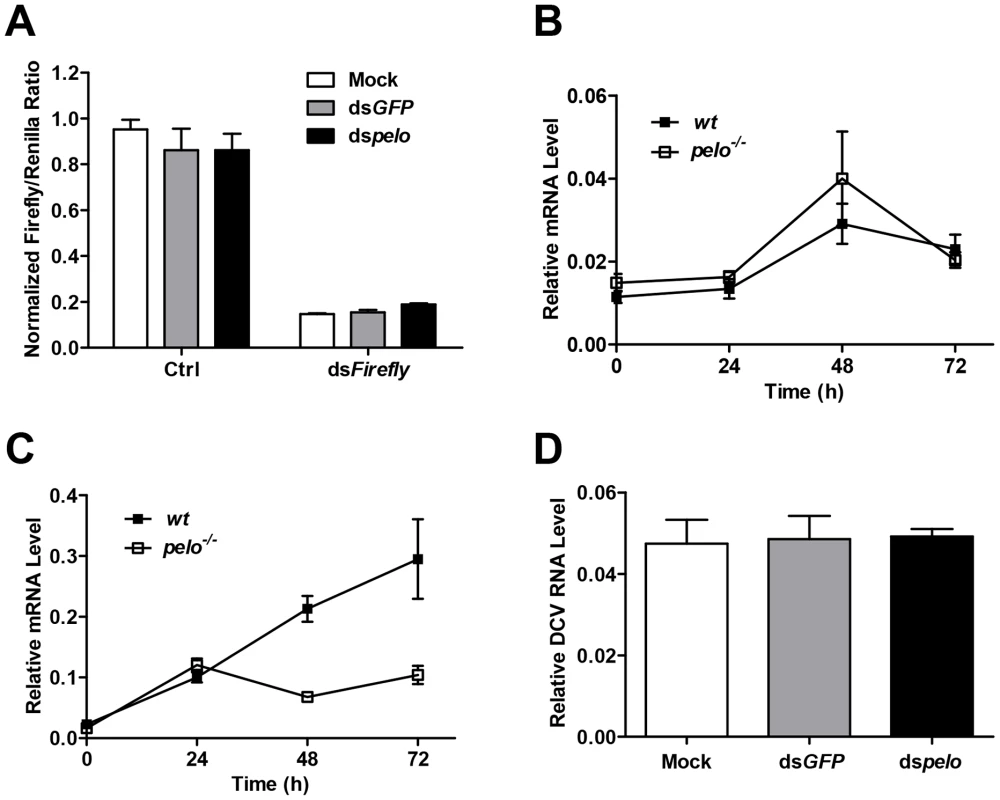
(A) Cells were treated with the indicated dsRNAs for 6 days and then co-transfected with reporter plasmids encoding Firefly and Renilla luciferase. 8 hours after transfection, cells were treated with dsRNA targeting the Firefly luciferase (dsFireflly) or were untreated as a control (Ctrl). Luciferase activities were assayed at 24 hours after dsRNA treatment. Ratios of Firefly to Renilla luciferase are shown. (B and C) Flies were infected with DCV and collected at the indicated time points post-infection. The inductions of the Dicer-2 mediated gene Vago (B) and JAK-STAT target gene Vir-1 (C) were analyzed by qRT-PCR. (D) Cells pretreated with the indicated dsRNAs were infected with DCV (MOI = 100) for 2 hours in the presence of cycloheximide (10 mg/ml) to prevent new synthesis of proteins. Viral uptake was measured by qRT-PCR analysis of internalized DCV genomic RNA. Data are the mean ± SD of triplicates. pelo deficiency selectively inhibits expression of DCV capsid proteins
The process of viral replication includes attachment, penetration, uncoating, biosynthesis of viral nucleic acids and proteins, assembly, and release [46]. To understand how pelo deficiency inhibits DCV replication, we ought to determine which step of DCV replication is affected by pelo. Since DCV is a positive-sense RNA virus, inhibition of protein synthesis by cycloheximide should block viral replication but not affect DCV entry. We therefore infected S2 cells with DCV in the presence of cycloheximide and analyzed the internalized DCV genomic RNA by qRT-PCR. The levels of DCV RNA in the control and pelo knockdown S2 cells were the same (Figure 3D), indicating that loss of pelo did not affect the efficiency of DCV entry.
Then we wanted to analyze the biosynthesis of viral nucleic acids and proteins. We measured the RNA level of DCV by qRT-PCR and labeled DCV-infected S2 cells with 35S-Met at different time points post-infection. We found that at 6 hour post-infection, there is no significant difference in viral RNA amounts between control and pelo knockdown S2 cells but the newly synthesized viral protein is less in pelo knockdown cells (Figure S5), suggesting that pelo plays a promoting role in viral protein synthesis.
DCV contains two open reading frames (ORFs). ORF1 encodes a ∼200 kDa polyprotein that includes the domains of helicase, protease, and RNA-dependent RNA polymerase. ORF2 encodes a ∼100 kDa polyprotein which is subsequently cleaved into the capsid proteins [47]. There is no subgenomic RNA transcription during the DCV lifecycle and translation of both ORFs proceeds from the genomic RNA [48]. To further analyze DCV protein synthesis, we labeled DCV-infected S2 cells with 35S-Met at 6 hours post-infection for 30 min. 7 proteins were detected in DCV-infected cells, but not in non-infected cells, and were named DCV-1 to 7 (Figure 4A). In order to make sure that these proteins were all viral proteins and not the host proteins induced by viral infection, we used actinomycin D, which can inhibit host transcription and thus inhibit protein synthesis, with the exception of DCV proteins. As expected, actinomycin D treatment greatly reduced the amount of host proteins but had no effect on DCV-1 to 7 (Figure S6A), demonstrating that all these proteins were encoded by viral RNA. pelo deficiency reduced the expression of DCV-2, DCV-4, DCV-5, DCV-6, and DCV-7, but had very little to no effect on the expression of DCV-1 and DCV-3 (Figures 4A and 4B). Analysis of viral RNA in the control and pelo knockdown cells revealed no significant difference at 6 hours post-infection (Figure 4C). These results suggested that pelo regulates the expression of some but not all DCV proteins at the translational level.
Fig. 4. pelo regulates the syntheses of some specific DCV proteins. 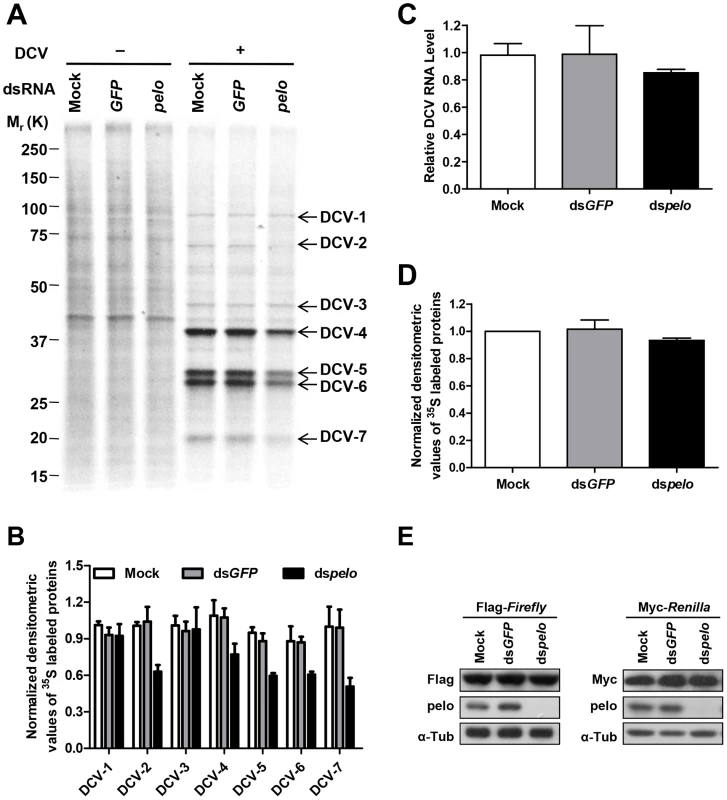
(A) Cells pretreated with the indicated dsRNAs were either uninfected (−) or infected (+) with DCV (MOI = 10) for 6 hours and then labeled with 35S-Met for 30 min. Labeled proteins were analyzed by Bis-Tris SDS-PAGE followed by autoradiography. The proteins that appeared after DCV infection are indicated by arrows and named DCV-1 to DCV-7. One 35S-Met labeled host cell protein between 37 and 50 KDa was used as loading control for protein analysis. (B) The quantities of DCV-1 to DCV-7 were determined by densitometry and normalized to mock control. The value of mock control was set at 1, and all other samples were normalized to it. (C) qRT-PCR analysis of DCV genomic RNA in different cells after 6 hours of DCV infection. Results were normalized to mock control and represent the mean ± SD of triplicates. (D) The quantities of metabolically labeled proteins (15–250 KD) were determined as decreased in B. (E) Immunoblot analysis of Flag-Firefly or Myc-Renilla expression in cells pretreated with indicated dsRNAs. The molecular weights of DCV-4 to 6 correspond to those of DCV capsid proteins [41]. More convincingly, antibodies against DCV capsid proteins recognized protein bands corresponding to the sizes of DCV-4 to 7 (Figure 2C and data not shown). The levels of these proteins are much higher than the levels of DCV-1 to 3, supporting the prediction that DCV-4 to 7 are in fact capsid proteins. To determine the identities of these proteins, we cut their bands from the Coomassie blue-stained SDS-PAGE gel and did mass spectrometry analysis (Table S1). Identified peptides of DCV-1 were located in the 975–1759 aa of ORF1, including the protease and RdRp domains. Peptides of DCV-3 were in the region of 297–679 aa, which includes the helicase domain in ORF1. Peptides of DCV-4 were in the 291–623 aa of ORF2, which corresponds to VP0. Because we could not separate DCV-5 and 6 well, the identified peptides of these two protein bands were almost the same. The peptides were distributed along the whole sequence of ORF2, consistent with the prediction that they are the mixture of VP1, VP2, and VP3. Peptides of DCV-7 were in the 647–850 aa of ORF2, indicating that DCV-7 is also encoded by ORF2. Interestingly, we could not find the corresponding band of DCV-2 in the Coomassie blue-stained gel despite its 35S-Met labeling level being equal to that of DCV-1 and DCV-3. We then analyzed the stability of the DCV proteins by a pulse-chase experiment and found that DCV-2, but not the other DCV proteins, had very short half-life (Figures S6B and S6C). pelo knockdown did not affect the turnover of these DCV proteins. The short half-life of DCV-2 explains why it is 35S-visible but Coomassie-invisible, because quick turnover make it rich in 35S-Met labeling (newly synthesized proteins) but its total amount is low. Because DCV-2 has almost equal 35S-Met labeling intensity as DCV-1 and DCV-3 but its half-life is much shorter than theirs, the instantaneous expression of DCV-2 should be higher than DCV-1 and DCV-3. Thus, DCV-2 may not be an ORF1 protein, but an ORF2 protein. Based on its size, DCV-2 cannot be the entire polyprotein of ORF2 or VP0. Since inhibition of either proteasomes or lysosomes by MG132 or chloroquine, respectively, could not increase the half-life of DCV-2 (data not shown), we predict that DCV-2 is an unprocessed precursor or intermediate product of capsids.
pelo deficiency apparently does not influence the synthesis of most, if not all kinds of cellular proteins in S2 cells based on the 35S-Met labeling experiments (Figures 4A, 4D and S6). The levels of proteins expressed by transiently transfected plasmids in the control and pelo knockdown cells were the same or almost the same (Figure 4E). Taken together, these data suggested that pelo is selectively involved in the expression of some DCV proteins such as capsids.
pelo does not target IRES or the termination sequence of ORF2
Since capsid proteins of DCV are encoded by ORF2, pelo may regulate the translation of DCV ORF2. The translation initiations of DCV ORFs are mediated by two different internal ribosomal entry sites (IRES). We constructed bicistronic luciferase reporters containing either the DCV IRES1 or IRES2 (Figure 5A), and neither of these two IRES-dependent translations was influenced by the depletion of pelo (Figure 5B). Because the structure of pelo is similar to eRF1 and pelo has functions in promoting ribosomal subunit dissociation, we tested whether the termination sequence of ORF2 is regulated by pelo. We fused termination regions of ORF1 or ORF2 to Firefly luciferase reporters (Figure 5C), and found that the expression of both reporters was not influenced by pelo knockdown (Figure 5D). Our reporter studies suggested that pelo does not function in the regulation of either IRES dependent translational initiation of ORF2 or termination of ORF2.
Fig. 5. pelo does not regulate IRES or the termination sequence of ORF2 of DCV. 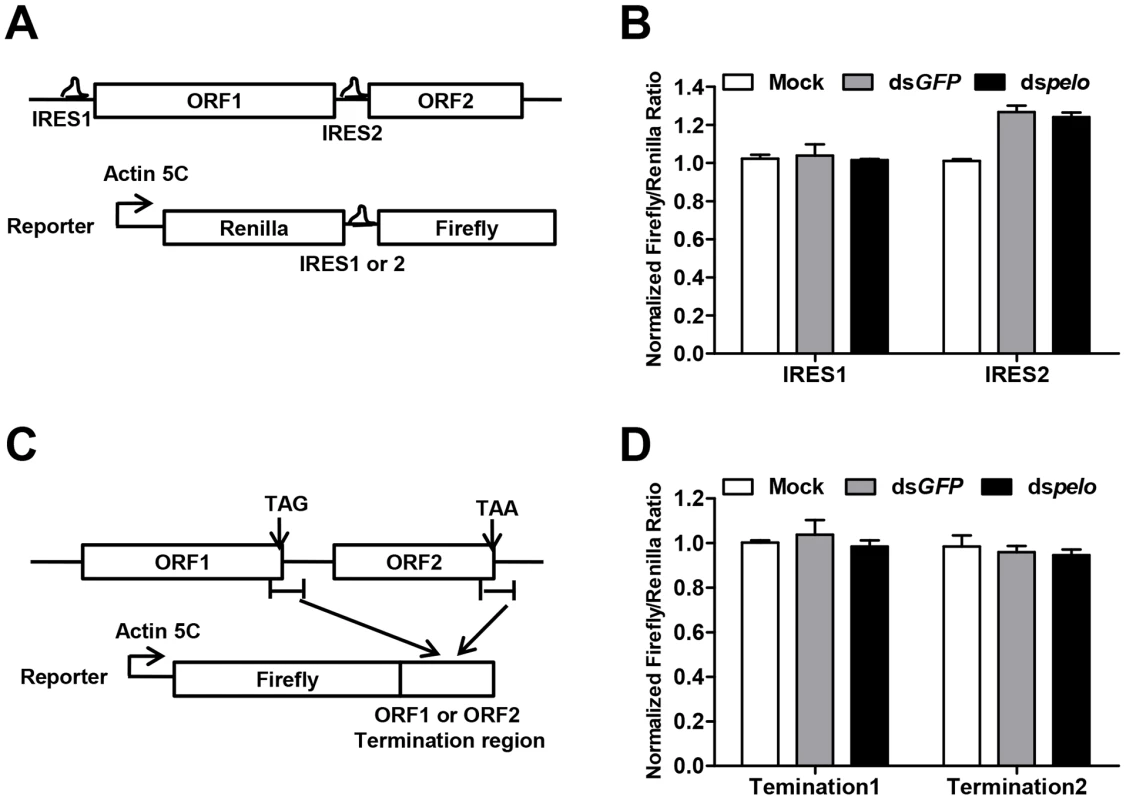
(A) Schematic diagram of the bicistronic reporter constructs. Translation from 5′cap (5C) is quantified by Renilla luciferase, while translation from the DCV IRES1 or IRES2 is measured by Firefly luciferase. (B) Luciferase activities were assayed at 36 hours after transfection of DCV IRES1 or IRES2 reporter plasmid. Ratios of Firefly to Renilla luciferase activity are shown after they were normalized to untreated cells (Mock), which was set at 1. Data are the mean ± SD of triplicates. (C) Schematic diagram of the Firefly luciferase reporter constructs. Termination region of DCV ORF1 or ORF2 (between ∼100 bp upstream and ∼200 bp downstream of stop codon) was fused to the c-terminal of Firefly luciferase. (D) Cells pretreated with indicated dsRNAs were transfected with each of the Firefly luciferase reporters shown in (C) together with a constitutively Renilla luciferase-expressing vector. Luciferase activities were assayed as described in (B). The increased non-functional 80S monoribosomes in pelo deficient cells may limit ribosome availability and contribute to the inhibition of DCV protein synthesis
Polysome profile analysis is frequently used to monitor the efficiency of translation [49]. We used it to examine whether there was any difference in the efficiency of DCV protein synthesis between control and pelo knockdown S2 cells. Cells were mock infected or infected with DCV for 6 hours. The 6 hours was chosen because at this time point the difference in DCV protein, but not DCV RNA, begins to appear between wild-type and pelo deficient cells (Figure S5). Cell lysates were resolved on a 10–50% continuous sucrose gradient (Figure 6A). Knockdown of pelo increased the amount of 80S monoribosomes, which is consistent with the result obtained by studying Dom34 deletion in yeast [37]. However, the distribution of DCV RNAs in ribosome profiling fractions is about the same in wild-type and pelo knockdown cells (Figure 6B). DCV infection also leads to increase of 80S monoribosomes in wild-type S2 cells over the time of infection (Figures 6A and S7). DCV infection does not affect the level of 80S monoribosome in pelo deficient cells much (Figure 6A), due to the already high level of 80S monoribosome and ineffective replication of DCV in pelo deficient S2 cells.
Fig. 6. The increased non-functional 80S monoribosomes in pelo-deficient cells may limit free ribosome availability for high level viral protein synthesis. 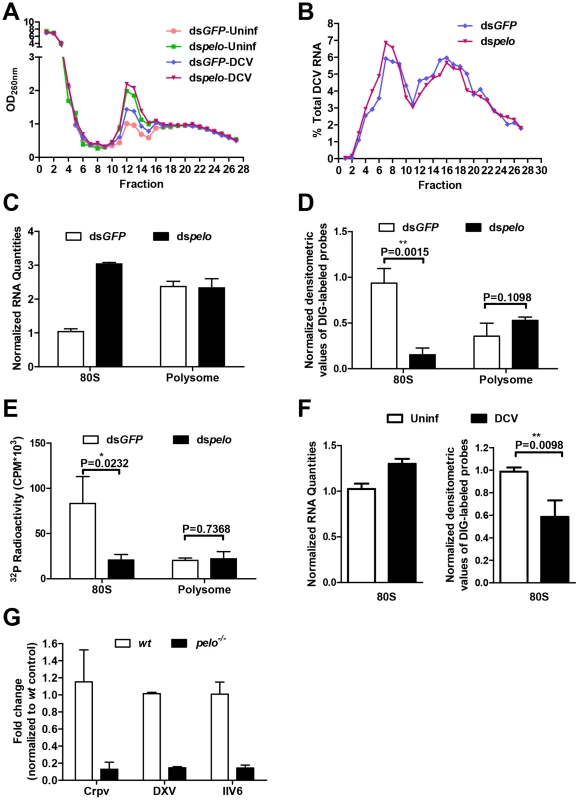
(A) Cells pretreated with the indicated dsRNAs were either uninfected (Uninf) or infected with DCV for 6 hours (MOI = 10) followed by polysome profile analysis. (B) qRT-PCR analysis of DCV RNA in individual fractions of sucrose gradients. The distribution of DCV RNA among the fractions is shown as a percentage of the total DCV RNA. (C) Total RNA of the 80S fraction and one of the polysome fractions were extracted. RNA quantities are shown as their relative ratios compared to that of 80S in dsGFP-treated cells. (D) Equal amounts of RNA from each sample were hybridized with digoxigenin (DIG)-labeled oligo (dT) probes and then applied to the nylon membrane. The membrane was probed with anti-DIG antibody and detected by chemiluminescence. The quantities of DIG-labeled probes were determined by densitometric analysis and normalized to the value of 80S in dsGFP-treated control cells. (E) Equal amounts of RNA from each sample were reverse transcribed with oligo (dT) in the presence of [α-32P] dTTP. The resulting cDNAs were recovered by using NucAway spin column. The radioactivity was measured with liquid scintillation counting. Results are the mean ± SD of triplicates. (F) Cells were uninfected (Uninf) or infected with DCV for 12 hours (MOI = 30) and then were followed by polysome profile analysis. Total RNA of 80S fractions of each sample were extracted and then analyzed as described in C and D. (G) pelo is generally involved in the replication of different viruses. wt and pelo−/− flies were infected with Crpv, DXV, or IIV6 for 72 hours. RNA was extracted from Crpv and DXV infected flies, and DNA was isolated from IIV6 infected flies. qRT-PCR or qPCR was used to analyze the amounts of these three different viruses. Results were normalized to wt and shown as the relative values. Data are the mean ± SD of triplicates. The 80S monoribosomes could associate with mRNA and also could be free. We extracted the total RNAs from 80S and polysome fractions prepared from non-DCV infected cells. The amount of RNA in the 80S fraction from the pelo-deficient cells was about three times of that from the control cells, while the quantities of RNA in the polysome fraction were about the same in both the control and pelo knockdown cells (Figure 6C). Because rRNA is the major component of the total RNA extracted from these fractions, this data confirms that pelo-deficient cells have more 80S monoribosomes. We then analyzed the amount of mRNA associated with 80S monoribosomes and polysomes. The samples with equal amounts of total RNA, which indicated that the RNA samples were extracted from an equal number of ribosomes, were hybridized with digoxigenin (DIG)-labeled oligo (dT) probes to detect mRNA. DIG was detected with alkaline phosphatase labeled anti-DIG antibody by chemiluminescence (Figure 6D). The amount of Poly (A) RNA in the 80S fraction of the pelo-deficient cells was less than that of the control cells, whereas poly (A) RNA amounts in the polysome fraction were not affected by pelo knockdown. The same result was obtained by reverse transcribing total RNA with oligo (dT) in the presence of [α-32P] dTTP (Figure 6E). The level of incorporated [α-32P] dTTP in cDNAs should correlate with the level of mRNA. Based on these data, we concluded that the increased portion of 80S monoribosomes in pelo-deficient S2 cells was primarily contributed to by mRNA free ribosomes. By using the same approach, we determined that DCV infection-induced 80S monoribosomes in wild-type S2 cells are also mainly poly (A) RNA free ribosomes (Figure 6F).
Since Dom34 forms a complex with HBS1 to function [29], [50], we knocked down HBS1 in S2 cells and found that DCV replication was indeed inhibited (Figure S8). It has been well established recently that Dom34-HBS1 plays an essential role in the quality control of protein translation by dissociation of stalled ribosomes at the 3′ end of aberrant mRNAs. The increase of 80S monoribosomes in pelo-deficient cells is most likely to be a result of those stalled ribosomes being processed to monomers. The increase in non-translating 80S ribosomes in DCV infected cells suggests that high level DCV protein synthesis produces more incidents of stalled ribosomes. Inefficient recycling of stalled 80S ribosomes in DCV RNA should result in the synthesis of truncated and mutated viral proteins and also could reduce the translation efficiency of viral RNA. Impairment of the dissociation of non-translating 80S ribosomes should reduce the availability of ribosomes for protein translation. It is believed that the relationship between free ribosome concentration and the rate of peptide formation follows the Michaelis-Menten equation [51] (Figure S9). Based on the Michaelis-Menten equation, the reduction of free ribosome concentration has a more inhibitory effect on the production of quickly synthesized proteins than on the production of slowly formed proteins (Figure S9). In the case of DCV, a single viral RNA encodes capsid proteins DCV-4 to 7 and non-capsid proteins DCV-1 and 3. Since a large amount of capsid proteins is required for efficient DCV replication, reduced free ribosomes in pelo knockdown cells are likely to be responsible for the phenomenon in which pelo knockdown had much more of an effect on the protein synthesis of DCV-4 to 7 than on that of DCV-1 and 3 (Figure 4).
High speed synthesis of capsid proteins is required for the replication of many different viruses. In order to further evaluate the notion that the availability of free ribosomes in pelo-deficient flies limits viral capsid protein synthesis, we examined whether pelo deficiency can influence the replication of three other viruses, single-stranded RNA virus Cricket Paralysis Virus (Crpv), double-stranded RNA virus Drosophila X virus (DXV), and a large dsDNA virus invertebrate iridescent virus 6 (IIV6). We used qRT-PCR as described by published studies [15], [17], [52] to measure the corresponding RNA or DNA levels of each virus in the infected flies, and found that the replications of these viruses were all slower in pelo−/− flies (Figure 6G). We also analyzed Crpv, DXV, and IIV6 at different time points after infection in wild-type and pelo−/− flies and measured viral titers in flies at 3 days post-infection (Figure S10). The data support the conclusion that the replications of these three different viruses are suppressed in pelo−/− flies. The pelo deficiency selectively resists viral infection as pelo−/− flies are not resistant but even more sensitive to bacterial infection (data not shown). These data support the idea that the defect in the recycling of stalled ribosomes in pelo-deficient cells limits viral replication and implies that pelo deficiency mediates a general antiviral activity.
Discussion
Through a forward genetic screen for Drosophila mutants with increased or decreased susceptibility to DCV-induced death, we have identified a virus-resistant line in which the pelo gene was mutated. We further demonstrated, by using this and other pelo-deficient Drosophila lines, and by rescuing pelo expression in pelo−/− flies, that pelo deficiency led to resistance to DCV infection (Figure 1). The pelo deficiency mediated DCV resistance can be phenotypically copied in S2 cells (Figure 2). Our mechanistic studies revealed that pelo deletion led to inefficient DCV replication (Figure 2). pelo protein appears to be involved in the high efficiency protein translation of DCV capsid proteins (Figure 4). Our data suggest that the role of pelo in the dissociation of 80S ribosomes links to pelo deficiency mediated resistance to DCV infection. Huge amounts of capsid protein synthesis in DCV infected cells could increase errors in protein translation. The increase in 80S ribosomes that were not associated with poly (A) mRNA in DCV infected cells suggests an increase of stalled ribosomes (Figure 6). Deletion of pelo impairs the recycling of the stalled ribosomes, which reduces the availability of free ribosomes and thus could be a limiting factor of DCV replication in pelo-deficient cells. Indeed, the effect of pelo deficiency on high speed translation of capsids is much greater than that on slowly translated DCV-1 and DCV-3 proteins from the same RNA (Figure 4), consistent with the kinetic prediction that a more quickly synthesized protein is limited more effectively than a more slowly translated protein by available free ribosome concentration (Figure S9). In addition, inefficient removal of stalled ribosomes from a given DCV RNA could also eliminate this DCV RNA from translation. It could create truncated or mutated DCV capsid proteins that might interfere with DCV assembling and further inhibit DCV replication. All these mechanistic predictions point to a general antiviral role of pelo deficiency. We have further confirmed that pelo−/− fly is indeed resistant to a broad panel of different viruses including a single-stranded RNA virus, a double-stranded RNA virus, and a DNA virus.
pelo may function beyond recycling stalled 80S ribosomes as a recent study showed that the formation of 80S-like complexes is an intermediate step in the process of 40S ribosome maturation, and pelo is involved in the dissociation of this 80S-like complex during the maturation of the 40S ribosome [53]. Regardless of whether pelo dissociates stalled 80S ribosomes or the 80S-like complex, it affects the supply of free ribosomes. pelo is known to form a complex with HBS1 to dissociate ribosomes that are vacant or stalled at the aberrant mRNA [27], [28], [29], [31], [32], [33], [54], which is supported by our data that HBS1 knockdown also inhibits DCV replication. pelo knockout is lethal in mice but not in yeast and Drosophila as we presented here [36], [38], [40], suggesting that pelo is not required for cell survival but is instead required for mouse development during the embryonic stage. pelo should also be important for the expression of certain genes since its knockout leads to male-sterility in flies (refs [38], [39]and data not shown). The selective inhibition of DCV capsid protein expression by pelo knockdown strongly suggests that pelo is not generally involved in the protein translation of all proteins. At present we know little about the identity of the cellular proteins whose translations are regulated by pelo. The current study revealed that at least some of the quickly synthesized proteins are subject to control by pelo. The regulation of the availability of free ribosomes should be at least part of the underlying mechanisms. The Michaelis-Menten equation-based prediction is certainly an over-simplified explanation since cellular processes are compartmentalized and the local concentration of free ribosomes in the synthesis site of a given protein is important. If certain protein synthesis sites are more dependent on recycled free ribosomes, pelo should have more influence on the synthesis of these proteins than those synthesized in other locations. Unfortunately, current technologies are unable to address this question.
It is known that multiple antiviral pathways including RNAi and antiviral gene expression are present in Drosophila [8]. Our data excluded the possibility that pelo deficiency mediated inhibition of viral replication resulted from the activation of these antiviral mechanisms (Figure 3). Without exception, viruses use cellular translational machinery to synthesize their proteins. pelo should belong to those proteins that are hijacked by infected virus to highly efficiently translate viral proteins. Because pelo is not required for cell survival, it is dispensable for protein synthesis of most cellular proteins. The general suppressive effect on virus replication imposed by pelo deficiency suggests that pelo is involved in a unique part of translational machinery used by viruses. Since viral replication requires high efficiency synthesis of a large amount of viral capsids, it is possible that viruses are able to utilize special cellular mechanisms for quickly synthesizing a large amount of proteins. If it is the case, pelo is involved in this special mechanism.
There is no doubt that viral recognition mechanisms in cells and the subsequent activation of antiviral responses are essential for host defense against viral infection. However, the cellular mechanisms that are preferentially hijacked by viruses are also important parts of host-virus interaction. Given the general antiviral effect of pelo deficiency, the factors that are dispensable for normal cell function but are involved in viral protein synthesis should draw more attention in our studies of host response to viral infection. Our work presented here identifies a new host factor that is essential for effective viral replication. Since pelo is a highly conserved protein, the function of pelo revealed in this study is very likely to be conserved in mammals as well. The discovery of the general antiviral activity by pelo deficiency may provide a new therapeutic target for broad-spectrum antiviral therapy.
Materials and Methods
Fly strains, screening, and viral infections
Flies were raised on a standard yeast-cornmeal-agar medium and all experiments were performed at 25°C. A large-scale P-element mutagenesis was performed by mobilizing a P {Mae-UAS.6.11} transposon (Bloomington stock #3025) from the X chromosome to random autosomal sites with the transposase P {Δ2-3} 99B (Bloomington stock #3664) by standard methodology. The insertion lines were infected with DCV and selected for more resistant or sensitive lines. We screened about 100 lines and selected a resistant line, R32. Identification of the P-element insertion site in R32 was performed by inverse PCR.
The pelo−/− line was generated by imprecise excision of the P-element from the line R32 and contained a deletion of about 1.1 kb including exon 1 and exon 2. A precise P-element excision line was used as a wild-type (wt) control. All other stocks were obtained from the Drosophila Bloomington stock center unless indicated otherwise.
For viral infection, 2–4 days old flies of the stated genotype were infected by septic injury with the indicated virus as described [55]. Briefly, the thorax of the fly was pricked with a thin needle previously dipped into the viral solution (DCV, 5×1011 TCID50/ml; Crpv, 4×107 TCID50/ml; DXV, 6×107 TCID50/ml; IIV6, 7×1010 TCID50/ml) and only the very tip of the needle is inserted into the fly. The infected flies were monitored for mortality or collected at indicated time points for different assays. Viral titers were determined in S2 cell culture and calculated according to the end-point method of Reed and Muench [56].
Cells, RNAi and transfection
Drosophila S2 cells (ATCC) were cultured at 25°C in SF900-II SFM medium (Gibco). dsRNA targeting pelo (nucleotides 304 bp–938 bp), HBS1 (nucleotides 706–2119 bp), Firefly (nucleotides 1144–1650 bp), and GFP (nucleotides 65–559 bp) were generated from T7-promoter-flanked PCR products by in vitro transcription using T7 transcription kit (Promega). RNAi treatments were performed as previously described [57]. In brief, S2 cells were incubated with SF900-II SFM medium containing 10 ug/ml dsRNA and then were replenished with fresh dsRNA-containing medium daily for 6 days.
For transfection, S2 cells were incubated in Schneider's medium (Lonza) supplemented with 10% FBS, and transfections were performed by the Calcium Phosphate precipitation method. After 12 hours, the medium was removed and replaced with SF900-II SFM medium. Luciferase expression was assayed using the Dual-Luciferase Reporter Assay System (Promega) and quantitated on the luminometer.
Quantitative RT-PCR
Total RNA was extracted from adult flies or S2 cells with RNAiso Plus (Takara) according to the manufacturer's instructions. cDNA was prepared with M-MLV reverse transcriptase and oligo-dT primers. Quantitative RT-PCR was performed using SYBR Green reagent along with gene-specific primers. All the results were analyzed by relative quantification, by normalizing to the Rp49 RNA level. Primer sequences are available on request.
Western blots
Cells or flies were lysed in Laemmli sample buffer and then boiled. Samples were separated by SDS-PAGE and blotted onto PVDF membrane. The blots were probed with indicated antibodies and detected by Chemiluminescent HRP Substrate (Millipore). Antibodies were obtained from the following sources: anti-α-tubulin (BGI), anti-Flag (Abmart), anti-Myc (Santa Cruz), and polyclonal antibodies to pelo and DCV were prepared in rabbits against bacterially expressed full-length pelo protein or purified DCV particles respectively.
Metabolic labeling
Cells were washed three times with PBS and then starved with methionine-free culture medium for 15 min. After methionine starvation, cells were incubated with 0.2 mCi/ml 35S-Methione in methionine-free medium for 30 min, washed three times with PBS and lysed with Laemmli sample buffer. Labeled proteins were resolved by Bis-Tris SDS-PAGE and then fixed. The dried gels were exposed to film. For protein stability analysis, cells were labeled with 35S-Met for 30 min, washed three times with PBS and then lysed (time point 0) or chased for the indicated time with excess cold methionine.
Polysome analysis
Cells were incubated with 100 µg/ml cycloheximide for 10 min and then washed twice with cold PBS containing cycloheximide (100 µg/ml). 5×108 cells were lysed with 1 ml polysome lysis buffer (10 mM HEPES-KOH pH 7.4, 5 mM MgCl2, 150 mM KCl, 0.5% NP-40, 0.5 mM DTT, 100 µg/ml cycloheximide, 100 U/ml RNAsin RNase inhibitor [Promega], and EDTA-free protease inhibitor cocktail Complete [Roche]). Cell debris was removed by centrifuging for 10 min at 16,000 g. The cytoplasmic supernatant was layered onto 10 ml of 10–50% continuous sucrose gradient and centrifuged at 4°C for 2 h at 36,000 rpm in a Beckman SW41 rotor. Then 0.4-ml fractions were collected from the top of the gradient and the polysome profile was monitored by RNA absorbance at 260 nm.
RNA of each fraction was extracted by using RNAiso Plus (Takara) and the distribution of DCV RNA was measured by quantitative RT-PCR. For the slot blot assay, RNA was incubated with digoxigenin (DIG)-labeled oligo (dT) probes. After hybridization, samples were applied to the nylon membrane by using a Bio-Dot apparatus (Bio-rad) and fixed by UV crosslinking. Following the washing and blocking steps, the membrane was probed with anti-DIG antibody that was coupled to alkaline phosphatase (Roche), and then detected by incubating with the chemiluminescent substrate CDP-star and exposing the blot to X-ray film. The quantities of DIG-labeled probes were determined by densitometric analysis. For radioactive assay, equal amounts of RNA from each sample were reverse transcribed with oligo (dT) in the presence of [α-32P] dTTP. The resulting cDNAs were recovered by using NucAway spin column (Ambion) while removing the salts and unincorporated [α-32P] dTTP. The radioactivity was measured with liquid scintillation counting (Beckman).
Statistical analysis
Statistical analysis was performed using the unpaired two-tailed Student's t-test with the Prism GraphPad software. P<0.05 was considered significantly different.
Accession numbers
NCBI gene ID numbers for Drosophila melanogaster genes mentioned in the text
pelo (34286); HBS1 (117365); Pka-C1 (34284); hoip (44173); CG31710 (318907); vir-1 (34652); vago (32040).
Genebank accession numbers for the genome sequences of the viruses
Drosophila C virus (DCV), NC_001834; Drosophila x virus (DXV), NC_004177, NC_004169; Cricket paralysis virus (Crpv), NC_003924; Invertebrate iridescent virus 6 (IIV6), NC_003038.
Supporting Information
Zdroje
1. OldstoneMB (1996) Principles of viral pathogenesis. Cell 87 : 799–801.
2. BeutlerB, EidenschenkC, CrozatK, ImlerJL, TakeuchiO, et al. (2007) Genetic analysis of resistance to viral infection. Nature reviews Immunology 7 : 753–766.
3. PichlmairA, Reis e SousaC (2007) Innate recognition of viruses. Immunity 27 : 370–383.
4. YanN, ChenZJ (2012) Intrinsic antiviral immunity. Nature immunology 13 : 214–222.
5. LemaitreB, HoffmannJ (2007) The host defense of Drosophila melanogaster. Annual review of immunology 25 : 697–743.
6. FerrandonD, ImlerJL, HetruC, HoffmannJA (2007) The Drosophila systemic immune response: sensing and signalling during bacterial and fungal infections. Nature reviews Immunology 7 : 862–874.
7. KempC, ImlerJL (2009) Antiviral immunity in drosophila. Current opinion in immunology 21 : 3–9.
8. SabinLR, HannaSL, CherryS (2010) Innate antiviral immunity in Drosophila. Current opinion in immunology 22 : 4–9.
9. WangXH, AliyariR, LiWX, LiHW, KimK, et al. (2006) RNA interference directs innate immunity against viruses in adult Drosophila. Science 312 : 452–454.
10. SalehMC, TassettoM, van RijRP, GoicB, GaussonV, et al. (2009) Antiviral immunity in Drosophila requires systemic RNA interference spread. Nature 458 : 346–350.
11. DingSW, VoinnetO (2007) Antiviral immunity directed by small RNAs. Cell 130 : 413–426.
12. LiH, LiWX, DingSW (2002) Induction and suppression of RNA silencing by an animal virus. Science 296 : 1319–1321.
13. van RijRP, SalehMC, BerryB, FooC, HoukA, et al. (2006) The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster. Genes & development 20 : 2985–2995.
14. NayakA, BerryB, TassettoM, KunitomiM, AcevedoA, et al. (2010) Cricket paralysis virus antagonizes Argonaute 2 to modulate antiviral defense in Drosophila. Nature structural & molecular biology 17 : 547–554.
15. DostertC, JouanguyE, IrvingP, TroxlerL, Galiana-ArnouxD, et al. (2005) The Jak-STAT signaling pathway is required but not sufficient for the antiviral response of drosophila. Nature immunology 6 : 946–953.
16. DeddoucheS, MattN, BuddA, MuellerS, KempC, et al. (2008) The DExD/H-box helicase Dicer-2 mediates the induction of antiviral activity in drosophila. Nature immunology 9 : 1425–1432.
17. ZambonRA, NandakumarM, VakhariaVN, WuLP (2005) The Toll pathway is important for an antiviral response in Drosophila. Proceedings of the National Academy of Sciences of the United States of America 102 : 7257–7262.
18. CostaA, JanE, SarnowP, SchneiderD (2009) The Imd pathway is involved in antiviral immune responses in Drosophila. PloS one 4: e7436.
19. ShellyS, LukinovaN, BambinaS, BermanA, CherryS (2009) Autophagy is an essential component of Drosophila immunity against vesicular stomatitis virus. Immunity 30 : 588–598.
20. NakamotoM, MoyRH, XuJ, BambinaS, YasunagaA, et al. (2012) Virus recognition by Toll-7 activates antiviral autophagy in Drosophila. Immunity 36 : 658–667.
21. PirieNW (1946) The viruses. Annual review of biochemistry 15 : 573–592.
22. CherryS, PerrimonN (2004) Entry is a rate-limiting step for viral infection in a Drosophila melanogaster model of pathogenesis. Nature immunology 5 : 81–87.
23. CherryS, DoukasT, ArmknechtS, WhelanS, WangH, et al. (2005) Genome-wide RNAi screen reveals a specific sensitivity of IRES-containing RNA viruses to host translation inhibition. Genes & development 19 : 445–452.
24. CherryS, KunteA, WangH, CoyneC, RawsonRB, et al. (2006) COPI activity coupled with fatty acid biosynthesis is required for viral replication. PLoS pathogens 2: e102.
25. BushellM, SarnowP (2002) Hijacking the translation apparatus by RNA viruses. The Journal of cell biology 158 : 395–399.
26. DeverTE, GreenR (2012) The elongation, termination, and recycling phases of translation in eukaryotes. Cold Spring Harbor perspectives in biology 4: a013706.
27. ShoemakerCJ, EylerDE, GreenR (2010) Dom34:Hbs1 promotes subunit dissociation and peptidyl-tRNA drop-off to initiate no-go decay. Science 330 : 369–372.
28. van den ElzenAM, HenriJ, LazarN, GasME, DurandD, et al. (2010) Dissection of Dom34-Hbs1 reveals independent functions in two RNA quality control pathways. Nature structural & molecular biology 17 : 1446–1452.
29. DomaMK, ParkerR (2006) Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature 440 : 561–564.
30. KurohaK, AkamatsuM, DimitrovaL, ItoT, KatoY, et al. (2010) Receptor for activated C kinase 1 stimulates nascent polypeptide-dependent translation arrest. EMBO reports 11 : 956–961.
31. TsuboiT, KurohaK, KudoK, MakinoS, InoueE, et al. (2012) Dom34:hbs1 plays a general role in quality-control systems by dissociation of a stalled ribosome at the 3′ end of aberrant mRNA. Molecular cell 46 : 518–529.
32. PisarevaVP, SkabkinMA, HellenCU, PestovaTV, PisarevAV (2011) Dissociation by Pelota, Hbs1 and ABCE1 of mammalian vacant 80S ribosomes and stalled elongation complexes. The EMBO journal 30 : 1804–1817.
33. IzawaT, TsuboiT, KurohaK, InadaT, NishikawaS, et al. (2012) Roles of dom34:hbs1 in nonstop protein clearance from translocators for normal organelle protein influx. Cell reports 2 : 447–453.
34. LeeHH, KimYS, KimKH, HeoI, KimSK, et al. (2007) Structural and functional insights into Dom34, a key component of no-go mRNA decay. Molecular cell 27 : 938–950.
35. GrailleM, ChailletM, van TilbeurghH (2008) Structure of yeast Dom34: a protein related to translation termination factor Erf1 and involved in No-Go decay. The Journal of biological chemistry 283 : 7145–7154.
36. DavisL, EngebrechtJ (1998) Yeast dom34 mutants are defective in multiple developmental pathways and exhibit decreased levels of polyribosomes. Genetics 149 : 45–56.
37. BhattacharyaA, McIntoshKB, WillisIM, WarnerJR (2010) Why Dom34 stimulates growth of cells with defects of 40S ribosomal subunit biosynthesis. Molecular and cellular biology 30 : 5562–5571.
38. EberhartCG, WassermanSA (1995) The pelota locus encodes a protein required for meiotic cell division: an analysis of G2/M arrest in Drosophila spermatogenesis. Development 121 : 3477–3486.
39. XiR, DoanC, LiuD, XieT (2005) Pelota controls self-renewal of germline stem cells by repressing a Bam-independent differentiation pathway. Development 132 : 5365–5374.
40. AdhamIM, SallamMA, StedingG, KorabiowskaM, BrinckU, et al. (2003) Disruption of the pelota gene causes early embryonic lethality and defects in cell cycle progression. Molecular and cellular biology 23 : 1470–1476.
41. ReavyB, MooreNF (1983) Cell-free translation of Drosophila C virus RNA: identification of a virus protease activity involved in capsid protein synthesis and further studies on in vitro processing of cricket paralysis virus specified proteins. Archives of virology 76 : 101–115.
42. JoussetFX, BergoinM, RevetB (1977) Characterization of the Drosophila C virus. The Journal of general virology 34 : 269–283.
43. KingLA, MooreNF (1988) Evidence for the presence of a genome-linked protein in two insect picornaviruses, cricket paralysis and DrosophilaC viruses. FEMS Microbiology Letters 50 : 41–44.
44. Lautie-HarivelN, Thomas-OrillardM (1990) Location of Drosophila C virus target organs in Drosophila host population by an immunofluorescence technique. Biology of the cell/under the auspices of the European Cell Biology Organization 69 : 35–39.
45. TeixeiraL, FerreiraA, AshburnerM (2008) The bacterial symbiont Wolbachia induces resistance to RNA viral infections in Drosophila melanogaster. PLoS biology 6: e2.
46. Fields BN, Knipe DM, Howley PM (2007) Fields virology. Philadelphia: Wolters Kluwer Health/Lippincott Williams & Wilkins.
47. JohnsonKN, ChristianPD (1999) Molecular characterization of Drosophila C virus isolates. Journal of invertebrate pathology 73 : 248–254.
48. JohnsonKN, ChristianPD (1998) The novel genome organization of the insect picorna-like virus Drosophila C virus suggests this virus belongs to a previously undescribed virus family. The Journal of general virology 79(Pt 1): 191–203.
49. MasekT, ValasekL, PospisekM (2011) Polysome analysis and RNA purification from sucrose gradients. Methods in molecular biology 703 : 293–309.
50. Carr-SchmidA, PfundC, CraigEA, KinzyTG (2002) Novel G-protein complex whose requirement is linked to the translational status of the cell. Molecular and cellular biology 22 : 2564–2574.
51. JohanssonM, BouakazE, LovmarM, EhrenbergM (2008) The kinetics of ribosomal peptidyl transfer revisited. Molecular cell 30 : 589–598.
52. KempC, MuellerS, GotoA, BarbierV, ParoS, et al. (2013) Broad RNA interference-mediated antiviral immunity and virus-specific inducible responses in Drosophila. Journal of immunology 190 : 650–658.
53. StrunkBS, NovakMN, YoungCL, KarbsteinK (2012) A translation-like cycle is a quality control checkpoint for maturing 40S ribosome subunits. Cell 150 : 111–121.
54. BeckerT, ArmacheJP, JaraschA, AngerAM, VillaE, et al. (2011) Structure of the no-go mRNA decay complex Dom34-Hbs1 bound to a stalled 80S ribosome. Nature structural & molecular biology 18 : 715–720.
55. TzouP, MeisterM, LemaitreB (2002) Methods for studying infection and immunity in Drosophila. Methods in Microbiology 31 : 507–529.
56. ReedLJ, MuenchH (1938) A simple method of estimating fifty per cent endpoints. Am J Epidemiol 27 : 493–497.
57. RogersSL, RogersGC (2008) Culture of Drosophila S2 cells and their use for RNAi-mediated loss-of-function studies and immunofluorescence microscopy. Nature protocols 3 : 606–611.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Affinity Proteomics Reveals Elevated Muscle Proteins in Plasma of Children with Cerebral MalariaČlánek The Transcriptional Activator LdtR from ‘ Liberibacter asiaticus’ Mediates Osmotic Stress ToleranceČlánek Complement-Related Proteins Control the Flavivirus Infection of by Inducing Antimicrobial PeptidesČlánek Fungal Chitin Dampens Inflammation through IL-10 Induction Mediated by NOD2 and TLR9 ActivationČlánek Parasite Fate and Involvement of Infected Cells in the Induction of CD4 and CD8 T Cell Responses to
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2014 Číslo 4- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- , , , Genetic Variability: Cryptic Biological Species or Clonal Near-Clades?
- Early Mortality Syndrome Outbreaks: A Microbial Management Issue in Shrimp Farming?
- Wormholes in Host Defense: How Helminths Manipulate Host Tissues to Survive and Reproduce
- Plastic Proteins and Monkey Blocks: How Lentiviruses Evolved to Replicate in the Presence of Primate Restriction Factors
- The 2010 Cholera Outbreak in Haiti: How Science Solved a Controversy
- Affinity Proteomics Reveals Elevated Muscle Proteins in Plasma of Children with Cerebral Malaria
- Noncanonical Role for the Host Vps4 AAA+ ATPase ESCRT Protein in the Formation of Replicase
- Efficient Parvovirus Replication Requires CRL4-Targeted Depletion of p21 to Prevent Its Inhibitory Interaction with PCNA
- Host-to-Pathogen Gene Transfer Facilitated Infection of Insects by a Pathogenic Fungus
- The Transcriptional Activator LdtR from ‘ Liberibacter asiaticus’ Mediates Osmotic Stress Tolerance
- Coxsackievirus B Exits the Host Cell in Shed Microvesicles Displaying Autophagosomal Markers
- TCR Affinity Associated with Functional Differences between Dominant and Subdominant SIV Epitope-Specific CD8 T Cells in Rhesus Monkeys
- Coxsackievirus-Induced miR-21 Disrupts Cardiomyocyte Interactions via the Downregulation of Intercalated Disk Components
- Ligands of MDA5 and RIG-I in Measles Virus-Infected Cells
- Kind Discrimination and Competitive Exclusion Mediated by Contact-Dependent Growth Inhibition Systems Shape Biofilm Community Structure
- Structural Differences Explain Diverse Functions of Actins
- HSCARG Negatively Regulates the Cellular Antiviral RIG-I Like Receptor Signaling Pathway by Inhibiting TRAF3 Ubiquitination Recruiting OTUB1
- Vaginitis: When Opportunism Knocks, the Host Responds
- Complement-Related Proteins Control the Flavivirus Infection of by Inducing Antimicrobial Peptides
- Fungal Chitin Dampens Inflammation through IL-10 Induction Mediated by NOD2 and TLR9 Activation
- Microbial Pathogens Trigger Host DNA Double-Strand Breaks Whose Abundance Is Reduced by Plant Defense Responses
- Alveolar Macrophages Are Essential for Protection from Respiratory Failure and Associated Morbidity following Influenza Virus Infection
- An Interaction between Glutathione and the Capsid Is Required for the Morphogenesis of C-Cluster Enteroviruses
- Concerted Spatio-Temporal Dynamics of Imported DNA and ComE DNA Uptake Protein during Gonococcal Transformation
- Potent Dengue Virus Neutralization by a Therapeutic Antibody with Low Monovalent Affinity Requires Bivalent Engagement
- Regulation of Human T-Lymphotropic Virus Type I Latency and Reactivation by HBZ and Rex
- Functionally Redundant RXLR Effectors from Act at Different Steps to Suppress Early flg22-Triggered Immunity
- The Pathogenic Mechanism of the Virulence Factor, Mycolactone, Depends on Blockade of Protein Translocation into the ER
- Role of Calmodulin-Calmodulin Kinase II, cAMP/Protein Kinase A and ERK 1/2 on -Induced Apoptosis of Head Kidney Macrophages
- An Overview of Respiratory Syncytial Virus
- First Experimental Model of Enhanced Dengue Disease Severity through Maternally Acquired Heterotypic Dengue Antibodies
- Binding of Glutathione to Enterovirus Capsids Is Essential for Virion Morphogenesis
- IFITM3 Restricts Influenza A Virus Entry by Blocking the Formation of Fusion Pores following Virus-Endosome Hemifusion
- Parasite Fate and Involvement of Infected Cells in the Induction of CD4 and CD8 T Cell Responses to
- Deficient IFN Signaling by Myeloid Cells Leads to MAVS-Dependent Virus-Induced Sepsis
- Pernicious Pathogens or Expedient Elements of Inheritance: The Significance of Yeast Prions
- The HMW1C-Like Glycosyltransferases—An Enzyme Family with a Sweet Tooth for Simple Sugars
- The Expanding Functions of Cellular Helicases: The Tombusvirus RNA Replication Enhancer Co-opts the Plant eIF4AIII-Like AtRH2 and the DDX5-Like AtRH5 DEAD-Box RNA Helicases to Promote Viral Asymmetric RNA Replication
- Mining Herbaria for Plant Pathogen Genomes: Back to the Future
- Inferring Influenza Infection Attack Rate from Seroprevalence Data
- A Human Lung Xenograft Mouse Model of Nipah Virus Infection
- Mast Cells Expedite Control of Pulmonary Murine Cytomegalovirus Infection by Enhancing the Recruitment of Protective CD8 T Cells to the Lungs
- Cytosolic Peroxidases Protect the Lysosome of Bloodstream African Trypanosomes from Iron-Mediated Membrane Damage
- Abortive T Follicular Helper Development Is Associated with a Defective Humoral Response in -Infected Macaques
- JC Polyomavirus Infection Is Strongly Controlled by Human Leucocyte Antigen Class II Variants
- Cationic Antimicrobial Peptides Promote Microbial Mutagenesis and Pathoadaptation in Chronic Infections
- Estimating the Fitness Advantage Conferred by Permissive Neuraminidase Mutations in Recent Oseltamivir-Resistant A(H1N1)pdm09 Influenza Viruses
- Progressive Accumulation of Activated ERK2 within Highly Stable ORF45-Containing Nuclear Complexes Promotes Lytic Gammaherpesvirus Infection
- Caspase-1-Like Regulation of the proPO-System and Role of ppA and Caspase-1-Like Cleaved Peptides from proPO in Innate Immunity
- Is Required for High Efficiency Viral Replication
- Modified Vaccinia Virus Ankara Triggers Type I IFN Production in Murine Conventional Dendritic Cells via a cGAS/STING-Mediated Cytosolic DNA-Sensing Pathway
- Evidence That Bank Vole PrP Is a Universal Acceptor for Prions
- Rapid Response to Selection, Competitive Release and Increased Transmission Potential of Artesunate-Selected Malaria Parasites
- Inactivation of Genes for Antigenic Variation in the Relapsing Fever Spirochete Reduces Infectivity in Mice and Transmission by Ticks
- Exposure-Dependent Control of Malaria-Induced Inflammation in Children
- A Neutralizing Anti-gH/gL Monoclonal Antibody Is Protective in the Guinea Pig Model of Congenital CMV Infection
- The Apical Complex Provides a Regulated Gateway for Secretion of Invasion Factors in
- A Highly Conserved Haplotype Directs Resistance to Toxoplasmosis and Its Associated Caspase-1 Dependent Killing of Parasite and Host Macrophage
- A Quantitative High-Resolution Genetic Profile Rapidly Identifies Sequence Determinants of Hepatitis C Viral Fitness and Drug Sensitivity
- Histone Deacetylase Inhibitor Romidepsin Induces HIV Expression in CD4 T Cells from Patients on Suppressive Antiretroviral Therapy at Concentrations Achieved by Clinical Dosing
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The 2010 Cholera Outbreak in Haiti: How Science Solved a Controversy
- , , , Genetic Variability: Cryptic Biological Species or Clonal Near-Clades?
- Efficient Parvovirus Replication Requires CRL4-Targeted Depletion of p21 to Prevent Its Inhibitory Interaction with PCNA
- An Overview of Respiratory Syncytial Virus
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



