-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaMethionine Biosynthesis in Is Tightly Controlled by a Hierarchical Network Involving an Initiator tRNA-Specific T-box Riboswitch
In line with the key role of methionine in protein biosynthesis initiation and many cellular processes most microorganisms have evolved mechanisms to synthesize methionine de novo. Here we demonstrate that, in the bacterial pathogen Staphylococcus aureus, a rare combination of stringent response-controlled CodY activity, T-box riboswitch and mRNA decay mechanisms regulate the synthesis and stability of methionine biosynthesis metICFE-mdh mRNA. In contrast to other Bacillales which employ S-box riboswitches to control methionine biosynthesis, the S. aureus metICFE-mdh mRNA is preceded by a 5′-untranslated met leader RNA harboring a T-box riboswitch. Interestingly, this T-box riboswitch is revealed to specifically interact with uncharged initiator formylmethionyl-tRNA (tRNAifMet) while binding of elongator tRNAMet proved to be weak, suggesting a putative additional function of the system in translation initiation control. met leader RNA/metICFE-mdh operon expression is under the control of the repressor CodY which binds upstream of the met leader RNA promoter. As part of the metabolic emergency circuit of the stringent response, methionine depletion activates RelA-dependent (p)ppGpp alarmone synthesis, releasing CodY from its binding site and thereby activating the met leader promoter. Our data further suggest that subsequent steps in metICFE-mdh transcription are tightly controlled by the 5′ met leader-associated T-box riboswitch which mediates premature transcription termination when methionine is present. If methionine supply is limited, and hence tRNAifMet becomes uncharged, full-length met leader/metICFE-mdh mRNA is transcribed which is rapidly degraded by nucleases involving RNase J2. Together, the data demonstrate that staphylococci have evolved special mechanisms to prevent the accumulation of excess methionine. We hypothesize that this strict control might reflect the limited metabolic capacities of staphylococci to reuse methionine as, other than Bacillus, staphylococci lack both the methionine salvage and polyamine synthesis pathways. Thus, methionine metabolism might represent a metabolic Achilles' heel making the pathway an interesting target for future anti-staphylococcal drug development.
Published in the journal: . PLoS Pathog 9(9): e32767. doi:10.1371/journal.ppat.1003606
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1003606Summary
In line with the key role of methionine in protein biosynthesis initiation and many cellular processes most microorganisms have evolved mechanisms to synthesize methionine de novo. Here we demonstrate that, in the bacterial pathogen Staphylococcus aureus, a rare combination of stringent response-controlled CodY activity, T-box riboswitch and mRNA decay mechanisms regulate the synthesis and stability of methionine biosynthesis metICFE-mdh mRNA. In contrast to other Bacillales which employ S-box riboswitches to control methionine biosynthesis, the S. aureus metICFE-mdh mRNA is preceded by a 5′-untranslated met leader RNA harboring a T-box riboswitch. Interestingly, this T-box riboswitch is revealed to specifically interact with uncharged initiator formylmethionyl-tRNA (tRNAifMet) while binding of elongator tRNAMet proved to be weak, suggesting a putative additional function of the system in translation initiation control. met leader RNA/metICFE-mdh operon expression is under the control of the repressor CodY which binds upstream of the met leader RNA promoter. As part of the metabolic emergency circuit of the stringent response, methionine depletion activates RelA-dependent (p)ppGpp alarmone synthesis, releasing CodY from its binding site and thereby activating the met leader promoter. Our data further suggest that subsequent steps in metICFE-mdh transcription are tightly controlled by the 5′ met leader-associated T-box riboswitch which mediates premature transcription termination when methionine is present. If methionine supply is limited, and hence tRNAifMet becomes uncharged, full-length met leader/metICFE-mdh mRNA is transcribed which is rapidly degraded by nucleases involving RNase J2. Together, the data demonstrate that staphylococci have evolved special mechanisms to prevent the accumulation of excess methionine. We hypothesize that this strict control might reflect the limited metabolic capacities of staphylococci to reuse methionine as, other than Bacillus, staphylococci lack both the methionine salvage and polyamine synthesis pathways. Thus, methionine metabolism might represent a metabolic Achilles' heel making the pathway an interesting target for future anti-staphylococcal drug development.
Introduction
Staphylococci are important skin and mucosa commensals but also major human pathogens. The most pathogenic species Staphylococcus aureus causes a wide range of diseases and, together with coagulase-negative staphylococci (CoNS), accounts for approximately 30 per cent of all hospital-acquired infections [1]. The development of antibiotic resistance in staphylococci increasingly limits therapeutic options and is a matter of major concern [2]. In recent years, studies into staphylococcal metabolism and its possible links to bacterial virulence have become a major focus of research but basic metabolic pathways remained largely unexploited in the development of new antibiotic drugs [3]. In this study, we investigate the regulation of methionine biosynthesis in staphylococci. Methionine and its chemical derivatives have important functions in the cell. For example, (formyl-)methionine is the universal N-terminal amino acid of nearly all proteins and therefore plays an eminent role in the initiation of protein biosynthesis. Moreover, the methionine derivative S-adenosylmethionine (SAM) serves as a methyl group donor in a variety of cellular processes and is the precursor molecule in polyamine synthesis [4]. Many microorganisms are able to synthesize methionine de novo and staphylococci employ the trans-sulfuration pathway to generate methionine [5]. Most bacteria from the order Bacillales are thought to control this pathway by SAM-binding S-box riboswitches [5], [6], [7]. Interestingly, in silico analysis predicts the presence of a T-box riboswitch in the 5′-untranslated region of the methionine biosynthesis operon (metICFE-mdh operon) in staphylococci [5], [6], [8], suggesting the use of alternative mechanisms to regulate methionine synthesis. T-box riboswitches are transcriptional control systems which have been extensively studied in Bacillus subtilis and other Firmicutes (reviewed in [6]). Their function is controlled by specific interactions and differential binding to charged and uncharged cognate tRNA, respectively, thus providing a means to “sense” the amino acid concentration in the cell [9]. T-box leader RNA/tRNA interaction essentially occurs at two sites: (i) the tRNA anticodon basepairs with the specifier-loop domain of the T-box leader RNA ensuring specific binding of the respective T-box element with its cognate tRNA; (ii) the free 3′-CCA end of an uncharged tRNA binds to the T-box motif, thereby triggering the formation and stabilization of an antiterminator which enables transcription of downstream genes [9]. In this study, we characterized interactions of the metICFE-mdh leader RNA with methionyl-tRNAs and demonstrate that they represent a functional T-box riboswitch that preferentially binds to initiator formylmethionyl-tRNA (tRNAifMet). We further show that, in staphylococci, T-box control of methionine biosynthesis has a key role in a complex regulatory network that also involves stringent response-mediated CodY regulation and RNA decay to tightly control this pathway.
Results
A 5′ met leader RNA harboring a T-box riboswitch precedes the methionine biosynthesis operon
The methionine biosynthesis genes (metI, metC, metF, metE, mdh (metal-dependent hydrolase)) are organized in an operon-like structure and are annotated as SACOL0431 - SACOL0427 in S. aureus COL and as NWMN_0351 - NWM_0347 in S. aureus Newman, respectively, with metF being named metH in the latter strain (Figure 1A). Northern blot analysis using a double-stranded DNA probe confirmed the expression of a stable transcript of approximately 400 nucleotides (nt) from the intergenic region (IGR) upstream of metI (Figure 1B, left panel). Hybridization employing in vitro-transcribed RNA probes revealed that the orientation of the transcript was identical to that of the metICFE-mdh operon (Figure 1B, middle and right panels). 5′ - and 3′-RACE experiments identified a single transcription start site and a transcript length of 439 nt (Figure 1C). Sequence analysis of various S. aureus and S. epidermidis strains demonstrated that the region is highly conserved (SI Figure S1) but lacks ribosomal binding sites and open reading frames, suggesting specific (non-coding) functions of the transcript, for example, as a 5′-untranslated region (5′-UTR) of the metICFE-mdh RNA. Interestingly, a putative binding site for the repressor protein CodY [10], [11] could be identified next to the 5′-UTR promoter region (Figure 1C). Most striking was the presence of a highly conserved canonical T-box sequence motif (5′-AAGGUGGUACCGCG-3′) which partially overlapped with a strong Rho-independent transcription termination signal in the 3′-portion of the transcript (Figure 1C). Overall, the transcript, which was named met leader RNA, harbored all the characteristics of previously characterized T-box riboswitches from Bacillus subtilis, and sequence alignments with Bacillus T-box systems led to a putative structural model of the Staphylococcus met leader RNA (SI Figure S2).
Fig. 1. Determination of a leader RNA transcript upstream of methionine biosynthesis genes. 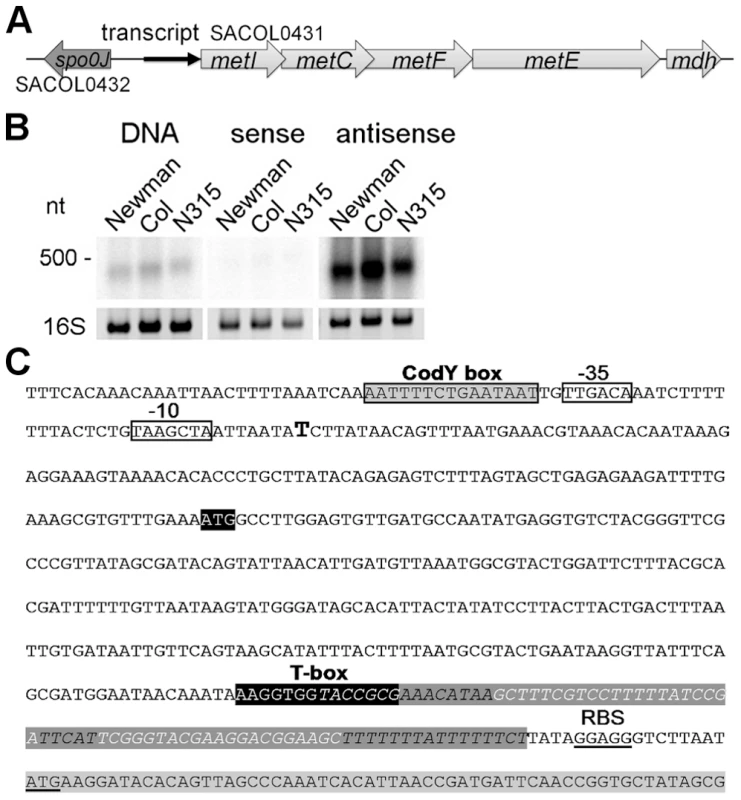
(A) Overview of the genomic organization of the methionine biosynthesis operon. A transcript (black arrow) was detected upstream of metI. (B) Northern blot analysis of total RNA from three S. aureus strains (Newmann, COL and N315) for the intergenic region (IGR) between spo0J and metI. Radiolabeled probes were either the PCR product (DNA) or in vitro transcribed RNA of each strand (sense and antisense) of the IGR. The experiment was done in duplicate. Fragment sizes correspond to a high-range RNA ladder (Fermentas). The 16S rRNA is shown as a loading control in the corresponding agarose gel. (C) Sequence of the IGR upstream of metI in S. aureus COL. Transcription start of the met leader RNA, as experimentally determined by 5′ RACE, is indicated by a bold ‘T’; putative -35 and -10 promoter sites are boxed; the CodY binding motif is boxed in grey. The potential specifier box and the highly conserved T-box motif are shown in black; overlapping with the T-box is a predicted Rho-independent transcription terminator (in italics and dark grey). The Shine-Dalgarno sequence (SD) and the start codon of the metI gene (light grey) are underlined. The met leader RNA specifically binds uncharged tRNAifMet
First, we tested if the Staphylococcus met leader RNA interacts specifically with methionyl-tRNAs (Figure 2A). met leader RNA was in vitro transcribed in the presence of the appropriate radioactively labeled tRNA species. Binding between radioactively labeled tRNA and in vitro-transcribed met leader RNA was determined by non-denaturing polyacrylamide gel electrophoresis and autoradiography. Staphylococci genomes harbor four methionyl-tRNA gene loci, two of which (tRNA-Met-1 and -2) being identical and representing the initiator tRNAifMet. Binding studies using tRNAifMet, tRNAMet3 and tRNAMet4, respectively, with free 3′-CCA ends revealed that the met leader RNA interacted strongly with tRNAifMet while interactions with tRNAMet3 and tRNAMet4 proved to be weak (Figure 2B). tRNAifMet binding to met leader RNA increased linearly within a 5-fold molar range (Figure 2C). In contrast, binding was abolished when the 3′-end of tRNAifMet included one additional cytosine, mimicking a charged tRNA molecule (3′-CCAC; AdC in Figure 2D). Also, no interaction was detectable in the presence of cysteinyl-tRNA, regardless of whether 3′-CCA or 3′-CCAC was present at the 3′ end (Figure 2C, D).
Fig. 2. In vitro binding of met leader RNA to tRNAs. 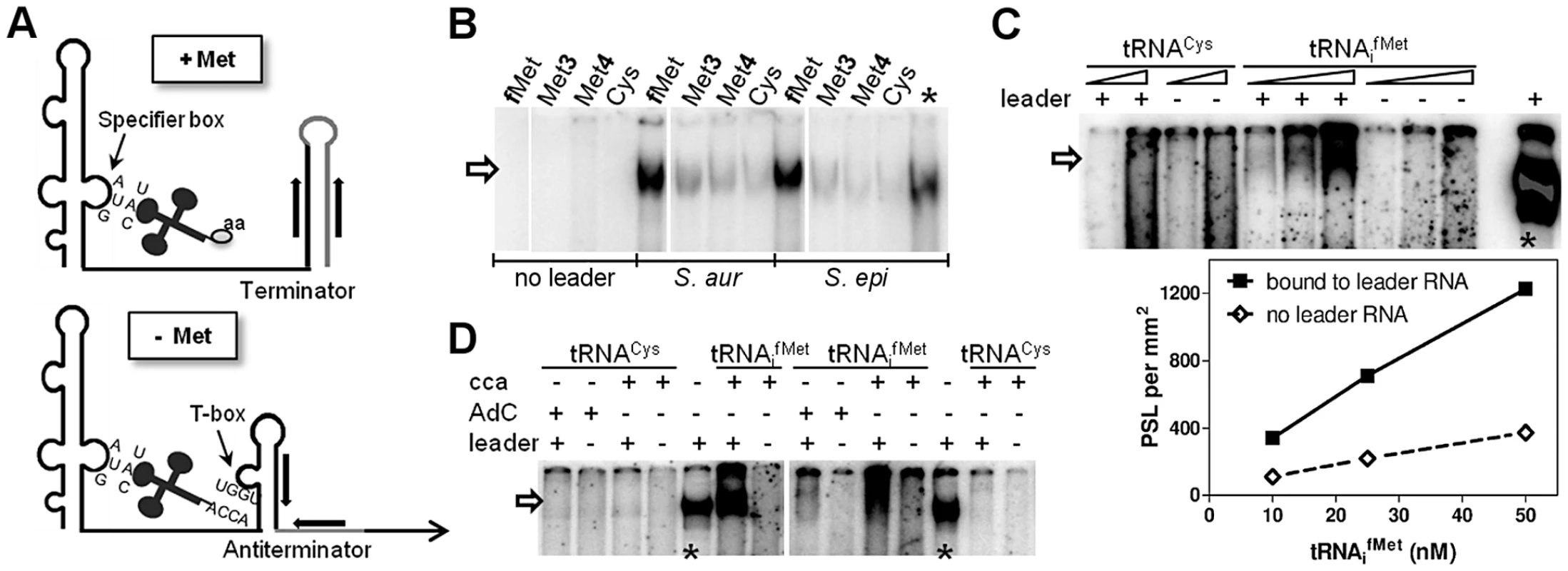
(A) Schematic of the binding interaction between tRNA and T-box leader RNA according to Bacillus T-box systems [9]. On the top (‘+ Met’), system under high intracellular levels of methionine, on the bottom (‘− Met’) under methionine starvation. Indicated are tRNAs with an amino acid (‘aa’, top) or a free 3′-CCA end (bottom) and the respective pairing sequences within the T-box leader RNA. (B–D) The met leader RNA was transcribed in vitro by T7 RNA polymerase from a defined DNA template. Where indicated, preformed and radiolabeled tRNA was present during the in vitro transcription (IVT) reaction. Samples were analyzed by a non-denaturing PAGE. Asterisks mark IVT met leader RNA reactions without tRNA, but with [α32P]-CTP present as an IVT efficiency control. The arrow indicates tRNAs bound to the met leader RNA. All experiments were performed at least twice. (B) Methionine-specific tRNA from the different genomic loci (tRNAifMet, tRNAMet3 and tRNAMet4) or tRNACys were present in the IVT. The leader RNA was transcribed from either the S. aureus COL or S. epidermidis RP62A sequence. (C) Increasing concentrations of each tRNA with 3′-CCA end (10 and 50 nM for tRNACys and 10, 25 and 50 nM for tRNAifMet, respectively) were present during IVT. Bound tRNAifMet was quantified by measuring the Photo-Stimulated Luminescence (PSL), which is proportional to the amount of radiation exposed to the IP plate. The PSL values are expressed per mm2 (y-axis) against the tRNA molarity (x-axis). (D) Either formylmethionine- (tRNAifMet) or cysteine- (tRNACys) specific tRNA was present during IVT. Two different tRNA species were used: with a free 3′-CCA end (‘cca’) or with an additional cytosine (‘AdC’) at the 3′-CCA end to mimic amino acid charging [31]. tRNAifMet binding requires the T-box motif
In classical T-box riboswitches, tRNA/leader RNA interaction is mediated by the T-box motif which forms a bulge that facilitates basepairing interactions with the tRNA 3′-CCA and supports antiterminator formation [9] (Figure 2A). We therefore studied whether the predicted T-box motif participates in tRNAifMet binding by generating a series of met leader RNAs carrying T-box mutations (SI Table S2, Figure 3A). tRNAifMet binding was clearly diminished in mutants SC2 and SC5, which are both likely to lack the putative T-box bulge for tRNA 3′-CCA interaction (Figure 3A). Also, in SC8, a single U to A exchange at position 363 was sufficient to reduce tRNAifMet binding, whereas other mutations within the putative T-box bulge, i.e. SC3, SC4, SC6 and SC7, enhanced tRNAifMet interactions with the met leader RNA (Figure 3B). Finally, alteration of a methionine-specific codon AUG to cysteine UGC in the putative specifier box of the met leader RNA in SC1 did not affect tRNAifMet binding efficiency (Figure 3B), nor did it confer tRNACys binding activity (SI Figure S3) suggesting that other components of this T-box system confer specificity to initiator tRNAifMet binding. Taken together, these data suggest that the met leader RNA upstream of the staphylococcal metICFE-mdh operon harbors a canonical T-box riboswitch that specifically binds uncharged initiator tRNAifMet.
Fig. 3. Effect of single nucleotide exchanges in the T-box on tRNA binding. 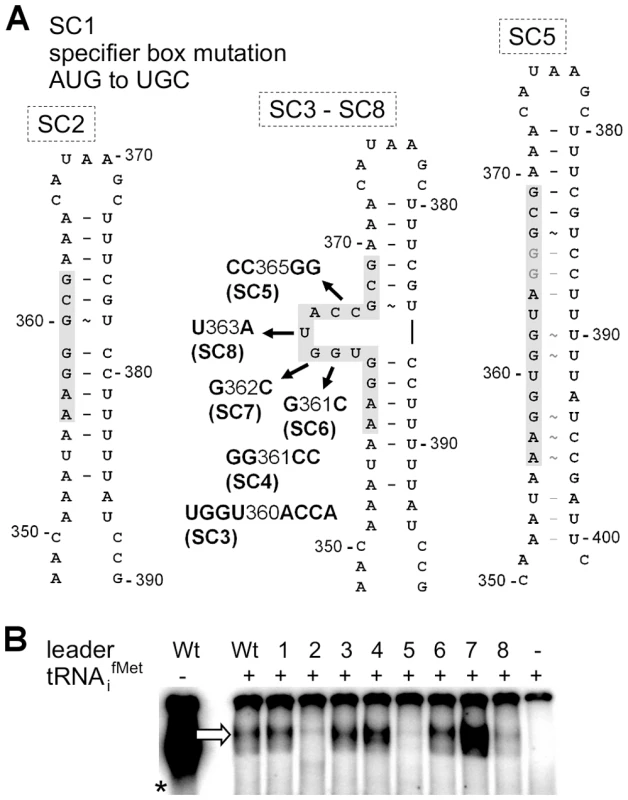
(A) Overview on mutations introduced into the met leader RNA. SI Table S2 lists all mutated constructs. The model of the antiterminator structure with the conserved T-box (in grey) demonstrates deletion of the side bulge in SC2, nucleotide substitutions of constructs SC3 to SC8 and the possible loss of the side bulge in SC5 through alternative base pairing. Nucleotide positions refer to the complete length of the met leader RNA of S. aureus COL. (B) IVT with WT or mutated constructs SC1 to SC8 (lanes 1–8) of the met leader RNA template with radiolabeled tRNAifMet present. The arrow indicates the bound tRNAifMet. The asterisk marks the control IVT reaction of the WT met leader RNA transcribed without tRNA, but in the presence of [α32P]-CTP. Results are representative of two independent experiments. Methionine starvation induces met leader RNA/metICFE-mdh transcription
Through interaction with either uncharged tRNAs (antiterminator formation) or charged tRNAs (terminator formation), T-box riboswitches indirectly sense amino acid levels in the bacterial cell [6], [9]. To determine whether or not met leader RNA/metICFE-mdh transcription is sensitive to methionine availability, S. aureus strain Newman was grown in chemically defined medium (CDM) in the presence or absence of methionine. RNA was isolated in the early exponential (E1), exponential (E2) and early stationary (S) growth phase and analyzed by Northern hybridization using met leader RNA - and metI-specific DNA probes, respectively (Figure 4A, B). In the presence of methionine, basal met leader RNA transcription could be detected. Methionine starvation induced the transcription of this RNA, especially during exponential growth (Figure 4A). In contrast, the metI mRNA signal was not detectable in the presence of methionine (Figure 4B). Upon methionine deprivation, however, metI mRNA transcription was activated with the strongest expression detected during exponential growth (Figure 4B). Interestingly, metI-specific Northern probing did not reveal a distinct fragment representing the full-length metICFE-mdh mRNA (Figure 4B). Instead, an RNA smear was detected in repeated experiments. As the RNA quality and integrity had been confirmed prior to the experiments, the Northern blot data suggest rapid degradation of the methionine starvation-induced metICFE-mdh transcript.
Fig. 4. Role of CodY in met leader RNA/metICFE-mdh transcription control. 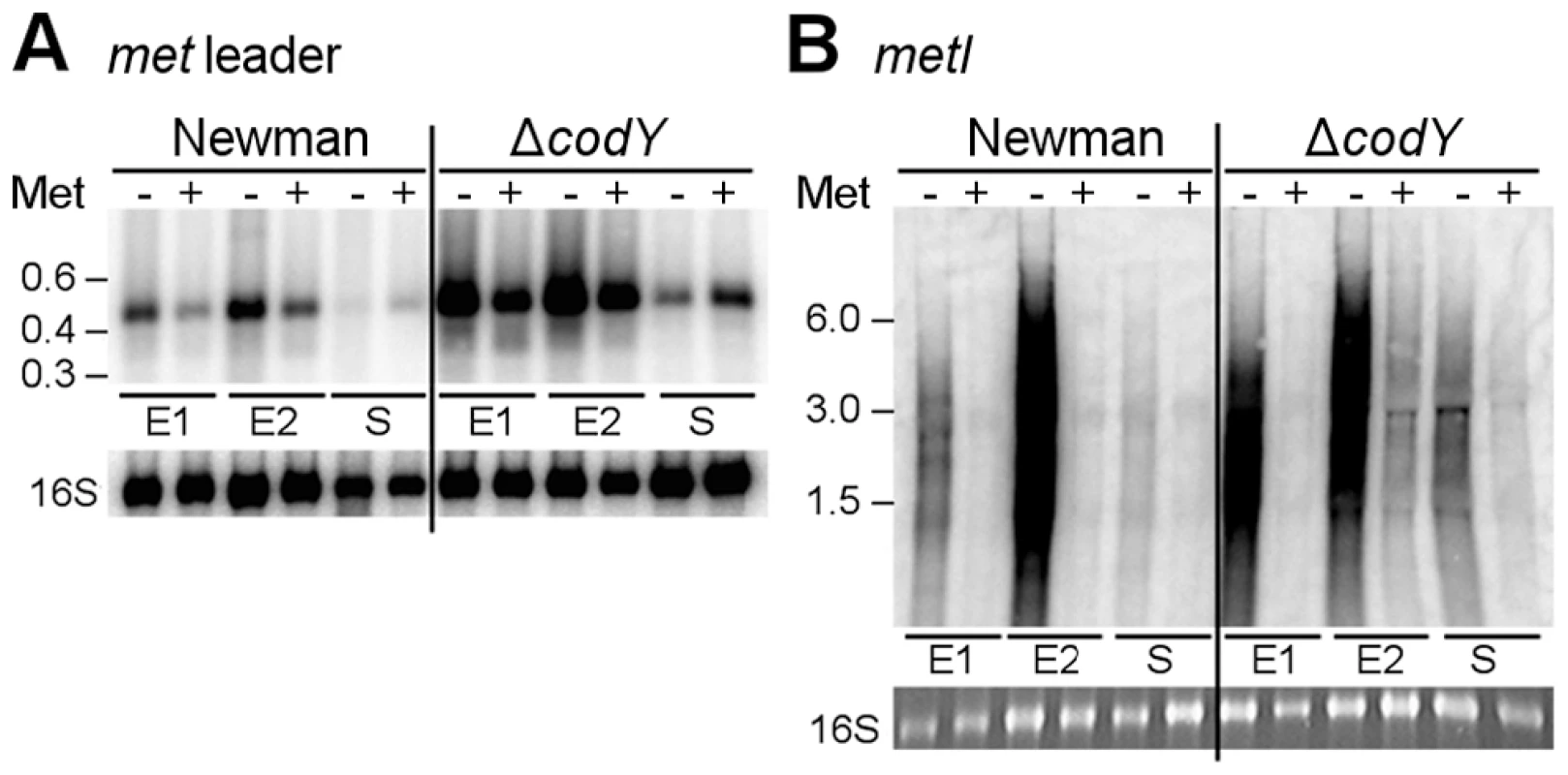
Northern blot analysis with radiolabeled probes. Total RNA from S. aureus strain Newman and its isogenic codY mutant was sampled at three different points of growth: early exponential (E1), exponential (E2) and early stationary phase (S). Cultures were grown in CDM without (‘−’) or supplemented with 1 mM L-methionine (‘+’). (A) Hybridization with the met leader RNA-specific probe (low range RNA ladder (Fermentas) indicated on the left). The blot was re-hybridized with a 16S rDNA-specific probe, for loading control. (B) Hybridization with metI-specific probe (high range RNA ladder (Fermentas) indicated on the left). The 16S rRNA signal on the gel is shown below. Results are representative of two independent experiments. CodY controls met leader RNA transcription in response to methionine supply
CodY is a global transcription repressor that controls the expression of a variety of genes in S. aureus, many of which being involved in amino acid biosynthesis and transport [10], [11]. Detection of a consensus sequence for CodY binding upstream of the met leader RNA and the recent identification of the methionine biosynthesis genes as direct CodY targets in S. aureus [10] prompted us to study the role of this factor for met leader/metICFE-mdh transcription in more detail. A S. aureus codY deletion mutant was grown in CDM with and without methionine and analyzed for met leader and metICFE-mdh transcription by Northern blot hybridization. In the absence of methionine, a stronger met leader RNA signal was detected in both the wildtype and the codY mutant (Figure 4A). However, compared to the wildtype, the codY mutant showed a generally enhanced met leader transcription, suggesting that met leader RNA expression was de-repressed in the absence of CodY irrespective of whether methionine was present or not (Figure 4A). In contrast, downstream metICFE-mdh operon expression remained sensitive to varying concentrations of methionine and was only activated both in the wildtype and the codY mutant in the absence of methionine (Figure 4B).
Methionine depletion is signaled to CodY via the stringent response
In many bacteria, nutrient limitation triggers the so-called stringent response to appropriately adjust gene expression patterns. Stringent response is characterized by the rapid synthesis of the alarmone (p)ppGpp involving bifunctional RelA/SpoT synthetases/hydrolases (RSHs) and affecting/modulating many cellular functions. Recently, a link between CodY and the stringent response of S. aureus has been demonstrated [12]. The CodY repressor function depends on its two effector molecules GTP and branched-chain amino acids (BCAA, valine, leucine, isoleucine) which enhance synergistically the affinity of CodY for its DNA targets [13], [14]. RSH-mediated (p)ppGpp synthesis lowers the GTP levels in the cell and eventually facilitates release of CodY from its DNA targets [12]. In a next set of experiments, we sought to identify possible regulatory links between methionine deficiency, stringent response and CodY. For this purpose, met leader RNA/metICFE-mdh transcription upon methionine depletion was studied in a rsh mutant carrying a deletion of the (p)ppGpp synthetase domain in strain S. aureus Newman [12]. As a marker for stringent response-controlled genes, brnQ-1, which encodes a CodY-repressed BCAA permease, was included in the analysis. In the S. aureus wildtype, methionine depletion led to brnQ-1 induction along with met leader RNA and metICFE-mdh expression (Figure 5A). In contrast, in the rsh mutant, induction of both brnQ-1 and met leader/metICFE-mdh transcription were significantly reduced upon methionine starvation in comparison to the wildtype, suggesting that RSH-mediated (p)ppGpp synthesis may be required for efficient activation of the system. Deletion of codY resulted in a generally higher basal transcription of both brnQ-1 and met leader RNA in the presence of methionine, whereas metICFE-mdh transcription remained tightly controlled and switched off under these conditions (Figure 5A). In the codY mutant, brnQ-1 expression was de-repressed and not further inducible by methionine deprivation, suggesting that CodY is responsible for the methionine-dependent brnQ-1 induction observed in the wildtype (Figure 5A).
Fig. 5. Stringent response relay and involvement of RNases. 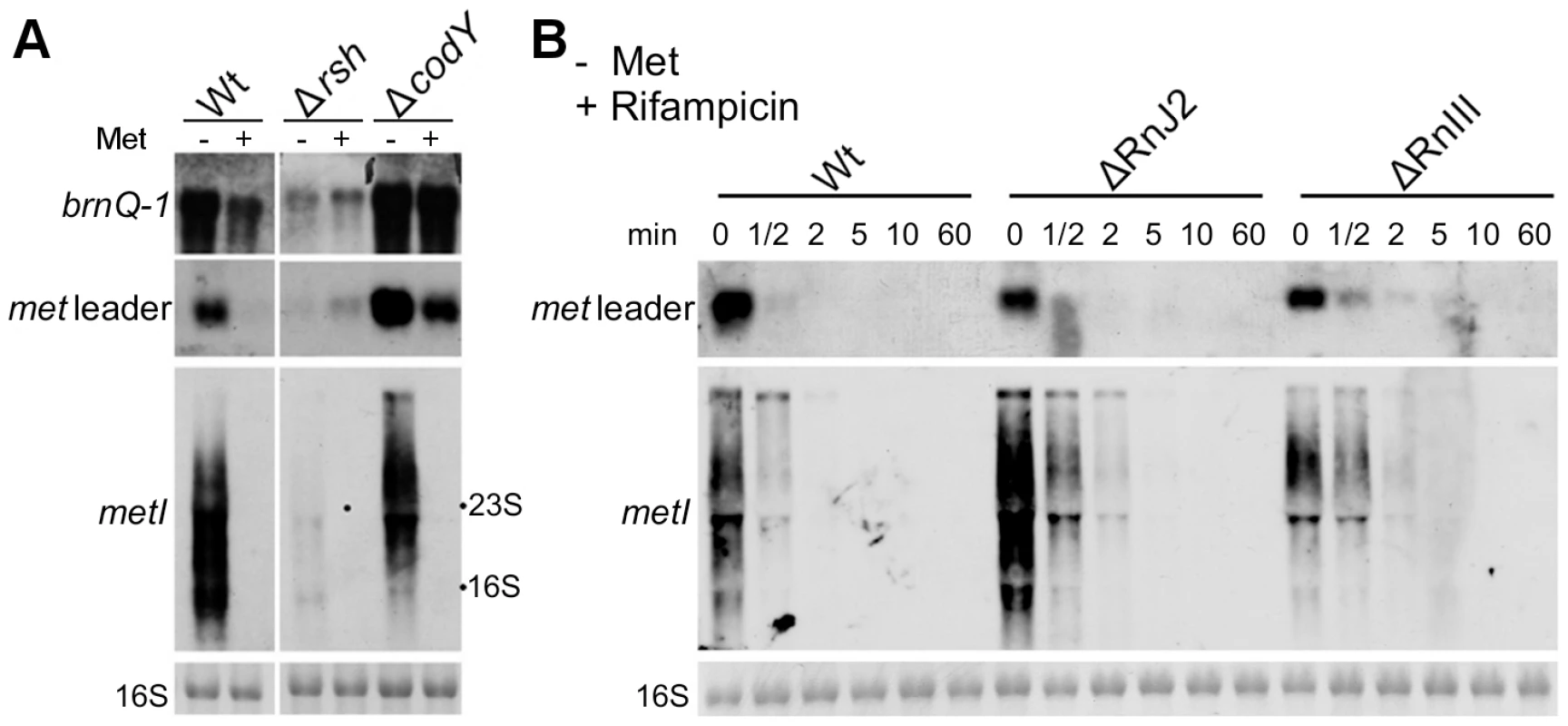
(A) Northern blot analysis of total RNA from S. aureus strain Newman (‘Wt’) and different isogenic mutants (‘Δ’) sampled at exponential growth phase. The cultures were grown in CDM without (‘−’) or supplemented with 1 mM L-methionine (‘+’). DIG-labeled DNA probes for brnQ-1 (branched-chain amino acid transport), the met leader RNA or metI were used to hybridize RNA from WT, the rsh (ppGpp synthetase) and a codY mutant. The data are representative of two independent experiments. (B) RNA stability assay. Northern blot analyses of met leader RNA (upper panel) and metICFE-mdh mRNA (lower panel) upon rifampicin exposure of S. aureus strain Newman (‘Wt’) and isogenic RNase J2 and RNase III deletion mutants (‘ΔRnJ2’, ‘ΔRnIII’), respectively. Bacteria were grown in CDM without methionine and total RNA was prepared from cells before (0 min) and after the addition of 500 µg ml−1 rifampicin at the time points indicated in the figure. Blots were hybridized with DIG-labeled met leader- and metI-specific DNA probes, respectively. Bottom panels in (A) and (B) show the 16S rRNA as loading controls in the corresponding agarose gels. RNase J2 and RNase III participate in met leader RNA and metICFE-mdh mRNA decay
The experiments described in Figure 4 suggest that metICFE-mdh mRNA is subject to rapid degradation. To investigate the possible involvement of specific RNases in this process, the stability of met leader RNA and metICFE-mdh was analyzed in S. aureus mutants that were deficient in RNase J2 and RNase III activity, respectively. For this purpose, de novo RNA synthesis was interrupted by the addition of the RNA polymerase inhibitor rifampicin to the cultures. Total RNA was isolated at different time points and subjected to Northern blot analysis (Figure 5B). Comparison of the wildtype and the RNase-deficient mutants revealed that the metICFE-mdh transcript was more stable in the RNase J2 mutant, suggesting that RNase J2 may be involved in metICFE-mdh degradation. In contrast, no significant effect on met leader RNA stability was detectable in the RNase J2 mutant (Figure 5B, upper panel). The RNase III mutant exhibited a slightly enhanced stability both of the met leader and metICFE-mdh RNA indicating a possible function of RNase III in met leader RNA and metICFE-mdh decay (Figure 5B).
Discussion
The staphylococcal met leader RNA interacts with initiator formylmethionyl-tRNA
In this study, we show that methionine biosynthesis control in S. aureus involves a T-box riboswitch. While the conservation of this T-box in staphylococci was predicted previously using bioinformatic tools [5], [6], [8], we now provide, for the first time, direct experimental proof for a specific interaction of the predicted T-box with initiator tRNAifMet. While methionyl-tRNA-specific T-box riboswitches (met-T-box) are rare among Bacillales they are more common in Lactobacillales. In both orders they are associated with methionine metabolism or transport (Table 1). Methionyl-tRNAs (tRNAMet) are encoded by four distinct gene loci in the genomes of S. aureus and S. epidermidis. Two of them are identical and represent the initiator tRNAifMet, while the other two tRNAMet loci differ in their nucleotide sequence from each other and from the tRNAifMet. Surprisingly, we found a clear preference for interaction of the met leader RNA T-box with the initiator tRNAifMet (Figure 2B). In prokaryotes, the first N-terminal methionine of newly synthesized proteins is N-formylated and, hence, N-formylmethionine (fMet) is indispensable for protein translation initiation and bacterial growth. fMet is carried to the ribosomal translation initiation complex by tRNAifMet which differs structurally from the elongator tRNAMet used for the incorporation of methionine residues into the growing polypeptide chain. Although all tRNAMet are charged with methionine by (the same) methionyl-tRNA synthetase, it is only tRNAifMet that is specifically recognized by the methionyl-tRNA-formyltransferase which then mediates N-formylation of methionine to produce fMet. At present, it is not clear how the observed specificity of the met leader RNA T-box for tRNAifMet is accomplished. An involvement of the putative specifier box in the 5′-region of the met leader RNA seems unlikely because all methionyl-tRNAs use the same anticodon, suggesting that other regions of the met leader RNA interact with structures that are unique to the initiator tRNAifMet. In line with this hypothesis, our data showed that tRNAifMet binding by the met leader RNA was not affected when the specifier box in the met leader was substituted with a cysteine-specific codon (Figure 3B). Also, these nucleotide replacements were insufficient to confer tRNACys binding activity (SI Figure S3). Our observation that the T-box riboswitch, shown in this study to be a key regulator of methionine biosynthesis in S. aureus, preferentially binds tRNAifMet points to an elegant mechanism by which protein translation initiation efficiency could both be sensed and, if necessary, adjusted by modulating fMet supply. It will be interesting to investigate if this tRNAifMet preference also applies to other met-T-box riboswitches that control the expression of genes not directly involved in methionine biosynthesis. Also, potential metabolic implications of the use of T-box-controlled fMet supply in staphylococci versus S-box-controlled methionine biosynthesis in other bacteria remain to be studied.
Tab. 1. Methionine metabolism and biosynthesis control among Bacillales and Lactobacillales. 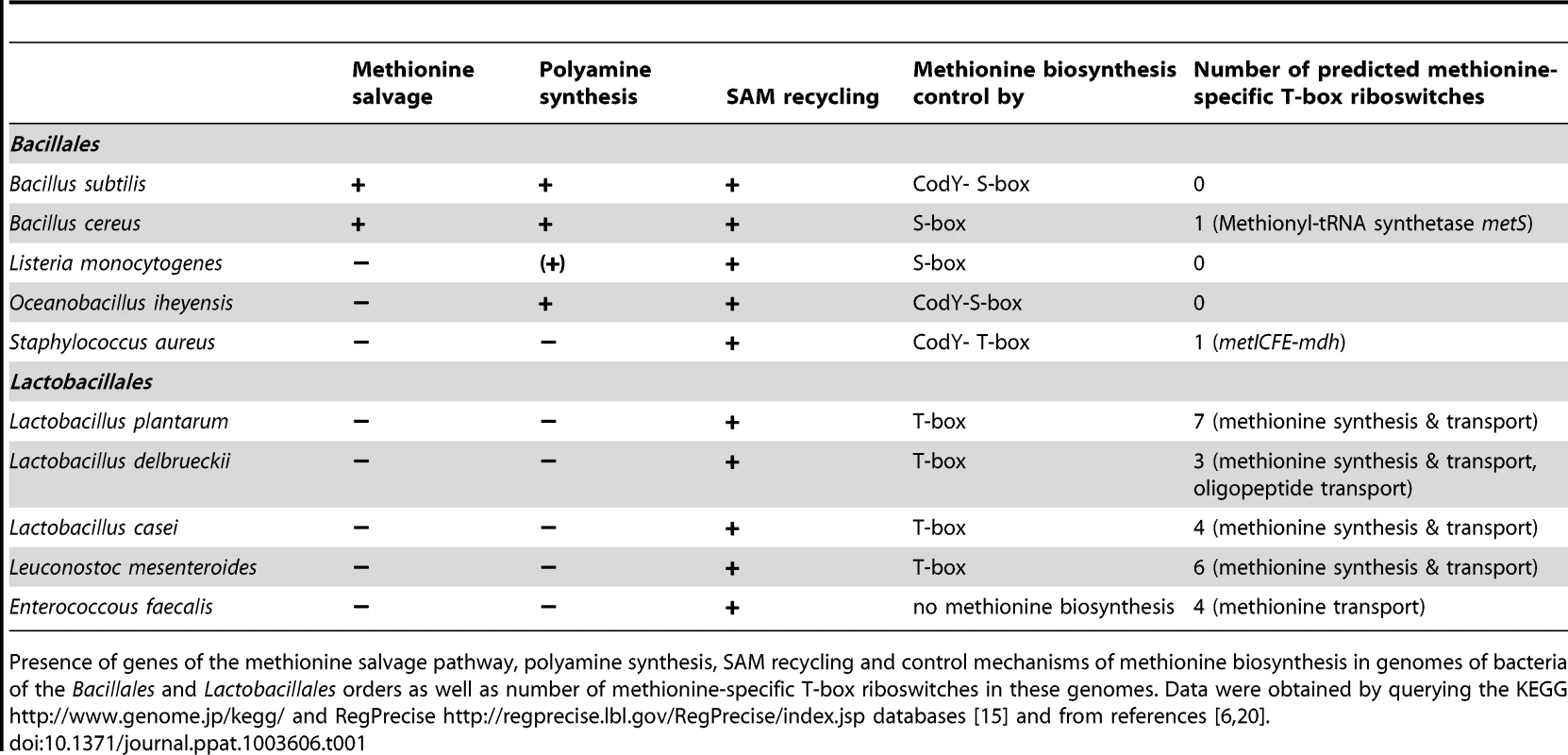
Presence of genes of the methionine salvage pathway, polyamine synthesis, SAM recycling and control mechanisms of methionine biosynthesis in genomes of bacteria of the Bacillales and Lactobacillales orders as well as number of methionine-specific T-box riboswitches in these genomes. Data were obtained by querying the KEGG http://www.genome.jp/kegg/ and RegPrecise http://regprecise.lbl.gov/RegPrecise/index.jsp databases [15] and from references [6], [20]. T-box-controlled methionine biosynthesis is linked to stringent response and CodY regulation
The data obtained in this study lead us to propose that a hierarchical regulatory network controls methionine biosynthesis in S. aureus, most likely, to minimize unnecessary de novo methionine biosynthesis. Centerpiece of this regulation turns out to be the tRNAifMet-specific T-box riboswitch located in the 5′-met leader that precedes the coding regions of the metICFE-mdh mRNA. Another important player is the global repressor CodY which drives met leader RNA transcription and links the system to the metabolic emergency circuit of the bacterial stringent response. Finally, staphylococcal RNases were implicated in this network by degrading both metICFE-mdh mRNA and met leader RNA, which may be considered a form of posttranscriptional control of metICFE-mdh gene expression. Figure 6 summarizes our major findings and suggests a model for the control of methionine biosynthesis in staphylococci. While stringent response-mediated CodY release and subsequent met leader RNA transcription are sensitive to general amino acid availability and the energy status of the cell, the T-box riboswitch is highly selective and ensures that downstream metICFE-mdh mRNA transcription only occurs if methionine concentration is low. The experiments also indicate that lack of methionine alone is sufficient to trigger the stringent response and, as a consequence, the release of CodY, thus securing efficient met leader RNA/metICFE-mdh transcription when needed (Figure 6). Interestingly, the regulatory cascade identified in this study seems to represent an exception rather than common rule. Thus, database searches of B. subtilis and S. aureus genomes revealed that combinations of CodY with T-box riboswitches are restricted to methionine and tryptophan biosynthesis in S. aureus and branched-chain amino acid (BCAA) biosynthesis in B. subtilis, respectively [15], [16] (Table 1).
Fig. 6. Model of a regulatory cascade for methionine biosynthesis operon control. 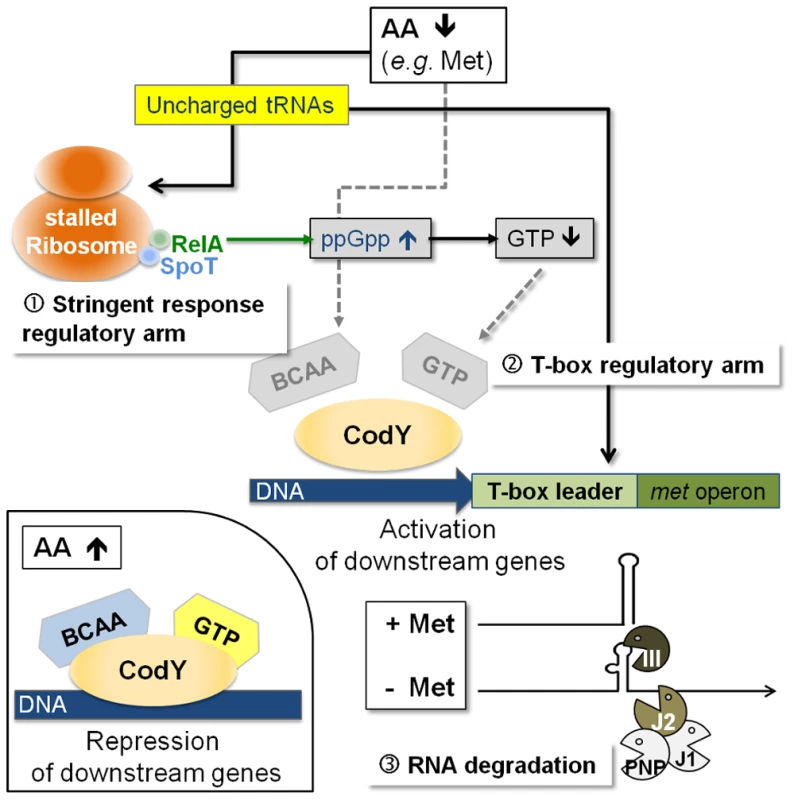
(i) With high amino acid levels, branched-chain amino acids (BCAA) and GTP are bound to the CodY repressor, increasing its affinity for target DNA binding; downstream genes are repressed (small picture, bottom left). (ii) Low amino acid levels will trigger the stringent response due to stalled ribosomes, which leads to an increase in RelA-mediated ppGpp alarmone synthesis resulting in less GTP. Subsequently, CodY dissociates from the DNA activating downstream transcription of the T-box leader RNA. The T-box acts as the crucial check-point sensing uncharged tRNAifMet levels and determines transcription of the met biosynthesis genes in a highly methionine-dependent manner. Rapid degradation of the met mRNA by the RNA degradosome is an additional mechanism to limit unnecessary translation of methionine biosynthesis mRNA. RNA decay is another checkpoint in methionine biosynthesis control
Methionine biosynthesis is a costly process consuming ATP and other resources. Riboswitches are generally regarded as fast and tight regulatory systems that do not depend on protein factors which, in many cases, react more slowly and/or are subject to complex regulation of protein expression, activation and degradation [5], [17]. Our study suggests that, in addition to regulation at the transcriptional level, S. aureus employs RNA decay mechanisms to quickly remove newly transcribed metICFE-mdh mRNA from the system, thus further limiting the risk of sustained (over)expression of genes involved in methionine synthesis. Although the complete mechanism and enzymes involved in the process still need to be established, the data give a first hint that RNase J2 participates in metICFE-mdh degradation (Figure 5B). This is in contrast to the situation in B. subtilis where mRNAs of the methionine biosynthesis genes, polyamine synthesis as well as the methionine salvage pathway (see below) were found to be not degraded by RNase J1/J2 [18]. The data further suggest that RNase III might be involved in met leader RNA degradation (Figure 5B). This observation is consistent with previously published data showing that RNase III co-immunoprecipitates with met leader RNA and targets also other S. aureus riboswitches for degradation [19]. Taken together, the data lead us to suggest that RNA decay is another mechanism involved in the control of methionine synthesis in staphylococci that merits further future investigation.
Why do staphylococci use a T-box riboswitch for methionine biosynthesis control?
T-box control of methionine biosynthesis genes in staphylococci is an exception among Bacillales which usually regulate this pathway by S-adenosylmethionine (SAM)-binding S-box riboswitches [5]. Apart from protein synthesis, most microorganisms use methionine to produce SAM which plays a central role in many cellular functions [4]. First, SAM serves as a methyl group donor for nucleic acid and protein methylation. Products of the methylation reaction are detoxified and recycled to homocysteine which is then reused for methionine/SAM synthesis. Second, SAM is used, following decarboxylation, to form polyamines. The remaining 5′-methylthioadenosine moiety is again metabolized to methionine by enzymes of the methionine salvage pathway. Comparative genomics using the KEGG database (http://www.genome.jp/kegg/) and experimental research [20], [21], [22] suggest that, unlike Bacillus, staphylococci may have only limited capacity to reuse or redirect methionine to other pathways because they lack both the methionine salvage and polyamine synthesis pathways (Table 1). Therefore, synthesis and recycling of SAM may be the only possibility to make use of excess methionine, implying that a more stringent control of de novo biosynthesis may be required in staphylococci. Interestingly, the Lactobacillales, which preferentially control their methionine biosynthesis genes by T-box riboswitches [5], also appear to lack polyamine synthesis and methionine salvage genes (Table 1). Based on these observations, we hypothesize that the lack of both polyamine synthesis and methionine salvage might favor control by a T-box rather than a S-box riboswitch, the major advantage being that the T-box riboswitch is able to sense methionine supply directly and then react immediately by switching off transcription of methionine biosynthesis genes, whereas S-box regulated systems would require an additional step (i.e. SAM synthesis) to produce the effector molecule required to stop methionine production. Alternatively, microorganisms that produce polyamines may need a larger SAM pool as precursor for the synthesis of these important compounds. Therefore, S-box control of methionine biosynthesis might be more effective in these organisms to ensure a constant SAM supply. More experimental work is needed to further substantiate these hypotheses.
The methionine pathway - a suitable anti-staphylococcal drug target?
The general life style of staphylococci provides easy access to methionine sources from the respective host they colonize or infect and, therefore, suggests that methionine supply may not be a limiting factor under normal conditions. The data presented in this paper provide first insight into the regulation of methionine synthesis gene expression in staphylococci and, more importantly, show that staphylococci have evolved special mechanisms to tightly restrict de novo methionine biosynthesis. It is tempting to speculate that overproduction (rather than lack) of methionine may be critical to staphylococci and, thus, this strict control of methionine de novo synthesis would not only save resources and energy but also meet the requirement to prevent methionine accumulation. While excess methionine biosynthesis might be critical for staphylococci under most conditions, de novo methionine biosynthesis becomes crucial for growth and survival when methionine supply is limited, for example, when entering the host during infection [23] or under specific external stress conditions like antibiotic and antimicrobial peptide exposure [24], [25]. It is therefore conceivable that both the efficient activation and shut-off of methionine biosynthesis might represent a metabolic “Achilles' heel” for staphylococci. Interestingly, successful inhibition of the S. aureus methionyl-tRNA synthetase by an experimental compound has already provided evidence that methionyl-tRNA metabolism is a suitable anti-staphylococcal target [26]. Also, structure-based drug design recently resulted in the identification of lead compounds that specifically interact with T-box structures in vitro, indicating that RNA-based drug targeting is a promising new avenue in medicinal chemistry [27], [28], [29]. The data presented in this paper may open perspectives for specifically targeting methionine metabolism and protein translation initiation in future efforts to develop novel Staphylococcus-specific antibiotics.
Materials and Methods
Bacterial strains, construction of mutants, growth conditions as well as RNA isolation and Northern blot procedures can be found in the SI Materials and Methods (Text S1, Table S1–S5).
3′ and 5′ RACE experiments
Total RNA was treated with recombinant DNase I (Ambion) and RNA quality was checked with the Agilent 2100 Bioanalyzer (Agilent Technologies). The 5′/3′ RACE Kit, 2nd Generation (Roche) was used for determination of transcript ends by synthesis of first-strand cDNA with reverse transcriptase (Fermentas) and the oligonucleotides listed in Table S4 (Text S1).
Site directed mutagenesis of the met leader RNA template
The met leader RNA sequence of S. aureus COL was amplified by PCR with oligonucleotides T7-F_met-sRNA and R_met-sRNA (Tables S5, Text S1). The product was inserted into the pGEM-T Easy vector (Promega) yielding plasmid pGEMmetCOL. For site directed mutagenesis, the oligonucleotides listed in Table S5 were used in PCR reactions with pGEMmetCOL as a template followed by DpnI treatment prior to transformation of the PCR products into Escherichia coli DH5α cells. The rescued plasmids were sequenced and constructs SC1 to SC8 (Table S2) with the appropriate mutations were used as met leader RNA templates in tRNA binding assays.
In vitro transcription (IVT) and tRNA binding assay
tRNA templates were generated by PCR from genomic DNA of S. aureus COL and oligonucleotides listed in Table S4 (Text S1). Four picomol DNA template were subsequently used for IVT in a 20 µl reaction volume consisting of T7 transcription buffer, 20 U RNase inhibitor, 20 U T7 RNA polymerase (Fermentas), 0.5 mM ATP, UTP, GTP, 12 µM CTP and 9 mM GMP as described [30]. [α32P]-CTP was added and the reaction was incubated at 37°C for six hours. For met leader RNA in vitro transcription, PCR templates were either generated from plasmid pGEMmetCOL or constructs SC1 to SC8 (Table S2) with oligonucleotides T7-F_met-sRNA and R_met-sRNA (Table S5). The products were used in IVT/tRNA binding assays in the presence of pre-formed tRNAs: 10 µl reaction volume consisted of T7 transcription buffer, 10 U RNase inhibitor, 6 U T7 RNA polymerase (Fermentas) and 0.5 mM NTPs. DNA templates of the met leader RNA and of pre-formed tRNA were adjusted to end concentrations of 8 nM and 50 nM, respectively, and the mix was incubated at 37°C for two hours. Samples were immediately separated on a non-denaturing 6% (w/v) polyacrylamide gel by electrophoresis at 4°C. Visualization was attained with the PhosphoImager (Fujifilm FLA-7000) and quantification of bands was achieved with the software Multi Gauge V2.2.
Rifampicin assay for determination of transcript stability
Bacteria from overnight cultures were diluted in 100-ml flasks in 40 ml CDM medium with methionine to an initial optical density at 600 nm (OD600) of 0.05 and grown with shaking at 220 rpm at 37°C to an OD of 0.5. The cultures were filtered over a 0.22 mm filter applying vacuum, washed twice with sterile phosphate buffered saline (PBS) and bacteria were resuspended in 15 ml CDM medium without methionine and grown for another 60 minutes in a 30-ml tube. Then rifampicin (500 µg ml−1) was added to the cultures. Before (0) and after 0.5, 2, 5, 10 and 60 minutes of rifampicin exposure, RNA was isolated and Northern blot analyses were performed as described in the Supporting Information (Text S1).
Supporting Information
Zdroje
1. HidronAI, EdwardsJR, PatelJ, HoranTC, SievertDM, et al. (2008) NHSN annual update: antimicrobial-resistant pathogens associated with healthcare-associated infections: annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006–2007. Infect Control Hosp Epidemiol 29 : 996–1011.
2. ChambersHF, DeleoFR (2009) Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat Rev Microbiol 7 : 629–641.
3. SomervilleGA, ProctorRA (2009) At the crossroads of bacterial metabolism and virulence factor synthesis in Staphylococci. Microbiol Mol Biol Rev 73 : 233–248.
4. ParveenN, CornellKA (2011) Methylthioadenosine/S-adenosylhomocysteine nucleosidase, a critical enzyme for bacterial metabolism. Mol Microbiol 79 : 7–20.
5. RodionovDA, VitreschakAG, MironovAA, GelfandMS (2004) Comparative genomics of the methionine metabolism in Gram-positive bacteria: a variety of regulatory systems. Nucleic Acids Res 32 : 3340–3353.
6. Gutierrez-PreciadoA, HenkinTM, GrundyFJ, YanofskyC, MerinoE (2009) Biochemical features and functional implications of the RNA-based T-box regulatory mechanism. Microbiol Mol Biol Rev 73 : 36–61.
7. GrundyFJ, HenkinTM (1998) The S box regulon: a new global transcription termination control system for methionine and cysteine biosynthesis genes in Gram-positive bacteria. Mol Microbiol 30 : 737–749.
8. VitreschakAG, MironovAA, LyubetskyVA, GelfandMS (2008) Comparative genomic analysis of T-box regulatory systems in bacteria. RNA 14 : 717–735.
9. GreenNJ, GrundyFJ, HenkinTM (2010) The T box mechanism: tRNA as a regulatory molecule. FEBS Lett 584 : 318–324.
10. MajerczykCD, DunmanPM, LuongTT, LeeCY, SadykovMR, et al. (2010) Direct targets of CodY in Staphylococcus aureus. J Bacteriol 192 : 2861–2877.
11. PohlK, FrancoisP, StenzL, SchlinkF, GeigerT, et al. (2009) CodY in Staphylococcus aureus: a regulatory link between metabolism and virulence gene expression. J Bacteriol 191 : 2953–2963.
12. GeigerT, GoerkeC, FritzM, SchäferT, OhlsenK, et al. (2010) Role of the (p)ppGpp synthase RSH, a RelA/SpoT homolog, in stringent response and virulence of Staphylococcus aureus. Infect Immun 78 : 1873–1883.
13. Ratnayake-LecamwasamM, SerrorP, WongKW, SonensheinAL (2001) Bacillus subtilis CodY represses early-stationary-phase genes by sensing GTP levels. Genes Dev 15 : 1093–1103.
14. ShiversRP, SonensheinAL (2004) Activation of the Bacillus subtilis global regulator CodY by direct interaction with branched-chain amino acids. Mol Microbiol 53 : 599–611.
15. NovichkovPS, LaikovaON, NovichkovaES, GelfandMS, ArkinAP, et al. (2010) RegPrecise: a database of curated genomic inferences of transcriptional regulatory interactions in prokaryotes. Nucleic Acids Res 38: D111–118.
16. BrinsmadeSR, KleijnRJ, SauerU, SonensheinAL (2010) Regulation of CodY activity through modulation of intracellular branched-chain amino acid pools. J Bacteriol 192 : 6357–6368.
17. SerganovA, PatelDJ (2007) Ribozymes, riboswitches and beyond: regulation of gene expression without proteins. Nat Rev Genet 8 : 776–790.
18. MäderU, ZigL, KretschmerJ, HomuthG, PutzerH (2008) mRNA processing by RNases J1 and J2 affects Bacillus subtilis gene expression on a global scale. Mol Microbiol 70 : 183–196.
19. LioliouE, SharmaCM, CaldelariI, HelferAC, FechterP, et al. (2012) Global regulatory functions of the Staphylococcus aureus endoribonuclease III in gene expression. PLoS Genet 8: e1002782.
20. JoshiGS, SpontakJS, KlapperDG, RichardsonAR (2011) Arginine catabolic mobile element encoded speG abrogates the unique hypersensitivity of Staphylococcus aureus to exogenous polyamines. Mol Microbiol 82 : 9–20.
21. SekowskaA, DenervaudV, AshidaH, MichoudK, HaasD, et al. (2004) Bacterial variations on the methionine salvage pathway. BMC Microbiol 4 : 9.
22. HulloMF, AugerS, SoutourinaO, BarzuO, YvonM, et al. (2007) Conversion of methionine to cysteine in Bacillus subtilis and its regulation. J Bacteriol 189 : 187–197.
23. ChaffinDO, TaylorD, SkerrettSJ, RubensCE (2012) Changes in the Staphylococcus aureus transcriptome during early adaptation to the lung. PLoS One 7: e41329.
24. PietiainenM, FrancoisP, HyyrylainenHL, TangomoM, SassV, et al. (2009) Transcriptome analysis of the responses of Staphylococcus aureus to antimicrobial peptides and characterization of the roles of vraDE and vraSR in antimicrobial resistance. BMC Genomics 10 : 429.
25. NagarajanV, ElasriMO (2007) SAMMD: Staphylococcus aureus microarray meta-database. BMC Genomics 8 : 351.
26. OchsnerUA, YoungCL, StoneKC, DeanFB, JanjicN, et al. (2005) Mode of action and biochemical characterization of REP8839, a novel inhibitor of methionyl-tRNA synthetase. Antimicrob Agents Chemother 49 : 4253–4262.
27. AnupamR, NayekA, GreenNJ, GrundyFJ, HenkinTM, et al. (2008) 4,5-Disubstituted oxazolidinones: High affinity molecular effectors of RNA function. Bioorg Med Chem Lett 18 : 3541–3544.
28. AnupamR, DenapoliL, MuchenditsiA, HinesJV (2008) Identification of neomycin B-binding site in T box antiterminator model RNA. Bioorg Med Chem 16 : 4466–4470.
29. LagojaIM, HerdewijnP (2007) Use of RNA in drug design. Expert Opin Drug Discov 2 : 889–903.
30. SampsonJR, UhlenbeckOC (1988) Biochemical and physical characterization of an unmodified yeast phenylalanine transfer RNA transcribed in vitro. Proc Natl Acad Sci U S A 85 : 1033–1037.
31. YousefMR, GrundyFJ, HenkinTM (2003) tRNA requirements for glyQS antitermination: a new twist on tRNA. RNA 9 : 1148–1156.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2013 Číslo 9- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Cross-Serotype Immunity Induced by Immunization with a Conserved Rhinovirus Capsid Protein
- The CLIP-Domain Serine Protease Homolog SPCLIP1 Regulates Complement Recruitment to Microbial Surfaces in the Malaria Mosquito
- Aggressive Chemotherapy and the Selection of Drug Resistant Pathogens
- Host Adaptation Is Contingent upon the Infection Route Taken by Pathogens
- Acute Neonatal Infections ‘Lock-In’ a Suboptimal CD8+ T Cell Repertoire with Impaired Recall Responses
- Lymph Node Colonization Dynamics after Oral Typhimurium Infection in Mice
- Highly Significant Antiviral Activity of HIV-1 LTR-Specific Tre-Recombinase in Humanized Mice
- Emerging and Emerged Pathogenic Species: Beyond the Paradigm
- Cross-Seeding of Misfolded Proteins: Implications for Etiology and Pathogenesis of Protein Misfolding Diseases
- Emergence of the Middle East Respiratory Syndrome Coronavirus
- Memory of Infections: An Emerging Role for Natural Killer Cells
- Death Be Not Proud—Cell Death Control in Plant Fungal Interactions
- Self and Non-self Discrimination of Intracellular Membranes by the Innate Immune System
- Innate Immune Sensing of Flaviviruses
- Bringing Culture to the Uncultured: and Lessons for Obligate Intracellular Bacterial Pathogens
- Atomic Force Microscopy: A New Look at Pathogens
- Methionine Biosynthesis in Is Tightly Controlled by a Hierarchical Network Involving an Initiator tRNA-Specific T-box Riboswitch
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Memory of Infections: An Emerging Role for Natural Killer Cells
- Emergence of the Middle East Respiratory Syndrome Coronavirus
- Emerging and Emerged Pathogenic Species: Beyond the Paradigm
- Death Be Not Proud—Cell Death Control in Plant Fungal Interactions
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



