-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaEmerging and Emerged Pathogenic Species: Beyond the Paradigm
article has not abstract
Published in the journal: . PLoS Pathog 9(9): e32767. doi:10.1371/journal.ppat.1003550
Category: Pearls
doi: https://doi.org/10.1371/journal.ppat.1003550Summary
article has not abstract
Candida albicans and Non-albicans Candida (NAC) Species Infections: General Information in Predisposing Conditions and Clinical Incidence
Many ascomycete yeast species from the Candida genus are widely distributed in nature and act as common saprophytic constituents of the normal human microflora. However, some of these fungal species can also become opportunistic pathogens following a transition from a commensal to a pathogenic phase, induced by alterations in the host environment. Candida species thereby rarely trigger infection in healthy people, but take advantage of a locally or systematically impaired immune system to proliferate in the host and cause diseases termed “candidiasis.” Such fungal infections can be subdivided into three major groups: cutaneous (skin and its appendages), mucosal (oropharyngeal, esophageal, and vulvovaginal) and systemic (bloodstream infections, i.e., candidemia and other forms of invasive candidiasis [IC]). While superficial candidiasis (cutaneous and mucosal) is often observed in AIDS patients, oropharyngeal thrush and vaginitis are more frequently seen in immunocompetent infants and adult women, respectively. Candidemia and IC are common in cancer patients or in transplant individuals following immunosuppression. Candidiasis currently represents the fourth leading cause of nosocomial infections, at 8% to 10%, and mortality due to systemic candidiasis remains high, ranging from 15% to 35% depending on the infecting Candida species [1].
Although Candida albicans remains the most frequently isolated agent of candidiasis, non-albicans Candida (NAC) species now account for a substantial part of clinical isolates collected worldwide in hospitals. NAC species of particular clinical importance include Candida glabrata, Candida tropicalis, Candida parapsilosis, and Candida krusei (synonym: Issatchenkia orientalis), as well as the less-prominent species Candida guilliermondii, Candida lusitaniae, Candida kefyr, Candida famata (synonym: Debaryomyces hansenii), Candida inconspicua, Candida rugosa, Candida dubliniensis, and Candida norvegensis (Table 1). A complementary set of about 20 opportunistic NAC species is also known, but exhibits lower isolation rates [2].
Tab. 1. Introducing characteristics of Candida species. 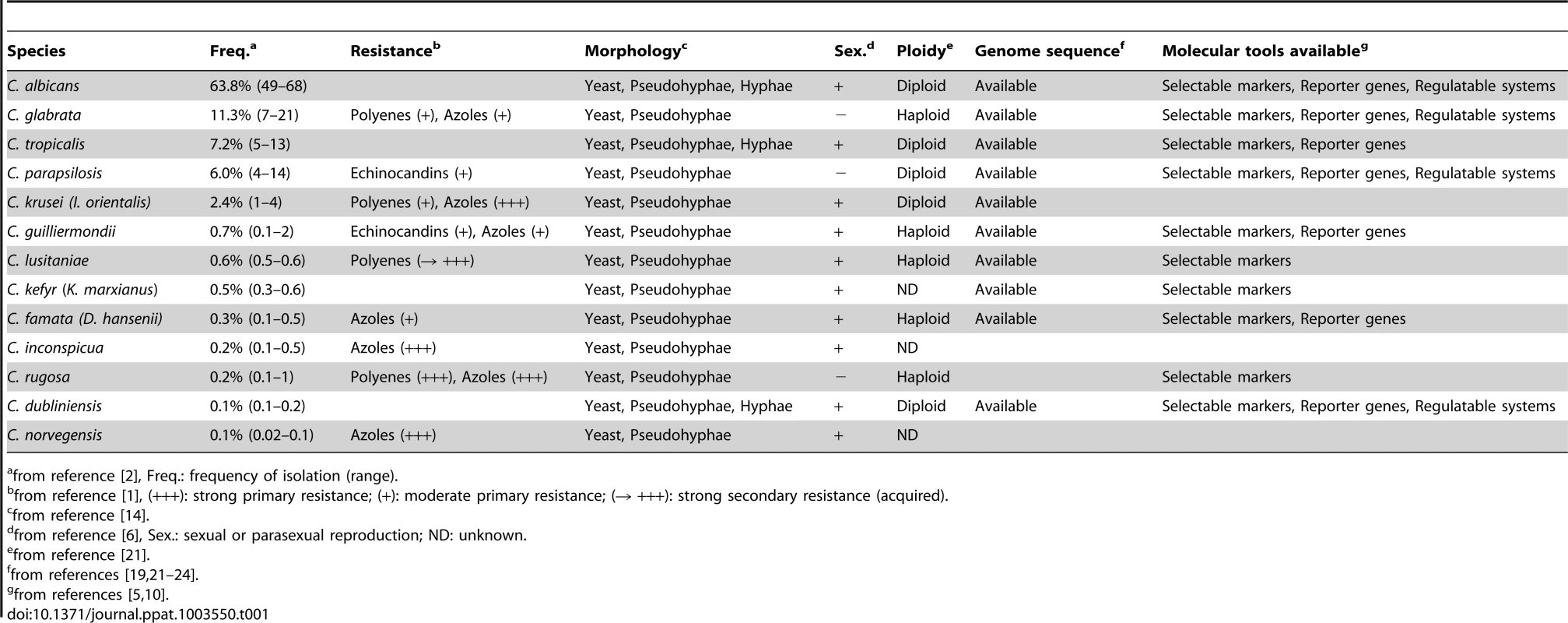
from reference [2], Freq.: frequency of isolation (range). Trends in Species Distribution and Antifungal Susceptibility of NAC Species
Global surveillance programs (e.g. SENTRY and ARTEMIS) provide a tremendous amount of data regarding global trends in various aspects of NAC candidiasis including geographical variation in the frequency of species, distribution by specimen type and patient age, as well as changes in the antifungal susceptibility of collected NAC isolates [2].
An overview of the literature from the last four decades highlights an important fact: Due to the growing size of the population at special risk (due to neutropenia, immunosuppression, metabolic dysfunction, and anticancer chemotherapy), candidiasis remains a persistent public health problem, and the proportion of NAC species among Candida isolates recovered from patients is increasing. Whereas NAC species accounted for 10%–40% of all systemic candidiasis from 1970 to 1990, this proportion reached 35%–65% in the last two decades [3]. A recent ten-year analysis of the worldwide distribution of NAC species indicated that C. glabrata remains the most common NAC species and that C. parapsilosis, C. tropicalis, and C. krusei are also frequently isolated (Table 1). C. guilliermondii and C. lusitaniae have shown gradual emergence as a cause of invasive candidiasis, while C. kefyr, C. famata, C. inconspicua, C. rugosa, C. dubliniensis, and C. norvegensis, although rarely isolated, are now considered emerging NAC species, as their isolation rate has increased between 2 - and 10-fold over the last 15 years [2].
Interestingly, significant geographic variation in the frequency of NAC species occurs. Among marked trends, C. glabrata is more prominent in North America than in Latin America. In addition, C. tropicalis is frequently isolated in Asia-Pacific and less often encountered in the rest of the world, whilst C. parapsilosis remains 3-fold more commonly recovered in North America than in Europe. Finally, C. guilliermondii and C. rugosa are more prominent in Latin America, and C. inconspicua and C. norvegensis in Europe [2] than in the rest of the world.
Antifungal compounds currently used to treat systemic candidiasis belong to three families: polyenes, azoles, and echinocandins. Most of the NAC species exhibit particular patterns of primary resistance or reduced susceptibility toward these antifungals (Table 1). For example, a high level of resistance toward azoles is well known for C. krusei, C. inconspicua, C. rugosa, and C. norvegensis, whereas C. parapsilosis and C. guilliermondii stand out due to their decreased susceptibility to echinocandins [4].
A Particular Codon Usage in Most NAC Species Delays Development of Genetic Tools
Since the end of the last century, the clinical importance of NAC species has promoted research aimed at identifying molecular events underlying pathogenicity and antifungal resistance in these emerging yeasts. However, the development of genetic approaches in NAC species has been hindered by three main factors: (i) most pioneering studies during the early stages of the “pathogenic yeast genetics” field were carried out in C. albicans; (ii) the particular codon usage of most of Candida species has precluded the direct use of S. cerevisiae or bacterial molecular tools in these NAC species [5]; (iii) most pathogenic Candida species have limited modes of sexual reproduction unlike S. cerevisiae [6].
Originally, the genus name Candida was attributed to yeast species able to form hyphae or pseudohyphae (Table 1) and for which no sexual spores were observed. Nevertheless, recent phylogenetic analysis has clarified that Candida species actually represent a polyphyletic group within the Saccharomycotina [7] (Figure 1). More precisely, C. tropicalis, C. parapsilosis, C. guilliermondii, C. lusitaniae, C. famata, C. rugosa, and C. dubliniensis form part of the Candida CTG clade and translate CTG codons as serine instead of leucine. In contrast, C. glabrata and C. kefyr belong to the Saccharomycetaceae, with C. glabrata and S. cerevisiae falling within the whole genome duplication (WGD) clade. The remaining species C. krusei, C. inconspicua, and C. norvegensis are probably closely related in the Saccharomycetaceae clade, which could give insights into their common resistance toward azole antifungals.
Fig. 1. Schematic representation illustrating the phylogeny of NAC species. 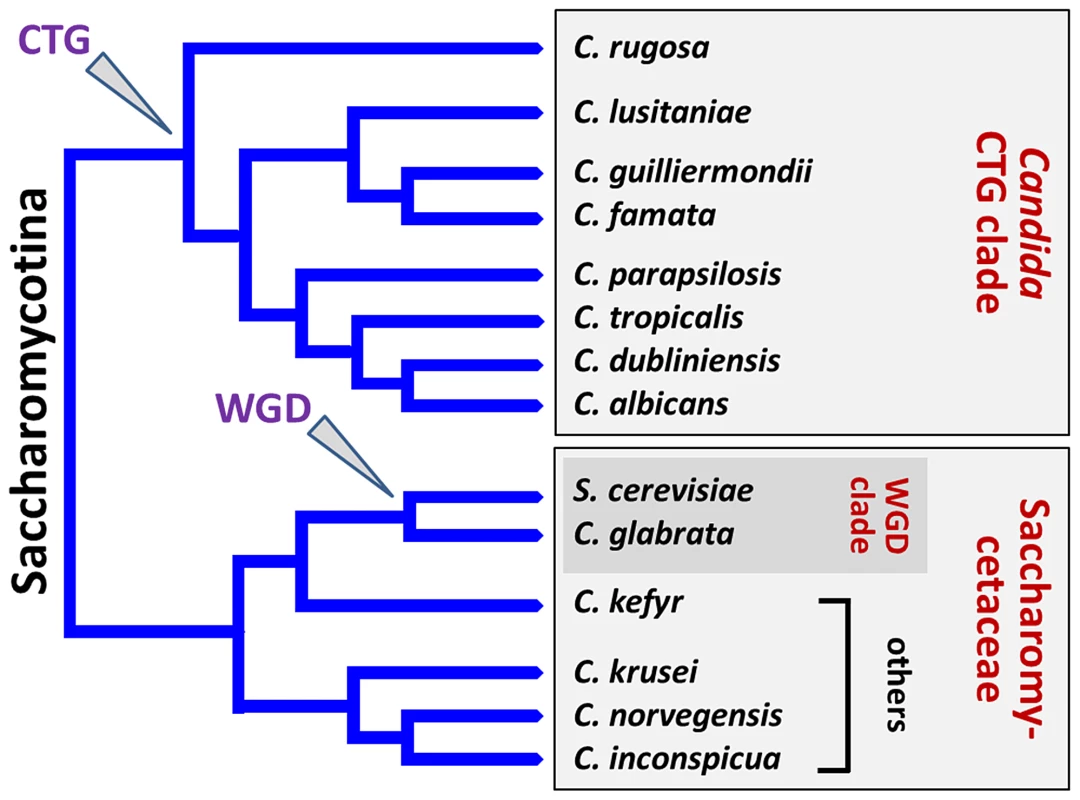
C. tropicalis, C. parapsilosis, C. guilliermondii, C. lusitaniae, C. famata (D. hansenii), C. rugosa, and C. dubliniensis form part of the Candida CTG clade and translate CTG codons as serine instead of leucine. In contrast, C. glabrata and C. kefyr (K. marxianus) belong to the Saccharomycetaceae, with C. glabrata and S. cerevisiae falling within the “whole genome duplication” (WGD) clade. The remaining species C. krusei (I. orientalis), C. inconspicua, and C. norvegensis are probably closely related in the Saccharomycetaceae clade. The branch lengths are arbitrary. During the late 1990s, C. glabrata genetics was by far the most advanced of the NAC species due to its haploid status, its classical codon usage (allowing the direct use of S. cerevisiae tools), and its high frequency of isolation in hospitals [8]. Genetic studies of CTG clade species expanded in the 2000s and focused on the development of molecular tools, as well as transformation procedures, due to the biotechnological potential of several Candida yeasts (C. guilliermondii, C. famata, C. tropicalis, and C. rugosa) as well as clinical incidence (C. dubliniensis and C. parapsilosis) [5], [9]. Specifically, drug-resistant markers and reporter genes (encoding fluorescent protein variants, luciferase, or beta-galactosidase) were adapted by changing CTG codons to allow their functionality in this particular clade [5] (Table 1).
Mechanisms Underlying Antifungal Resistance, Virulence, and Morphological Transitions in NAC Species: Is Candida albicans the Rule or the Exception?
C. albicans genetics, with the construction and phenotypical analysis of targeted mutant strains since 1994, has provided a foundation for understanding fundamental processes in pathogenic yeasts [10]. Intense research in C. albicans from the end of the 20th century shed light on the molecular mechanisms involved in drug resistance [11], biofilm formation [12], adherence [13], yeast-hyphal switching and its role in virulence [14], and sexual mating [15], [16]. C. albicans has therefore become the model yeast for investigating the multiple factors controlling the host–pathogen interaction. As a result, C. albicans biology is now the paradigm for Candida research in the medical mycology community.
In response to the clinical emergence of NAC species, research programs were initiated to further understand these opportunistic yeasts. The first studies highlighted marked differences in behavior between different Candida species. This included stress adaptation [17], which may come from the fact that each species has independently evolved to promote survival in their respective natural niches and their specific host. It must also be kept in mind that each Candida species displays specific traits such as ploidy, sexual behavior (if any) [6], and morphology [14] (Table 1). These could directly impact their ability to adapt to the host's response, to disseminate in the organism, and to develop resistance mechanisms to antifungals during treatments.
Due to the lack of genetic and molecular resources, researchers have often assumed that if a yeast species is related to another yeast species, the underlying molecular and cellular mechanisms must also be closely related. However, even within a clade, the genetic distance between any two NAC species is often larger than the genetic distance between humans and some fishes [18]. Therefore, in no way should it be argued that C. albicans makes the rules for all NAC species. As a corollary, in future investigations, the biology of each Candida species should continue to be addressed on a case-by-case basis.
Perspectives: Genome Resources and Postgenomic Technologies Dedicated to NAC
A large range of rapidly evolving genomic and postgenomic approaches, including genome sequences and gene expression data, have recently enhanced the understanding of Candida yeasts pathogenicity.
The first published genomes of Candida species were C. glabrata in 2003 (alongside the C. famata genome sequence) [19], followed by C. albicans [20] in 2004, which has further strengthened the prominent role of C. albicans and C. glabrata in the field. In January 2005, the Broad Institute Fungal Genome Initiative, in collaboration with the Wellcome Trust Sanger Institute, made available the sequences of five CTG clade genomes, including C. tropicalis, C. parapsilosis, C. dubliniensis, C. guilliermondii, and C. lusitaniae [21], [22]. Finally, genome sequences of C. kefyr (teleomorph Kluyveromyces marxianus) [23] and C. krusei [24] were recently published. These genome resources have provided new insights into gene family evolution within Candida species and identified gene families enriched in the most common pathogenic NAC species [21]. This area of research is further supported by the creation of databases dedicated to genome annotation, including gene ontology browsers specializing in metabolic pathways, virulence, and morphogenesis [25]. These bioinformatics tools provide an accurate annotation of NAC genome sequences and give precious help to future Candida gene evolutionary analyses.
Postgenomic technologies have also emerged to support the Candida research field. Quantitative transcriptional profiling strategies (e.g. RNA-Seq, microarray) currently allow the active screening of genes commonly or specifically required for pathogenicity, morphogenesis, and antifungal resistance in multiple Candida species [26]–[28].
Thanks to the growing number of yeast genome sequences available, as well as the utilization of postgenomic approaches, the palette of newly identified pathogenicity-related genes in NAC species is now predicted to increase rapidly. However, efforts need to continue toward the development of classical molecular tools dedicated to each pathogenic NAC species to further analyze the function of large numbers of uncharacterized genes. This is an essential prerequisite for the identification of new fungal targets and the subsequent development of novel antifungal drugs.
Zdroje
1. PfallerMA, DiekemaDJ (2007) Epidemiology of invasive candidiasis: a persistent public health problem. Clin Microbiol Rev 20 : 133–163 doi:10.1128/CMR.00029-06
2. PfallerMA, DiekemaDJ, GibbsDL, NewellVA, EllisD, et al. (2010) Results from the ARTEMIS DISK Global Antifungal Surveillance Study, 1997 to 2007: a 10.5-year analysis of susceptibilities of Candida Species to fluconazole and voriconazole as determined by CLSI standardized disk diffusion. J Clin Microbiol 48 : 1366–1377 doi:10.1128/JCM.02117-09
3. KrcmeryV, BarnesAJ (2002) Non-albicans Candida spp. causing fungaemia: pathogenicity and antifungal resistance. J Hosp Infect 50 : 243–260 doi:10.1053/jhin.2001.1151
4. WalkerLA, GowNA, MunroCA (2013) Elevated chitin content reduces the susceptibility of Candida species to caspofungin. Antimicrob Agents Chemother 57 : 146–154 doi:10.1128/AAC.01486-12
5. PaponN, CourdavaultV, ClastreM, SimkinAJ, CrècheJ, et al. (2012) Deus ex Candida genetics: overcoming the hurdles for the development of a molecular toolbox in the CTG clade. Microbiology 158 : 585–600 doi:10.1099/mic.0.055244-0
6. BennettRJ (2010) Coming of age–sexual reproduction in Candida species. PLoS Pathog 6: e1001155 doi:10.1371/journal.ppat.1001155
7. FitzpatrickDA, LogueME, StajichJE, ButlerG (2006) A fungal phylogeny based on 42 complete genomes derived from supertree and combined gene analysis. BMC Evol Biol 6 : 99 doi:10.1186/1471-2148-6-99
8. SilvaS, NegriM, HenriquesM, OliveiraR, WilliamsDW, et al. (2012) Candida glabrata, Candida parapsilosis and Candida tropicalis: biology, epidemiology, pathogenicity and antifungal resistance. FEMS Microbiol Rev 36 : 288–305 doi:10.1111/j.1574-6976.2011.00278.x
9. PaponN, SaviniV, LanoueA, SimkinAJ, CrècheJ, et al. (2013) Candida guilliermondii: biotechnological applications, perspectives for biological control, emerging clinical importance and recent advances in genetics. Curr Genet 59 : 73–90 doi:10.1007/s00294-013-0391-0
10. SamaranayakeDP, HanesSD (2011) Milestones in Candida albicans gene manipulation. Fungal Genet Biol 48 : 858–865 doi:10.1016/j.fgb.2011.04.003
11. PfallerMA (2012) Antifungal drug resistance: mechanisms, epidemiology, and consequences for treatment. Am J Med 125: S3–13 doi:10.1016/j.amjmed.2011.11.001
12. FanningS, MitchellAP (2012) Fungal biofilms. PLoS Pathog 8: e1002585 doi:10.1371/journal.ppat.1002585
13. FinkelJS, XuW, HuangD, HillEM, DesaiJV, et al. (2012) Portrait of Candida albicans adherence regulators. PLoS Pathog 8: e1002525 doi:10.1371/journal.ppat.1002525
14. ThompsonDS, CarlislePL, KadoshD Coevolution of morphology and virulence in Candida species. Eukaryot Cell 10 : 1173–1182 doi:10.1128/EC.05085-11
15. HullCM, RaisnerRM, JohnsonAD (2000) Evidence for mating of the “asexual” yeast Candida albicans in a mammalian host. Science 289 : 307–310 doi:10.1126/science.289.5477.307
16. MageeBB, MageePT (2000) Induction of mating in Candida albicans by construction of MTLa and MTLalpha strains. Science 289 : 310–313 doi:10.5580/1f5e
17. LiD, AgrellosOA, CalderoneR (2010) Histidine kinases keep fungi safe and vigorous. Curr Opin Microbiol 13 : 424–430 doi:10.1016/j.mib.2010.04.007
18. DujonB (2010) Yeast evolutionary genomics. Nat Rev Genet 11 : 512–524 doi:10.1038/nrg2811
19. WongS, FaresMA, ZimmermannW, ButlerG, WolfeKH (2003) Evidence from comparative genomics for a complete sexual cycle in the ‘asexual’ pathogenic yeast Candida glabrata. Genome Biol 4: R10 doi:10.1186/gb-2003-4-2-r10
20. JonesT, FederspielNA, ChibanaH, DunganJ, KalmanS, et al. (2004) The diploid genome sequence of Candida albicans. Proc Natl Acad Sci USA 101 : 7329–7334 doi:10.1073/pnas.0401648101
21. ButlerG, RasmussenMD, LinMF, SantosMA, SakthikumarS, et al. (2009) Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature 459 : 657–662 doi:10.1038/nature08064
22. JacksonAP, GambleJA, YeomansT, MoranGP, SaundersD, et al. (2009) Comparative genomics of the fungal pathogens Candida dubliniensis and Candida albicans. Genome Res 19 : 2231–2244 doi:10.1101/gr.097501.109
23. JeongH, LeeDH, KimSH, KimHJ, LeeK, et al. (2012) Genome sequence of the thermotolerant yeast Kluyveromyces marxianus var. marxianus KCTC 17555. Eukaryot Cell 11 : 1584–1585 doi:10.1128/EC.00260-12
24. ChanGF, GanHM, LingHL, RashidNA (2012) Genome sequence of Pichia kudriavzevii M12, a potential producer of bioethanol and phytase. Eukaryot Cell 11 : 1300–1301 doi:10.1128/EC.00229-12
25. MaguireSL, OhéigeartaighSS, ByrneKP, SchröderMS, O'GaoraP, et al. (2013) Comparative Genome Analysis and Gene Finding in Candida Species Using CGOB. Mol Biol Evol 30 : 1281–1291 doi:10.1093/molbev/mst042
26. GrumazC, LorenzS, StevensP, LindemannE, SchöckU, et al. (2013) Species and condition specific adaptation of the transcriptional landscapes in Candida albicans and Candida dubliniensis. BMC Genomics 14 : 212 doi:10.1186/1471-2164-14-212
27. SilvaAP, MirandaIM, GuidaA, SynnottJ, RochaR, et al. (2011) Transcriptional profiling of azole-resistant Candida parapsilosis strains. Antimicrob Agents Chemother 55 : 3546–3556 doi:10.1128/AAC.01127-10
28. PormanAM, HirakawaMP, JonesSK, WangN, BennettRJ (2013) MTL-independent phenotypic switching in Candida tropicalis and a dual role for Wor1 in regulating switching and filamentation. PLoS Genet 9: e1003369 doi:10.1371/journal.pgen.1003369
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2013 Číslo 9- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Cross-Serotype Immunity Induced by Immunization with a Conserved Rhinovirus Capsid Protein
- The CLIP-Domain Serine Protease Homolog SPCLIP1 Regulates Complement Recruitment to Microbial Surfaces in the Malaria Mosquito
- Aggressive Chemotherapy and the Selection of Drug Resistant Pathogens
- Host Adaptation Is Contingent upon the Infection Route Taken by Pathogens
- Acute Neonatal Infections ‘Lock-In’ a Suboptimal CD8+ T Cell Repertoire with Impaired Recall Responses
- Lymph Node Colonization Dynamics after Oral Typhimurium Infection in Mice
- Highly Significant Antiviral Activity of HIV-1 LTR-Specific Tre-Recombinase in Humanized Mice
- Emerging and Emerged Pathogenic Species: Beyond the Paradigm
- Cross-Seeding of Misfolded Proteins: Implications for Etiology and Pathogenesis of Protein Misfolding Diseases
- Emergence of the Middle East Respiratory Syndrome Coronavirus
- Memory of Infections: An Emerging Role for Natural Killer Cells
- Death Be Not Proud—Cell Death Control in Plant Fungal Interactions
- Self and Non-self Discrimination of Intracellular Membranes by the Innate Immune System
- Innate Immune Sensing of Flaviviruses
- Bringing Culture to the Uncultured: and Lessons for Obligate Intracellular Bacterial Pathogens
- Atomic Force Microscopy: A New Look at Pathogens
- Methionine Biosynthesis in Is Tightly Controlled by a Hierarchical Network Involving an Initiator tRNA-Specific T-box Riboswitch
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Memory of Infections: An Emerging Role for Natural Killer Cells
- Emergence of the Middle East Respiratory Syndrome Coronavirus
- Emerging and Emerged Pathogenic Species: Beyond the Paradigm
- Death Be Not Proud—Cell Death Control in Plant Fungal Interactions
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



