-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaA Type IV Pilus Mediates DNA Binding during Natural Transformation in
Natural genetic transformation is widely distributed in bacteria and generally occurs during a genetically programmed differentiated state called competence. This process promotes genome plasticity and adaptability in Gram-negative and Gram-positive bacteria. Transformation requires the binding and internalization of exogenous DNA, the mechanisms of which are unclear. Here, we report the discovery of a transformation pilus at the surface of competent Streptococcus pneumoniae cells. This Type IV-like pilus, which is primarily composed of the ComGC pilin, is required for transformation. We provide evidence that it directly binds DNA and propose that the transformation pilus is the primary DNA receptor on the bacterial cell during transformation in S. pneumoniae. Being a central component of the transformation apparatus, the transformation pilus enables S. pneumoniae, a major Gram-positive human pathogen, to acquire resistance to antibiotics and to escape vaccines through the binding and incorporation of new genetic material.
Published in the journal: . PLoS Pathog 9(6): e32767. doi:10.1371/journal.ppat.1003473
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1003473Summary
Natural genetic transformation is widely distributed in bacteria and generally occurs during a genetically programmed differentiated state called competence. This process promotes genome plasticity and adaptability in Gram-negative and Gram-positive bacteria. Transformation requires the binding and internalization of exogenous DNA, the mechanisms of which are unclear. Here, we report the discovery of a transformation pilus at the surface of competent Streptococcus pneumoniae cells. This Type IV-like pilus, which is primarily composed of the ComGC pilin, is required for transformation. We provide evidence that it directly binds DNA and propose that the transformation pilus is the primary DNA receptor on the bacterial cell during transformation in S. pneumoniae. Being a central component of the transformation apparatus, the transformation pilus enables S. pneumoniae, a major Gram-positive human pathogen, to acquire resistance to antibiotics and to escape vaccines through the binding and incorporation of new genetic material.
Introduction
Natural transformation, first discovered in Streptococcus pneumoniae [1], is observed in many Gram-negative and Gram-positive bacteria [2]. It increases bacterial adaptability by promoting genome plasticity through intra - and inter-species genetic exchange [3]. In S. pneumoniae, a major human pathogen responsible for severe diseases such as pneumonia, meningitis and septicemia, transformation is presumably responsible for capsular serotype switching and could therefore reduce the efficiency of capsule-based vaccines after a short period [4]. In this species, it occurs during a genetically programmed and differentiated state called competence that is briefly induced at the beginning of exponential growth. During this competent state, pneumococci secrete a peptide pheromone called Competence-Stimulating-Peptide (CSP) [5], which spreads competence in the pneumococcal population. Interestingly, in S. pneumoniae, some antibiotics and DNA-damaging agents induce competence, which would act as an alternative SOS response and ultimately increases bacterial resistance to external stresses [6].
During transformation, environmental DNA is bound at the surface of competent cells and transported through the cell envelope to the cytosolic compartment. This process has been mostly studied in the Gram-positive bacterium Bacillus subtilis with additional information coming from studies in S. pneumoniae [7], [8]. In both species, a DNA translocation apparatus mediates the transfer of DNA through the cellular membrane. In S. pneumoniae, it is composed of ComEA, EndA, ComEC and ComFA. Incoming double-stranded DNA would bind the membrane receptor ComEA. One DNA strand crosses the membrane through ComEC while the endonuclease EndA degrades the other strand. On the cytoplasmic side, ComFA, an ATPase that contains a helicase-like domain, would facilitate DNA internalization through ComEC. Once inside the bacterium, single-stranded DNA is either integrated into the chromosome by RecA-mediated homologous recombination or entirely degraded.
Strikingly, all transformable Gram-positive bacteria also carry a comG operon that resembles operons encoding Type IV pili and Type II secretion pseudopili in Gram-negative bacteria, as well as a gene encoding a prepilin peptidase homolog, pilD [7]. In B. subtilis and S. pneumoniae, comG and pilD genes are exclusively expressed in competent cells and are essential for transformation [9], [10], [11]. In S. pneumoniae, the comG operon encodes a putative ATPase (ComGA), a polytopic membrane protein (ComGB) and five prepilin candidates named ComGC, ComGD, ComGE, ComGF and ComGG (Figure 1A and B and table S1). By homology with Type IV pili, it is generally proposed that these proteins could be involved in the assembly of a transformation pseudo-pilus at the surface of competent cells [7], [8], [12]. So far, two studies show that a large macromolecular complex containing ComGC can be found at the surface of competent B. subtilis cells [9], [12]. In this complex, ComGC subunits appear to be linked together by disulfide bridges [9]. All the other ComG proteins and the PilD homolog, ComC, are necessary for the formation of this complex [9]. It was proposed that this complex could correspond to the transformation pseudo-pilus.
Fig. 1. Genes potentially essential for transformation pilus assembly in S. pneumonia and prepilin sequences. 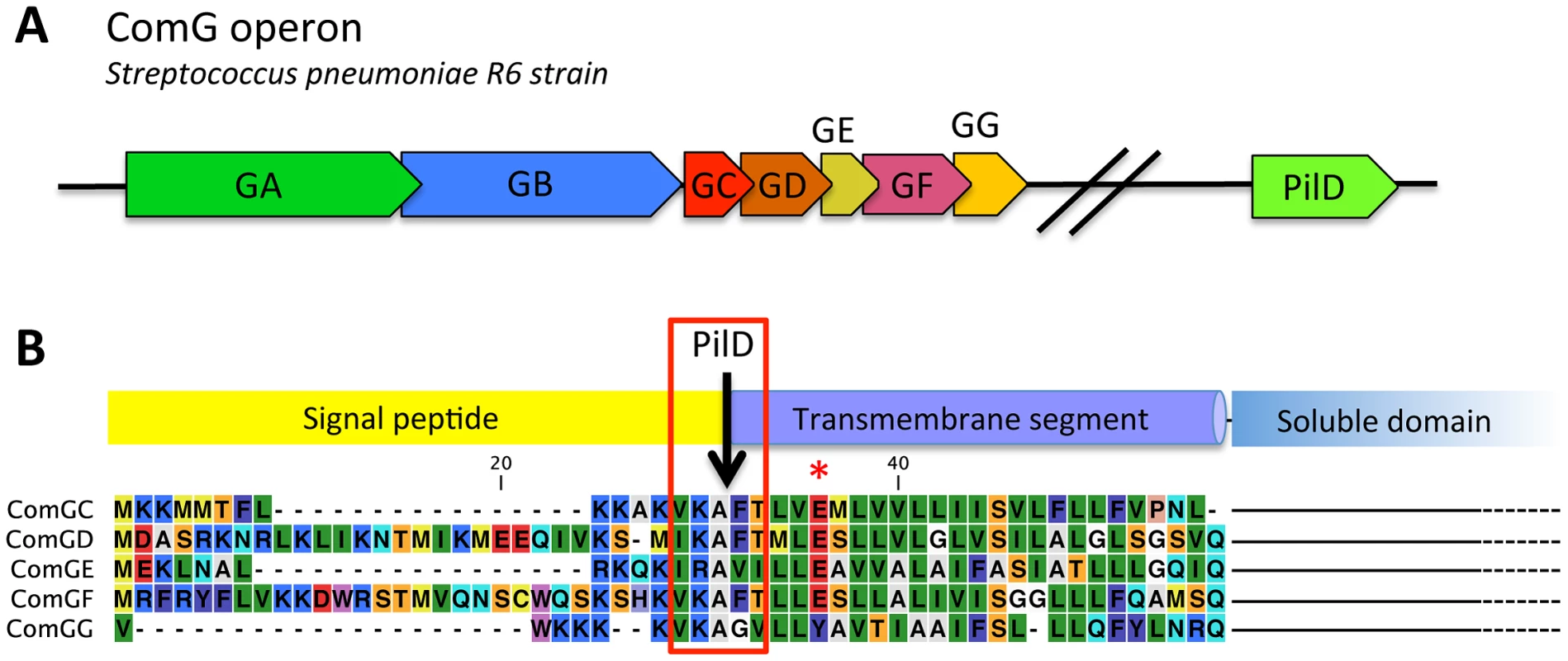
(A) Genes that are potentially essential in transformation pilus assembly. The comG operon contains 7 genes named comGA to comGG. A gene that encodes a pre-pilin peptidase homolog, pilD, is found elsewhere on the chromosome. The name used to designate the comG and pilD genes varies in different pneumoccocal strains. For clarity, we refer to the comG nomenclature used in B. subtilis. Names of the corresponding genes in different S. pneumoniae strains are found in the Table S1. (B) Alignment of the N-terminal extremity of the five possible prepilins show that ComGC, ComGD, ComGE and comGF contain a canonical prepilin cleavage motif (red rectangle) [20]. All four proteins also contain a conserved glutamic acid residue in position 5 after the cleavage site (red asterisk), a conserved feature of all Type IV pilins [43]. ComGG has a degenerated peptidase motif and does not contain a conserved glutamic acid in position 5. Secondary structure predictions for the five proteins propose a N-terminal hydrophobic helix and a C-terminal soluble domain of variable size (not shown in the figure), the typical topology found in Type IV pilins. Despite these first clues, no transformation appendage could be directly visualized at the surface of any competent Gram-positive bacterium. In addition, the function of the ComG proteins during transformation remains unclear. Mutations in the cytosolic ComGA protein abolish DNA binding at the surface of both B. subtilis and S. pneumoniae [13], [14], [15]. This strongly suggests that the ComGC-containing macromolecular complex detected at the surface of competent B. subtilis cells could bind DNA. However, it was recently shown that ComGA is the only ComG protein essential to the initial DNA binding at the surface of competent B. subtilis cells [14]. This protein would interact with an unknown DNA receptor at the surface of competent cells while the other ComG proteins would only be required at a later stage during transformation.
In this study, we provide the first direct evidence for the existence of a transformation pilus in a Gram-positive bacterium. We discovered a new appendage at the surface of competent pneumococci that we could visualize using immuno-fluorescence and electron microscopy. Competent cells harbor one or a few appendages that are morphologically similar to Type IV pili found in Gram-negative bacteria. We were able to purify this pilus and showed that it is essentially composed of the ComGC pilin. We also demonstrate that pilus assembly is required for transformation. As we provide direct evidence that the transformation pilus binds extracellular DNA, we propose it is the primary DNA receptor at the surface of competent pneumococci.
Results
An extracellular appendage containing ComGC is detected at the surface of competent S. pneumoniae
Mechanical shearing is frequently used to release bacterial surface appendages and to study their protein composition [16]. To see if ComGC was part of a macromolecular complex at the surface of competent S. pneumoniae, we adapted the method to this bacterium and raised antibodies against the purified soluble domain of ComGC. Using this antibody, we showed that ComGC could be detected by immunoblotting in the sheared fraction of competent bacteria (Figure 2A). While ComGC level in the cell fraction was not affected, no ComGC could be found in the sheared fraction in a comGA knockout mutant (Figure 2A). These data strongly suggest that ComGC is part of an extra-cellular appendage and that ComGA is necessary to its assembly.
Fig. 2. Competence-induced appendages assembly and transformation efficiency. 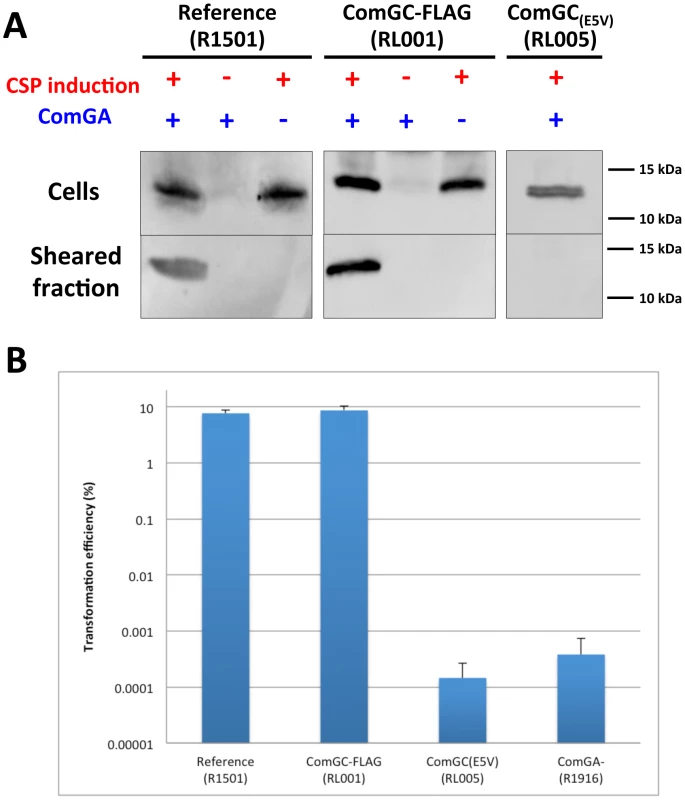
(A) Detection of ComGC in the sheared and cellular fractions by immunoblot. ComGC was detected in the sheared fraction of reference (R1501) and ComGC-FLAG (RL001) competent strains. ComGC was not detected in the sheared fraction of cultures that were not competence-induced. Deletion of comGA completely abolished detection of the ComGC pilin in the sheared fraction in both reference and ComGC-FLAG strains. Substitution of the conserved glutamic acid in position 5 of the mature ComGC pilin (Figure 1) by alanine (ComGC(E5V)) also completely abolished detection of ComGC in the sheared fraction. For the reference and ComGC(E5V) strains, detection of ComGC was performed with a polyclonal rabbit antibody raised against the soluble domain of ComGC. With the ComGC-FLAG strain, the FLAG-tagged pilin was detected by an anti-FLAG monoclonal antibody. (B) Transformation assay using the reference strain (R1501), comGC-FLAG strain (RL001), comGC(E5V) mutant (RL005) and comGA deletion mutant (R1916). Both comGC(E5V) and comGA mutants were defective for transformation. A ComGC-containing appendage is visualized at the surface of competent pneumococci
We inserted a FLAG tag at the C-terminus of ComGC to directly visualize the competence-induced appendages by immuno-fluorescence. It was not possible to insert the sequence encoding the tag at the comGC locus on the chromosome because comGC and comGD genes overlap in the comG operon. Therefore, a copy of comGC encoding a C-terminally FLAG-tagged ComGC (ComGC-FLAG) was integrated ectopically into the chromosome of S. pneumoniae under the control of a competence-induced promoter [17]. The transformation efficiency was not affected in this strain (Figure 2B). Using anti-FLAG antibodies, we could show by immuno-fluorescence that almost all the cells appeared to harbour one or a few ComGC foci or distinct fluorescent appendages (Figure 3A and B; Figure S1A). Due to sample preparation, many broken appendages were also found in the background. No preferential location of the foci/appendages at the cell surface was observed. They are absent in comGA knockout cells (Figure 3A). Note that anti-ComGC antibodies were not able to label the competent cells. They probably recognize epitopes that are masked when ComGC is included in the appendages.
Fig. 3. Direct visualization of competence-induced appendages by Immuno-fluorescence. 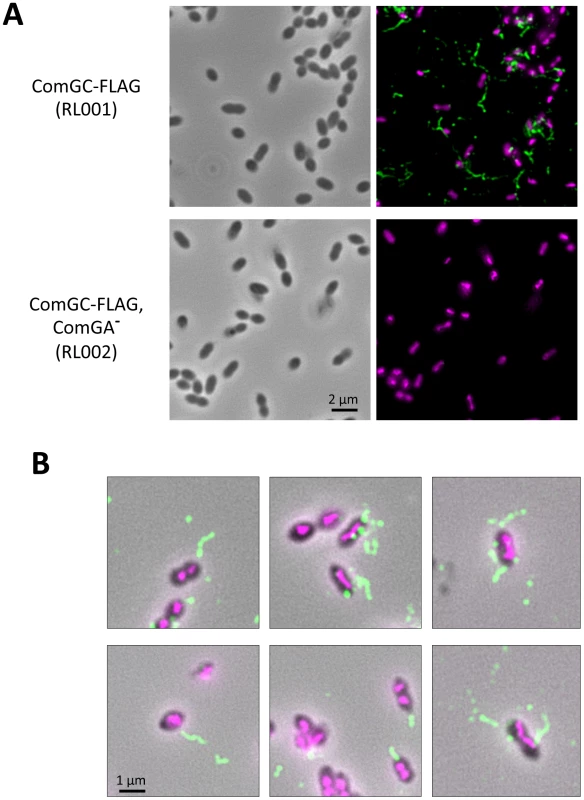
(A) Immuno-fluorescence microscopy showing intact competent cells expressing a FLAG-tagged ComGC pilin in presence (top row) or absence of ComGA (bottom row). Left column correspond to bright field image, right column to overlay between anti-FLAG antibody fluorescence (green) and DAPI fluorescence (magenta). (B) Zoom on several bacterial cells visualized by immuno-fluorescence. Overlay of bright filed image, anti-FLAG antibody fluorescence (green) and DAPI fluorescence (magenta). Distinct appendages are visible on competent cells. Competence-induced appendage morphology and composition
Using electron microscopy, we observed filaments attached to the cell surface of negatively stained competent pneumococci (Figure 4A and B). These flexible filaments are 5–6 nm in diameter. Their length could reach up to 2–3 micrometers (Figure 4A). A maximum of 2–3 filaments per cell could be observed. Their average length was difficult to assess because they break easily into smaller fragments during sample preparation. Using the ComGC-FLAG expressing strain, we confirmed by immunogold-labelling that they contain ComGC (Figure 4C).
Fig. 4. Direct visualization of the competence-induced appendage. 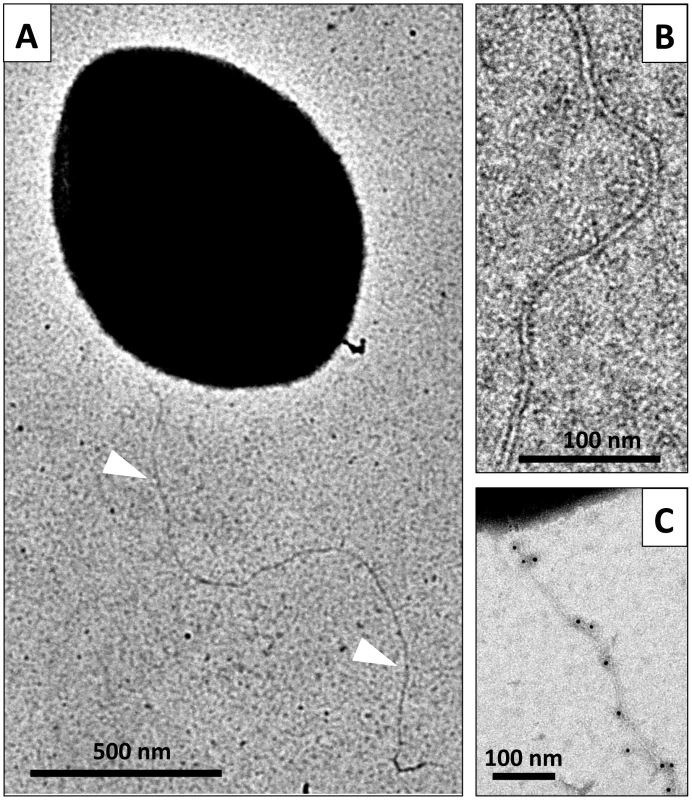
Competent reference bacteria observed by transmission electron microscopy. Single appendages, 5–6 nm wide, were observed at the surface most of the competent cells in the culture. Many long filaments were observed, reaching up to several micrometers in length. (A) a competent S. pneumoniae cell with a long pilus (white triangle). (B) closer view of a transformation pilus. (C) A pilus observed by transmission electron microscopy after immunogold labeling with anti-FLAG antibody (5 nm gold beads) using the ComGC-FLAG strain. ComGC-FLAG proteins are detected within the appendages. Appendages were then purified using anti-FLAG affinity chromatography after mechanical shearing. Appendage fragments of between 50 and 500 nm in length were observed by electron microscopy (Figure 5A), showing that these filamentous structures do not disassemble during purification. SDS-PAGE analysis of the purified fraction showed that ComGC is the major component of the appendages (Figure 5B). Using whole protein mass profiling by high-resolution mass spectrometry [18], we could only detect ComGC and ComGC-FLAG in the purified material (Figure 5C), confirming that ComGC is the major constituent of these appendages. Monoisotopic mass measurements of intact proteins and top-down fragmentation using a variety of activation techniques confirmed that the ComGC prepilin is cleaved after the alanine residue in position 15 and that the first amino acid of the mature protein is methylated, presumably by PilD (Figure S2). Indeed, PilD homologs in Gram-negative bacteria catalyze this post-translational modification of the Type IV pilins [19]. No other post-translational modification was detected in ComGC. Other proteins, including other ComG proteins, were not detected in the purified material by the methods used in this study. This suggests that these proteins are either absent, present in very low amount within the appendage or weakly bound to it and lost during sample preparation. These morphological and biochemical features are typical of Gram-negative Type IV pili. Therefore, we propose that the competence-induced appendage observed in S. pneumoniae belongs to the Type IV pilus family.
Fig. 5. Nature of the transformation pilus. 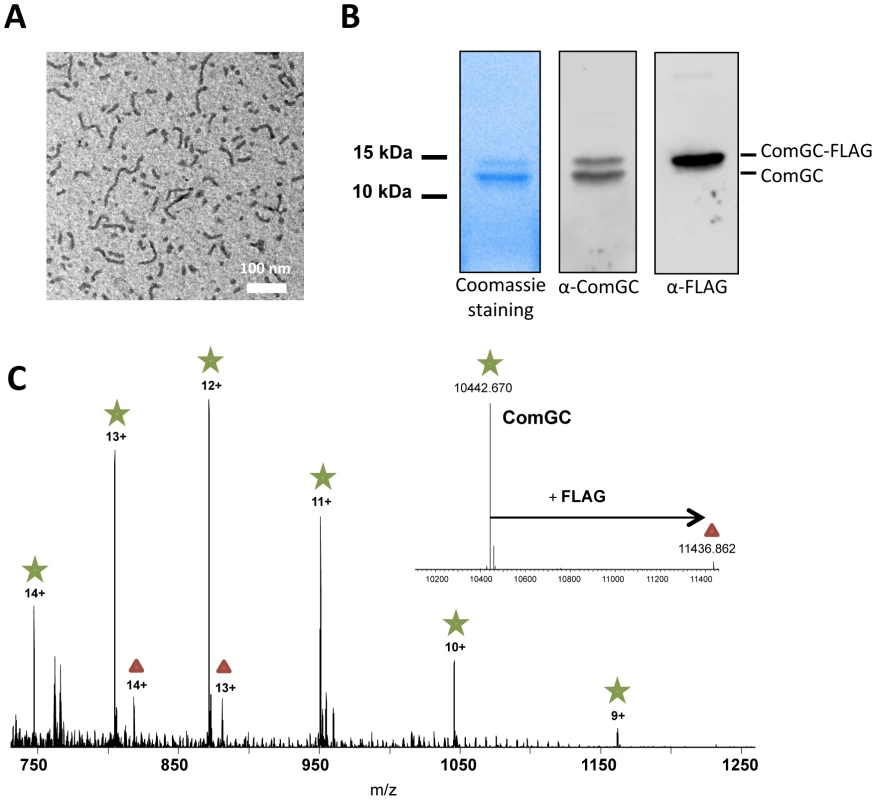
(A) Purified pili visualized by negative stain electron microscopy. Short pilus fragments ranging from 50 to 500 nm were observed. (B) SDS-PAGE analysis of the purified fraction. Left lane, Coommassie blue staining. Middle lane, Western blot using anti-ComGC antibody. Right lane, Western blot analysis using anti-FLAG analysis. (C) Nano-ESI FT-MS spectrum of purified pili. Peaks corresponding to ComGC are labelled with a green star those corresponding to ComGC-FLAG with a red triangle. ComGC-FLAG is present at approximately 6+/−2% abundance of the native form. The deconvoluted spectrum showing monoisotopic masses of the neutral protein forms is presented in the inset. The measured mass of methylated comGC (10,442.670 Da) compares very well to the calculated theoretical mass (10,442.636 Da) with an error of +3 ppm. The competence-induced pili are required for transformation
It was important to determine whether these competence-induced pili were involved in transformation. Indeed, it was previously shown that S. pneumoniae and B. subtilis comGA knockout could not be transformed (Figure 2B) [9] [13]. In this study, we were able to show in S. pneumoniae that comGA mutant cells lack pili (Figure 2A and 3A). It was enticing to conclude that competence-induced pili assembly is essential for transformation. However, it was recently shown that a comGA mutation could have a pleiotropic effect on transformation in B. subtilis [14]. Therefore, we generated a comGC mutant in S. pneumoniae in which the conserved glutamic acid in position 5 was substituted by an alanine (Figure 1B). Such a substitution was shown to impair Type IV pilus assembly in Gram-negative bacteria [20]. ComGC cellular level was not affected by this point mutation (Figure 2A). Our results show that this mutant strain could not assemble any pilus and that it was defective for transformation (Figure 2A and B). Therefore we conclude from the analysis of both comGA and comGC(E5A) mutants that the assembly of the competence-induced pilus is required for transformation.
Transformation pili bind extracellular DNA
The nature of the primary DNA receptor at the surface of transformable Gram-positive bacteria is not known. It is generally proposed that the transformation pseudopilus would bind extracellular DNA at the surface of competent Gram-positive bacteria [8], [21]. However, this hypothesis has never been confirmed experimentally. Using affinity purification, we show that DNA naturally released in the culture medium co-fractionates with the purified pili. No DNA could be found in the purified fraction in absence of the pilus (Figure 6A). These data were a first hint suggesting that DNA present in the environment could bind to the transformation pilus. However, it was not clear if this binding was related to the transformation process or fortuitous. By using specific electron microscopy methods [22], we visualized DNA directly bound to the transformation appendage after adding linear double stranded DNA (dsDNA) to competent bacteria. Long stretches of dsDNA interacting with the transformation pilus were observed with clearly visible multiple contact points (Figure 6 B–E). Interestingly, it was extremely difficult to see DNA bound on the pilus in the reference bacteria (R1501 strain), which are known to internalize exogenous DNA quickly [23]. On the other hand, in ComEC and comFA mutants, we could easily observe bound DNA on transformation pili. These strains are defective for DNA uptake and accumulate bound DNA at their surface [13]. Given that the dsDNA was added in large excess, no difference between the reference and mutant strains should be observed if DNA binding on the pili was a coincidental event. The fact that the uncoupling of DNA binding and uptake processes facilitates the observation of the DNA/pilus interaction is a strong indication that DNA binding on the transformation pilus is related to the transformation process.
Fig. 6. Interactions between transformation pili and DNA. 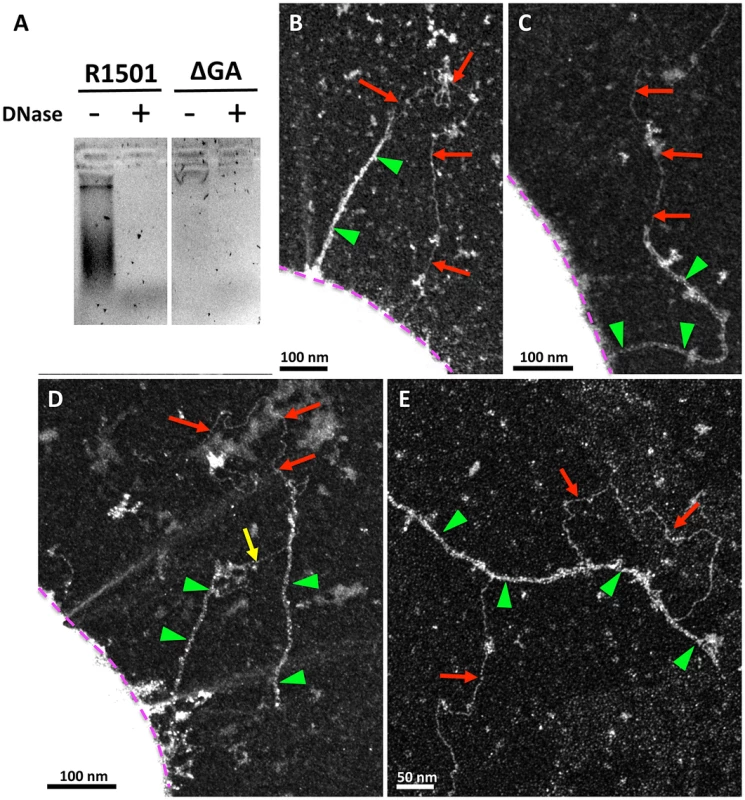
(A) DNA co-purified with the pili by affinity chromatography was revealed after migration on agarose gel and Gel Green staining. DNA was present in the ComGC-FLAG strain. In the absence of ComGA, DNA was not detected. DNase treatment unambiguously showed that the bands detected on the gel are DNA. Linear DNA (red arrow) bound to the pilus (green triangle) in ΔcomFA (B) and ΔcomEC (C), (D) and (E) strains. (B) and (C), linear DNA interacting with a single pilus. The DNA molecule contacts the pilus at several points. D, a DNA molecule maintains a broken pilus close to the bacterium (yellow arrow), demonstrating the existence of several DNA binding sites on the pilus. (E) Detached pilus found in the medium with bound DNA. In (B), (C) and (D), bacterial cell envelope is highlighted by a purple dotted line. Discussion
The comG operon is conserved in all transformable Gram-positive bacteria. This operon encodes proteins that are homologous to proteins involved in Type IV pilus assembly in Gram-negative bacteria. Therefore it has been proposed that a pilus (or pseudopilus) could be assembled at the surface of competent Gram-positive bacteria. Since all comG genes are essential for transformation, this pilus could be directly involved in transformation. The first biochemical clues for the existence of a transformation pilus were found in B. subtilis although decisive observational support was lacking. In addition, it was not clear if the ComGC-containing macromolecular complex found in B. subtilis was a common feature of competent Gram-positive bacteria or specific to this species. Finally, the function of this putative transformation pilus, and in general of the ComG proteins, was unclear.
Discovery of a new pneumococcal appendage
The pneumococcal transformation pilus represents a newly discovered pneumococcal surface structure. For a long time, no external appendage could be found at the surface of S. pneumoniae cells while many electron microscopy images were published in the literature. Recently, sortase-mediated pili have been discovered in some pathogenic S. pneumoniae strains [24]. To our knowledge, no specific ultrastructural study of competent S. pneumoniae has ever been described. Here, we analysed a laboratory strain that is commonly used to study the transformation process in S. pneumoniae [10] [13]. In this strain, competence can be induced in a rapid and synchronous manner upon addition of synthetic CSP in the medium of an exponentially growing culture [5], [25]. To make sure that the appearance of the transformation pilus is a common feature of competent pneumococci and not a mere one-off property of our reference strain, we observed negatively stained G54 and CP strains by electron microscopy. The G54 strain is a wild-type clinical strain. The CP strain is a laboratory strain that has a different genetic background than our reference strain [26]. In both cases, transformation pili were observed at the surface of competent cells (Figure S3). Therefore, we think that transformation pili are found at the surface of most, if not all, pneumoccocal strains, including clinical strains.
The transformation pilus is a Type IV pilus
The pneumococcal transformation pilus is morphologically very similar to Type IV pili found in many Gram-negative bacteria. Its major component, the ComGC pilin, is cleaved and probably methylated by a PilD homolog. We therefore propose that the transformation pilus is a bona fide Type IV pilus. Since its length can reach up to 2–3 µm, we think that the “pseudo-pilus” appellation does not apply to the pneumococcal transformation appendage. By comparison, the type II secretion pseudo-pilus is just 50–100 nm long [27]. The transformation pilus is the first Type IV pilus clearly observed in a Gram-positive bacterium. So far, Type IV pilus-dependent gliding motility had been described in Clostridium species [28]. However, no clear picture of this pilus was provided. A recent genomic study show the existence of numerous and diverse Type IV pilus-like operons in a wide range of Gram-positive bacteria [29]. This suggests that many other Type IV-like pili remain to be discovered in these bacteria. The conservation of comG operons argues in favor of the presence of a transformation pilus in all naturally transformable Gram-positive bacteria. However, species-specific variations in pilus length can be anticipated because of variations in thickness of the capsule and/or the cell wall.
Pilus function
We envision the transformation pilus to act as a “DNA-trap” to capture DNA in the environment. In Gram-negative bacteria, Type IV pilus assembly is also essential for natural transformation [30]. It is proposed that this pilus interacts directly with DNA but the molecular details of this interaction remains enigmatic [31], [32], [33]. No DNA binding protein could be identified in the pilus [34]. Recently a minor pilin, ComP, has been identified as a DNA receptor specific of genus-specific DNA uptake sequence (DUS) motifs in Neisseria meningitidis [35]. Our data explicitly show that the pneumoccocal transformation pilus binds DNA. At this stage, it is not clear if this DNA binding ability is due to the physicochemical properties of the pilus and/or due to a yet undetected minor pilin. Interestingly, only ComGA homolog was found indispensable for initial DNA binding at the surface of B. subtilis [14]. These bacteria could assemble only short transformation pili that are not sufficient to bind DNA at the surface of the competent cells. An unknown DNA receptor that interacts with ComGA ATPase could be required for efficient DNA binding in this species. It is possible that the mechanism of initial DNA binding at the surface of competent Gram-positive bacteria vary.
Pilus assembly and retraction
In Gram-negative bacteria, 12 to 15 proteins are necessary for Type IV pilus biogenesis [36], [37]. The assembly of the pneumococcal transformation pilus could require the simplest Type IV pilus assembly apparatus discovered so far in bacteria. Indeed, only 7 ComG proteins might be sufficient to assemble this pilus. Strikingly, Type IV pili are retractile appendages in Gram-negative bacteria [38]. It is therefore appealing to propose that the transformation pilus could retract to guide bound DNA through the cell wall and the polysaccharide capsule that is often present in bacteria (as in S. pneumoniae), and, ultimately deliver it to the ComEA/EndA complex located in the membrane [7]. It is unlikely that this DNA would find its way along several hundreds of nanometers through the cell wall and capsule without pilus retraction. However, a dedicated ATPase called PilT powers pilus retraction in Gram-negative bacteria and S. pneumoniae genome does not encode any PilT homolog. All the comG operons that have been identified so far in Gram-positive bacteria encode only one ATPase, ComGA, that is required for the pilus assembly. Therefore, the possible retraction of the transformation pilus and role of ComGA in this process should be assessed.
Our data clearly establish the existence and function of a transformation pilus at the surface of competent pneumococci. As an essential component of the transformation apparatus, it enables this major human Gram-positive pathogen to acquire resistance to antibiotics and to escape vaccines through the binding and incorporation of new genetic material. Future work should establish whether transformation pili exist and play similar roles in other transformable pathogens. Intriguingly, ComG proteins also appear to play an important and direct role in phagosomal escape and virulence in Listeria monocytogenes [39]. It would be interesting to determine whether they also assemble into a pilus to play this role.
Materials and Methods
Strain construction and growth
Cells were grown at 37°C under anaerobic condition, without agitation, in a Casamino Acid Tryptone medium (CAT) up to OD600 = 0.3 for stock cultures [40]. After addition of 15% glycerol, stocks were kept frozen at −80°C. For competence induction, cells were grown in CAT supplemented with BSA (2 g/L), calcium chloride (1 mM) and adjusted to pH = 7.8. Competence was triggered by incubating cells with the Competence Stimulating Peptide (CSP) at OD600 = 0.1 for 12 min as described previously [40]. For transformation, DNA was then added and transformants were selected on CAT agar plates [17]. Competence was induced following the same protocol in G54 and TCP1251 strains.
For transformation efficiency assays, 100 µL of competent bacteria were transformed by the addition of 100 ng of S. pneumoniae R304 genomic DNA (contains the streptomycin resistance gene str41). Bacteria were plated in presence and absence of streptomycin (100 µg/mL final concentration) and incubated at 37°C overnight before colony counting.
The annotated names of the comG genes in different strains of S. pneumoniae are listed in Table S1. The S. pneumoniae strains used derived from the non-capsulated R6 strain and are listed in Table S2. The comGC-FLAG gene was cloned by PCR using genomic DNA of pneumococcal R6 strain (ATCC BAA-255) as template. The resulting fragment was digested with NcoI and BamHI and inserted into the same sites of the pCEPx vector [17]. RL001 strain was constructed by transformation of R1501 cells with the pCEPx plasmid containing comGC-FLAG, followed by selection with kanamycin (Kan). RL002 was obtained by transformation of RL001 with R1062 chromosomal DNA and selection with spectinomycin (Spc). For RL003, a 2 kb genomic fragment of R6 genome containing comGC in the middle was amplified, and the codon 20 was changed from GAG to GTG by cross-over PCR. R1501 was transformed with this modified genomic fragment, and clones were screened by sequencing the comGC gene.
Chemically competent Escherichia coli BL21 Star (Life Technologies) were used for heterologous production of ComGC soluble domain. The corresponding DNA sequence was amplified from genomic DNA of strain R800 and cloned into pET15b expression vector (Novagen), using NdeI/XhoI. The protein was purified from the soluble fraction using IMAC affinity and gel filtration in 50 mM Tris/HCl pH = 8, 200 mM NaCl. The anti-ComGC were raised against the purified protein (Eurogentec).
Detection and purification of cell surface appendages
Shearing experiments were adapted from Sauvonnet et al. [41]. Competence was induced exactly as described above in a 50 mL culture. Cells were harvested by centrifugation 15 min at 4,500 g, 4°C. The pellet was suspended in 1 mL LB and immediately vortexed for 1 min to apply mechanical pressure. The suspension was then centrifuged twice at 13,000 g for 5 min to separate the bacteria (pellet fraction) from the pilus-enriched supernatant (sheared fraction). The supernatant was then precipitated with 10% trichloroacetic acid for 30 min on ice. Both fractions were loaded on SDS 15% polyacrylamide gels and subjected to electrophoresis and immunoblot with rabbit polyclonal antibodies raised against ComGC soluble domain (38–108) or anti-FLAG M2 antibody (Sigma-Aldrich F1804).
The pili containing ComGC-FLAG were purified from the sheared fraction of a 1 L culture. Shearing was performed in 2 mL Tris Buffered Saline (TBS, Tris pH 7.6 0.05 M, NaCl 0.15 M, protease inhibitor cocktail Roche 11873580001) and incubated overnight on a rotating wheel at 4°C with ANTI-FLAG M2 affinity resin (Sigma-Aldrich A2220). After washing with TBS, the pili were eluted by adding 3×FLAG-peptide at 100 µg/mL (Sigma Aldrich F4799) 30 min at room temperature under agitation.
To prevent DNA aspecific binding on the ANTI-FLAG M2 affinity resin, the resin was saturated 2 h at 4°C with a 1.5 kb PCR fragment (20 ng/µL). For DNA detection, 20 µL of the eluted pili were run on a 1% agarose gel and stained with SYBR safe (Life technologies S33102).
Visualization of pili and immunogold labelling
Competence was induced exactly as described above in a 10 mL culture. Cells were harvested by centrifugation 15 min at 4,500 g, 4°C. The pellet was suspended in 60 µL phosphate-buffered saline (PBS) (Sigma-Aldrich P4417). A drop of this suspension was placed on a glow discharged carbon coated grid (EMS, USA) for 1 min. The grid was then placed on a drop of PBS-3% formaldehyde, 0.2% glutaraldehyde for 10 min, and washed on drops of distilled water. The grids were then treated with 2% uranyl acetate in water. Specimens were examined using a Philips CM12 transmission electron microscope operated at 120 kV. Pictures were recorded using a camera KeenView (SIS, Germany) and ITEM software. For immunogold labelling, additional steps were applied after fixation: 3 washes with PBS, PBS–50 mM NH4Cl (10 min), 3 washes with PBS, PBS with 1% BSA (5 min), 1 hour incubation with ANTI-FLAG M2 antibody (Sigma-Aldrich F1804) diluted 1/100 in PBS with 1% BSA, 3 washes with PBS-BSA 1% (5 min), 1 hour incubation with goat anti-mouse antibody (5 nm gold particles, BritishBioCell, UK) diluted 1/25 in PBS containing 1% BSA.
Fluorescence microscopy
S. pneumoniae cells were grown in the same conditions as above for visualization by electron microscopy. Cells were harvested by centrifugation for 15 min at 4,500 g, 4°C. The pellet was suspended in 500 µL PBS and directly immobilized on poly-L-lysine-coated coverslips. Samples were fixed for 30 min with 3.7% formaldehyde, washed 3 times with PBS containing 1% BSA and incubated on a 100 µL drop of anti-FLAG antibodies (1∶300) and secondary Alexa Fluor 488 - coupled anti-mouse IgG (Invitrogen). Samples were examined with an Axio Imager.A2 microscope (Zeiss). Images were taken with AxioVision (Zeiss) and processed in ImageJ [42].
Mass spectrometry
Protein samples were desalted and eluted directly into a 10 µL spray solution of methanol∶water∶formic acid (75∶25∶3). Approximately 4 µL was loaded into a coated, medium sized, nano-ESI capillary (Proxeon) and introduced into an Orbitrap Velos mass spectrometer, equipped with ETD module (Thermo Fisher Scientific, Bremen, Germany) using the off-line nanospray source in positive ion mode. A full set of automated positive ion calibrations was performed immediately prior to mass measurement. The transfer capillary temperature was lowered to 100°C, sheath and axillary gasses switched off and source transfer parameters optimised using the auto tune feature. Helium was used as the collision gas in the linear ion trap. For MSn experiments, ions were selected with a 3 Da window and both CID and HCD were performed at normalised collision energies of 15–25%, with the appropriate HCD charge state set and other activation parameters left as default. For ETD the reagent gas was fluoranthene and the interaction time 10 ms. Supplemental activation was used as noted. The FT automatic gain control (AGC) was set at 1×106 for MS and 2×105 for MSn experiments. Spectra were acquired in the FTMS over several minutes with one microscan and a resolution of 60,000 @ m/z 400 before being summed using Qualbrowser in Thermo Xcalibur 2.1. Summed spectra were then deconvoluted using Xtract and a, b, c−1, y, z, z+1 ions assigned using in house software at a tolerance of 5 ppm. N-terminal ions were verified manually.
Positive staining electron microscopy
Five microliters of bacterial culture (wild-type, ΔcomFA or ΔcomEC) were diluted in 45 µL of Tris 10 mM, pH 8, NaCl 150 mM. Bacteriophage lambda DNA (0,1 mg/ml final) was then added to bacteria. Five µL of mix were immediately adsorbed onto a 600 mesh copper grid coated with a thin carbon film, activated by glow-discharge. After 1 min, grids were washed with 0,02% (w/vol) uranyl acetate solution (Merck, France) and then dried with filter paper. TEM observations were carried out with a Zeiss 912AB transmission electron microscope in filtered crystallographic dark field mode. Electron micrographs were obtained using a ProScan 1024 HSC digital camera and Soft Imaging Software system.
Supporting Information
Zdroje
1. GriffithF (1928) The Significance of Pneumococcal Types. The Journal of hygiene 27 : 113–159.
2. JohnsborgO, EldholmV, HavarsteinLS (2007) Natural genetic transformation: prevalence, mechanisms and function. Research in microbiology 158 : 767–778.
3. PopaO, DaganT (2011) Trends and barriers to lateral gene transfer in prokaryotes. Current opinion in microbiology 14 : 615–623.
4. HillerNL, AhmedA, PowellE, MartinDP, EutseyR, et al. (2010) Generation of genic diversity among Streptococcus pneumoniae strains via horizontal gene transfer during a chronic polyclonal pediatric infection. PLoS pathogens 6: e1001108.
5. HavarsteinLS, CoomaraswamyG, MorrisonDA (1995) An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proceedings of the National Academy of Sciences of the United States of America 92 : 11140–11144.
6. PrudhommeM, AttaiechL, SanchezG, MartinB, ClaverysJP (2006) Antibiotic stress induces genetic transformability in the human pathogen Streptococcus pneumoniae. Science 313 : 89–92.
7. ChenI, DubnauD (2004) DNA uptake during bacterial transformation. Nature reviews Microbiology 2 : 241–249.
8. ClaverysJP, MartinB, PolardP (2009) The genetic transformation machinery: composition, localization, and mechanism. FEMS microbiology reviews 33 : 643–656.
9. ChenI, ProvvediR, DubnauD (2006) A macromolecular complex formed by a pilin-like protein in competent Bacillus subtilis. The Journal of biological chemistry 281 : 21720–21727.
10. DagkessamanskaiaA, MoscosoM, HenardV, GuiralS, OverwegK, et al. (2004) Interconnection of competence, stress and CiaR regulons in Streptococcus pneumoniae: competence triggers stationary phase autolysis of ciaR mutant cells. Molecular microbiology 51 : 1071–1086.
11. PetersonSN, SungCK, ClineR, DesaiBV, SnesrudEC, et al. (2004) Identification of competence pheromone responsive genes in Streptococcus pneumoniae by use of DNA microarrays. Molecular microbiology 51 : 1051–1070.
12. KaufensteinM, van der LaanM, GraumannPL (2011) The three-layered DNA uptake machinery at the cell pole in competent Bacillus subtilis cells is a stable complex. Journal of bacteriology 193 : 1633–1642.
13. BergeM, MoscosoM, PrudhommeM, MartinB, ClaverysJP (2002) Uptake of transforming DNA in Gram-positive bacteria: a view from Streptococcus pneumoniae. Molecular microbiology 45 : 411–421.
14. BrileyKJr, Dorsey-OrestoA, PrepiakP, DiasMJ, MannJM, et al. (2011) The secretion ATPase ComGA is required for the binding and transport of transforming DNA. Molecular microbiology 81 : 818–830.
15. ChungYS, DubnauD (1998) All seven comG open reading frames are required for DNA binding during transformation of competent Bacillus subtilis. Journal of bacteriology 180 : 41–45.
16. NunnD, BergmanS, LoryS (1990) Products of three accessory genes, pilB, pilC, and pilD, are required for biogenesis of Pseudomonas aeruginosa pili. Journal of bacteriology 172 : 2911–2919.
17. MartinB, GranadelC, CampoN, HenardV, PrudhommeM, et al. (2010) Expression and maintenance of ComD-ComE, the two-component signal-transduction system that controls competence of Streptococcus pneumoniae. Molecular microbiology 75 : 1513–1528.
18. Chamot-RookeJ, MikatyG, MalosseC, SoyerM, DumontA, et al. (2011) Posttranslational modification of pili upon cell contact triggers N. meningitidis dissemination. Science 331 : 778–782.
19. StromMS, NunnDN, LoryS (1993) A single bifunctional enzyme, PilD, catalyzes cleavage and N-methylation of proteins belonging to the type IV pilin family. Proceedings of the National Academy of Sciences of the United States of America 90 : 2404–2408.
20. StromMS, LoryS (1991) Amino acid substitutions in pilin of Pseudomonas aeruginosa. Effect on leader peptide cleavage, amino-terminal methylation, and pilus assembly. The Journal of biological chemistry 266 : 1656–1664.
21. BurtonB, DubnauD (2010) Membrane-associated DNA transport machines. Cold Spring Harbor perspectives in biology 2: a000406.
22. DupaigneP, Le BretonC, FabreF, GangloffS, Le CamE, et al. (2008) The Srs2 helicase activity is stimulated by Rad51 filaments on dsDNA: implications for crossover incidence during mitotic recombination. Molecular cell 29 : 243–254.
23. MejeanV, ClaverysJP (1993) DNA processing during entry in transformation of Streptococcus pneumoniae. The Journal of biological chemistry 268 : 5594–5599.
24. BarocchiMA, RiesJ, ZogajX, HemsleyC, AlbigerB, et al. (2006) A pneumococcal pilus influences virulence and host inflammatory responses. Proceedings of the National Academy of Sciences of the United States of America 103 : 2857–2862.
25. AlloingG, MartinB, GranadelC, ClaverysJP (1998) Development of competence in Streptococcus pneumonaie: pheromone autoinduction and control of quorum sensing by the oligopeptide permease. Molecular microbiology 29 : 75–83.
26. PestovaEV, HavarsteinLS, MorrisonDA (1996) Regulation of competence for genetic transformation in Streptococcus pneumoniae by an auto-induced peptide pheromone and a two-component regulatory system. Molecular microbiology 21 : 853–862.
27. CamposM, CisnerosDA, NivaskumarM, FranceticO (2013) The type II secretion system - a dynamic fiber assembly nanomachine. Research in microbiology [Epub ahead of print] doi:10.1016/j.resmic.2013.03.013
28. VargaJJ, NguyenV, O'BrienDK, RodgersK, WalkerRA, et al. (2006) Type IV pili-dependent gliding motility in the Gram-positive pathogen Clostridium perfringens and other Clostridia. Molecular microbiology 62 : 680–694.
29. ImamS, ChenZ, RoosDS, PohlschroderM (2011) Identification of surprisingly diverse type IV pili, across a broad range of gram-positive bacteria. PloS one 6: e28919.
30. CraigL, PiqueME, TainerJA (2004) Type IV pilus structure and bacterial pathogenicity. Nature reviews Microbiology 2 : 363–378.
31. BiswasGD, SoxT, BlackmanE, SparlingPF (1977) Factors affecting genetic transformation of Neisseria gonorrhoeae. Journal of bacteriology 129 : 983–992.
32. DoughertyTJ, AsmusA, TomaszA (1979) Specificity of DNA uptake in genetic transformation of gonococci. Biochemical and biophysical research communications 86 : 97–104.
33. van SchaikEJ, GiltnerCL, AudetteGF, KeizerDW, BautistaDL, et al. (2005) DNA binding: a novel function of Pseudomonas aeruginosa type IV pili. Journal of bacteriology 187 : 1455–1464.
34. LangE, HaugenK, FleckensteinB, HombersetH, FryeSA, et al. (2009) Identification of neisserial DNA binding components. Microbiology 155 : 852–862.
35. CehovinA, SimpsonPJ, McDowellMA, BrownDR, NoscheseR, et al. (2013) Specific DNA recognition mediated by a type IV pilin. Proceedings of the National Academy of Sciences of the United States of America 110 : 3065–3070.
36. CarbonnelleE, HelaineS, NassifX, PelicicV (2006) A systematic genetic analysis in Neisseria meningitidis defines the Pil proteins required for assembly, functionality, stabilization and export of type IV pili. Molecular microbiology 61 : 1510–1522.
37. GeorgiadouM, CastagniniM, KarimovaG, LadantD, PelicicV (2012) Large-scale study of the interactions between proteins involved in type IV pilus biology in Neisseria meningitidis: characterization of a subcomplex involved in pilus assembly. Molecular microbiology 84 : 857–873.
38. MerzAJ, SoM, SheetzMP (2000) Pilus retraction powers bacterial twitching motility. Nature 407 : 98–102.
39. RabinovichL, SigalN, BorovokI, Nir-PazR, HerskovitsAA (2012) Prophage excision activates Listeria competence genes that promote phagosomal escape and virulence. Cell 150 : 792–802.
40. MartinB, PrudhommeM, AlloingG, GranadelC, ClaverysJP (2000) Cross-regulation of competence pheromone production and export in the early control of transformation in Streptococcus pneumoniae. Molecular microbiology 38 : 867–878.
41. SauvonnetN, VignonG, PugsleyAP, GounonP (2000) Pilus formation and protein secretion by the same machinery in Escherichia coli. The EMBO journal 19 : 2221–2228.
42. SchneiderCA, RasbandWS, EliceiriKW (2012) NIH Image to ImageJ: 25 years of image analysis. Nature methods 9 : 671–675.
43. HansenJK, ForestKT (2006) Type IV pilin structures: insights on shared architecture, fiber assembly, receptor binding and type II secretion. Journal of molecular microbiology and biotechnology 11 : 192–207.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2013 Číslo 6- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Asthma and the Diversity of Fungal Spores in Air
- The Spatial Organization of DNA Virus Genomes in the Nucleus
- Novel Polyomaviruses of Nonhuman Primates: Genetic and Serological Predictors for the Existence of Multiple Unknown Polyomaviruses within the Human Population
- Introducing the Outbreak Threshold in Epidemiology
- An Apicoplast Localized Ubiquitylation System Is Required for the Import of Nuclear-encoded Plastid Proteins
- Streptolysin O and its Co-Toxin NAD-glycohydrolase Protect Group A from Xenophagic Killing
- A Type IV Pilus Mediates DNA Binding during Natural Transformation in
- Location of the CD8 T Cell Epitope within the Antigenic Precursor Determines Immunogenicity and Protection against the Parasite
- Dietary Cholesterol Modulates Pathogen Blocking by
- Extreme Genetic Fragility of the HIV-1 Capsid
- Novel Immunomodulators from Hard Ticks Selectively Reprogramme Human Dendritic Cell Responses
- Extramedullary Myelopoiesis in Malaria Depends on Mobilization of Myeloid-Restricted Progenitors by IFN-γ Induced Chemokines
- Developing Models of Disease Transmission: Insights from Ecological Studies of Insects and Their Baculoviruses
- Cryotomography of Budding Influenza A Virus Reveals Filaments with Diverse Morphologies that Mostly Do Not Bear a Genome at Their Distal End
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Asthma and the Diversity of Fungal Spores in Air
- Streptolysin O and its Co-Toxin NAD-glycohydrolase Protect Group A from Xenophagic Killing
- Cryotomography of Budding Influenza A Virus Reveals Filaments with Diverse Morphologies that Mostly Do Not Bear a Genome at Their Distal End
- A Type IV Pilus Mediates DNA Binding during Natural Transformation in
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



