-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaThe Spatial Organization of DNA Virus Genomes in the Nucleus
article has not abstract
Published in the journal: . PLoS Pathog 9(6): e32767. doi:10.1371/journal.ppat.1003386
Category: Pearls
doi: https://doi.org/10.1371/journal.ppat.1003386Summary
article has not abstract
Where Do the Genomes of DNA Viruses Go When They Enter the Nucleus?
During lytic infection, DNA viruses that replicate in the nucleus of cells establish an environment that favors viral replication while evading cellular defenses. Consequently, these viruses must engage with cellular factors that are necessary for efficient viral transcription and DNA replication, while disarming restrictive cellular responses to the presence of the invading pathogenic DNA. It is important to consider these events not only at the biochemical level of protein-protein and protein-nucleic acid interactions, but also in terms of spatial organization within the nucleus. The nucleus contains many distinct substructures, including nucleoli, chromosome territories, splicing speckles, PML Nuclear Bodies (PML NBs, also known as ND10), and transcription sites, all of which are in a dynamic environment involving the exchange of protein molecules between the structures themselves and the general nucleoplasm [1]. DNA viruses establish their own transcription sites and replication compartments within the nucleus, and a topic that has been of interest for many years is how these virus-specific loci relate to the preexisting cellular nuclear substructures, particularly during the early stages of infection. Is viral genome localization random, or is it regulated in some way?
Gerd Maul made a breakthrough discovery when he found that after their delivery into the nucleus, the genomes of many different DNA viruses are frequently associated with PML NBs [2], [3]. These intriguing findings posed many questions: How does this association occur? What are the viral and cellular signals involved? Does the association have a positive or negative influence on infection?
Viral Genome Association with PML NBs—A Cellular Response?
Initially it was thought that viral genomes might migrate through the nucleoplasm until they engaged with preexisting PML NBs. While this has not been excluded, particularly for the smaller DNA viruses, this model presents some practical problems for large DNA virus genomes. For example, the genomes of herpesviruses, in excess of 150 kbp, are likely to have limited mobility in the nucleoplasm, especially as they will rapidly accumulate chromatin-related and other binding partners and thus achieve significant bulk and mass. Indeed, there is strong evidence that HSV-1 genomes do not move far from the inner nuclear membrane after their entry into the nucleus through the nuclear pore [4], [5]. PML NBs are also of substantial size (of the order of 1 µm diameter), but while the structures themselves have limited mobility, the component proteins undergo rapid exchange with the general nucleoplasm [6]. Therefore an alternative explanation of viral genome–PML NB association is that it occurs not because either entity migrates through the nucleoplasm, but because new PML NBs are formed at the sites of the viral genomes due to the deposition of PML NB protein molecules. In the case of HSV-1, a chance observation provided very strong evidence that this was indeed the case—the viral genomes became very rapidly associated with novel PML NB–like structures in the earliest stages of infection through recruitment of PML NB proteins [4] (Figure 1). While for technical reasons it is more difficult to confirm that this is also the case in other DNA virus infections, it is likely that analogous events occur more generally. If so, the association of viral genomes and PML NB–like structures can be viewed as a cellular response to the entry of a viral genome into the nucleus.
Fig. 1. The association of PML and DNA damage response foci with HSV-1 genomes. 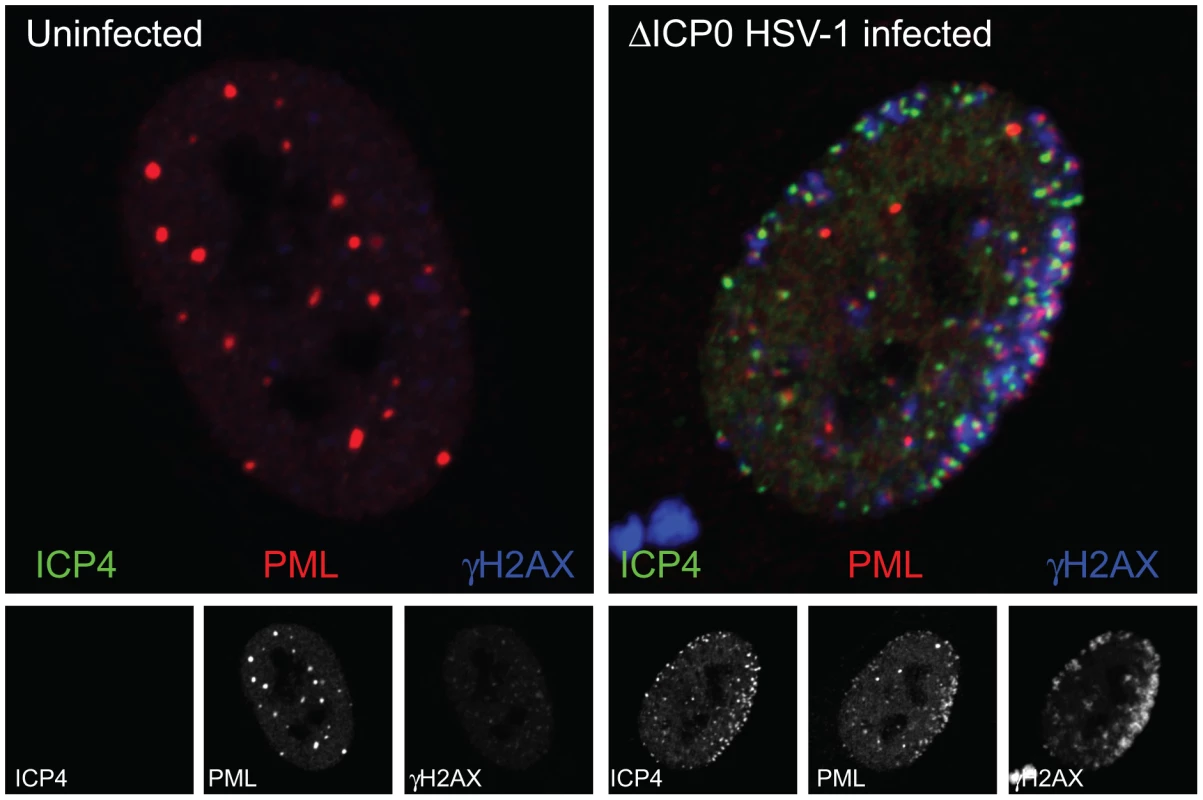
The uninfected human diploid fibroblast nucleus on the left shows a typical distribution of PML NBs (PML, red) with only faint staining of the DNA damage response protein γH2AX (blue), with the separated channels shown in grayscale below. The infected cell nucleus on the right is of a cell at the edge of a developing ICP0-null mutant HSV-1 plaque, which has been infected with a very high number of virus particles by spread from neighboring heavily infected cells. The viral genomes can be detected by the presence of the viral transcriptional activator ICP4 (green), which binds efficiently to viral DNA. PML (red) has been redistributed from its normal locations to sites that closely associated with the viral genomes. This association is stabilized by the absence of ICP0, which efficiently inhibits the formation of these foci in a normal wild type HSV-1 infection. A DNA damage response has been initiated to produce a marked increase in γH2AX (blue) in regions close to the viral genomes. What Factors Are Involved in the Association between DNA Virus Genomes and PML NBs?
The issues that influence the association between DNA virus genomes and PML NBs include the virus factors that are required for the response, the properties of PML NB proteins required for their recruitment, and the actions of viral regulatory proteins that affect the stability of the recruitment. Because the association of adenovirus and HSV-1 genomes with PML NBs occurs when viral transcription and protein expression are inhibited [2]–[4], it appears that the viral genome itself may be sufficient to trigger the response. It is possible that the initial signal involves unchromatinized “foreign” DNA, but note that PML NB components can also be recruited to unusual chromosomal locations such as integrated transgene arrays with multiple regulatory factor binding sites [7].
From the cellular aspect, although PML is absolutely required for the assembly of PML NBs in uninfected cells [8], a number of other PML NB proteins (such as Sp100, hDaxx, ATRX, and SUMO-2/3) become associated with HSV-1 genomes even in cells highly depleted of PML [9]–[12]. There is evidence that similar events occur in HCMV-infected cells [13]. The assembly and dynamics of PML NBs are strongly linked to modification by the ubiquitin-like SUMO family of proteins, and this also holds true for the viral genome–associated structures. Recruitment of PML requires both its sumoylation and its ability to interact noncovalently with SUMO through its SUMO interaction motif (SIM) [12], and recruitment of both Sp100 and hDaxx requires their SIMs [12]. ATRX on the other hand is recruited through its interaction with hDaxx [11]. Therefore the association requires viral genomes on the one hand, and a cascade of events involving both sumoylation-dependent and -independent events on responsive cellular proteins on the other.
An important factor involved in the association of DNA viral genomes and PML NBs is the effect of viral regulatory proteins on their stability. The recruitment is very short-lived in normal wild type HSV-1 infection because the viral immediate-early protein ICP0, an E3 ubiquitin ligase, induces degradation of PML and disruption of PML NBs [14], [15]. However, in the absence of ICP0, the recruitment is stable and easily visualized [4] (Figure 1). The association between PML NBs and the genomes of HCMV and adenovirus is also most readily observed at early times of infection or with mutant viruses that fail to express regulatory proteins that would normally disrupt PML NBs [2], [16]. In contrast, in cultured cells harboring quiescent HSV-1 [9] and in latently infected mouse neurons [17] (conditions in which ICP0 is not expressed), HSV-1 genomes are surrounded by a shell of PML NB proteins. It is possible that this reflects sequestration of the viral genomes in an inactive, repressed environment.
Is the Recruitment of PML NB Proteins to Viral Genomes Linked to a DNA Damage Response?
The initial stages of the recruitment process are of obvious interest. Because DNA viral genomes in the initial stages of infection are in a state quite distinct from cellular chromatin, it is possible that a DNA damage response (DDR) is involved. Many DNA viruses induce a DDR [18], which in some cases involves the accumulation of DNA repair proteins in close proximity to viral DNA [19], [20], or within developing viral replication compartments (reviewed in [18]) (see also Figure 1). Furthermore, PML has been observed to accumulate at sites of DNA damage [21]. PML NBs have been suggested to act as sensors for damaged DNA [22], they include proteins involved in both DNA repair and chromatin modification [8], and sumoylation events are involved in both the recruitment of PML NB proteins to HSV-1 genomes [12] and the DDR [23]. Despite these parallels, it remains unclear whether PML NB and DNA damage responses during the initial stages of DNA virus infection are related. Although ICP0 of HSV-1 inhibits both responses, this occurs through distinct mechanisms [19]. Furthermore, the formation of DDR foci close to HSV-1 genomes does not require PML or other major PML NB components, and is also atypical of classical DDR because it does not require Mre11 or ATM [19]. Therefore the association of DNA viral genomes with PML NBs and DDR foci may occur through distinct mechanisms. Whether other DNA sensors, such as IFI16, are involved in the PML NB response remains to be determined. It is noteworthy that IFI16 becomes associated with herpesvirus genomes during infection [24], [25].
What Are the Consequences of Viral Genome Association with Cellular Nuclear Substructures and Protein Complexes?
Whether the association between DNA virus genomes and PML NBs has a positive or negative influence on the outcome of infection has long been debated. On the one hand, viral transcription and replication sites develop initially in association with PML NB–like structures [16], which could indicate a beneficial effect. On the other hand, the defective phenotype of virus mutants unable to disrupt PML NBs suggests a negative influence, and it is this view that dominates current thinking. RNAi-mediated depletion of PML NB proteins has revealed several examples of herpesvirus infections that are restricted by one or more PML NB components, and in these cases the viruses express regulatory proteins that overcome the restriction [26]. There is evidence in the case of HSV-1 that the DDR also contributes to initial restrictive responses to viral DNA [19], although in general DDR proteins play both positive and negative roles during DNA virus infection [18]. The degree of restriction imparted by any one PML NB protein may be modest, but there is evidence that PML NB–mediated inhibition of herpesvirus infections involves the cooperative actions of several PML NB components [27], [28].
In conclusion, the current information is consistent with the hypothesis that entry of DNA virus genomes into the nucleus stimulates a cellular response that leads to the accumulation of PML NB proteins, and probably other restrictive proteins yet to be identified, in close association with the viral genomes. In turn, viruses express regulatory proteins that inactivate this defensive response and thus stimulate lytic infection. Thus the outcome of DNA virus infection is governed not only by the biochemical properties of cellular and viral regulatory proteins, but also by the dynamic manner in which they are spatially organized in the nucleus.
Zdroje
1. LamondAI, EarnshawWC (1998) Structure and function in the nucleus. Science 280 : 547–553.
2. IshovAM, MaulGG (1996) The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J Cell Biol 134 : 815–826.
3. MaulGG, IshovAM, EverettRD (1996) Nuclear domain 10 as preexisting potential replication start sites of herpes simplex virus type-1. Virology 217 : 67–75.
4. EverettRD, MurrayJ (2005) ND10 components relocate to sites associated with herpes simplex virus type 1 nucleoprotein complexes during virus infection. J Virol 79 : 5078–5089.
5. SilvaL, CliffeA, ChangL, KnipeDM (2008) Role for A-type lamins in herpesviral DNA targeting and heterochromatin modulation. PLoS Pathog 4: e1000071 doi:10.1371/journal.ppat.1000071
6. Weidtkamp-PetersS, LenserT, NegorevD, GerstnerN, HofmannTG, et al. (2008) Dynamics of component exchange at PML nuclear bodies. J Cell Sci 121 : 2731–2743.
7. NewhartA, Rafalska-MetcalfIU, YangT, NegorevDG, JanickiSM (2012) Single-cell analysis of Daxx and ATRX-dependent transcriptional repression. J Cell Sci 125 : 5489–5501.
8. BernardiR, PandolfiPP (2007) Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat Rev Mol Cell Biol 8 : 1006–1016.
9. EverettRD, MurrayJ, OrrA, PrestonCM (2007) Herpes simplex virus type 1 genomes are associated with ND10 nuclear substructures in quiescently infected human fibroblasts. J Virol 81 : 10991–11004.
10. EverettRD, RechterS, PapiorP, TavalaiN, StammingerT, et al. (2006) PML contributes to a cellular mechanism of repression of herpes simplex virus type 1 infection that is inactivated by ICP0. J Virol 80 : 7995–8005.
11. LukashchukV, OrrA, EverettRD (2010) Regulation of ICP0 null mutant HSV-1 infection by ND10 components ATRX and hDaxx. J Virol 84 : 4026–4040.
12. Cuchet-LourençoD, BoutellC, LukashchukV, GrantK, SykesA, et al. (2011) SUMO pathway dependent recruitment of cellular repressors to herpes simplex virus type 1 genomes. PLoS Pathog 7: e1002123 doi:10.1371/journal.ppat.1002123
13. TavalaiN, PapiorP, RechterS, LeisM, StammingerT (2006) Evidence for a role of the cellular ND10 protein PML in mediating intrinsic immunity against human cytomegalovirus infections. J Virol 80 : 8006–8018.
14. EverettRD, FreemontP, SaitohH, DassoM, OrrA, et al. (1998) The disruption of ND10 during herpes simplex virus infection correlates with the Vmw110 - and proteasome-dependent loss of several PML isoforms. J Virol 72 : 6581–6591.
15. BoutellC, Cuchet-LourençoD, VanniE, OrrA, GlassM, et al. (2011) A viral ubiquitin ligase has substrate preferential SUMO targeted ubiquitin ligase activity that counteracts intrinsic antiviral defence. PLoS Pathog 7: e1002245 doi:10.1371/journal.ppat.1002245
16. IshovAM, StenbergRM, MaulGG (1997) Human cytomegalovirus immediate early interaction with host nuclear structures: definition of an immediate transcript environment. J Cell Biol 138 : 5–16.
17. CatezF, PicardC, HeldK, GrossS, RousseauA, et al. (2012) HSV-1 genome subnuclear positioning and associations with host-cell PML-NBs and centromeres regulate LAT locus transcription during latency in neurons. PLoS Pathog 8: e1002852 doi:10.1371/journal.ppat.1002852
18. WeitzmanMD, LilleyCE, ChaurushiyaMS (2010) Genomes in conflict: maintaining genome integrity during virus infection. Annu Rev Microbiol 64 : 61–81.
19. LilleyCE, ChaurushiyaMS, BoutellC, EverettRD, WeitzmanMD (2011) The intrinsic antiviral defense to incoming HSV-1 genomes includes specific DNA repair proteins and is counteracted by the viral protein ICP0. PLoS Pathog 7: e1002084 doi:10.1371/journal.ppat.1002084
20. MohniKN, MastrocolaAS, BaiP, WellerSK, HeinenCD (2011) DNA mismatch repair proteins are required for efficient herpes simplex virus 1 replication. J Virol 85 : 12241–12253.
21. CarboneR, PearsonM, MinucciS, PelicciPG (2002) PML NBs associate with the hMre11 complex and p53 at sites of irradiation induced DNA damage. Oncogene 21 : 1633–1640.
22. DellaireG, ChingRW, AhmedK, JalaliF, TseKC, et al. (2006) Promyelocytic leukemia nuclear bodies behave as DNA damage sensors whose response to DNA double-strand breaks is regulated by NBS1 and the kinases ATM, Chk2, and ATR. J Cell Biol 175 : 55–66.
23. JacksonSP, DurocherD (2013) Regulation of DNA Damage Responses by Ubiquitin and SUMO. Mol Cell 49 : 795–807.
24. KerurN, VeettilMV, Sharma-WaliaN, BotteroV, SadagopanS, et al. (2011) IFI16 acts as a nuclear pathogen sensor to induce the inflammasome in response to Kaposi Sarcoma-associated herpesvirus infection. Cell Host Microbe 9 : 363–375.
25. OrzalliMH, DelucaNA, KnipeDM (2012) Nuclear IFI16 induction of IRF-3 signaling during herpesviral infection and degradation of IFI16 by the viral ICP0 protein. Proc Natl Acad Sci U S A 109: E3008–3017.
26. TavalaiN, StammingerT (2008) New insights into the role of the subnuclear structure ND10 for viral infection. Biochim Biophys Acta 1783 : 2207–2221.
27. GlassM, EverettRD (2013) Components of PML Nuclear Bodies (ND10) act cooperatively to repress herpesvirus infection. J Virol 87 : 2174–2185.
28. TavalaiN, PapiorP, RechterS, StammingerT (2008) Nuclear domain 10 components promyelocytic leukemia protein and hDaxx independently contribute to an intrinsic antiviral defense against human cytomegalovirus infection. J Virol 82 : 126–137.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2013 Číslo 6- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Asthma and the Diversity of Fungal Spores in Air
- The Spatial Organization of DNA Virus Genomes in the Nucleus
- Novel Polyomaviruses of Nonhuman Primates: Genetic and Serological Predictors for the Existence of Multiple Unknown Polyomaviruses within the Human Population
- Introducing the Outbreak Threshold in Epidemiology
- An Apicoplast Localized Ubiquitylation System Is Required for the Import of Nuclear-encoded Plastid Proteins
- Streptolysin O and its Co-Toxin NAD-glycohydrolase Protect Group A from Xenophagic Killing
- A Type IV Pilus Mediates DNA Binding during Natural Transformation in
- Location of the CD8 T Cell Epitope within the Antigenic Precursor Determines Immunogenicity and Protection against the Parasite
- Dietary Cholesterol Modulates Pathogen Blocking by
- Extreme Genetic Fragility of the HIV-1 Capsid
- Novel Immunomodulators from Hard Ticks Selectively Reprogramme Human Dendritic Cell Responses
- Extramedullary Myelopoiesis in Malaria Depends on Mobilization of Myeloid-Restricted Progenitors by IFN-γ Induced Chemokines
- Developing Models of Disease Transmission: Insights from Ecological Studies of Insects and Their Baculoviruses
- Cryotomography of Budding Influenza A Virus Reveals Filaments with Diverse Morphologies that Mostly Do Not Bear a Genome at Their Distal End
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Asthma and the Diversity of Fungal Spores in Air
- Streptolysin O and its Co-Toxin NAD-glycohydrolase Protect Group A from Xenophagic Killing
- Cryotomography of Budding Influenza A Virus Reveals Filaments with Diverse Morphologies that Mostly Do Not Bear a Genome at Their Distal End
- A Type IV Pilus Mediates DNA Binding during Natural Transformation in
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



