-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaStructural Determinants for Activity and Specificity of the Bacterial Toxin LlpA
Lectin-like bacteriotoxic proteins, identified in several plant-associated bacteria, are able to selectively kill closely related species, including several phytopathogens, such as Pseudomonas syringae and Xanthomonas species, but so far their mode of action remains unrevealed. The crystal structure of LlpABW, the prototype lectin-like bacteriocin from Pseudomonas putida, reveals an architecture of two monocot mannose-binding lectin (MMBL) domains and a C-terminal β-hairpin extension. The C-terminal MMBL domain (C-domain) adopts a fold very similar to MMBL domains from plant lectins and contains a binding site for mannose and oligomannosides. Mutational analysis indicates that an intact sugar-binding pocket in this domain is crucial for bactericidal activity. The N-terminal MMBL domain (N-domain) adopts the same fold but is structurally more divergent and lacks a functional mannose-binding site. Differential activity of engineered N/C-domain chimers derived from two LlpA homologues with different killing spectra, disclosed that the N-domain determines target specificity. Apparently this bacteriocin is assembled from two structurally similar domains that evolved separately towards dedicated functions in target recognition and bacteriotoxicity.
Published in the journal: . PLoS Pathog 9(2): e32767. doi:10.1371/journal.ppat.1003199
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1003199Summary
Lectin-like bacteriotoxic proteins, identified in several plant-associated bacteria, are able to selectively kill closely related species, including several phytopathogens, such as Pseudomonas syringae and Xanthomonas species, but so far their mode of action remains unrevealed. The crystal structure of LlpABW, the prototype lectin-like bacteriocin from Pseudomonas putida, reveals an architecture of two monocot mannose-binding lectin (MMBL) domains and a C-terminal β-hairpin extension. The C-terminal MMBL domain (C-domain) adopts a fold very similar to MMBL domains from plant lectins and contains a binding site for mannose and oligomannosides. Mutational analysis indicates that an intact sugar-binding pocket in this domain is crucial for bactericidal activity. The N-terminal MMBL domain (N-domain) adopts the same fold but is structurally more divergent and lacks a functional mannose-binding site. Differential activity of engineered N/C-domain chimers derived from two LlpA homologues with different killing spectra, disclosed that the N-domain determines target specificity. Apparently this bacteriocin is assembled from two structurally similar domains that evolved separately towards dedicated functions in target recognition and bacteriotoxicity.
Introduction
In most natural settings, complex interactions occur among microorganisms, ranging from nutritional co-operation to warfare among competitors. Examples of such interplay have been reported not only between unrelated microorganisms (e.g. fungi and bacteria [1], [2]), but also between distant relatives (e.g. members of different bacterial genera [3]), and even between close relatives (e.g. at inter - and intra-species levels [4], [5]). A major strategy in niche colonization is the production of growth inhibitors or toxins directed at microbial competitors [6]. While a huge variety of secondary metabolites is used to target phylogenetically-distant competitors, ribosome-synthesized peptides or proteins are typically active against close relatives. These protein toxins are collectively referred to as bacteriocins, and may either be released into the environment or transferred to the host via specialized contact-dependent delivery systems [7]–[9].
Bacteriocins are structurally and mechanistically very diverse. This is reflected in the bacteriocinogenic potential of the genus Pseudomonas [10]. Their R - and F-type pyocins are multi-subunit protein complexes evolutionarily related to contractile tails of bacteriophages [11]–[13]. R-pyocins attach to specific lipopolysaccharide moieties at the cell surface of susceptible cells and insert their core structure through the cell envelope, causing depolarization of the cytoplasmic membrane [14]. The S-type pyocins of Pseudomonas aeruginosa share structural and functional features with Escherichia coli colicins [15]. Following docking onto surface-exposed targets such as siderophore receptors [16], [17], S-pyocins kill cells by nucleic acid degradation [10], [17], cytoplasmic membrane damage [18], or inhibition of peptidoglycan synthesis [19], [20]. Putidacin A (or LlpABW), first identified in Pseudomonas putida BW11M1 [21], represents a class of Pseudomonas-specific antibacterial proteins not related to any known bacteriocin. Additional llpA-like genes encoding functional bacteriocins were identified by genome mining in the biocontrol strain Pseudomonas fluorescens Pf-5 [22] and in the phytopathogen Pseudomonas syringae pv. syringae 642 [23]. Identification of this type of protein in two Xanthomonas pathovars extended its occurrence as a genus-specific killer protein [23]. The Xanthomonas LlpA precursor is proteolytically processed by removal of a characteristic Type II secretion signal peptide, whereas such N-terminal sequence is lacking in Pseudomonas homologues, indicating that secretory routes may differ among LlpA producers.
The amino acid sequence of LlpA suggests the presence of two related domains belonging to the ‘monocot mannose-binding lectin’ (MMBL) family [24]. The MMBL domain consists of a β-prism fold containing three potential carbohydrate-binding pockets, each generated by a QxDxNxVxY sequence (with x, any amino acid), but some sites may be inactive due to degeneracy of the signature motif [25]. This domain (Pfam domain: B_lectin - PF01453) was initially identified in lectins of monocot plants [26], [27], but a more widespread occurrence of MMBL lectins has become evident and includes representatives in fungi [28], [29], slime molds [30], sponges [31], and fishes [32]–[34]. The LlpA branch occupies a unique position among MMBL-domain proteins, harboring non-eukaryotic representatives and being equipped with the capacity to kill bacterial cells with bacteriocin-like specificity, a property not yet demonstrated for other family members [25]. Next to proteins with the LlpA-type tandem-MMBL organization, many other predicted MMBL proteins are encoded by bacterial genomes. Often the MMBL module is embedded in a larger protein. For one such protein, bacteriocin-like activity among Ruminococcus species, Gram-positive bacteria colonizing the rumen, was demonstrated [35].
Here we report on the crystal structure of LlpABW as the prototype of a novel family of antibacterial proteins and explore how domain architecture and specific structural elements contribute to its activity and specificity.
Results
LlpA forms a rigid MMBL tandem
The crystal structure of LlpABW from P. putida BW11M1 (LlpABW) shows it contains two β-prism MMBL domains, referred to as the N-domain and the C-domain following their position in the amino acid sequence (Figure 1A,B; Figure S1). The N-domain spans residues Arg4-Pro135 while the C-domain encompasses residues Ala136-Gln253. Each domain exhibits pseudo-threefold symmetry and the corresponding subdomains will be referred to as IN, IIN, IIIN, IC, IIC and IIIC, respectively (Figure 1A and Figure S1). Following these two domains, a β-hairpin extension is formed by residues Pro254-His275 (the numbering used in this paper corresponds to that of the wild-type protein without His-tag [21]).
Fig. 1. Overall structure of LlpABW. 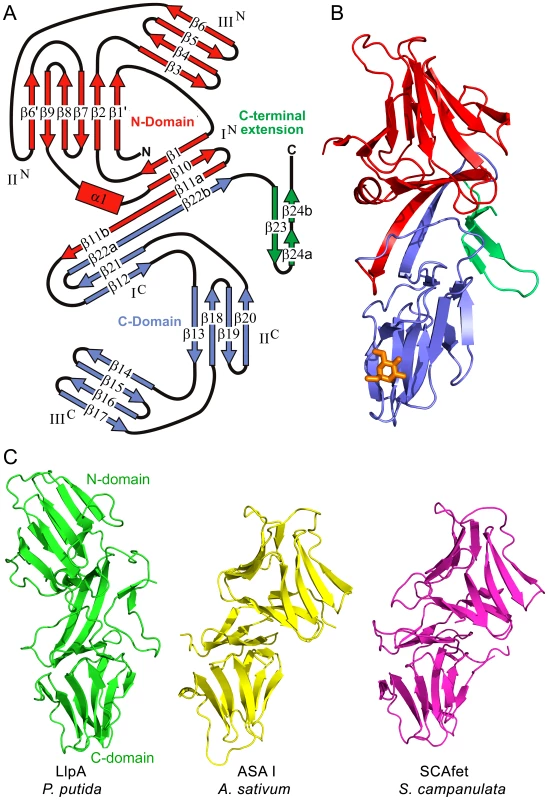
(A) Topology diagram of LlpABW. The N-domain is shown in red, the C-domain in blue and the C-terminal extension in green. The different strands and subdomains are labeled. Domain swapping involves β-strand segments β11b and β22b, which together with β-strand segments β11a and β22a link both MMBL domains. (B) Cartoon representation of LlpABW with the different domains colored as in panel A. The bound Me-Man residue is shown as an orange stick representation. (C) Domain orientations of LlpABW compared with the heterodimeric MMBL ASA I (Allium sativum agglutinin, PDB entry 1KJ1) and tandem MMBL SCAfet (Scilla campanulata fetuin-binding lectin, PDB entry 1DLP). In each case, the C-domain is shown in the same orientation, highlighting the different relative orientation of the N-domain in LlpABW. Domain-swapped dimers in homo-oligomeric plant MMBL lectins such as snowdrop lectin have their domain orientation similar to ASA I and SCAfet. The two-domain architecture reflects the β-strand swapping that is typical in dimers of single-domain mannose-binding monocot lectins (Figure 1A,B) [36] and which apparently is retained after the ancestral fusion or duplication of the two domains, as is also the case in certain MMBL tandems or heterodimers from monocots [37], [38]. Thus, residues Asp126-Pro135 from the first MMBL sequence complement the fold of the C-domain while residues Pro245-Gln253 from the second MMBL sequence complement the fold of the N-domain. However, in LlpABW, the relative orientation of both domains is different compared to what is observed in a canonical MMBL lectin dimer, such as snowdrop lectin [36], in the heterodimeric MMBL lectin ASA I from Allium sativum [38], or in the tandem MMBL SCAfet from Scilla campanulata [37] (Figure 1C and Figure S2). In contrast to these plant MMBL proteins, the resulting architecture of LlpABW does not obey pseudo-twofold symmetry (Figure 1C).
LlpABW is a very rigid molecule. The two monomers present in the asymmetric unit are essentially identical with a root-mean-square deviation (RMSD) of 0.34 Å for 270 Cα atoms. This RMSD value does not change significantly when the individual domains are fitted separately (0.32 Å for 120 Cα's of the N-domain and 0.22 Å for 115 Cα's of the C-domain), indicating that the inter-domain orientation is fixed. This stems from three sets of interactions (Figure 2). Both domains are connected by a two-stranded anti-parallel β-sheet that is involved in the β-strand swapping mentioned above and that links both domains. The C-terminal β-hairpin extension makes extensive contacts, through hydrophobic and hydrogen bonds, with both domains. Finally, the stretch Val140-Asp145 of the C-domain makes extensive contacts with stretch Val115-Asp118 and with the side chains of Ser15 and Pro32 of the N-domain.
Fig. 2. Domain interactions within LlpABW. 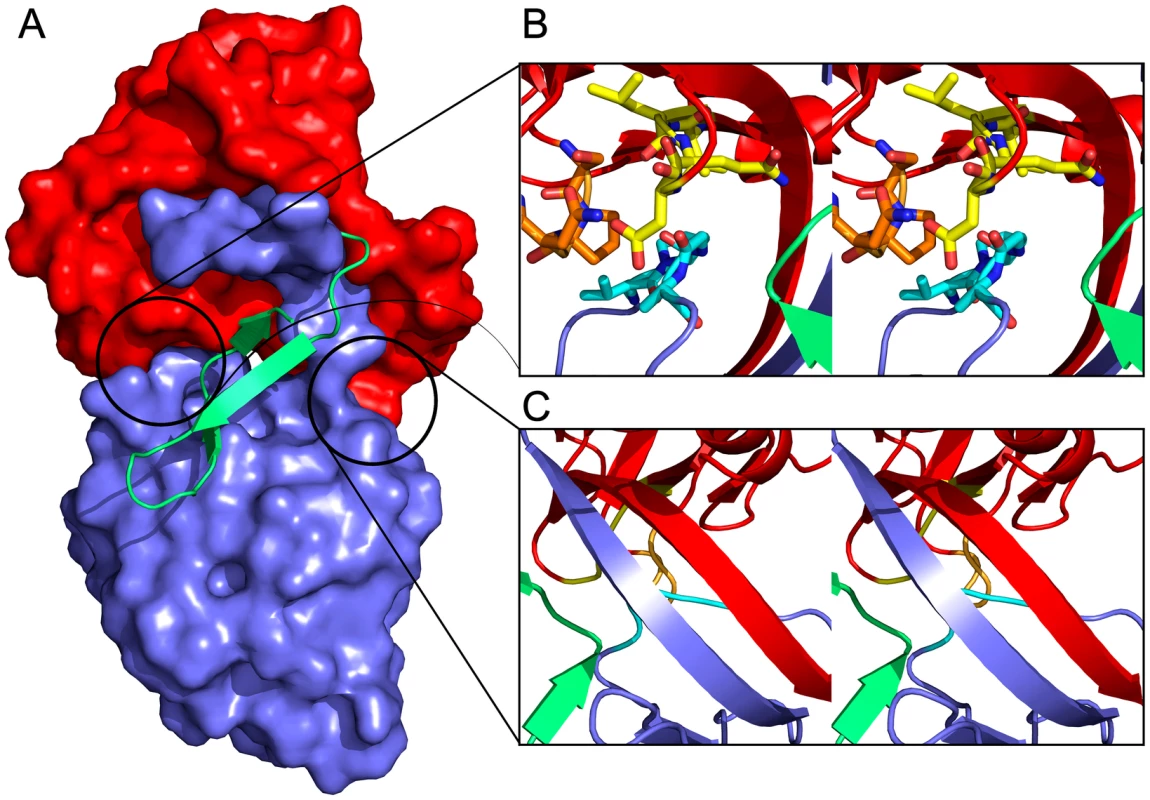
(A) The C-terminal hairpin extension (green cartoon) covers the interface between the N-domain (red surface representation) and the C-domain (blue surface representation). (B) Stereo view of the interactions between loop segments Val140-Asp145 (cyan) of the C-domain and Val115-Asp118 (yellow) and Ser31-Gln34 (orange) of the N-domain. Other structural elements are colored according to panel A. (C) Stereo view of the two-stranded β-sheet formed by strands β11a,b and β22a,b that links the N- and the C-domains and gives rise to domain swapping. Colors according to panel A and B. Domains of LlpABW are shaped by differential evolutionary pressure
A superposition of the Cα-trace of the N - and C-domain of LlpABW as well as the MMBL domain of snowdrop lectin is shown in Figure S3. Based on 79 Cα atoms that form the common β-sheet core of the MMBL domains, the RMSD between the N - and C-domains of LlpABW is 1.84 Å. While the secondary structure elements of the C-domain are restricted to the three four-stranded β-sheets of the β-prism fold, the N-domain contains three additional secondary structure elements (Figure 1A). A three-turn α-helix (α1) is inserted in the loop between strands β9 and β10, and sheet IIN contains two additional strands. Strand β6′ is inserted in the loop between strands β6 and β7 and provides an anti-parallel extension to sheet II (hydrogen bonding to strand β9). Strand β1′ is a short piece of β-strand that is part of the long N-terminus and forms a parallel extension on the opposite site of sheet IIN (hydrogen bonding to strand β2), making this β-sheet a mixed type six-stranded one rather than the canonical four-stranded anti-parallel sheet.
Despite these additions to the β-prism fold, the common core of the N-domain more closely resembles that of the well-studied and highly conserved monocot lectins (e.g. RMSD of 1.35 Å with snowdrop lectin compared to 1.82 Å for the C-domain). This structural divergence is in contrast with the degree of conservation of the carbohydrate-binding motif characteristic of the monocot lectins (QxDxNxVxY) in each of the three subdomains. In the N-domains of LlpA homologues, the surface-exposed motifs III and II are not well conserved and likely lost their function during evolution. In contrast they seem to be better conserved in the C-domains (Figure S4). Apparently, the two MMBL domains of LlpA experienced a differential evolutionary pressure resulting in different degrees of global and local (carbohydrate-binding motif) conservation, suggesting distinct functional roles for each domain.
The C-domain of LlpABW further extends into a β-hairpin that helps to define the relative orientations of its two MMBL domains. This β-hairpin is highly bent due to a β-bulge inserted into its second β-strand (Figure 1B). It is absent in all plant representatives including tandem MMBL proteins such as SCAfet (Figure 1C). In bacteria it represents the most divergent part of LlpA homologues, both in primary sequence and in length (Figure S5). Most of these C-terminal extensions terminate with a phenylalanine residue. This is reminiscent of the conserved terminal phenylalanine of outer membrane proteins from Gram-negative bacteria such as PhoE, required for their translocation to the cell envelope [39]. An equivalent extension appears to be absent in the Xanthomonas and Arthrobacter sequences (Figure S5).
LlpA is capable of binding mannose-containing carbohydrates
Subdomains IIC and IIICof LlpABW contain the typical sugar-binding signature (QxDxNxVxY) of an active MMBL mannose-binding site (Figure S1 and S4). Soaking crystals of LlpABW with 200 mM methyl-α-D-mannopyranoside (Me-Man) led to clear electron density of a single Me-Man in site IIIC of each of the two LlpABW monomers in the asymmetric unit (Figure S6A). This site comprises the side chains from Gln171, Asp173, Asn175 and Tyr179, which contribute to hydrogen bond interactions and the side chains of residues Val177, Asn188, Gln192 and Ala185, which contribute to van der Waals contacts with the carbohydrate ligand (Figure 3A, Figure S7A,C). This architecture is very similar to what is observed for mannose bound to other MMBL-type lectins such as snowdrop and garlic lectin (Figure S7B).
Fig. 3. Carbohydrate binding in site IIIC of LlpABW. 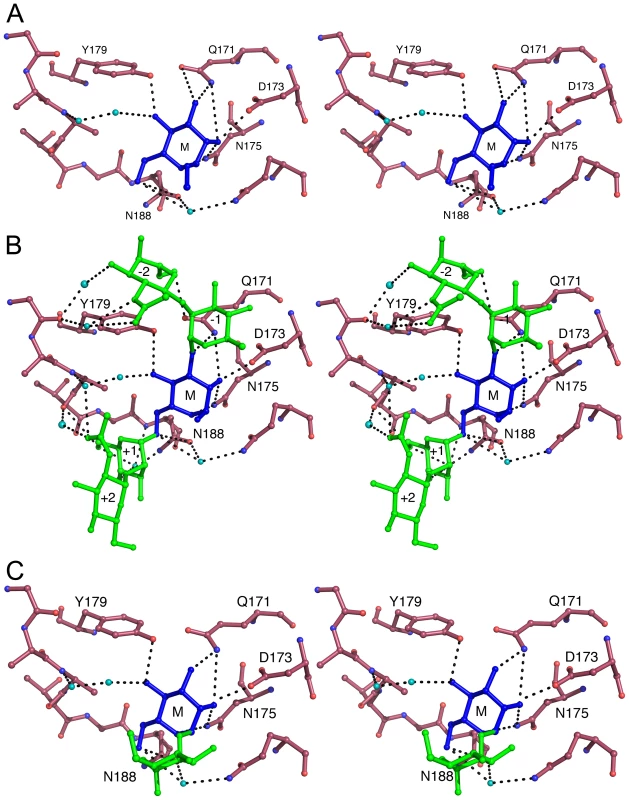
(A) Stereoview of methyl-α-D-mannopyranoside bound to subdomain IIIC. Methyl-α-D-mannopyranoside is shown in blue and indicated by M. Residues belonging to the QxDxNxVxY motif and hydrogen bonding to the sugar as well as Asn188 are labeled. Water molecules bridging protein and carbohydrate are shown in cyan (B) Similar view of the pentasaccharide GlcNAcβ(1–2)Manα(1–3)[GlcNAcβ(1–2)Manα(1–6)]Man. The mannose residue occupying the primary binding site is shown in blue and labeled M. The additional two mannoses (labeled +1 and −1) and two N-acetyl glucosamine residues (labeled +2 and −2) are shown in green. Other colors are as in panel A. (C) Binding of the disaccharide Manα(1–2)Man. The non-reducing mannose residue occupying the primary binding site is shown in blue and labeled M. The second, reducing mannose is shown in green. Other colors are as in panel A. Soaks with oligomannoses revealed additional sugar-binding subsites. Binding site IIIC accommodates the disaccharide Manα(1–2)Man and the pentasaccharide GlcNAcβ(1–2)Manα(1–3)[GlcNAcβ(1–2)Manα(1–6)]Man (Figure S6B,C). In the case of the pentasaccharide, the central reducing mannose is located in the shallow Me-Man binding site and the two GlcNAcβ(1–2)Man moieties stretch out over the surface making only a few additional hydrogen bonds or van der Waals contacts (Figure 3B). In the bound disaccharide, the non-reducing mannose is located in the Man-Me binding site while the reducing mannose faces the solvent and does not interact directly with the protein (Figure 3C).
Site IIC of both LlpABW molecules in the asymmetric unit is involved in crystal packing interactions and the presence of Me-Man is therefore sterically excluded. All residues that form specific hydrogen bonds with Me-Man are retained but substitutions occur for three side chains that provide van der Waals contacts (Figure S4 and S8A). In contrast, site IC lost the conserved QxDxNxVxY motif (Figure S4) and is involved in inter-domain contacts and therefore inaccessible to ligands (Figure S8B).
The putative carbohydrate-binding sites in the N-domain of LlpABW are less conserved. Similar to the C-domain, site IN is inaccessible and involved in inter-domain interactions (Figure S9A). In the IIN subdomain, the canonical mannose-binding motif QxDxNxVxY is essentially absent, with only the Gln residue of the motif being conserved as Gln82 (Figure S4). All other donors or acceptors required for hydrogen bonds with a mannose ligand are missing. In addition, the presence of Phe86 at the equivalent position of the expected Val sterically hinders the binding of mannose (Figure S9B). The potential carbohydrate-binding site on subdomain IIIN is only partially conserved (Figure S9C) and contains two relevant substitutions from the canonical signature: (1) the Tyr residue of the QxDxNxVxY motif is replaced by the shorter Gln49, thereby removing the canonical hydrogen bond between Man O4 and Tyr OH, and (2) a threonine at position 54 which may compensate the hydrogen bond lost due to the Tyr-to-Gln substitution in the canonical motif. The lack of electron density at this site in our Me-Man soak nevertheless indicates that this site does not recognize this ligand or that its affinity is so low that recognition would only be achieved in the context of a larger and as yet unidentified mannose-containing ligand. Alternatively, this putative site may possess specificity for a different monosaccharide. In order to evaluate this hypothesis, we soaked LlpABW crystals with D-galactose, N-acetyl-D-glucosamine and L-fucose. No electron density was observed for any of these sugars, suggesting that the N-domain has a function distinct from carbohydrate recognition (data not shown).
Carbohydrate-binding capacity is required for LlpA toxicity
The LlpABW motifs IIIN, IIIC and IIC create potential carbohydrate binding sites that may be involved in bacteriotoxicity of the protein. We therefore examined the role of carbohydrate binding in the bactericidal function of LlpABW. The presence of methyl-α-D-mannopyranoside up to 500 mM in the medium did not influence the activity of LlpABW on P. syringae GR12-2R3. Glycan array profiling did not highlight any specific oligosaccharide structure that could represent a natural ligand of LlpABW (Table S1). This could be due to the array design that is principally based on eukaryotic glycans and may therefore lack an appropriate carbohydrate for this prokaryotic toxin. Previously, it was observed that LlpABW from concentrated culture supernatant does not agglutinate rabbit red blood cells, nor binds to a mannose-agarose affinity matrix [21].
To assess whether the mannose-recognizing QxDxNxVxY motifs in LlpABW are nevertheless relevant for bactericidal activity, the conserved valine residue was mutated to tyrosine in subdomains IIIN, IIIC, and IIC. These mutations sterically preclude mannose or any other ligand to enter the binding sites (Figure S7C). Semi-quantitative activity assays with permeabilized E. coli cells expressing the LlpA variants in motifs IIIN, IIIC and IIC were used to assess the relationship between carbohydrate binding and bactericidal activity. Modification of the IIIN site, for which no mannose binding was observed, does not affect the antibacterial activity against P. syringae GR12-2R3 (Figure 4). In contrast, the altered IIIC pocket strongly diminishes activity, either alone or in pairwise combination with the other mutated sites (IIIN or IIC). A minor negative effect of the IIC mutation is only apparent in a double mutant, when combined with a modified IIIN motif.
Fig. 4. Inhibitory activity of wild-type LlpABW and selected mutants with modified (potential) mannose-binding sites. 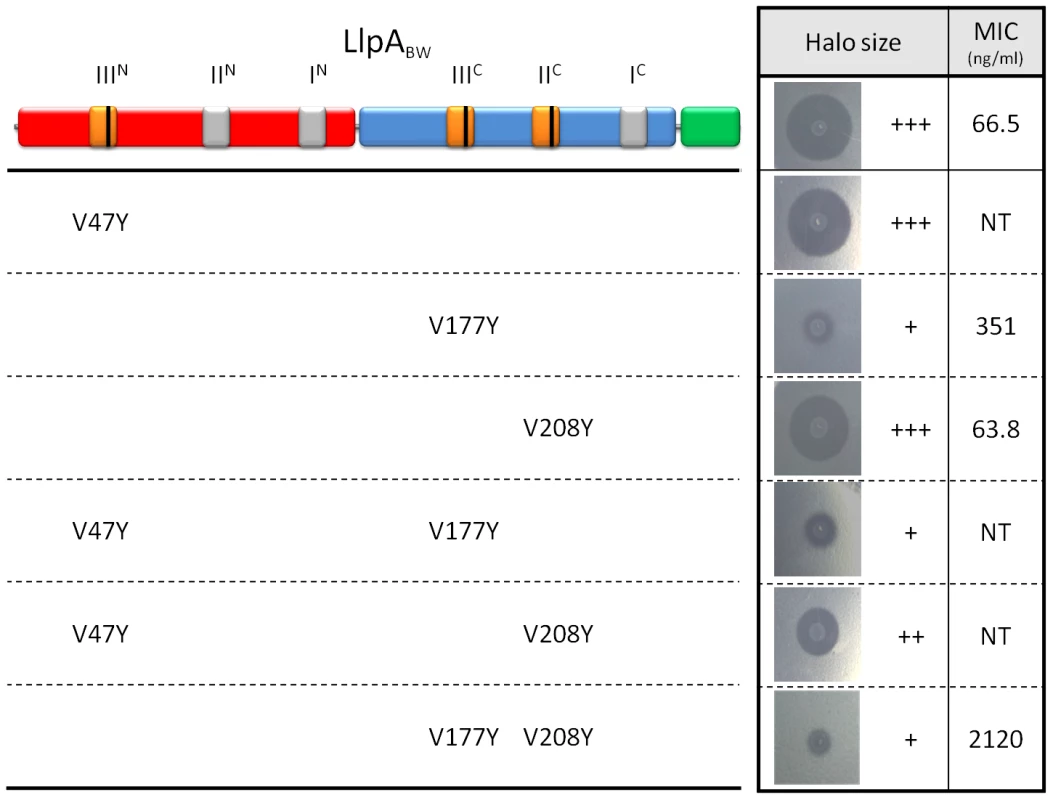
The domain structure (N-domain in red, C-domain in blue and C-terminal extension in green) and the position of the MMBL motifs (potentially active binding sites in orange, inactive ones in grey) are shown. The positions of conserved valine residues converted to tyrosine residues by site-directed mutagenesis are indicated with a black bar. Inhibitory activity of E. coli strains expressing mutant LlpABW forms was assayed against P. syringae GR12-2R3 and semi-quantified according to the size (inner zone radius) of the growth inhibition halo relative to LlpABW (+++, native LlpABW; ++, halo size reduced; + halo size strongly reduced; −, no halo; NT, not tested). For wild-type LlpABW and three purified His-tagged mutant forms (LlpAV177Y, LlpAV208Y and LlpAV177Y-V208Y) the MIC values were determined with indicator P. syringae GR12-2R3. Molar minimal inhibitory concentrations of recombinant proteins (with standard deviations): LlpA, 2.08 nM (±0.58 nM); LlpAV177Y, 10.9 nM (±0.66 nM); LlpAV208Y, 1.98 nM (±0.066 nM); 65.72 nM (±2.80 nM). Purified proteins were prepared to further quantify these effects. Far UV CD spectra of these mutant forms are identical to that of native protein LlpABW, indicating that the mutations do not affect the overall structure of the protein. Isothermal titration calorimetry (ITC) showed that LlpABW has an affinity of 2.1 mM for the pentasaccharide GlcNAcβ(1–2)Manα(1–3)[GlcNAcβ(1–2)Manα(1–6)]Man, the highest among all the tested oligo-mannosides (See Figure 5 and Table 1 for a summary of the experimentally validated LlpABW-carbohydrate interactions). This is in agreement with the crystal structures of the different complexes since this sugar is the one with the largest binding interface (Figure 3). Titrations of LlpABW, of the mutants LlpAV177Y (a site IIIC knockout), LlpAV208Y (a site IIC knockout) and of the double mutant LlpAV177Y-V208Y with α-methyl mannoside clearly pinpoint site IIIC as the only responsible for the sugar binding activity. Point mutations in both sites or IIIC (V177Y) alone, completely abrogate sugar binding. However knocking out site IIC (V208Y) has little effect in binding and the affinities of LlpAV208Y for α-methyl mannoside and Manα(1–3)Man are very close to the ones measured for the wild-type protein (See Table 1 and Figure 5B).
Fig. 5. ITC analysis of carbohydrate binding to LlpABW and mutants. 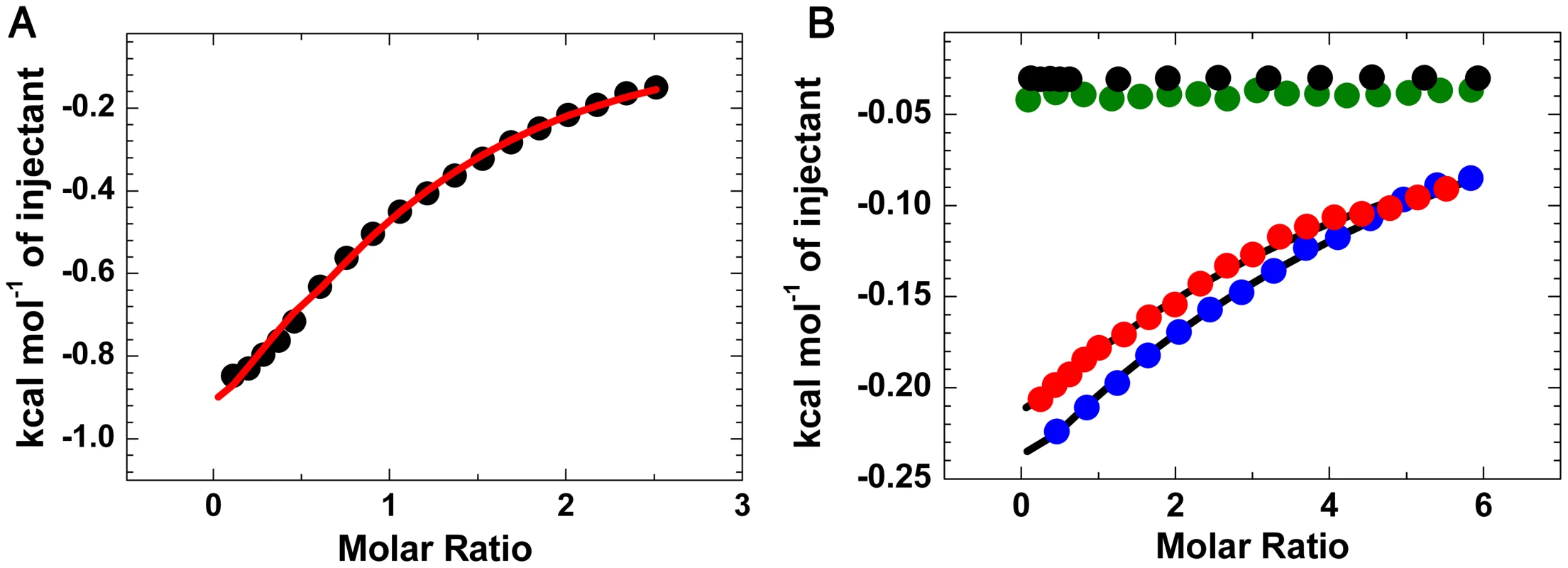
(A) Binding of LlpABW to the pentasaccharide GlcNAcβ(1–2)Manα(1–3)[GlcNAcβ(1–2)Manα(1–6)]Man. (B) Binding of LlpABW (blue circles, wild type) and the mutants LlpAV177Y (green circles, site IIIC knockout), LlpAV208Y (red circles, site IIC knockout) and LlpAV177Y-V208Y (black circles, site IIC and IIIC knockout) to α-methyl mannoside. There is no heat exchanged in the titration of the double mutant or the site IIIC knockout LlpAV177Y, whereas the site IIC knockout LlpAV208Y, binds the monosaccharide in a “wildtype”-like fashion, showing that only site IIIC is involved in sugar binding. Tab. 1. Binding affinities and thermodynamic parameters obtained from ITC titrations. 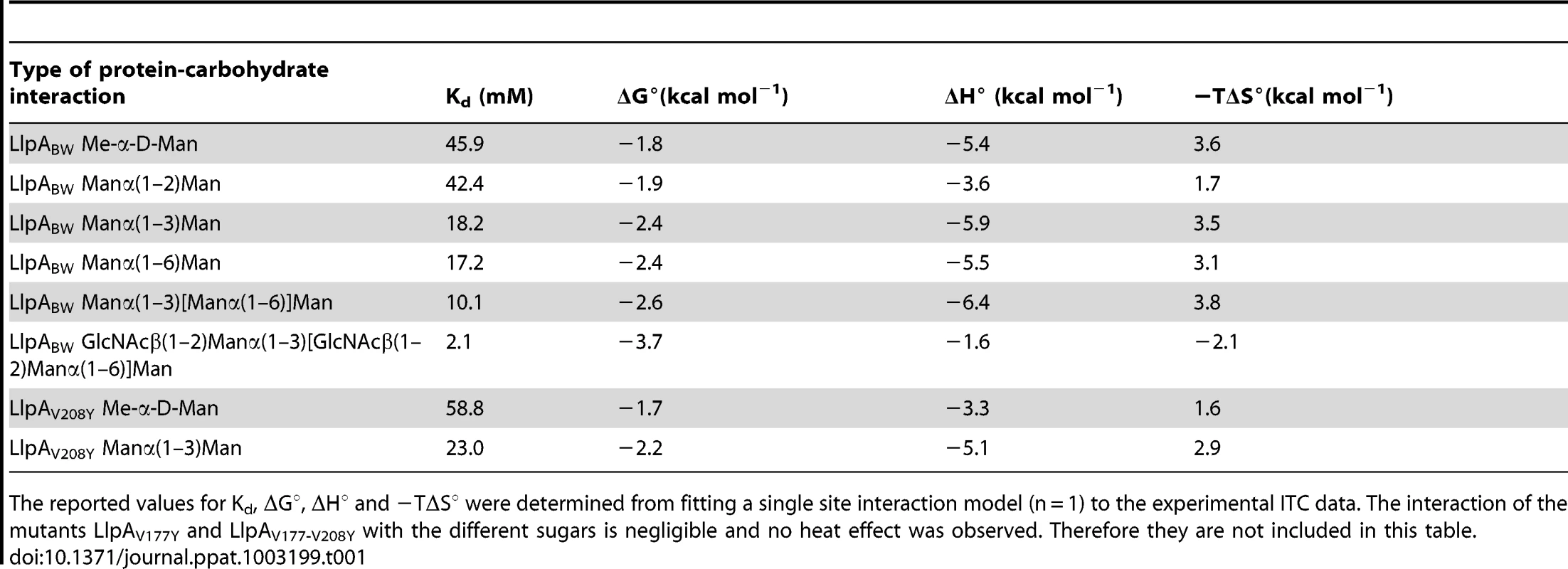
The reported values for Kd, ΔG°, ΔH° and −TΔS° were determined from fitting a single site interaction model (n = 1) to the experimental ITC data. The interaction of the mutants LlpAV177Y and LlpAV177-V208Y with the different sugars is negligible and no heat effect was observed. Therefore they are not included in this table. While the V208Y mutation in the IIC site has no observable effect on the MIC value for P. syringae GR12-2R3, the altered IIIC motif engenders a 5.2-fold increase in MIC (Figure 4). The mutant protein LlpAV177Y-V208Y suffers a further reduction in activity, yielding a 31.6-fold increased MIC compared to native LlpABW. The biological activities of LlpA and its mutants were further assessed by live/dead staining and subsequent flow cytometry analysis (Figure 6, Figure S10). Proportions of dead cells after 1 hour of exposure to LlpA or LlpAV208Y were comparable (10.1% and 9.7%, respectively). For LlpAV177Y this value was reduced to 6.1%, significantly lower than for LlpA. Killing activity was even further reduced for LlpAV177Y-V208Y (3.7%). These results are consistent with the MIC determination and ITC data, indicating that an active site IIIC is required to generate a fully active LlpA bacteriocin. The difference in bacteriotoxicity between LlpAV177Y and LlpAV177Y-V208Y suggests that site IIC has a supporting role in the LlpABW bacteriotoxicity.
Fig. 6. Killing activity of LlpABW and mutant proteins. 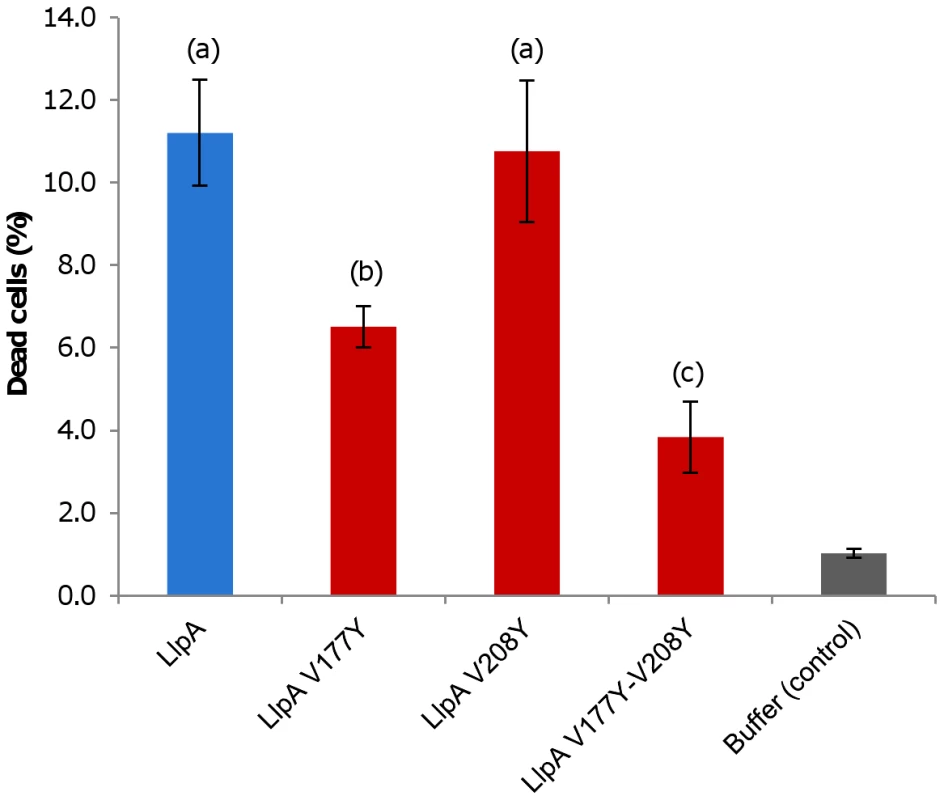
Percentages of dead cells after live/dead staining as quantified by flow cytometry analysis (Figure S10). P. syringae GR12-2R3 was used as indicator strain and treated at a final concentration of 50 µg/ml for 1 h. Average values (with standard deviations; indicated by error bars): LlpA, 10.1 (±1.04); LlpAV177Y, 6.1 (±0.44); LlpAV208Y, 9.7 (±1.39); LlpAV177Y-V208Y, 3.7 (±0.90); buffer (control), 1.0 (±0.11).Values are significantly different for (a) and (b), (b) and (c) (p<0.01). All domains are necessary for LlpABW functionality
The site-directed mutagenesis approach revealed an important role for the C-domain's carbohydrate-binding capacity in LlpABW toxicity. Considering the increased binding motif degeneration in the N-domain and the fact that a Ruminococcus bacteriocin composed of only a single MMBL domain fused to an unknown domain has been identified [35], the N-domain may fulfill a distinct function, different from that of the C-domain. In order to scrutinize the contribution of individual domains to overall activity, six domain deletion constructs of llpABW were engineered to potentially encode proteins lacking the first or second MMBL domain, a gene product devoid of the C-terminal hairpin, or a protein retaining only an individual domain (N-domain, C-domain, or hairpin) (Figure S11). To take the domain swapping into account, the constructs containing only a single MMBL domain were designed with a fusion of the swapped C-terminal β-strands to the corresponding domain via a short linker.
None of these deletion constructs resulted in the production of an active protein, indicating that none of the domains are dispensable. Removal of the terminal phenylalanine residue still allows expression of a functional bacteriocin in E. coli (Figure 7), but a further C-terminal truncation (deletion of Trp-His-Phe tail) resulted in a negative bacteriocin assay (data not shown). From these data we conclude that both MMBL domains as well as the C-terminal hairpin extension are required for activity of LlpA. Whether the role of the C-terminal hairpin is any other than simply stabilization of the C-domain cannot be concluded.
Fig. 7. Differential inhibitory activity of wild-type LlpABW and LlpA/LlpA1 domain chimers. 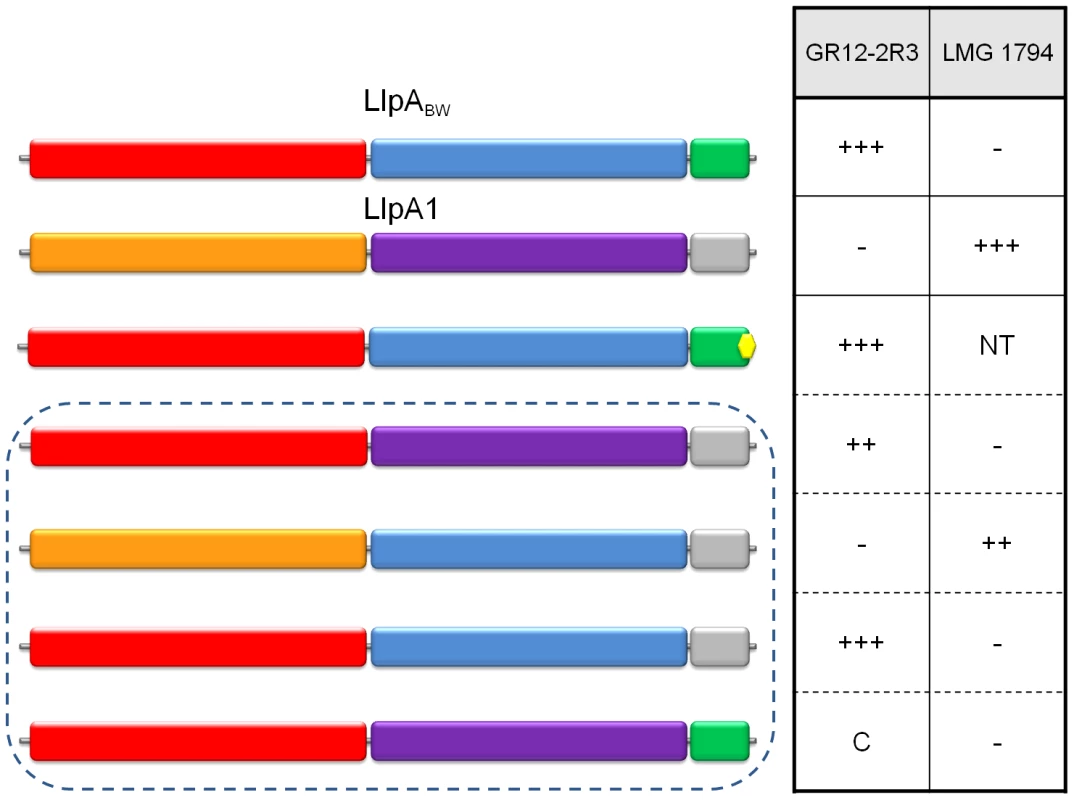
The domain structures of LlpABW (as in Figure 4) and of LlpA1 (inferred by pairwise alignment; N-domain in orange, C-domain in purple and C-terminal extension in grey) are depicted, along with those of chimeric forms (in dashed box). The LlpA variant lacking the terminal phenylalanine residue is marked with a yellow hexagon. Inhibitory activity of the respective E. coli recombinants was tested with diagnostic indicators for LlpABW (P. syringae GR12-2R3) and LlpA1 (P. fluorescens LMG 1794). Halo sizes are semi-quantified according to size of the growth inhibition halo (+++, native halo size of LlpABW and LlpA1; ++, halo size reduced; C, local clearing confined to producer colony spot; −, no halo or clearing; NT, not tested). Additional chimeric and domain deletion constructs not conferring bacteriocin activity against one of the indicator strains are specified in Figure S11. Target specificity of LlpA is hosted by the N-domain
In order to investigate the role of the different domains in target specificity, we created hybrid LlpA proteins using the domains of LlpABW from P. putida BW11M1 and LlpA1 from P. fluorescens Pf-5. These two LlpA proteins share 45% sequence identity and differ in their target spectra. Strains P. syringae GR12-2R3 and P. fluorescens LMG1794 were identified as specific indicators for LlpABW [21] and LlpA1 [22], respectively. Six constructs carrying llpABW/llpA1chimeric genes were made with domain exchanges involving the N-domain, C-domain, and hairpin region (Figure 7 and Figure S11). For four of these constructs activity against one of both indicators was detected. Only constructs retaining the original N-domain give rise to inhibition of the cognate indicator strain. The C-domain or the hairpin of LlpABW could be replaced with the corresponding LlpA1 domains without changing target specificity. Conversely, the original specificity of LlpA1 is retained upon replacement of its C-domain by the LlpABW equivalent.
Discussion
Structure elucidation of LlpABW from P. putida BW11M1 unequivocally assigns this bacteriocin to the MMBL lectin family, in which it constitutes the first prokaryotic member, representative for a group of bacterial proteins composed of two MMBL domains [21]–[23]. Systematic inactivation of the three potential carbohydrate-binding sites present in the N-domain (IIIN) and in the C-domain (IIIC and IIC) of LlpABW, revealed that a non-occluded IIIC pocket is required to obtain a fully active LlpABW molecule. A negative co-operative effect on activity resulted when the IIC site was additionally modified. Although mannose-containing carbohydrates can bind to the IIIC pocket of LlpABW, it remains unclear if these are part of or mimic the natural ligand required for biological activity since bacteriocin activity is not impaired in the presence of excess mannose.
A mutated IIIN site did not provoke a negative effect on antibacterial activity. However, the N-domain appears to play a major role in target selection. This was demonstrated by assessing the differential activity of domain chimers against two target strains, diagnostic for the LlpABW - and LlpA1-specific killing.
The β-hairpin does not appear to be a specificity determinant, although it constitutes the most variable region among LlpA-like bacteriocins. Possibly, it is required for thermodynamic stability since it needs to be intact in LlpABW. An equivalent C-terminal stretch is absent from the Xanthomonas citri LlpA-like bacteriocin [23]. From our results relying on heterologous expression in E. coli and a bacteriocin assay with permeabilized cells, it cannot be excluded that this structural element may play a role in the way an LlpA protein is exported by its native producer cells.
In general, a defensive role has been proposed for the (oligo)mannose-binding MMBL lectins based on insecticidal, nematicidal, antifungal, or even antiviral activities demonstrated for several of these proteins that are abundantly found in monocot plants [40]–[46]. Some of these plant lectins trigger apoptosis in cancer cells [47]. Also their identification in fish mucus and epithelial cells is in line with a general protective (antimicrobial) function for MMBL domains [32]. LlpABW as a bactericidal protein fits within this picture of MMBL domains being involved in general defense mechanisms. Since no antibacterial activity has been assigned to the eukaryotic MMBL proteins, it is challenging to identify structural features that confer the intragenus-specific bacteriocin activity of LlpA, as shown for proteins from P. putida [21], P. fluorescens [22], P. syringae [23], and Xanthomonas citri [23]. Their target spectra are narrower than reported for the mammalian antibacterial C-type lectins of the RegIII family, such as mouse RegIIIγ and its human homolog HIP/PAP that bind to the surface-exposed peptidoglycan layer of Gram-positive bacteria [48], and RegIIIβ that also binds to the lipid A moiety of lipopolysaccharides on the cell envelope of Gram-negative bacteria [49].
The absence of any known secretory signal sequence in LlpABW and its homologues in other Pseudomonas species is intriguing in view of their extracellular location [21]. The translocation of the outer membrane-associated mannose/fucose-specific lectin LecB of P. aeruginosa, that also lacks such signal sequence [50], is dependent on its glycosylation [51]. Contrary to LlpA that is exported to the culture supernatant to exert its antagonistic activity, LecB remains associated with the cell envelope through interaction with the major outer membrane protein OprF [52], in line with its role in biofilm formation.
Materials and Methods
Strains and culture conditions
Bacterial strains and plasmids used in this study are listed in Table S2. Escherichia coli was routinely grown in shaken Luria-Bertani (LB, MP Biomedicals) broth at 37°C. Pseudomonas strains were grown in Tryptic Soy Broth (BD Biosciences) at 30°C with shaking. Media were solidified with 1.5% agar (Invitrogen) and supplemented with filter-sterilized antibiotics as required at following concentrations: ampicillin (Sigma-Aldrich), 100 µg/ml or kanamycin (Sigma-Aldrich), 50 µg/ml. Isopropyl β-D-thiogalactoside (IPTG 40 µg/ml, ForMedium) and 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-Gal 40 µg/ml, ForMedium) were added for blue/white screening of pUC18-derived plasmids in E. coli.
Plasmids used for antibacterial testing and sequencing were propagated in E. coli TOP10F′ (Invitrogen). E. coli BL21(DE3) (Novagen) was used as a host for plasmids driving recombinant protein expression. Genomic DNA from P. putida BW11M1 was isolated using the Puregene Yeast/Bact. Kit B (Qiagen). Plasmid DNA was extracted using the QIAprep Spin Miniprep Kit (Qiagen). Stocks were stored at −80°C in the appropriate medium in 25% (v/v) glycerol.
Recombinant DNA methods
Standard methods were used for preparation of competent E. coli cells, heat shock transformation of E. coli and DNA electrophoresis [53]. Restriction enzymes were used according to the supplier's specifications (Roche Diagnostics and BIOKÉ). DNA ligation was performed using T4 DNA ligase (Invitrogen). Plasmid sequencing was performed by GATC Biotech (Constance, Germany). Constructs that were generated are listed in Table S2 and primers are listed in Table S3.
A 921-bp fragment containing llpABW was amplified by PCR with Platinum Pfx DNA polymerase (Invitrogen), using a C1000 Thermal Cycler (Bio-Rad). P. putida BW11M1 genomic DNA was taken as a template, and combined with primers PGPRB-3155 and PGPRB-3156. The amplicon was purified using the QIAquick PCR Purification Kit (Qiagen), digested with KpnI and BamHI, ligated in pUC18, and transformed to E. coli TOP10F′. Transformants were verified for the presence of the insert by PCR using Taq Polymerase (BIOKÉ) with primers PGPRB-2545 and PGPRB-2546. The cloned construct (pCMPG6129) was purified and its insert confirmed by sequencing.
DpnI-mediated site-directed mutagenesis was performed to construct valine-to-tyrosine mutant forms of llpABW in pUC18 (pCMPG6129) and N-terminal His-tagged llpABW in pET28a (pCMPG6056 [54]). PCR conditions were: 2 min initial denaturation, followed by 16 cycles of denaturation (1 min), annealing (1 min, primer-dependent temperature) and elongation at 68°C (1 min./kb). Final elongation was for 8 min at 68°C. After PCR, samples were immediately treated with DpnI at 37°C for 2 h and transformed into E. coli TOP10F′ and selected on the appropriate medium. Plasmid inserts of selected transformants were verified by sequence analysis. Double mutants were constructed using plasmids with a single point mutation as a template.
Domain deletants of llpABW were constructed using pCMPG6129 as a template (llpABW from P. putida BW11M1). Chimeric constructs were obtained using pCMPG6129 and pCMPG6053 (llpA1 from P. fluorescens Pf-5 [22]) as templates. Artificial ligation of gene fragments, generated with the PCR primers specified in Table S3, was performed by using splicing by overlap extension (SOE). The resulting recombinant amino acid sequences are listed in Table S4.
Recombinant protein expression and purification
Protein isolation and purification of N-terminal His6-tagged LlpABW, LlpAV177Y, LlpAV208Y, and LlpAV177Y-V208Y from E. coli BL21(DE3), carrying expression constructs pCMPG6056, pCMPG6149, pCMPG6150 and pCMPG6151 respectively, were performed as described by Parret and collaborators [54]. The presence of His-tagged protein was observed via immunodetection by Western blot, using monoclonal anti-His6 (IgG1 from mouse; Roche Diagnostics) as primary antibody. Fractions free of other proteins, as verified by SDS-PAGE and subsequent Coomassie Blue staining, were dialyzed against bis-TRIS propane buffer (20 mM, 200 mM NaCl, pH 7.0). Concentrations of purified proteins were determined by absorbance measurement at 280 nm using molar extinction coefficients of 62910 M−1 cm−1 for LlpABW, 64400 M−1 cm−1 for LlpAV177Y and LlpAV208Y, and 65890 M−1 cm−1 for LlpAV177Y-V208Y. Extinction coefficients were calculated according to Pace and collaborators [55].
Antibacterial assays
Bacteriocin production by E. coli cells carrying pUC18-derived constructs was assayed as follows: 2-µl drops of an overnight stationary-phase culture were spotted onto LB agar plates and incubated for 8 h at 37°C. Next, plates were exposed to chloroform vapor (30 min), aerated and subsequently overlaid with 5 ml of soft agar (0.5%), seeded with 200 µl of a cell culture of an indicator strain (∼108 CFU/ml), followed by overnight incubation at 30°C. Next day, plates were scored for the presence of halos in the confluently grown overlay.
Antibacterial activity assays with purified recombinant His6-tagged proteins were performed as described by Ghequire and collaborators [23]. To assess the influence of added sugar, the same assay was carried out on agar medium supplemented with D-mannose (Sigma-Aldrich) or methyl-α-D-mannopyranoside (Sigma-Aldrich) to a final concentration of 0.01 M to 0.5 M.
A Bioscreen C apparatus (Oy Growth Curves Ab Ltd, Finland) was used to determine the minimum inhibitory concentration (MIC). An overnight culture (16 h) of the indicator strain was diluted to 104–105 CFU/ml and incubated at 30°C, with a two-fold dilution series of recombinant His6-tagged LlpABW or mutant LlpABW. Bis-TRIS propane buffer was used as control. The MIC value was determined as the minimum concentration of protein at which no growth of the indicator strain (OD600<0.2) occurred after 24 h. At least three independent repeats, each with three replicates, were carried out.
Glycan array
His6-tagged LlpABW was lyophilized and verified for antibacterial activity. After re-dissolving in MilliQ water, recombinant LlpABW was diluted to 200 µg/ml with binding buffer (20 mM TRIS-HCl pH 7.4, 150 mM NaCl, 2 mM CaCl2, 2 mM MgCl2, 0.05% Tween 20, 1% BSA), and used to probe the printed glycan arrays [56] following the standard procedures of Core H of the Consortium for Functional Glycomics (http://www.functionalglycomics.org/). Monoclonal anti-His6 antibodies (Roche Diagnostics) were used as primary antibodies, and fluorescently labeled anti-mouse IgG as secondary antibodies.
Circular dichroism
CD spectra were acquired on a Jasco J-715 spectropolarimeter. Curves were averaged over 8 scans, taken at 25°C using a 1 mm cuvette. Samples were dialyzed against bis-TRIS propane buffer (20 mM, NaCl 200 mM, pH 7.0), filtered and degassed prior to data acquisition. All proteins were assayed at 0.4 mg/ml.
Isothermal titration calorimetry
ITC titrations were carried out on an ITC200 apparatus (MicroCal). Prior to the measurement, LlpABW, LlpAV177Y, LlpAV208Y and LlpAV177Y-V208Y was dialyzed to bis-TRIS propane buffer. Sugars were directly dissolved into the same buffer. The samples were filtered and degassed for 10 min at 25°C before being examined in the calorimeter. The titrations were carried out at 25°C, injecting the sugars (methyl-α-D-mannoside, Manα(1–2)Man, Manα(1–3)Man, Manα(1–6)Man, Manα(1–3)[Manα(1–6)]Man and GlcNAcβ(1–2)Manα(1–3)[GlcNAcβ(1–2)Manα(1–6)]Man) into a protein solution (protein concentrations ranged from 2 mM to 4 mM depending on protein availability). All data were analyzed using the MicroCal Origin ITC 7.0 software. Binding affinities and thermodynamic parameters from all ITC titrations are reported in Table 1.
X-ray data collection and structure determination
Expression, purification and crystallization of recombinant His-tagged LlpABW have been described [54]. X-ray data for native and derivative crystals were collected on EMBL beamline BW7A of the DESY synchrotron (Hamburg, Germany). For each potential derivative, the wavelength was chosen to be at the high-energy side of the absorption edge in order to ensure a usable anomalous signal. All data were scaled and merged with the HKL package of programs. Data collection statistics are given in Table S5.
The crystal structure of free LlpABW was solved combining single isomorphous replacement with anomalous scattering (SIRAS strategy) from a p-chloromercurybenzoate derivative. The heavy-atom substructure was determined with SHELXD [57] using a resolution cutoff of 4.0 Å. Heavy-atom refinement and phasing were performed with SHARP [58]. Phase improvement by solvent flattening was performed with SOLOMON [59]. Non-crystallographic symmetry averaging with density modification [60] further improved the electron density. A partial model (94% of the residues comprising the asymmetric unit) was automatically built with ARP/wARP [61] and the remainder was built manually over several cycles of model building with Coot [62], alternated with refinement using phenix.refine [63], [64]. Phasing and refinement statistics are shown in Table S5.
Carbohydrate soaks
Crystals of LlpABW were transferred to artificial mother liquor (0.1 M imidazole pH 6.5, 1.3 M sodium acetate) enriched with either 200 mM methyl-α-D-mannopyranoside (Me-Man), Manα(1–2)Man, GlcNAcβ(1–2)Manα(1–3)[GlcNAcβ(1–2)Manα(1–6)]Man (M592), D-galactose, L-fucose or N-acetyl-D-glucosamine and allowed to equilibrate overnight (all carbohydrates obtained from Dextra Laboratories, Reading, U.K.). Data were collected at room temperature on EMBL beamline X13, except for the N-acetyl-D-glucosamine soak collected at the PROXIMA-1 beamline of the SOLEIL synchrotron (Gif-sur-Yvette, France) and the D-galactose soak collected at ESRF beam line ID14-1 (Grenoble, France). All data were scaled and merged using the HKL package. Refinement was started from the coordinates of the ligand-free structure using phenix.refine. Manual rebuilding, including the introduction of the carbohydrate ligand if present, was done with Coot [62]. Crystal structures of LlpABW from P. putida BW11M1 (PDB entry 3M7H) and LlpABW in complex with methyl-α-D-mannoside (PDB entry 3M7J), with Manα(1–2)Man (PDB entry 4GC1), and with GlcNAcβ(1–2)Manα(1–3)[GlcNAcβ(1–2)Manα(1–6)]Man (PDB entry 4GC2) have been deposited at the PDB.
Flow cytometry
Overnight cultures of P. syringae GR12-2R3 (16 h) were diluted to OD600 0.5 and washed twice with phosphate-buffered saline (PBS). Cells were treated with LlpA, mutant proteins or buffer (bis-TRIS propane buffer, negative control), at a final concentration of 50 µg/ml for 1 h, at 20°C. Next, PBS-washed bacteria were stained using the Live/Dead BacLight bacterial viability kit (Invitrogen), incubated for 15 minutes, and analyzed on a BD Influx (BD Biosciences). Excitation of the dyes was done at 488 nm, and fluorescence measured at 530 nm for SYTO 9 and at 610 nm for propidium iodide. Results were processed with FlowJo 10.0.4 software (Figure S10). Measurements were done independently and based on six biological repeats. Results are expressed as percentages of dead cells [dead/(live+dead) * 100)].
Supporting Information
Zdroje
1. Frey-KlettP, BurlinsonP, DeveauA, BarretM, TarkkaM, et al. (2011) Bacterial-fungal interactions: hyphens between agricultural, clinical, environmental, and food microbiologists. Microbiol Mol Biol Rev 75 : 583–609.
2. MelaF, FritscheK, de BoerW, van VeenJA, de GraaffLH, et al. (2011) Dual transcriptional profiling of a bacterial/fungal confrontation: Collimonas fungivorans versus Aspergillus niger. ISME J 5 : 1494–1504.
3. GarbevaP, SilbyMW, RaaijmakersJM, LevySB, de BoerW (2011) Transcriptional and antagonistic responses of Pseudomonas fluorescens Pf0-1 to phylogenetically different bacterial competitors. ISME J 5 : 973–985.
4. ShankEA, Klepac-CerajV, Collado-TorresL, PowersGE, LosickR, et al. (2011) Interspecies interactions that result in Bacillus subtilis forming biofilms are mediated mainly by members of its own genus. Proc Natl Acad Sci U S A 108: E1236–E1243.
5. RosenthalAZ, MatsonEG, EldarA, LeadbetterJR (2011) RNA-seq reveals cooperative metabolic interactions between two termite-gut spirochete species in co-culture. ISME J 5 : 1133–1142.
6. HibbingME, FuquaC, ParsekMR, PetersonSB (2010) Bacterial competition: surviving and thriving in the microbial jungle. Nat Rev Microbiol 8 : 15–25.
7. AokiSK, DinerEJ, de RoodenbekeCT, BurgessBR, PooleSJ, et al. (2010) A widespread family of polymorphic contact-dependent toxin delivery systems in bacteria. Nature 468 : 439–442.
8. PooleSJ, DinerEJ, AokiSK, BraatenBA, t'Kint de RoodenbekeC, et al. (2011) Identification of functional toxin/immunity genes linked to contact-dependent growth inhibition (CDI) and rearrangement hotspot (Rhs) systems. PLoS Genet 7: e1002217.
9. RussellAB, HoodRD, BuiNK, LeRouxM, VollmerW, et al. (2011) Type VI secretion delivers bacteriolytic effectors to target cells. Nature 475 : 343–347.
10. Michel-BriandY, BaysseC (2002) The pyocins of Pseudomonas aeruginosa. Biochimie 84 : 499–510.
11. NakayamaK, TakashimaK, IshiharaH, ShinomiyaT, KageyamaM, et al. (2000) The R-type pyocin of Pseudomonas aeruginosa is related to P2 phage, and the F-type is related to lambda phage. Mol Microbiol 38 : 213–231.
12. WilliamsSR, GebhartD, MartinDW, SchollD (2008) Retargeting R-type pyocins to generate novel bactericidal protein complexes. Appl Environ Microbiol 74 : 3868–3876.
13. FischerS, GodinoA, QuesadaJM, CorderoP, JofréE, et al. (2012) Characterization of a phage-like pyocin from the plant growth-promoting rhizobacterium Pseudomonas fluorescens SF4c. Microbiology 158 : 1493–1503.
14. KöhlerT, DonnerV, van DeldenC (2010) Lipopolysaccharide as shield and receptor for R-pyocin-mediated killing in Pseudomonas aeruginosa. J Bacteriol 192 : 1921–1928.
15. CascalesE, BuchananSK, DuchéD, KleanthousC, LloubèsR, et al. (2007) Colicin biology. Microbiol Mol Biol Rev 71 : 158–229.
16. DenayerS, MatthijsS, CornelisP (2007) Pyocin S2 (Sa) kills Pseudomonas aeruginosa strains via the FpvA type I ferripyoverdine receptor. J Bacteriol 189 : 7663–7668.
17. ElfarashA, WeiQ, CornelisP (2012) The soluble pyocins S2 and S4 from Pseudomonas aeruginosa bind to the same FpvAI receptor. Microbiologyopen 1 : 268–275.
18. LingH, SaeidiN, RasoulihaBH, ChangMW (2010) A predicted S-type pyocin shows a bactericidal activity against clinical Pseudomonas aeruginosa isolates through membrane damage. FEBS Lett 584 : 3354–3358.
19. BarreteauH, TiouajniM, GrailleM, JosseaumeN, BouhssA, et al. (2012) Functional and structural characterization of PaeM, a colicin M-like bacteriocin produced by Pseudomonas aeruginosa. J Biol Chem 287 : 37395–37405.
20. GrinterR, RoszakAW, CogdellRJ, MilnerJJ, WalkerD (2012) The crystal structure of the lipid II-degrading bacteriocin syringacin M suggests unexpected evolutionary relationships between colicin M-like bacteriocins. J Biol Chem 287 : 38876–38888.
21. ParretAHA, SchoofsG, ProostP, De MotR (2003) Plant lectin-like bacteriocin from a rhizosphere-colonizing Pseudomonas isolate. J Bacteriol 185 : 897–908.
22. ParretAHA, TemmermanK, De MotR (2005) Novel lectin-like bacteriocins of biocontrol strain Pseudomonas fluorescens Pf-5. Appl Environ Microbiol 71 : 5197–5207.
23. GhequireMGK, LiW, ProostP, LorisR, De MotR (2012) Plant lectin-like antibacterial proteins from phytopathogens Pseudomonas syringae and Xanthomonas citri. Environ Microbiol Rep 4 : 373–380.
24. Van DammeEJM, LannooN, PeumansWJ (2008) Plant lectins. Adv Bot Res 48 : 107–209.
25. GhequireMGK, LorisR, De MotR (2012) MMBL proteins: from lectin to bacteriocin. Biochem Soc Trans 40 : 1553–1559.
26. Van DammeEJM, KakuH, PeriniF, GoldsteinIJ, PeetersB, et al. (1991) Biosynthesis, primary structure and molecular cloning of snowdrop (Galanthus nivalis L.) lectin. Eur J Biochem 202 : 23–30.
27. HesterG, WrightCS (1996) The mannose-specific bulb lectin from Galanthus nivalis (snowdrop) binds mono - and dimannosides at distinct sites. Structure analysis of refined complexes at 2.3 Å and 3.0 Å resolution. J Mol Biol 262 : 516–531.
28. FouquaertE, PeumansWJ, GheysenG, Van DammeEJM (2011) Identical homologs of the Galanthus nivalis agglutinin in Zea mays and Fusarium verticillioides. Plant Physiol Biochem 49 : 46–54.
29. ShimokawaM, FukudomeA, YamashitaR, MinamiY, YagiF, et al. (2012) Characterization and cloning of GNA-like lectin from the mushroom Marasmius oreades. Glycoconj J 29 : 457–465.
30. JungE, FuciniP, StewartM, NoegelAA, SchleicherM (1996) Linking microfilaments to intracellular membranes: the actin-binding and vesicle-associated protein comitin exhibits a mannose-specific lectin activity. EMBO J 15 : 1238–1246.
31. WiensM, BelikovSI, KaluzhnayaOV, KraskoA, SchröderHC, et al. (2006) Molecular control of serial module formation along the apical-basal axis in the sponge Lubomirskia baicalensis: silicateins, mannose-binding lectin and mago nashi. Dev Genes Evol 216 : 229–242.
32. TsutsuiS, TasumiS, SuetakeH, SuzukiY (2003) Lectins homologous to those of monocotyledonous plants in the skin mucus and intestine of pufferfish, Fugu rubripes. J Biol Chem 278 : 20882–20889.
33. de Santana EvangelistaK, AndrichF, Figueiredo de RezendeF, NilandS, CordeiroMN, et al. (2009) Plumieribetin, a fish lectin homologous to mannose-binding B-type lectins, inhibits the collagen-binding α1β1 integrin. J Biol Chem 284 : 34747–34759.
34. RajanB, FernandesJM, CaipangCM, KironV, RomboutJH, et al. (2011) Proteome reference map of the skin mucus of Atlantic cod (Gadus morhua) revealing immune competent molecules. Fish Shellfish Immunol 31 : 224–231.
35. ChenJ, StevensonDM, WeimerPJ (2004) Albusin B, a bacteriocin from the ruminal bacterium Ruminococcus albus 7 that inhibits growth of Ruminococcus flavefaciens. Appl Environ Microbiol 70 : 3167–3170.
36. WrightCS, HesterG (1996) The 2.0 Å structure of a cross-linked complex between snowdrop lectin and a branched mannopentaose: evidence for two unique binding modes. Structure 4 : 1339–1352.
37. WrightLM, ReynoldsCD, RizkallahPJ, AllenAK, Van DammeEJM, et al. (2000) Structural characterisation of the native fetuin-binding protein Scilla campanulata agglutinin: a novel two-domain lectin. FEBS Lett 468 : 19–22.
38. RamachandraiahG, ChandraNR, SuroliaA, VijayanM (2002) Re-refinement using reprocessed data to improve the quality of the structure: a case study involving garlic lectin. Acta Crystallogr D Biol Crystallogr 58 : 414–420.
39. RobertV, VolokhinaEB, SenfF, BosMP, Van GelderP, et al. (2006) Assembly factor Omp85 recognizes its outer membrane protein substrates by a species-specific C-terminal motif. PLoS Biol 4: e377.
40. WangX, BauwG, Van DammeEJM, PeumansWJ, ChenZL, et al. (2001) Gastrodianin-like mannose-binding proteins: a novel class of plant proteins with antifungal properties. Plant J 25 : 651–661.
41. BalzariniJ (2007) Targeting the glycans of glycoproteins: a novel paradigm for antiviral therapy. Nat Rev Microbiol 5 : 583–597.
42. TianQ, WangW, MiaoC, PengH, LiuB, et al. (2008) Purification, characterization and molecular cloning of a novel mannose-binding lectin from rhizomes of Ophiopogon japonicus with antiviral and antifungal activities. Plant Sci 175 : 877–884.
43. BharathiY, Vijaya KumarS, PasaluIC, BalachandranSM, ReddyVD, et al. (2011) Pyramided rice lines harbouring Allium sativum (asaI) and Galanthus nivalis (gna) lectin genes impart enhanced resistance against major sap-sucking pests. J Biotechnol 152 : 63–71.
44. HoorelbekeB, Van DammeEJM, RougéP, ScholsD, Van LaethemK, et al. (2011) Differences in the mannose oligomer specificities of the closely related lectins from Galanthus nivalis and Zea mays strongly determine their eventual anti-HIV activity. Retrovirology 8 : 10.
45. VandenborreG, SmaggheG, Van DammeEJM (2011) Plant lectins as defense proteins against phytophagous insects. Phytochemistry 72 : 1538–1550.
46. YangY, XuHL, ZhangZT, LiuJJ, LiWW, et al. (2011) Characterization, molecular cloning, and in silico analysis of a novel mannose-binding lectin from Polygonatum odoratum (Mill.) with anti-HSV-II and apoptosis-inducing activities. Phytomedicine 18 : 748–755.
47. FuLL, ZhouCC, YaoS, YuJY, LiuB, et al. (2011) Plant lectins: targeting programmed cell death pathways as antitumor agents. Int J Biochem Cell Biol 43 : 1442–1449.
48. LehotzkyRE, PartchCL, MukherjeeS, CashHL, GoldmanWE, et al. (2010) Molecular basis for peptidoglycan recognition by a bactericidal lectin. Proc Natl Acad Sci U S A 107 : 7722–7727.
49. MikiT, HolstO, HardtWD (2012) The bactericidal activity of the C-type lectin RegIIIβ against Gram-negative bacteria involves binding to Lipid A. J Biol Chem 287 : 34844–34855.
50. TielkerD, HackerS, LorisR, StrathmannM, WingenderJ, et al. (2005) Pseudomonas aeruginosa lectin LecB is located in the outer membrane and is involved in biofilm formation. Microbiology 151 : 1313–1323.
51. BartelsKM, FunkenH, KnappA, BrockerM, WilhelmS, et al. (2011) Glycosylation is required for outer membrane localization of the lectin LecB in Pseudomonas aeruginosa. J Bacteriol 193 : 1107–1113..
52. FunkenH, BartelsKM, WilhelmS, BrockerM, BottM, et al. (2012) Specific association of lectin LecB with the surface of Pseudomonas aeruginosa: role of outer membrane protein OprF. PLoS One 7: e46857..
53. Green MR, Sambrook JR (2012) Molecular cloning: a laboratory manual. 4th edition. New York: Cold Spring Harbor Laboratory Press. 2028 p.
54. ParretAHA, WynsL, De MotR, LorisR (2004) Overexpression, purification and crystallization of bacteriocin LlpA from Pseudomonas sp. BW11M1. Acta Crystallogr D Biol Crystallogr 60 : 1922–1924.
55. PaceCN, VajdosF, FeeL, GrimsleyG, GrayT (1995) How to measure and predict the molar absorption coefficient of a protein. Protein Sci 4 : 2411–2423.
56. BlixtO, HeadS, MondalaT, ScanlanC, HuflejtME, et al. (2004) Printed covalent glycan array for ligand profiling of diverse glycan binding proteins. Proc Natl Acad Sci U S A 101 : 17033–17038.
57. SchneiderTR, SheldrickGM (2002) Substructure solution with SHELXD. Acta Crystallogr D Biol Crystallogr 58 : 1772–1779.
58. de la Fortelle E, Bricogne G (1997) Maximum likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. In: Carter CW, Sweet RM, editors. Methods in Enzymology. Volume 276, Macromolecular Crystallography Part A. New York: Academic Press. 472–494.
59. AbrahamsJP, LeslieAG (1996) Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr D Biol Crystallogr 52 : 30–42.
60. CowtanK (2010) Recent developments in classical density modification. Acta Crystallogr D Biol Crystallogr 66 : 470–478.
61. PerrakisA, MorrisR, LamzinVS (1999) Automated protein model building combined with iterative structure refinement. Nat Struct Biol 6 : 458–463.
62. EmsleyP, CowtanK (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60 : 2126–2132.
63. AdamsPD, GopalK, Grosse-KunstleveRW, HungLW, IoergerTR, et al. (2004) Recent developments in the PHENIX software for automated crystallographic structure determination. J Synchrotron Radiat 11 : 53–55.
64. AfoninePV, Grosse-KunstleveRW, AdamsPD (2005) A robust bulk-solvent correction and anisotropic scaling procedure. Acta Crystallogr D Biol Crystallogr 61 : 850–855.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2013 Číslo 2- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- The Apoptogenic Toxin AIP56 Is a Metalloprotease A-B Toxin that Cleaves NF-κb P65
- Phylodynamic Analysis of the Emergence and Epidemiological Impact of Transmissible Defective Dengue Viruses
- Isolation of a Novel Swine Influenza Virus from Oklahoma in 2011 Which Is Distantly Related to Human Influenza C Viruses
- Using Existing Drugs as Leads for Broad Spectrum Anthelmintics Targeting Protein Kinases
- MCMV-mediated Inhibition of the Pro-apoptotic Bak Protein Is Required for Optimal Replication
- Neutrophils Exert a Suppressive Effect on Th1 Responses to Intracellular Pathogen
- -32 Ligand/Receptor Silencing Phenocopy Faster Plant Pathogenic Nematodes
- Targeted and Random Mutagenesis of for the Identification of Genes Required for Infection
- Noncanonical Inflammasomes: Caspase-11 Activation and Effector Mechanisms
- A Roadmap to the Human Virome
- Bacterial Survival Amidst an Immune Onslaught: The Contribution of the Leukotoxins
- Fifty Shades of Immune Defense
- Proteins Secreted via the Type II Secretion System: Smart Strategies of to Maintain Fitness in Different Ecological Niches
- Structural Determinants for Activity and Specificity of the Bacterial Toxin LlpA
- Protein Complexes and Proteolytic Activation of the Cell Wall Hydrolase RipA Regulate Septal Resolution in Mycobacteria
- Programmed Protection of Foreign DNA from Restriction Allows Pathogenicity Island Exchange during Pneumococcal Transformation
- Behavior of Prions in the Environment: Implications for Prion Biology
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Isolation of a Novel Swine Influenza Virus from Oklahoma in 2011 Which Is Distantly Related to Human Influenza C Viruses
- A Roadmap to the Human Virome
- Neutrophils Exert a Suppressive Effect on Th1 Responses to Intracellular Pathogen
- -32 Ligand/Receptor Silencing Phenocopy Faster Plant Pathogenic Nematodes
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



