-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaFive Mechanisms of Manipulation by Bacterial Effectors: A Ubiquitous Theme
article has not abstract
Published in the journal: . PLoS Pathog 8(8): e32767. doi:10.1371/journal.ppat.1002823
Category: Pearls
doi: https://doi.org/10.1371/journal.ppat.1002823Summary
article has not abstract
Ubiquitin, a highly conserved polypeptide of 76 amino acids, participates in a vast range of eukaryotic cell processes through its role as a reversible post-translational modifier (see review [1], Figure 1A). Such extensive utilization of a single protein within a host cell lends itself to be an ideal target for microbial manipulation. Host-pathogen co-evolution has endowed present-day pathogens with an ever-expanding repertoire of proteins that function to modulate this system. The majority of these proteins are effectors of type III secretion (T3S) or type IV secretion (T4S) pathways, which are major virulence determinants of many Gram-negative pathogens [2], [3]. This review is focused on five distinct mechanisms in which secreted bacterial effector proteins exploit the host ubiquitylation system (Figure 2).
Fig. 1. The three classes of host enzymes (E1, purple; E2, red; E3, yellow) involved in ubiquitin modification of target host proteins. 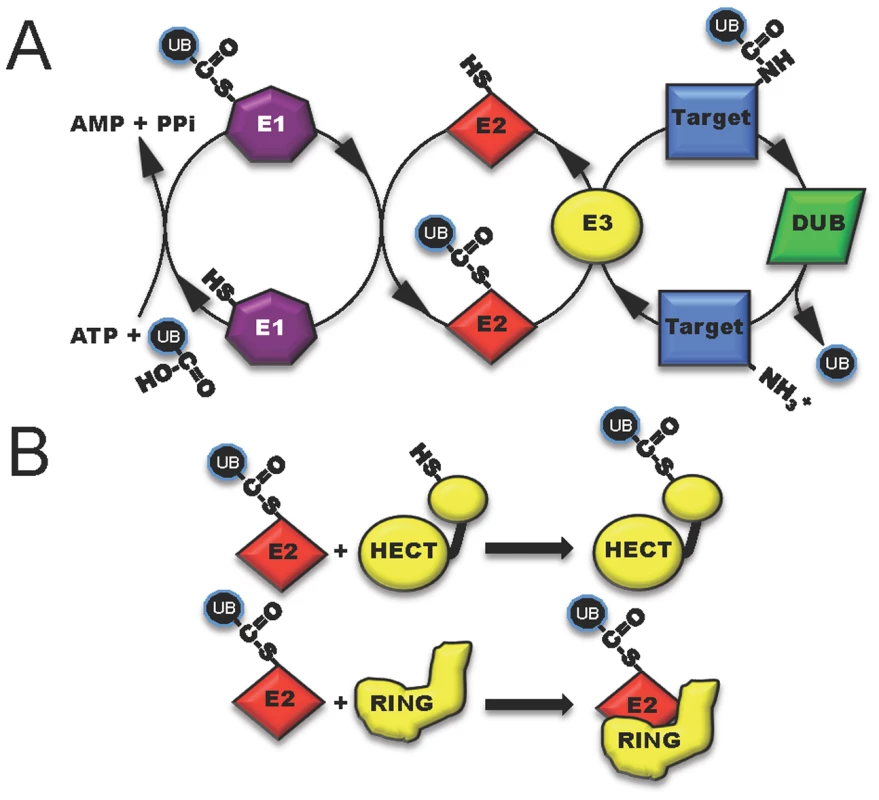
(A) E1 (activating enzyme, purple) charges ubiquitin in an ATP-dependent manner to form an E1-ubiquitin thioester intermediate. Activated ubiquitin is then transferred to the conjugating enzyme E2 (red). Target specificity (blue) is determined by E3 ligating enzymes. Linkage to the final modified target protein is by an isopeptide linkage. Deubiquitylase enzymes (DUB, green) can remove ubiquitin for recycling. (B) The two classes of host E3 ligases are illustrated in yellow as HECT and RING. HECT enzymes have a conserved cysteine residue and participate in catalysis. RING enzymes serve as adaptor-like proteins. Fig. 2. Five distinct mechanisms in which bacterial proteins manipulate the host ubiquitin system. 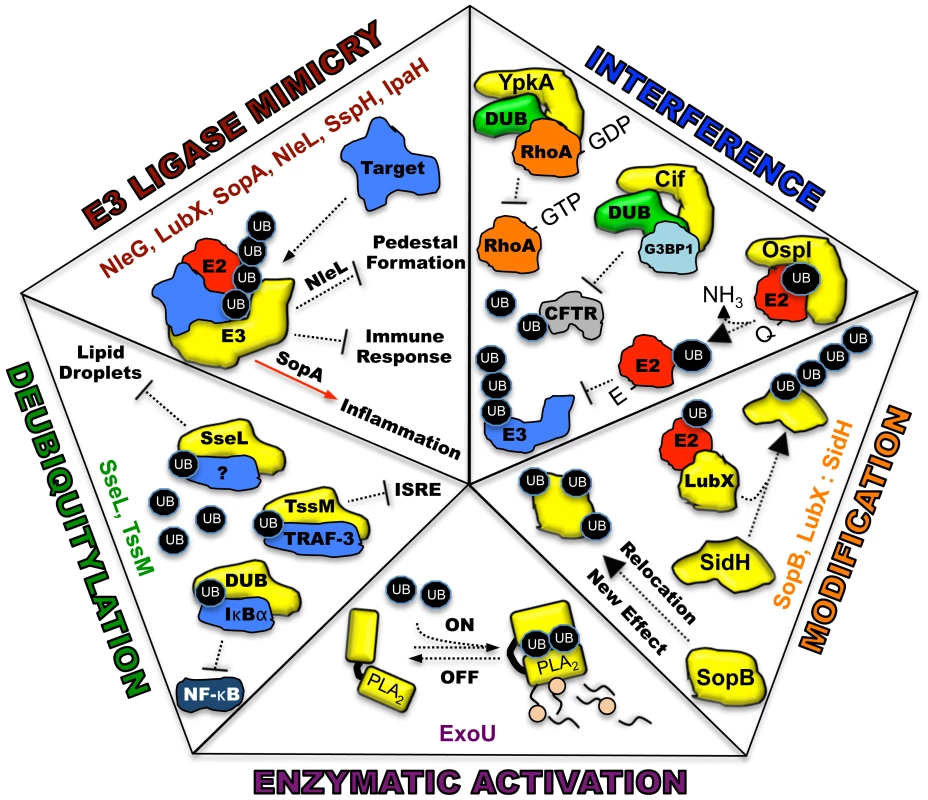
Each piece of the pentagon illustrates a known mechanism for the intersection between pathogen effectors and the host ubiquitin system. Bacterial proteins utilizing each specific mechanism are listed. All bacterial effectors are depicted in yellow. Host target proteins are identified when known and labeled as (?) when unknown. E2, E2 conjugating enzyme, red; E3, E3 ligating enzyme; DUB, deubiquitylating activity, green; UB, monoubiquituin or ubiquitin chains, black ovals; NF-κB, nuclear factor kappa B; IκBα, I kappa B alpha; PLA2, phospholipase activity with A2 specificity. E3 Ubiquitin Ligase Mimicry
One method to disrupt host cell physiology involves the injection of bacterial effectors that mimic proteins in the final step of ubiquitylation involving the E3 ligases [4]. Eukaryotic cells possess two main, functionally distinct classes of E3 ligases: RING-finger (really interesting new gene) and HECT-type (homologous to the E6-associated protein C-terminus) enzymes. RING (and related U-box) ligases act as bridging partners between E2-ubiquitin conjugates and target proteins, while HECT ligases directly participate in the chemistry of ubiquitylation (Figure 1B) [5], [6]. Bacterial effectors mimic both RING and HECT-class ligases and include a newly discovered third mechanistic class, appropriately named novel E3 ligases or NEL [7].
Recently characterized examples of bacterial ligases resembling the RING-finger/U-box enzymes include NleG, encoded by enterohaemorrhagic Escherichia coli, and LubX, encoded by Legionella pneumophila. The NleG family contains a conserved C-terminal domain that interacts with human E2 enzymes in a similar fashion as their E3 eukaryotic counterparts. The intracellular targets for NleG ligase activity, however, are unknown [8]. LubX is a novel, double U-box-containing enzyme. The first U-box is critical for ubiquitin ligase activity, while the second is necessary for targeting substrates such as the kinase, Clk1 [9]. A recent report revealed an additional target for LubX (see Effector Modification). Bacterial HECT-like ligases such as SopA (Salmonella) and NleL (E. coli O157:H7) each contain a conserved catalytic cysteine residue and interact with the E2 enzyme, UbcH7, on the same surface as eukaryotic ligases [10], [11]. The structural flexibility of their C-terminal subdomains, which are required for transthiolation of ubiquitin, appears similar between bacterial and eukaryotic enzymes as well [12]. NEL family members are related to HECT E3s because of the formation of a thioester bond with ubiquitin via a conserved cysteine in the catalytic domain. However, they differ in their mechanisms of interacting with substrates. SopA/NleL HECT-like enzymes possess a flexible, bilobed catalytic domain while novel ligases IpaH3 (Shigella) and SspH2 (Salmonella) likely undergo a dramatic reorientation between their N-terminal leucine-rich repeat and C-terminal NEL domains upon substrate recognition and before catalysis [7], [12]–[14]. Collectively, these mimics are profound examples of convergent evolution and the utilization of different strategies for solving a similar biological problem.
Deubiquitylation
Many bacteria synthesize effectors that interfere with host ubiquitylation by mimicking host deubiquitylases (DUBs). Eukaryotic DUBs exhibit exquisite substrate specificity and are important regulatory components within the host ubiquitin system [15]. Bacterial DUBs are generally modeled after eukaryotic cysteine proteases and are commonly used to attenuate NF-κB-related inflammatory responses by deubiquitylating and stabilizing IκBα. The Burkholderia pseudomallei DUB, TssM, additionally targets lysine 63-linked TNFR-associated factor-3 (TRAF-3) and TRAF-6, affecting interferon stimulated response element (ISRE) signaling and IKK activation, respectively [16]. Other mechanisms include the alteration of the intracellular environment. The DUB activity of Salmonella SseL was recently shown to affect lipid metabolism in infected gallbladder epithelial cells to prevent the accumulation of lipid droplets. These results suggest that in addition to IκBα, SseL possesses other target substrates [17]. Such disruption of complex cellular processes by the activity of a single enzyme provides both merit to the vulnerability of the host ubiquitin system to manipulation and insight into eukaryotic cell physiology.
Effector Modification
Bacterial pathogens have also evolved effectors that allow the host ubiquitin system to fine-tune their function, localization, or temporal regulation. One fascinating example of temporal regulation was revealed for two Salmonella Rho GTPase-modulating enzymes, SopE and SptP. SopE, a guanine exchange factor (GEF), is rapidly degraded by the host proteasome via ubiquitylation. SptP plays the opposing role of a GTPase activating protein (GAP) and differs in amino acid sequence resulting in a longer half-life compared to SopE. Thus, bacterial internalization is facilitated through membrane ruffling induced by SopE GTPase stimulation. Membrane and cytoskeletal homeostasis is then achieved though SopE degradation and the opposing actions of the longer-lived SptP [18]. During the intracellular replication of Legionella pneumophila, the bacterial E3 ligase mimic, LubX, regulates the stability of a second effector, SidH, via ubiquitylation. In the absence of LubX, SidH is stabilized, which leads to a hyper-virulent phenotype in a Drosophila infection model. LubX was thus coined to function as a “metaeffector,” a remarkable testament of pathogen co-evolution with its host [19].
Diversification of effector function and localization through ubiquitylation has been demonstrated for the Salmonella phosphoinositide phosphatase, SopB. In this case, an enzyme with a single catalytic function can act to modulate multiple cellular processes depending on its ubiquitylation state. SopB first traffics to the host plasma membrane, affecting actin reorganization and bacterial entry, macropinocytosis, and Akt activation. Post-multimonoubiquitylation by host enzymes, these activities are downregulated and SopB relocalizes to the Salmonella-containing vacuole to alter vesicular trafficking for the promotion of bacterial replication [20]. In sum, each of these strategies uses the host ubiquitin system to minimize the genetic cost of maintaining multiple virulence factors by simplifying the number of effectors required to alter host cell function.
Signaling Interference
There is a growing array of effectors that function to interfere with host ubiquitylation by mechanisms that differ from direct ubiquitylation/deubiquitylation. For instance, the Shigella flexneri protein, OspI, inhibits NF-κB signaling through the E3 ligase, TRAF6. Structural and mass spectrometry-based investigations indicate that OspI deamidase activity modifies an E2 enzyme (UBC13) recognizing TRAF6. Deamidation of glutamine 100 to glutamate on the E2 prevents polyubiquitylation of the E3 and activation of downstream innate immune response [21].
Host enzymes such as DUBS can also play scaffolding roles for bacterial effectors to enhance virulence. Pseudomonas aeruginosa uses outer membrane vesicles to export a protein known as Cif. Cif interferes with the endosomal recycling of the cystic fibrosis transmembrane conductance regulator (CFTR) to ultimately inhibit chloride secretion. Functionally, Cif stabilizes an intracellular complex of a DUB (USP10) and G3BP1, preventing deubiquitylation of CFTR, which is required for recycling the receptor to the plasma membrane [22]. Lastly, YpkA, a multifunctional serine/threonine kinase of Yersinia, has been postulated to sequester a phosphorylated DUB (OTUB1) and GDP-bound RhoA together in a complex. As OTUB1 activity and GTP-RhoA are implicated in enhancing bacterial uptake, inhibition of each may serve a role in preventing bacterial internalization and cell death [23]. The finesse of each of these effectors greatly enhances their specificity and prevents unintended collateral damage that might stimulate further inflammatory responses.
Enzymatic Activation
Our group has recently discovered a fifth mechanism of pathogen manipulation of host ubiquitylation. Pseudomonas aeruginosa possess a suite of T3S effectors known as ExoS, ExoT, ExoU, and ExoY [24]. Each enzyme contains domains that are catalytically inactive until injection into a target cell and association with a specific eukaryotic cofactor. Mechanistically this strategy ensures that each protein can be safely synthesized within Pseudomonas. We identified ubiquitin as such a cofactor for ExoU, a patatin-like A2 phospholipase with potent cytotoxic activity [25]. This appears to be the first example of a bacterial enzyme specifically requiring ubiquitin for catalysis. Co-expression of ExoU and ubiquitin in a prokaryotic system results in membrane damage and cell lysis, suggesting that no host ubiquitylation components, other than monoubiquitin, are required for phospholipase activity. Analysis of the enzyme activity in vitro indicates that multiple isoforms of ubiquitin activate ExoU, including ubiquitylated proteins and purified polyubiquitin chains with a variety of linkages [26]. Interestingly, a diubiquitin chain modifies ExoU soon after injection, an event with potential trafficking repercussions but not affecting overall toxicity [27], [28]. It is unclear whether the ubiquitylation of ExoU is part of the intracellular activation mechanism. Bioinformatic searches suggest as many as 4,400 bacterial proteins with typical patatin domains reside within sequenced bacterial genomes [29]. As many of these proteins remain uncharacterized, it is tempting to speculate that some of them may also require eukaryotic proteins such as ubiquitin or specific ubiquitylated proteins for activation.
Conclusion
Ubiquitin is an extremely conserved protein used for an ever-increasing array signaling cascades and regulatory events within all eukaryotic cells. During host-pathogen co-evolution, microbes have exploited these essential pathways to diversify and regulate the function of their effectors. The absence of ubiquitin and its related machinery from prokaryotes makes it a safe and effective target. Continued research into the host-pathogen relationship will no doubt reveal additional mechanisms by which bacterial effectors usurp the host ubiquitin system. Discovery of such mechanisms will provide a basis for therapeutic intervention as well as reveal new aspects of eukaryotic physiology.
Zdroje
1. HusnjakK, DikicI (2012) Ubiquitin-binding proteins: decoders of ubiquitin-mediated cellular functions. Annu Rev Biochem 81 : 291–322.
2. DeanP (2011) Functional domains and motifs of bacterial type III effector proteins and their roles in infection. FEMS Microbiol Rev 35 : 1100–1125.
3. ThanassiDG, BliskaJB, ChristiePJ (2012) Surface organelles assembled by secretion systems of gram-negative bacteria: diversity in structure and function. FEMS Microbiol Rev Epub ahead of print 30 April 2012. doi:10.1111/j.1574-6976.2012.00342.x.
4. HicksSW, GalanJE (2010) Hijacking the host ubiquitin pathway: structural strategies of bacterial E3 ubiquitin ligases. Curr Opin Microbiol 13 : 41–46.
5. DeshaiesRJ, JoazeiroCA (2009) RING domain E3 ubiquitin ligases. Annu Rev Biochem 78 : 399–434.
6. RotinD, KumarS (2009) Physiological functions of the HECT family of ubiquitin ligases. Nat Rev Mol Cell Biol 10 : 398–409.
7. QuezadaCM, HicksSW, GalanJE, StebbinsCE (2009) A family of Salmonella virulence factors functions as a distinct class of autoregulated E3 ubiquitin ligases. Proc Natl Acad Sci U S A 106 : 4864–4869.
8. WuB, SkarinaT, YeeA, JobinMC, DileoR, et al. (2010) NleG type 3 effectors from enterohaemorrhagic Escherichia coli are U-box E3 ubiquitin ligases. PLoS Pathog 6: e1000960 doi:10.1371/journal.ppat.1000960.
9. KuboriT, HyakutakeA, NagaiH (2008) Legionella translocates an E3 ubiquitin ligase that has multiple U-boxes with distinct functions. Mol Microbiol 67 : 1307–1319.
10. DiaoJ, ZhangY, HuibregtseJM, ZhouD, ChenJ (2008) Crystal structure of SopA, a Salmonella effector protein mimicking a eukaryotic ubiquitin ligase. Nat Struct Mol Biol 15 : 65–70.
11. PiscatelliH, KotkarSA, McBeeME, MuthupalaniS, SchauerDB, et al. (2011) The EHEC type III effector NleL is an E3 ubiquitin ligase that modulates pedestal formation. PLoS ONE 6: e19331 doi:10.1371/journal.pone.0019331.
12. LinDY, DiaoJ, ChenJ (2012) Crystal structures of two bacterial HECT-like E3 ligases in complex with a human E2 reveal atomic details of pathogen-host interactions. Proc Natl Acad Sci U S A 109 : 1925–1930.
13. SingerAU, RohdeJR, LamR, SkarinaT, KaganO, et al. (2008) Structure of the Shigella T3SS effector IpaH defines a new class of E3 ubiquitin ligases. Nat Struct Mol Biol 15 : 1293–1301.
14. ZhuY, LiH, HuL, WangJ, ZhouY, et al. (2008) Structure of a Shigella effector reveals a new class of ubiquitin ligases. Nat Struct Mol Biol 15 : 1302–1308.
15. KomanderD (2010) Mechanism, specificity and structure of the deubiquitinases. Subcell Biochem 54 : 69–87.
16. TanKS, ChenY, LimYC, TanGY, LiuY, et al. (2010) Suppression of host innate immune response by Burkholderia pseudomallei through the virulence factor TssM. J Immunol 184 : 5160–5171.
17. ArenaET, AuweterSD, AntunesLC, VoglAW, HanJ, et al. (2011) The deubiquitinase activity of the Salmonella pathogenicity island 2 effector, SseL, prevents accumulation of cellular lipid droplets. Infect Immun 79 : 4392–4400.
18. KuboriT, GalanJE (2003) Temporal regulation of Salmonella virulence effector function by proteasome-dependent protein degradation. Cell 115 : 333–342.
19. KuboriT, ShinzawaN, KanukaH, NagaiH (2010) Legionella metaeffector exploits host proteasome to temporally regulate cognate effector. PLoS Pathog 6: e1001216 doi:10.1371/journal.ppat.1001216.
20. PatelJC, HuefferK, LamTT, GalanJE (2009) Diversification of a Salmonella virulence protein function by ubiquitin-dependent differential localization. Cell 137 : 283–294.
21. SanadaT, KimM, MimuroH, SuzukiM, OgawaM, et al. (2012) The Shigella flexneri effector OspI deamidates UBC13 to dampen the inflammatory response. Nature 483 : 623–626.
22. BombergerJM, YeS, MaceachranDP, KoeppenK, BarnabyRL, et al. (2011) A Pseudomonas aeruginosa toxin that hijacks the host ubiquitin proteolytic system. PLoS Pathog 7: e1001325 doi:10.1371/journal.ppat.1001325.
23. EdelmannMJ, KramerHB, AltunM, KesslerBM (2010) Post-translational modification of the deubiquitinating enzyme otubain 1 modulates active RhoA levels and susceptibility to Yersinia invasion. FEBS J 277 : 2515–2530.
24. EngelJ, BalachandranP (2009) Role of Pseudomonas aeruginosa type III effectors in disease. Curr Opin Microbiol 12 : 61–66.
25. SatoH, FrankDW (2004) ExoU is a potent intracellular phospholipase. Mol Microbiol 53 : 1279–1290.
26. AndersonDM, SchmalzerKM, SatoH, CaseyM, TerhuneSS, et al. (2011) Ubiquitin and ubiquitin-modified proteins activate the Pseudomonas aeruginosa T3SS cytotoxin, ExoU. Mol Microbiol 82 : 1454–1467.
27. StirlingFR, CuzickA, KellySM, OxleyD, EvansTJ (2006) Eukaryotic localization, activation and ubiquitinylation of a bacterial type III secreted toxin. Cell Microbiol 8 : 1294–1309.
28. GendrinC, Contreras-MartelC, BouillotS, ElsenS, LemaireD, et al. (2012) Structural basis of cytotoxicity mediated by the type III secretion toxin ExoU from Pseudomonas aeruginosa. PLoS Pathog 8: e1002637 doi:10.1371/journal.ppat.1002637.
29. LangC, FliegerA (2011) Characterization of Legionella pneumophila phospholipases and their impact on host cells. Eur J Cell Biol 90 : 903–912.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2012 Číslo 8- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Exon Level Transcriptomic Profiling of HIV-1-Infected CD4 T Cells Reveals Virus-Induced Genes and Host Environment Favorable for Viral Replication
- Five Mechanisms of Manipulation by Bacterial Effectors: A Ubiquitous Theme
- Nonhuman Primate Models for HIV Cure Research
- The Ebola Virus Glycoprotein Contributes to but Is Not Sufficient for Virulence
- Trichomonosis, a Common Curable STI, and Prostate Carcinogenesis—A Proposed Molecular Mechanism
- Host Defense and Tolerance: Unique Challenges in the Placenta
- CPAF: A Chlamydial Protease in Search of an Authentic Substrate
- Small Protease Sensitive Oligomers of PrP in Distinct Human Prions Determine Conversion Rate of PrP
- Invariant NKT Cells: Regulation and Function during Viral Infection
- Human Monoclonal Antibody HCV1 Effectively Prevents and Treats HCV Infection in Chimpanzees
- Chemokine Receptor Ccr1 Drives Neutrophil-Mediated Kidney Immunopathology and Mortality in Invasive Candidiasis
- Phagocyte Responses to Protozoan Infection and How Meets the Challenge
- Telomere Length Affects the Frequency and Mechanism of Antigenic Variation in
- A Biofilm-Induced Pathway for Matrix Glucan Delivery: Implications for Drug Resistance
- The HSV-1 Exonuclease, UL12, Stimulates Recombination by a Single Strand Annealing Mechanism
- Inhibition of Fatty Acid Synthase (Fas2) Induces Mitochondrial Cell Death in Serum
- Interferon-alpha Subtype 11 Activates NK Cells and Enables Control of Retroviral Infection
- Transposon-mediated Chromosomal Integration of Transgenes in the Parasitic Nematode and Establishment of Stable Transgenic Lines
- Structural and Biochemical Basis for Development of Influenza Virus Inhibitors Targeting the PA Endonuclease
- Upregulation of Retinal Dehydrogenase 2 in Alternatively Activated Macrophages during Retinoid-dependent Type-2 Immunity to Helminth Infection in Mice
- Measles Immune Suppression: Lessons from the Macaque Model
- Fungi and the Rise of Mammals
- Bacterial Cell Surface Heterogeneity: A Pathogen's Disguise
- Cytoplasmic Entry Induces Fetal Wastage by Disrupting Maternal Foxp3 Regulatory T Cell-Sustained Fetal Tolerance
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Invariant NKT Cells: Regulation and Function during Viral Infection
- Host Defense and Tolerance: Unique Challenges in the Placenta
- Nonhuman Primate Models for HIV Cure Research
- Exon Level Transcriptomic Profiling of HIV-1-Infected CD4 T Cells Reveals Virus-Induced Genes and Host Environment Favorable for Viral Replication
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



