-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaStructural Basis of Cytotoxicity Mediated by the Type III Secretion Toxin ExoU from
The type III secretion system (T3SS) is a complex macromolecular machinery employed by a number of Gram-negative pathogens to inject effectors directly into the cytoplasm of eukaryotic cells. ExoU from the opportunistic pathogen Pseudomonas aeruginosa is one of the most aggressive toxins injected by a T3SS, leading to rapid cell necrosis. Here we report the crystal structure of ExoU in complex with its chaperone, SpcU. ExoU folds into membrane-binding, bridging, and phospholipase domains. SpcU maintains the N-terminus of ExoU in an unfolded state, required for secretion. The phospholipase domain carries an embedded catalytic site whose position within ExoU does not permit direct interaction with the bilayer, which suggests that ExoU must undergo a conformational rearrangement in order to access lipids within the target membrane. The bridging domain connects catalytic domain and membrane-binding domains, the latter of which displays specificity to PI(4,5)P2. Both transfection experiments and infection of eukaryotic cells with ExoU-secreting bacteria show that ExoU ubiquitination results in its co-localization with endosomal markers. This could reflect an attempt of the infected cell to target ExoU for degradation in order to protect itself from its aggressive cytotoxic action.
Published in the journal: . PLoS Pathog 8(4): e32767. doi:10.1371/journal.ppat.1002637
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1002637Summary
The type III secretion system (T3SS) is a complex macromolecular machinery employed by a number of Gram-negative pathogens to inject effectors directly into the cytoplasm of eukaryotic cells. ExoU from the opportunistic pathogen Pseudomonas aeruginosa is one of the most aggressive toxins injected by a T3SS, leading to rapid cell necrosis. Here we report the crystal structure of ExoU in complex with its chaperone, SpcU. ExoU folds into membrane-binding, bridging, and phospholipase domains. SpcU maintains the N-terminus of ExoU in an unfolded state, required for secretion. The phospholipase domain carries an embedded catalytic site whose position within ExoU does not permit direct interaction with the bilayer, which suggests that ExoU must undergo a conformational rearrangement in order to access lipids within the target membrane. The bridging domain connects catalytic domain and membrane-binding domains, the latter of which displays specificity to PI(4,5)P2. Both transfection experiments and infection of eukaryotic cells with ExoU-secreting bacteria show that ExoU ubiquitination results in its co-localization with endosomal markers. This could reflect an attempt of the infected cell to target ExoU for degradation in order to protect itself from its aggressive cytotoxic action.
Introduction
Type III secretion systems (T3SS) are needle-like, membrane-anchored, multi-component complexes that enable a number of pathogenic bacteria to inject effectors from the cytosol directly into the cytoplasm of eukaryotic cells [1]–[6]. T3SS are widespread among Gram-negative bacteria, and although the structure of the T3SS apparatus itself can display notable similarities amongst different bacterial species [5], [7], the nature of translocated effectors are widely different. Many T3SS-translocated proteins have been shown to modulate cellular functions, i.e. by mimicking protein kinases, phosphatases, GTPase activating proteins, or ubiquitin ligases, or by covalently modifying target proteins through phosphorylation or acetylation. The consequences to the target cell may range from modifications of the cytoskeleton to membrane disruption and apoptosis [8], underlining the key nature of T3SS for extreme pathogenesis in a number of systems.
Pseudomonas aeruginosa is a leading cause of nosocomial infections and is a major threat to cystic fibrosis patients and others with impaired immune defenses. It carries a T3SS whose upregulation during acute phases of infection is directly related to poor patient prognosis [9], [10], and which translocates four effectors, namely exoenzymes S, T, U, and Y [11]–[13]. ExoS and ExoT are bifunctional molecules with GTPase-activating (GAP) and ADP-ribosyltransferase activities essential for the inhibition of bacterial internalization and epithelial cell migration [14]–[19]. ExoY is an adenylate cyclase reported to play a role in actin cytoskeleton disruption and cause cell rounding [20]–[22]. However, it is ExoU which is the most detrimental toxin injected by the T3SS of P. aeruginosa. ExoU is expressed by approximately 30% of clinical strains, 90% of which cause acute illness [10], [11]. It is encoded on a pathogenicity island together with its cognate chaperone SpcU, which is required for ExoU's efficient secretion from the bacterial cytoplasm [23], [24]. ExoU is a 687-residue protein that, once translocated through the T3SS, induces cytotoxic effects leading to rapid necrotic cell death; exoU knockout P. aeruginosa strains display greatly decreased virulence in mouse models of acute infection [25], [26]. In clinical settings, ExoU-expressing P. aeruginosa strains lead to poor patient prognosis, since the toxin causes acute lung epithelial injury and is linked to the development of septic shock [10], [18], [27]–[30]. To date, the precise mechanism underlying ExoU's significant potency for cellular destruction has remained unclear.
Notably, ExoU has been shown to carry phospholipase activity with broad substrate specificity which relies on an essential catalytic dyad (Ser142/Asp344) [24], [31]–[33], as is the case for other phospholipases. Enzymes with PLA2 activity hydrolyze the sn-2 ester bond of phospholipids, and thus play a role in membrane disruption, fatty acid release, and in many cases, signal transduction [34]–[36]. Interestingly, expression of ExoU in yeast causes rapid fragmentation of the vacuolar compartment, suggesting that eukaryotic membranes are the major targets for the toxin [33]. It is of note that the C-terminus of ExoU is critical for its in vitro phospholipase activity and cellular cytotoxicity [32], [37]–[40]. In addition, pre-incubation with cellular extracts has been shown to be essential for detection of phospholipase activity in vitro [33], [41], indicating the requirement for host eukaryotic cofactors in order for ExoU to exert its phospholipase activity on lipidic substrates. Eukaryotic Cu/Zn superoxide dismutase (SOD1) and other ubiquitinated proteins, as well as ubiquitin itself, have been suggested as being potential activators of the toxin [42]–[44].
The extreme toxicity of ExoU has limited functional studies of the wild type form, necessitating the employment of a catalytically-inactive mutant in cellular assays [31]–[33]. Trafficking studies of the ExoU-Ser142Ala mutant in eukaryotic cells have revealed that, upon translocation through the T3SS, ExoU is initially targeted to the plasma membrane (PM). The membrane localization potential of ExoU resides in its C-terminal domain (specifically, in residues 679–683). ExoU trafficking to the PM enables its ubiquitination on Lys178 [39], but this modification has only a modest effect on ExoU turnover and an ExoU-K178R-expressing mutant strain displays cytotoxicity levels that are similar to those of the wild-type. These observations attest to the complex fate of ExoU in target cells, which implies the coordination between membrane targeting, phospholipase activity, activation by cofactors, and ubiquitination.
In order to understand the multi-faceted behavior of ExoU, we solved its crystal structure, which was achieved in the presence of its chaperone, SpcU. ExoU forms a 1∶1 complex with SpcU, and folds into three distinct domains, which fulfill catalytic, bridging, and membrane-binding functions. The active site of ExoU is localized within an α/β hydrolase fold in a cleft sheltered by flexible loop regions, thus suggesting an inactive conformation that would require a structural modification to allow access of the nucleophilic serine to the lipidic substrates on the plasma membrane. The localization of Lys178 in proximity to the active site region implies that such structural rearrangements could also facilitate ubiquitination of ExoU by PM-localized E3 ligases. Ubiquitinated ExoU co-localizes with markers of endosomal compartments both upon transfection of cultured cells and infection of eukaryotic cells by an ExoU-expressing P. aeruginosa strain. This reflects a potential attempt of the cell to eliminate the toxin by targeting it for lysosomal destruction.
Results
SpcU's type IA fold stabilizes the N-terminus of ExoU
The structure of the ExoU(30–687)∶SpcU(1–127) complex was solved to a resolution of 2.94 Å by performing a SAD experiment on the selenium edge at the ESRF synchrotron in Grenoble (Supplementary Table I), and harbors a 1∶1 complex in the asymmetric unit. This stoichiometry is unusual, since SpcU has a typical type IA T3SS chaperone fold (Figure 1), and complexes between T3SS effectors and type I chaperones typically associate with stoichiometries of 1∶2 [3], [5], [45]. In order to determine the precise stoichiometry of ExoU∶SpcU in vitro, we performed native mass spectrometry measurements of the purified complex, which allowed us to identify a mixture of two distinct stoichiometric species, 1∶1 and 1∶2. Interestingly, close analysis of the structure reveals how both stoichiometries are possible in the crystal lattice.
Fig. 1. The crystal structure of ExoU in complex with its chaperone SpcU. 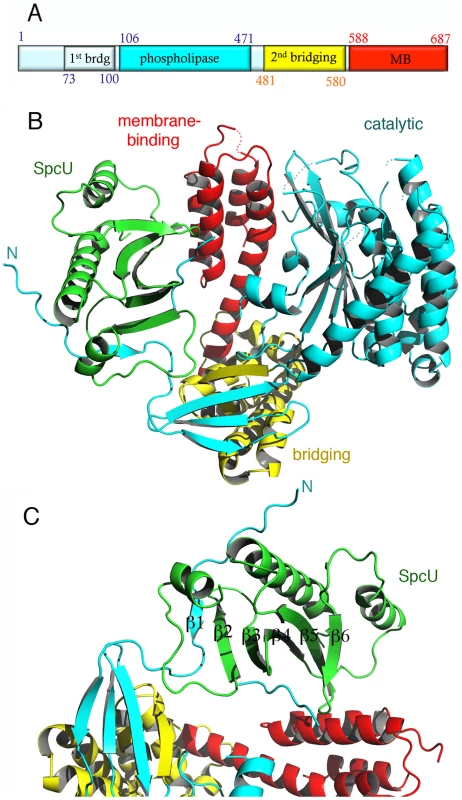
(A) Schematic diagram of the ExoU construct used in this work and limitations of different domains. The first and last ExoU residues observed in the electron density map are Gly53 and Glu685. The catalytic domain is inserted within the first and second bridging subdomains. (B) ExoU folds into three distinct domains and its N-terminal β-strand is paired with SpcU's β-sheet. The bridging domain (yellow) contains elements that are both N-terminal and C-terminal to the catalytic domain (blue). The membrane-binding domain makes few contacts with the other domains of ExoU. (C) SpcU (green) is a type IA T3SS chaperone. The N-terminus of ExoU, packs as a β-strand against the central 5-stranded SpcU sheet. For clarity, only the asymmetric unit is shown. SpcU is a typical type IA chaperone, being composed of a curved, 5-stranded sheet flanked by α-helices. SpcU displays two hydrophobic patches that recognize the effector. One involves the extremity of the 5-stranded β-sheet, which binds a short β-strand located at the N-terminus of ExoU (residues 62–64), thus generating a 6-stranded sheet (Figures 1c and S1). This binding region corresponds to the β motif that is also present in SipA∶InvB, SptP∶SicP, YopH∶SycH, and YopE∶SycE complexes [46]–[48]. An additional hydrophobic region of SpcU, located at the center of the β-sheet, binds a region of ExoU that is partly disordered in the electron density map. It is conceivable that this region corresponds to the flexible N-terminus of ExoU that is stabilized in a symmetry-related SpcU molecule. This becomes more evident upon the generation of an ExoU∶SpcU dimer (Figure S1c) based on the possible biological assemblies calculated by PISA (http://www.ebi.ac.uk/msd-srv/prot_int/picite.html), as well as by symmetry within the C2 cell, which suggests that the flexible N-terminus of ExoU in fact wraps around the crystallographic SpcU dimer by employing two different hydrophobic patches on the chaperone (the “front" and the “back", Figures S1b, S1c). It is of interest that the effectors in the abovementioned 1∶2 complexes display N-termini that surround their respective chaperone dimers in comparable fashion. Thus, it is conceivable that in vitro, both ExoU∶SpcU stoichiometric forms exist in equilibrium, but the 1∶1 form, presented here, is the one that crystallizes in the asymmetric unit of a C2 cell. Notably, the complex between the Yersinia pestis chaperone SycH and a fragment from the effector YscM2 also crystallized with 1∶1 stoichiometry, with the effector wrapped around the chaperone in a similarly extended conformation [49].
This association presumably ensures that the N-terminus of ExoU is ready for translocation through the T3SS, and is also in agreement with data indicating that the minimal domain required for cytotoxicity begins in a region residing between residues 52 and 100 [50]; in fact, the last residue in the N-terminal region of ExoU that makes contacts with SpcU is Ser65.
ExoU folds into three independent domains
Immediately following the SpcU-bound N-terminus of ExoU is the first part of the bridging domain, which precedes the catalytic region. The bridging domain of ExoU consists of two subdomains: an N-terminal 4-stranded region (cyan and yellow in Fig. 1b) and an all-helical section (yellow in Figures 1b and S2). Notably, the ExoU sequence that immediately follows the N-terminal, SpcU-associated region contributes 3 β-strands to the 4-stranded sheet of the first subdomain (β2–β4). The catalytic domain is intercalated at this point; β11 then complements the first subdomain, and this is followed by the all-helical C-terminal subdomain (residues 480–580). The bridging domain itself has no significant structural similarity with any known folds, as determined by the DALI server (http://ekhidna.biocenter.helsinki.fi/dali_server/). Due to its intercalated position between catalytic and membrane-binding domains, it is possible that its function is uniquely to serve as a ‘platform’ to place both functionally critical domains in close proximity.
Residues 106–471 delimit the catalytic, phospholipase domain, which is composed of an α/β hydrolase fold with a 6-stranded central β-sheet surrounded by 10 α-helices (Figures 1 and 2a). The sheet displays a superhelical twist with β strands 8 and 10 lying at approximately 90° to each other. The catalytic serine (Ser142) is located at an elbow between β6 and the following helix, in a funnel-like region surrounded by a number of flexible loops. A triple glycine sequence (Gly111, 112, 113), in close proximity, provides its backbone atoms as an oxyanion hole that potentially stabilizes the charge of the transition state intermediate in the phospholipase reaction (Figure 2a). This structural arrangement is highly similar to that of a well-studied and potent phospholipase, human cPLA2, whose structure also carries a twisted β-sheet that prominently displays the catalytic serine in proximity to the oxyanion hole [51] (Figure 2b). Additionally, the structure of cPLA2 harbors an active site ‘cap’ – a presumably flexible region that must be displaced in order to enable substrate access into the active site. It is precisely in this ‘cap’ region that is located the catalytic aspartate residue, essential for phospholipase activity [51]. In the structure of ExoU, this region is also found to be flexible, and the catalytic aspartate (Asp344) is not traceable in the electron density map (Figure 2a), suggesting not only that ExoU could also harbor a ‘cap’ region, but that the structure presented here is in the ‘open ’, and potentially inactive, conformation.
Fig. 2. The catalytic domain of ExoU displays an α/β hydrolase fold. 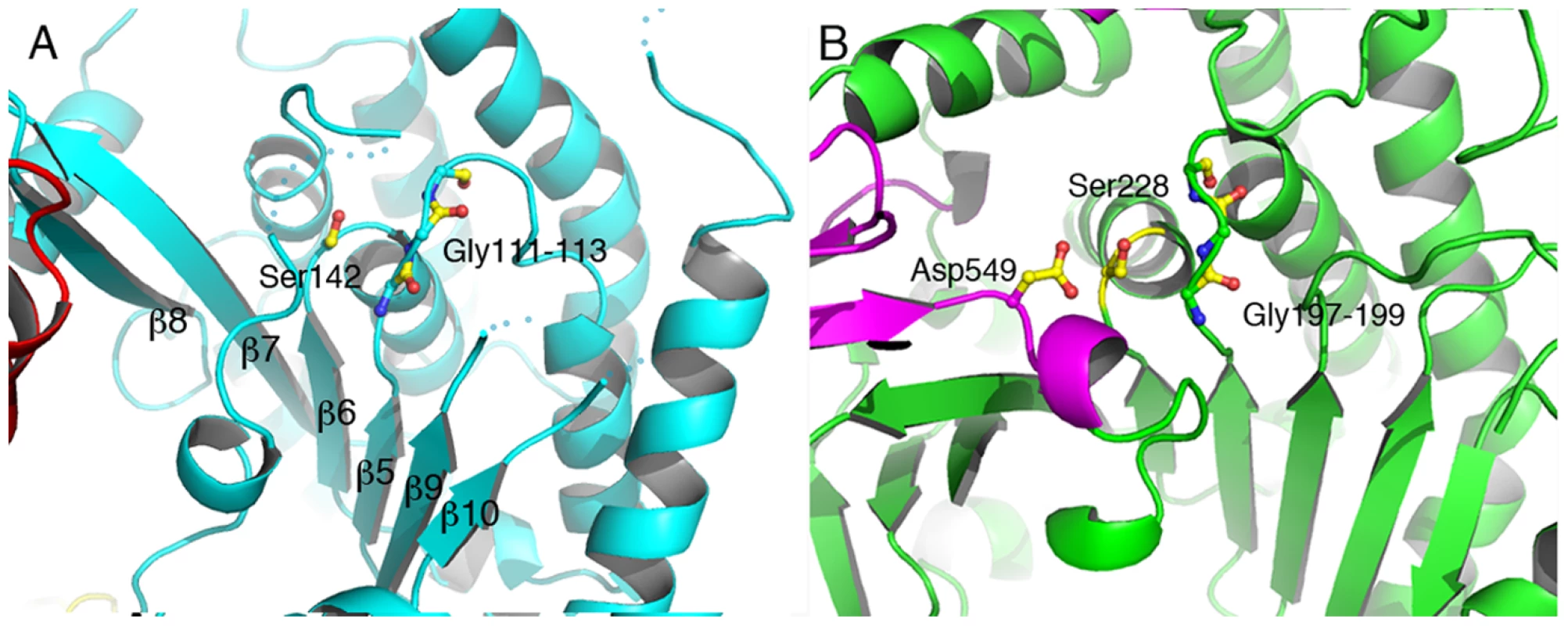
(A) Ser142 is in close proximity to the Gly111–113 elbow, which acts as an oxyanion hole for the phospholipase-catalyzed reaction. The second member of the Ser-Asp catalytic dyad, Asp344, is located in a flexible region that is not traceable in the electron density map. The overall arrangement of the core region of ExoU's catalytic domain is reminiscent of that of human cPLA2 (B), which also harbors the catalytic aspartate in a moveable ‘cap’ (in violet). Interestingly, Lys178, shown to be the target for ubiquitination once ExoU upon membrane localization, is located within the phospholipase domain, in close proximity to the active site (Figures 3a, 3b). Its side chain is located approximately 7 Å away from the active site serine, in a region that displays high flexibility. This strategic localization could favor the ability of this particular lysine residue to become ubiquitinated (see below).
Fig. 3. Membrane-binding, ubiquitination, and catalysis are structurally intertwined. 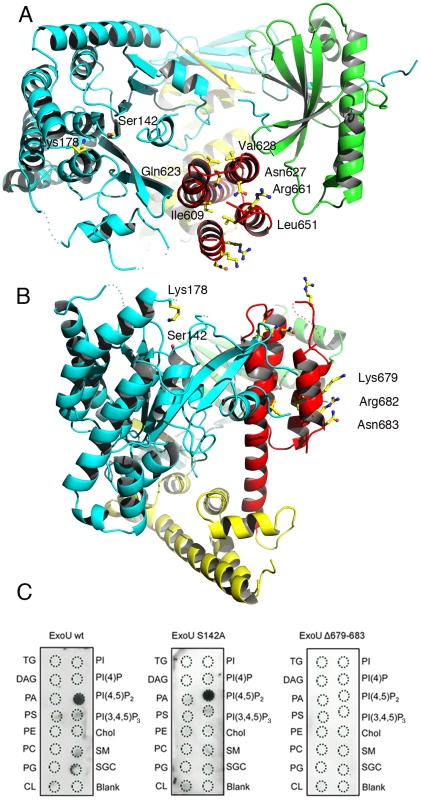
(A) Mutations in the C-terminus of ExoU [31], [32], [37]–[40] that abrogate membrane binding and cytotoxicity are all located within the four-helical bundle (red). This region is located on the same face of the molecule as the catalytic region, as well as Lys178 (left). (B) Residues 679–683 are exposed to solvent, in a potential appropriate orientation for bilayer recognition. (C) Protein-lipid overlays of wt ExoU, ExoU-S142A and ExoU-Δ679–683 reveal that the C-terminal region is essential for a strong, specific interaction with PI(4,5)P2. TG, triglyceride; DAG, diacylglycerol; PA, phosphatidic acid; PS, phosphatidylserine; PE, phosphatidylethanolamine; PC, phosphatidylcholine; PG, phosphatidylglycerol; CL, cardiolipin; PI, phosphatidyl-inositol; PI(4)P, phosphatidylinositol 4 phosphate; PI(4,5)P2, phosphatidylinositol (4,5) di-phosphate; PI(3,4,5)P3, phosphatidylinositol (3,4,5) triphosphate; chol, cholesterol; SM, sphingomyelin; SGC, 3 sulfogalactosylceramide. Blank=no lipid spotted. The C-terminus of ExoU has been shown to play a key role in membrane targeting, and numerous modifications of this region that have an impact ExoU's cytotoxicity have been described, including the deletion of residues 679–683, which results in P. aeruginosa strains whose cytotoxicity potential is greatly diminished [31], [37]–[40], [50]. Our structure reveals that the C-terminus of ExoU forms a distinct domain (encompassing residues 588–687) that folds into a 4-helical bundle (red in Figures 1 and 3a). One group of residues whose mutation affects ExoU intracellular functionality (Ile609, Ile654, Leu651, Ala678; [38], [40]) makes hydrophobic interactions within the bundle itself, and their mutation into polar residues could destabilize the non-polar character of the interior of the bundle. A second group (Arg661, Lys679, Arg682, Asn683 [38], [39]) is exposed on the surface of ExoU, with Arg661 located on a loop that precedes the C-terminal α-helix that harbors amino acids 679–683 (Figures 3a and 3b). Interestingly, residues 679, 682, and 683 form a polar, mostly basic ‘backbone’ that could bind to negatively charged eukaryotic phospholipids (Figure S3), and facilitate lipid bilayer recognition. To explore this idea, we performed protein-lipid overlay assays and demonstrated that ExoU strongly interacts with PI(4,5)P2 (Figure 3c), a negatively-charged polyvalent phospholipid that is abundant in the cytosolic leaflet of the PM. The non-catalytic mutant ExoU-S142A also displays a strong affinity for PI(4,5)P2, but this interaction is completely abolished in the case of a mutant lacking residues 679–683 (Figure 3c), which suggests a direct role of PI(4,5)P2 interaction for ExoU PM localization. Our results thus provide a molecular explanation for ExoU's PM targeting, which constitutes the toxin's premier localization upon T3SS translocation.
Ubiquitinated ExoU is targeted to endosomes
Upon localization to the PM, ExoU becomes ubiquitinated on Lys178, a modification which has a small effect on its intracellular turnover rate [39]. In order to explore the fate of translocated ExoU, we adapted the Ubiquitin-mediated Fluorescence Complementation (UbFC) method [52] to directly visualize ubiquitinated ExoU (Ub-ExoU) in eukaryotic cells. Catalytically-inactive ExoU-S142A was fused to the C-terminal fragment of the Venus fluorescent protein (generating ExoU-S142A-VC), and Ub to its complementary N-terminal fragment (generating VN-Ub; Figure 4a). Co-transfection of the two plasmids, but not single transfections, led to a fluorescent signal located at the cell periphery, but also revealed punctate structures throughout the cytoplasm (Figure 4b upper panel). To ascertain that the observed signal was specific, we tested the fluorescence complementation potential of a mutated, non-conjugatable Ub protein, in which the 7 endogenous lysine residues and the two C-terminal glycine residues were modified (VN-Ub Mut). No fluorescence complementation was observed when co-transfecting ExoU-S142A-VC and VN-Ub Mut, whereas both proteins were successfully expressed as evidenced by immunofluorescence detection of tagged proteins (Figure 4b, bottom panels). These observations suggested that the punctate pattern observed by Phillips et al upon transfection of GFP-ExoU-S142A into eukaryotic cells [32] could reflect targeting of the ubiquitinated form of ExoU to specific intracellular compartments.
Fig. 4. Direct visualization of Ub-ExoU by UbFC. 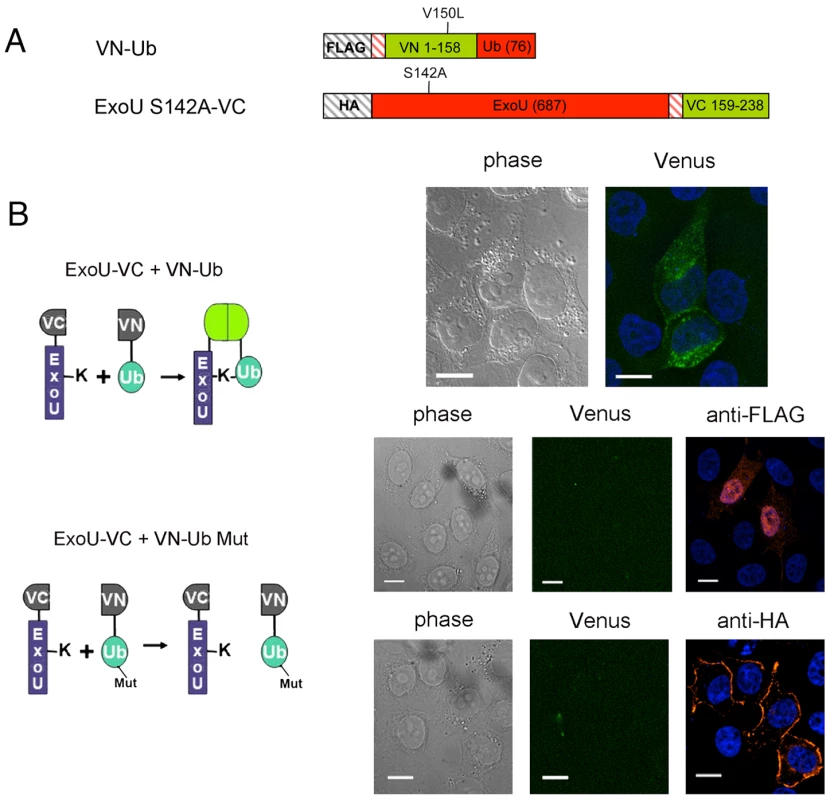
(A) Schematic representation of the plasmid constructs used for UbFC. The genes from which the different fragments were amplified are indicated within the boxes, and the number of residues of each fragment is indicated in parentheses. (B) ExoU-S142A-VC and VN-Ub (upper panel) or ExoU-S142A-VC and VN-Ub Mut (lower panels) were co-expressed in HeLa cells, and fluorescence complementation was observed 36 h after transfection. Immunodetection of fusion proteins with the anti-HA and the anti-FLAG antibodies was performed to confirm protein expression Bars, 12 µm. Pearson's correlation coefficient for Fig. 4B top=0.727. In order to identify this intracellular location, we compared the fluorescence complementation signal with the localization of proteins of known distribution. Ub-ExoU-S142A did not co-localize with endoplasmic reticulum nor with Golgi markers (Figure S4), but was targeted to acidic organelles, as evidenced by its co-localization with the early endosome marker EEA1 and with lysotracker-stained compartments (Figure 5a, top panels). Since ubiquitination of ExoU has been proposed to involve Lys63-linked Ub chains [39], we tested a Venus-fusion of a Ub mutant in which all Lys residues, except Lys63, were mutated to Arg (VN-Ub-K63). ExoU-S142A-Ub K63 co-localized with lysotracker (Figure 5a, bottom panel), confirming that K63-linked chains of Ub are most likely responsible for ExoU targeting to endosomal compartments.
Fig. 5. Ub-ExoU is targeted to endosomes. 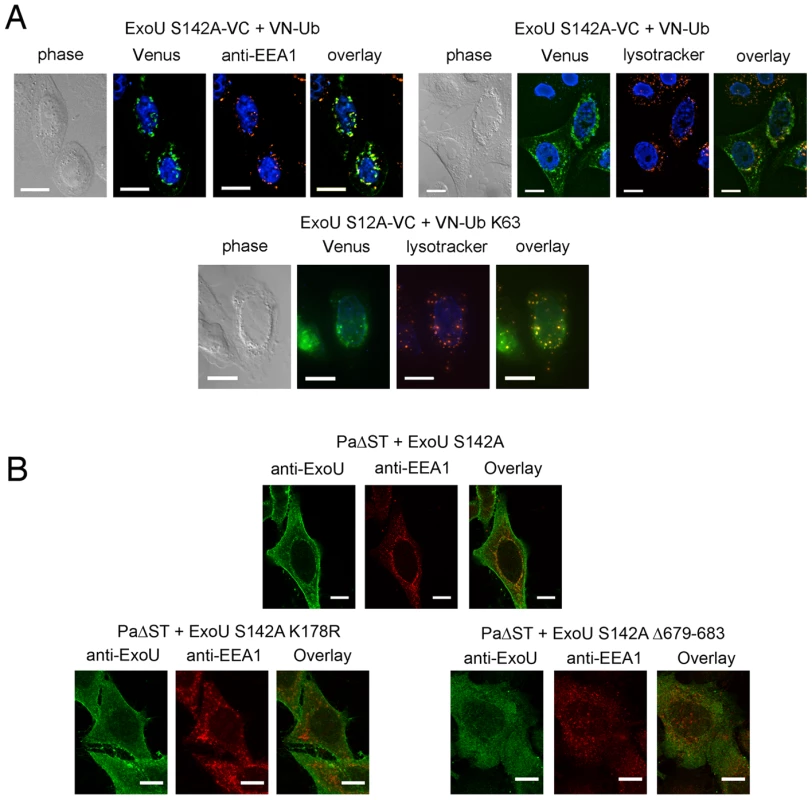
(A) Co-localization of Ub-ExoU with endosomal markers. ExoU S142A-VC was co-transfected with VN-Ub (upper panel) or with VN-Ub K63 (bottom panel). 36 h later, cells were stained with the anti-EEA1, or treated with lysotracker red. Pearson's correlation coefficients correspond to 0.692, 0.570, and 0.801 the top left, top right, and bottom panels, respectively. (B). HeLa cells were infected for 3 h with PaΔST+ExoU S142A (upper panel), PaΔST+ExoU S142A-K178R or PaΔST+ExoU S142A Δ679–683 (bottom panel), fixed and immunolabeled with anti-ExoU and anti-EEA1 antibodies. Single two-channel confocal images are presented. The Pearson's correlation coefficient for EEA1-ExoU=0.213. Bars, 12 µm. In order to study ExoU targeting in the natural context of infection, we engineered a P. aeruginosa strain that translocates catalytically inactive ExoU-S142A upon induction of the T3SS into HeLa cells (PaΔST+ExoU). Confocal microscopy analyses of infected cells revealed the presence of ExoU at the PM, as previously identified [32], [37], [39], but also revealed that translocated ExoU co-localizes with EEA1 (Figure 5b top panel and Video S1). In order to relate this localization to the ubiquitination of ExoU, we generated a P. aeruginosa strain that expresses ExoU-S142A-K178R, which cannot be ubiqutinated [39]. Confocal microscopy analyses of HeLa cells infected with this strain did not indicate any co-localization of this mutant with endosomal compartments, but a prominent PM staining. These observations thus indicate that ExoU's co-localization with endosomes is directly dependent on its ubiquitination of Lys178.
Stirling et al. showed that residues 679–683, located at the extremity of the C-terminal helical bundle (Figure 3b) are required for ubiquitination [39]. The intracellular fate of this mutant was investigated by constructing a P. aeruginosa strain that expresses ExoU-S142A-Δ679–683. Confocal microscopy images of infected HeLa cells indicated that this mutant form of ExoU not only failed to associate with the PM, but was also not able to co-localize with EEA1 (Figure 5b, bottom right panel). These observations point clearly to the interdependence of the membrane-binding bundle, of Lys178 ubiquitination, and of phospholipase activity in mediating the cellular functionality of ExoU.
Discussion
The acute cytotoxic potential of ExoU-carrying P. aeruginosa strains is evidenced by the significant damage suffered by a variety of cell lineages and the toxin's ability to rapidly destroy lung tissue in vivo [24], [25]. The structural and functional characterization of this unique T3SS toxin, reported here, indicates how this multi-domain molecule can intertwine functionalities, and sheds light on its intracellular fate upon translocation.
In the bacterial cytosol and prior to translocation, ExoU is bound to SpcU [23], a small, acidic Type IA T3SS chaperone. Type IA chaperones are typically dimeric, and contact their cognate effectors mostly through the latter's N-terminus, which is ‘wrapped around’ the chaperone dimer [3], [45]–[48], [53]. Here, we identify the two key recognition sites between SpcU and ExoU as the β motif and a hydrophobic cavity at the center of the chaperone, both of which have been described for other Type IA chaperone∶effector complexes, such as InvB∶SipA from Salmonella [46], SycE∶YopE (23–78) from Yersinia [47], and SicP∶SptP (35–139) from Salmonella [48]. In all of these structures, the chaperone-binding domain (CBD) is localized within the effector's N-terminal region. SpcU maintains the N-terminal residues of ExoU in a non-globular conformation (albeit still maintaining the structure of β1). This could allow ExoU to be ready for secretion and rapid unfolding upon recognition by the base of the T3SS, as suggested for other systems [5], [54]. This hypothesis is also supported by mutagenesis studies performed on the β motifs of SicP∶SptP and SycE∶YopE, which reveal that modification of a few strategic residues suffices for total disruption of chaperone∶effector interactions [46].
The structure of the phospholipase domain of ExoU reveals that the catalytic Ser142 is not exposed on the surface of the molecule, but is located within a cleft surrounded by flexible regions. This suggests that the active site does not have unobstructed access to its phospholipidic substrates. Local conformational changes could thus be required to occur upon membrane targeting, leading to active site exposure, which could constitute a potential mechanism of protection of membranes (bacterial or host) that are not originally intended for targeting. Notably, the localization of a phospholipase active site serine residue within a funnel-like region protected by a flexible lid carrying the catalytic aspartate has been observed in the structure of the eukaryotic enzyme cPLA2 [51], which undergoes interfacial activation upon binding to a membranous interface [55], [56]. In cPLA2, closure of the lid region is necessary not only for formation of a ‘complete’ active site, but also for masking of a large hydrophobic region that surrounds the catalytic serine [51]. In ExoU, the localization of Asp344, the second member of the catalytic dyad, in a flexible region in close proximity to Ser142, provides additional supportive evidence for the requirement for local conformational changes in the vicinity of Ser142 in order to place Asp344 in position for catalysis. This suggests that the structure presented here is in an ‘open’, inactive conformation, and local conformational changes are required to ‘close’ the flexible loops and define the placement of Asp344.
How could such conformational modifications be initiated? The data presented here, including our structural analysis and phospholipid binding studies, together with mutagenesis results, spectroscopic studies, and cofactor identification experiments performed by other laboratories [31]–[33], [37], [38], [40]–[44] provide a working hypothesis. ExoU binds specifically to PI(4,5)P2, the major phosphoinositide of the eukaryotic plasma membrane, and the C-terminal α-helical bundle plays a key role in this association (Figure 3c). Notably, this C-terminal region is of central importance for ExoU's phospholipase activity [31], [38]–[40], an observation that reveals the functional interdependence of membrane-binding and catalytic domains. Thus, upon translocation of ExoU into target cells and PM recognition by the C-terminal domain, ExoU could undergo a conformational change that allows partial insertion of the nearby catalytic domain, and thus the active site, into the bilayer. The proximity of the substrate, and the interaction with a eukaryotic cofactor, could suffice to locally modify the flexible regions that surround the active site region, allowing the catalytic serine to have access to phospholipids. The distance between the C-terminus of ExoU and the active site is of approximately 25 Å, and the lack of many contact points between these two domains suggests that some level of rotation could occur between them in order to place the active site in an optimal position. This suggestion is also in agreement with recent, elegant studies by Benson et al., who used spin-labeling electron paramagnetic resonance spectroscopy to identify conformational changes in the C-terminus domain and within the active site of ExoU, in the presence of both liposomes and of a potential eukaryotic cofactor [43]. Strikingly, Benson et al. also identified that in the absence of cofactor or lipids, ExoU displays multiple conformations, which is in particular agreement with the considerable flexibility observed in the region of the active site in our apo structure. In another recent study, ubiquitin and ubiquitinated proteins were also identified as cofactors, and potential activating agents, for ExoU [44]. Considering that many ubiquitination processes occur in the vicinity of the PM, these observations are also in agreement with the model discussed above.
ExoU has been reported to be ubiquitinated by the host machinery on its Lys178 upon translocation and binding to the PM [39]. Our structure reveals that Lys178 is located within the catalytic domain, on the same side of the molecule as both the active site and the membrane-binding domain (Figures 3a and 3b). Following the model described above, the partial imbedding of the catalytic domain of ExoU into the bilayer (upon recognition of the PM through the C-terminal region and binding to cofactor(s)) would place Lys178 in close proximity to ubiquitin ligase complexes located in the vicinity [57], allowing for rapid ubiquitination. Notably, we show that ubiquitinated ExoU is observed at the level of the organelles of the endo-lysosomal pathway, both by transfection studies using fluorescence complementation, and confocal microscopy analyses of P. aeruginosa-infected eukaryotic cells. It has been previously shown that a P. aeruginosa strain carrying the non-ubiquitinated variant ExoU-K178R is as cytotoxic to eukaryotic cells as the wild type strain [39], suggesting that endosomal localization is not linked to ExoU's cytotoxic potential. Rather, the effects described here could represent an attempt by the eukaryotic cell to protect itself from ExoU's phospholipase action by targeting it to lysosomes, viewing its potential destruction. Given the rapid effect of ExoU on target cells, however, this protection mechanism does not seem to be efficient enough to offer protection from the toxin's ability to efficiently disrupt membranes, which is at the basis for the extremely aggressive nature of ExoU-expressing P. aeruginosa strains.
In summary, the structure of ExoU presented here reveals the toxin in its chaperone-bound, inactive, pre-secretion conformation. Since ExoU has broad substrate specificity, this conformation most likely prevents the toxin from binding to bacterial membranes, enabling the bacterium to be protected from ExoU's phospholipase action. Thus, our structural data provide a basis for the study of conformational changes that must accompany ExoU's activation upon binding to cellular factors, which in turn lead to phospholipase activity and cytotoxicity. Finally, the structure of the ExoU∶SpcU complex could serve as a basis for the development of ligands designed to stabilize this conformation, which in turn could prevent ExoU's secretion and thus diminish bacterial pathogenicity.
Methods and Materials
Protein expression, crystallization, and structure solution
ExoU (1–687) and SpcU (1–137) were first amplified from a clinical P. aeruginosa strain (GESPA 1999 collection). Both full-length forms of ExoU and SpcU were initially cloned into pETDueT-1, co-expressed in E. coli BL21/DE3, and expression was performed with 1 mM isopropyl 1-thio-β-D-thiogalactopyranoside for 3 h at 37°C. The complex was purified by nickel affinity chromatography (Ni-NTA, Qiagen) and gel filtration (HR 10/60 column, GE Healthcare) techniques. This strategy yielded thin crystals that diffracted poorly. Limited proteolysis experiments with thrombin identified that removal of the first 29 residues of ExoU and the last 10 residues of SpcU generated better quality crystals. Thus a thrombin site was introduced between residues 29 and 30 of ExoU, and a stop codon after residue 127 of SpcU, in the same pETDueT vector. The complex was expressed as described above. After Ni affinity chromatography and treatment with thrombin, ExoU∶SpcU was further purified a MonoQ column (GE Healthcare) in 50 mM Tris-HCl pH 8.8, 1 mM EDTA (elution gradient, 0–1 M NaCl). Samples for crystallization were gel filtered in 25 mM Tris-HCl pH 8.0, 100 mM NaCl, 1 mM EDTA. Crystals of the ExoU (29–687)∶SpcU (1–127) complex were obtained in 0.1 M KNO3, 20% PEG 3350. A SAD data set on the selenium edge was collected at the European Synchrotron Radiation Facility (ESRF) ID29 beamline (Grenoble, France) to a resolution of 2.94 Å. Data collection and refinement statistics are included in Table S1.
Diffraction images were indexed and scaled with XDS [58]. Initial selenomethionine sites were identified with PHENIX AutoSol [59]. Initial experimental density maps were improved after model building and phase extension using PHENIX. PARROT [60] and PIRATE [61]were used in density modification-phase improvement steps interspersed with cycles of automatic model-building using BUCCANEER [62]. The ExoU∶SpcU structure was completed by cycles of manual model-building using COOT 0.6.2 [63]. Water molecules were added to the residual electron density map using ARP/wARP 7.1.1 [64]. Cycles of restrained refinement were performed with REFMAC 5.6 [65] as implemented in the CCP4 program suite (Table S1). Stereochemical verification was performed by PROCHECK [66] and secondary structure assignment by DSSP [67]. Figures were generated with PyMol (www.pymol.org).
Mass spectrometry
Native mass spectrometry measurements of ExoU∶SpcU were carried out with a Micro-TOF-Q Bruker mass spectrometer (Wissembourg, France) with an electrospray ion source. Mass spectra were recorded in the 500–7000 mass-to-charge (m/z) range. Sample concentration was 15 µm in 20 mm ammonium acetate and continuously infused at a flow rate of 7 µl/min. Data were acquired in the positive mode and calibration was performed using a solution of 0.1 mg/ml CsI in water/isopropyl alcohol (1∶1, v/v). The system was controlled with the MicrOTOF Control software package and data were processed with DataAnalysis.
Construction of plasmids for transfection of HeLa cells
Plasmids encoding ExoU S142A-VC and VN-Ub originate from Addgene plasmids 22011 and 22010, initially provided by Dr. C.-D. Hu, Purdue Univ. ExoU was cloned N-terminally to the VC155 fragment, whereas Ub was cloned C-terminally to the VN173 fragment. The S142A mutation was inserted into the ExoU sequence via site-directed mutagenesis (Stratagene). To limit background fluorescence [68], we deleted specific regions of the Venus fragments to generate VN158-Ub and ExoU-S142A-VC159, and introduced the V150L mutation in the VN fragment, to obtain plasmids ExoU-S142A-VC and VN-Ub as described in [69]. We amplified Ub K63 from Addgene plasmid 17606 (donated by Dr T. Dawson), and cloned it in our modified pBiFC-VN plasmid. Insertion of the Lys63Arg, Gly75Lys and Gly76Leu substitutions led to the control plasmid VN-Ub Mut.
Construction of mutant ExoU-expressing P. aeruginosa strains
A 3-kb-long DNA fragment containing the promoter and the sequence of the exoUspcU operon from the strain of the GESPA collection [70] was cloned into the replicative pUCP20 plasmid. The S124A and K178R mutations, as well as the 679–683 deletion were introduced into the exou sequence using Quick Change Mutagenesis kit (Stratagene). The different plasmids were transferred by transformation [71] into the P. aeruginosa CHAΔSTlox strain kindly provided by Prof B. Polack.
Antibodies and probes
Antibodies used for immunofluorescence co-staining include an anti-HA from Covance (AFC-101P), an anti-FLAG from Euromedex (EL1B11), an anti-EEA1 from BD transduction (610457) and an anti-GORASP2 from Euromedex (10598-1-AP). The anti-ExoU antibodies were raised against His6-ExoU in rabbits by Eurogentec, as described by the manufacturer. Specific anti-ExoU antibodies were immunopurified. Lysotracker Red (Molecular probes) was added at 500 nM and incubated for 1 h at 37°C before fixation.
Mammalian cell transfections and immunofluorescence microscopy
One day before transfection, Hela cells were seeded on 12-mm coverslips. Transient transfections were performed using Jetprime (Polyplus transfection), following the manufacturer's instructions. For bifluorescence complementation assays, both plasmids were added in equal quantities. On the following day, cells were fixed with 2% paraformaldehyde and permeabilized with 0.2% TX-100. Primary antibodies were added for 30 min at 37°C, washed away and the Alexa-fluor 594 conjugated anti-mouse antibody (Molecular Probes) was incubated for 1 h at room temperature. Nuclei were counterstained with DAPI. Images were collected with an inverted Olympus IX81 epifluorescence motorized microscope equipped with a motorized piezo stage (Ludl Electronic Products, USA) and a Retiga-SRV CCD camera (QImaging) driven by VOLOCITY software (Improvision) with a binning of 1, using a PlanApo 60× NA1.42 objective (Olympus). Images were deconvoluted using the Volocity 5.5 software.
P. aeruginosa infection and analysis by confocal microscopy
Sparse HeLa cells were infected with bacteria in exponential growth at an MOI value of 10 and incubated for 3 h at 37°C in RPMI. At this point, approximately 10% of the total amount of ExoU is ubiquitinated. Subsequently, cells were washed and fixed in 4% paraformaldehyde and 0.5% Triton X-100 and labeled with anti-ExoU and anti-EEA1 antibodies. After incubation with secondary fluorescent antibodies, cells were observed with a confocal fluorescent microscope (Leica TCS SP2). Images were treated with the ImageJ software for 3D reconstruction.
Detection of protein-lipid interactions
Membrane lipid strips (Echelon Biosciences) were blocked in PBS+4% milk, and incubated overnight with proteins at 0.5 µg/mL. Detection of bound proteins was performed as recommended by the manufacturer.
Supporting Information
Zdroje
1. WorrallLJLameignereEStrynadkaNC 2011 Structural overview of the bacteria injectisome. Curr Opin Microbiol 14 3 8
2. CornelisGR 2006 The type III secretion injectisome. Nat Rev Microbiol 4 811 825
3. IzoréTJobVDessenA 2011 Biogenesis, regulation, and targeting of the type III secretion system. Structure 19 603 612
4. HayesCSAokiSKLowDA 2010 Bacterial contact-dependent delivery systems. Annu Rev Genet 44 71 90
5. GalánJEWolf-WatzH 2006 Protein delivery into eukaryotic cells by type III secretion machines. Nature 444 567 573
6. MarlovitsTCStebbinsCE 2009 Type III secretion systems shape up as they ship out. Curr Opin Microbiol 13 1 6
7. BlockerAKomoriyaKAizawaS 2003 Type III secretion systems and bacterial flagella: insights into their function from structural similarities. Proc Natl Acad Sci USA 100 3027 3030
8. GalánJE 2009 Common themes in the design and function of bacterial effectors. Cell Host Microbe 5 571 579
9. El SolhAAAkinnusiMEWiener-KronishJPLynchSVPinedaLA 2008 Persistent infection with Pseudomonas aeruginosa in ventilator-associated pneumonia. Am J Respir Crit Care Med 178 513 519
10. HauserARCobbEBodiMMariscalDVallesJ 2002 Type III protein secretion is associated with poor clinical outcomes in patients with ventilator-associated pneumonia caused by Pseudomonas aeruginosa. Crit Care Med 30 521 528
11. EngelJBalachandranP 2009 Role of Pseudomonas aeruginosa type III effectors in disease. Curr Opin Microbiol 12 61 66
12. HauserAR 2009 The type III secretion system of Pseudomonas aeruginosa: infection by injection. Nat Rev Microbiol 7 654 665
13. FrankDW 1997 The enxoenzyme S regulon of Pseudomonas aeruginosa. Mol Microbiol 26 621 629
14. DengQBarbieriJT 2008 Modulation of host cell endocytosis by the type III cytotoxin, Pseudomonas ExoS. Traffic 9 1948 1957
15. Garrity-RyanLShafikhaniSBalachandranPNguyenLOzaJ 2004 The ADP ribosyltransferase domain of Pseudomonas aeruginosa ExoT contributes to its biological activities. Infect Immun 72 546 558
16. ShafikhaniSHMoralesCEngelJ 2008 The Pseudomonas aeruginosa type III secreted toxin ExoT is necessary and sufficient to induce apoptosis in epithelial cells. Cell Microbiol 10 994 1007
17. PielageJFPowellKRKalmanDEngelJN 2008 RNAi screen reveals an Abl kinase-dependent host cell pathway involved in Pseudomonas aeruginosa internalization. PLoS Pathog 4 e1000031
18. ShaverCMHauserAR 2004 Relative contributions of Pseudomonas aeruginosa ExoU, ExoS, and ExoT to virulence in the lung. Infect Immun 72 6969 6977
19. Frithz-LindstenEDuYRosqvistRForsbergA 1997 Intracellular targeting of exoenzyme S of Pseudomonas aeruginosa via type III-dependent translocation induces phagocytosis resistance, cytotoxicity and disruption of actin microfilaments. Mol Microbiol 25 1125 1139
20. CowellBAEvansDJFleiszigSM 2005 Actin cytoskeleton disruption by ExoY and its effects on Pseudomonas aeruginosa invasion. FEMS Microbiol Lett 250 71 76
21. YahrTLVallisAJHancockMKBarbieriJTFrankDW 1998 ExoY, an adenylate cyclase secreted by the Pseudomonas aeruginosa type III system. Proc Natl Acad Sci U S A 95 13899 13904
22. HritonenkoVMunJJTamCSimonNCBarbieriJT 2011 Adenylate cyclase activity of Pseudomonas aeruginosa ExoY can mediate bleb-niche formation in epithelial cells and contributes to virulence. Microb Pathog 51 305 312
23. Finck-BarbançonVYahrTLFrankDW 1998 Identification and characterization of SpcU, a chaperone required for efficient secretion of the ExoU cytotoxin. J Bacteriol 180 6224 6231
24. SatoHFrankDW 2004 ExoU is a potent intracellular phospholipase. Mol Microbiol 53 1279 1290
25. Finck-BarbançonVGoransonJZhuLSawaTWiener-KronishJP 1997 ExoU expression by Pseudomonas aeruginosa correlates with acute cytotoxicity and epithelial injury. Mol Microbiol 25 547 557
26. AlleweltMColemanFTGroutMPriebeGPPierGB 2000 Acquisition of expression of the Pseudomonas aeruginosa ExoU cytotoxin leads to increased bacterial virulence in a murine model of acute pneumoniae and systemic spread. Infect Immun 68 3998 4004
27. MachadoGBde AssisMCLeãoRSalibaAMSilvaMC 2010 ExoU-induced vascular hyperpermeability and platelet activation in the course of experimental Pseudomonas aeruginosa pneumosepsis. Shock 33 315 321
28. KurahashiKKajikawaOSawaTOharaMGropperM 1999 Pathogenesis of septic shock in Pseudomonas aeruginosa pneumonia. J Clin Invest 104 743 750
29. PankhaniyaRRTamuraMAllmondLRMoriyamaKAjayiT 2004 Pseudomonas aeruginosa causes acute lung injury via the catalytic activity of the patatin-like phospholipase domain of ExoU. Crit Care Med 32 2293 2299
30. Roy-BurmanASavelRHRacineSSwansonBLRevadigarNS 2001 Type III protein secretion is associated with death in lower respiratory and systemic Pseudomonas aeruginosa infections. J Infect Dis 183 1767 1774
31. RabinSDPHauserAR 2005 Functional regions of the Pseudomonas aeruginosa cytotoxin ExoU. Infect Immun 73 573 582
32. PhillipsRMSixDADennisEAGhoshP 2003 In vivo phospholipase activity of the Pseudomonas aeruginosa cytotoxin ExoU and protection of mammalian cells with phospholipase A2 inhibitors. J Biol Chem 278 41326 41332
33. SatoHFrankDWHillardCJFeixJBPankhaniyaRR 2003 The mechanism of action of the Pseudomonas aeruginosa-encoded type III cytotoxin, ExoU. EMBO J 22 2959 2969
34. MurakamiMTaketomiYSatoHYamamotoK 2011 Secreted phospholipase A2 revisited. J Biochem 150 233 255
35. GhoshMTuckerDEBurchettSALeslieCC 2006 Properties of the group IV phospholipase A2 family. Prog Lipid Res 45 487 510
36. BurkeJEDennisEA 2009 Phospholipase A2 structure/function, mechanism, and signaling. J Lipid Res 50 S237 S242
37. RabinSDPVeesenmeyerJLBiegingKTHauserAR 2006 A C-terminal domain targets the Pseudomonas aeruginosa cytotoxin ExoU to the plasma membrane of host cells. Infect Immun 74 2552 2561
38. VeesenmeyerJLHowellHHalavatyASAhrensSAndersonWF 2010 Role of the membrane localization domain of the Pseudomonas aeruginosa effector protein ExoU in cytotoxicity. Infect Immun 78 3346 3357
39. StirlingFRCuzickAKellySMOxleyDEvansTJE 2006 Eukaryotic localization, activation and ubiquitinylation of a bacterial type III secreted toxin. Cell Microbiol 8 1294 1309
40. SchmalzerKBensonMAFrankDW 2010 Activation of ExoU phospholipase activity requires specific C-terminal regions. J Bacteriol 192 1801 1812
41. SatoHFeixJBHillardCJ 2005 Characterization of phospholipase activity of the Pseudomonas aeruginosa type III cytotoxin, ExoU. J Bacteriol 187 1192 1195
42. SatoHFeixJBFrankDW 2006 Identification of superoxide dismutase as a cofactor for the Pseudomonas type III toxin, ExoU. Biochemistry 45 10368 10375
43. BensonMAKomasSMSchmalzerKCaseyMSFrankDW 2011 Induced conformational changes in the activation of the Pseudomonas aeruginosa type III toxin, ExoU. Biophys J 100 1335 1343
44. AndersonDMSchmalzerKMSatoHCaseyMTerhuneSS 2011 Ubiquitin and ubiquitin-modified proteins activate the Pseudomonas aeruginosa T3SS cytotoxin, ExoU. Mol Microbiol 82 1452 67
45. ParsotCHamiauxCPageA-L 2003 The various and varying roles of specific chaperones in type III secretion systems. Curr Opin Microbiol 6 7 14
46. LilicMVujanacMStebbinsCE 2006 A common structural motif in the binding of virulence factors to bacterial secretion chaperones. Mol Cell 21 653 664
47. BirtalanSCPhillipsRMGhoshP 2002 Three-dimensional secretional signals in chaperone-effector complexes of bacterial pathogens. Mol Cell 9 971 980
48. StebbinsCEGalánJE 2001 Maintenance of an unfolded polypeptide by a cognate chaperone in bacterial type III secretion. Nature 414 77 81
49. PhanJTropeaJEWaughDS 2004 Structure of the Yersinia pestis type III secretion chaperone SycH in complex with a stable fragment of YscM2. Acta Cryst Sect D 60 1591 1599
50. Finck-BarbançonVFrankDW 2001 Multiple domains are required for the toxic activity of Pseudomonas aeruginosa ExoU. J Bacteriol 183 4330 4344
51. DessenATangJSchmidtHStahlMClarkJD 1999 Crystal structure of human cytosolic phospholipase A2 reveals a novel topology and catalytic mechanism. Cell 97 349 360
52. KerppolaTK 2009 Visualization of molecular interactions using bimolecular fluorescence complementation analysis: characteristics of protein fragment complementation. Chem Soc Rev 38 2876 2886
53. LuoYBerteroMGFreyEAPfuetznerRAWenkMR 2001 Structural and biochemical characterization of the type III secretion chaperones CesT and SigE. Nat Struct Biol 8 1031 1036
54. AkedaYGalánJE 2005 Chaperone release and unfolding of substrates in type III secretion. Nature 437 911 915
55. JainMKBergOG 2006 Coupling of the i-face and the active site of phospholipase A2 for interfacial activation. Curr Opin Chem Biol 10 473 479
56. DennisEACaoJHsuY-HMagriotiVKokotosG 2011 Phospholipase A2 enzymes: physical structure, biological function, disease implication, chemical inhibition, and therapeutic intervention. Chem Rev 111 6130 6185
57. PiperRCLehnerPJ 2011 Endosomal transport via ubiquitination. Trends Cell Biol 21 647 655
58. KabschW 1993 Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Cryst 26 795 800
59. AdamsPDAfoninePVBunkocziGChenVBDavisIW 2010 PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D 66 213 221
60. CowtanK 2010 Recent developments in classical density modification. Acta Crystallogr D 66 470 478
61. CowtanK 2000 General quadratic functions in real and reciprocal space and their application to likelihood phasing. Acta Crystallogr D 56 1612 1621
62. CowtanK 2006 The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr D 62 1002 1111
63. EmsleyPCowtanK 2004 Coot: model-building tools for molecular graphics. Acta Crystallogr sect D 60 2126 2132
64. PerrakisAMorrisRMLamzinVS 1999 Automated protein model building combined with iterative structure refinement. Nat Struct Biol 6 458 463
65. MurshudovGVaginADodsonE 1997 Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr sect D 53 240 255
66. LaskowskiRAMacArthurMWMossDSThorntonJM 1993 PROCHECK: a program to check the stereo chemical quality of protein structures. J Appl Crystallog 26 283 291
67. KabschWSanderC 1983 Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22 2577 2637
68. ShyuYJHuCD 2008 Fluorescence complementation: an emerging tool for biological research. Trends Biotechnol 26 622 630
69. LinJWangNLiYTianSZhaoL 2011 LEC-BIFC: a new method for rapid assay of protein interaction. Biotech Histochem 86 272 279
70. BerthelotPAttreeIPlesiatPChabertJde BentzmannS 2003 Genotypic and phenotypic analysis of type III secretion system in a cohort of Pseudomonas aeruginosa bacteremia isolates: evidence for a possible association between O serotypes and exo genes. J Infect Dis 188 512 518
71. ChuanchuenRNarasakiCTSchweizerHP 2002 Benchtop and microcentrifuge preparation of Pseudomonas aeruginosa competent cells. Biotechniques 33 762 763
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Fungal Biofilms
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2012 Číslo 4- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Systematic Review of Mucosal Immunity Induced by Oral and Inactivated Poliovirus Vaccines against Virus Shedding following Oral Poliovirus Challenge
- The Arbuscular Mycorrhizal Symbiosis: Origin and Evolution of a Beneficial Plant Infection
- Modelling the Evolutionary Dynamics of Viruses within Their Hosts: A Case Study Using High-Throughput Sequencing
- Structural Basis of Cytotoxicity Mediated by the Type III Secretion Toxin ExoU from
- Fungal Biofilms
- The Problem of Auto-Correlation in Parasitology
- T Regulatory Cells Control Susceptibility to Invasive Pneumococcal Pneumonia in Mice
- Small-Molecule Inhibitors of Dengue-Virus Entry
- The Baculovirus Uses a Captured Host Phosphatase to Induce Enhanced Locomotory Activity in Host Caterpillars
- Matrix Metalloprotease 9 Mediates Neutrophil Migration into the Airways in Response to Influenza Virus-Induced Toll-Like Receptor Signaling
- Necrotrophism Is a Quorum-Sensing-Regulated Lifestyle in
- Modeling of the N-Glycosylated Transferrin Receptor Suggests How Transferrin Binding Can Occur within the Surface Coat of
- The Role of Cofactors in Prion Propagation and Infectivity
- The Role of Mast Cells in the Defence against Pathogens
- The Accessory Genome as a Cradle for Adaptive Evolution in Pathogens
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The Accessory Genome as a Cradle for Adaptive Evolution in Pathogens
- Systematic Review of Mucosal Immunity Induced by Oral and Inactivated Poliovirus Vaccines against Virus Shedding following Oral Poliovirus Challenge
- The Arbuscular Mycorrhizal Symbiosis: Origin and Evolution of a Beneficial Plant Infection
- Modelling the Evolutionary Dynamics of Viruses within Their Hosts: A Case Study Using High-Throughput Sequencing
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



