-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaNecrotrophism Is a Quorum-Sensing-Regulated Lifestyle in
How pathogenic bacteria infect and kill their host is currently widely investigated. In comparison, the fate of pathogens after the death of their host receives less attention. We studied Bacillus thuringiensis (Bt) infection of an insect host, and show that NprR, a quorum sensor, is active after death of the insect and allows Bt to survive in the cadavers as vegetative cells. Transcriptomic analysis revealed that NprR regulates at least 41 genes, including many encoding degradative enzymes or proteins involved in the synthesis of a nonribosomal peptide named kurstakin. These degradative enzymes are essential in vitro to degrade several substrates and are specifically expressed after host death suggesting that Bt has an active necrotrophic lifestyle in the cadaver. We show that kurstakin is essential for Bt survival during necrotrophic development. It is required for swarming mobility and biofilm formation, presumably through a pore forming activity. A nprR deficient mutant does not develop necrotrophically and does not sporulate efficiently in the cadaver. We report that necrotrophism is a highly regulated mechanism essential for the Bt infectious cycle, contributing to spore spreading.
Published in the journal: . PLoS Pathog 8(4): e32767. doi:10.1371/journal.ppat.1002629
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1002629Summary
How pathogenic bacteria infect and kill their host is currently widely investigated. In comparison, the fate of pathogens after the death of their host receives less attention. We studied Bacillus thuringiensis (Bt) infection of an insect host, and show that NprR, a quorum sensor, is active after death of the insect and allows Bt to survive in the cadavers as vegetative cells. Transcriptomic analysis revealed that NprR regulates at least 41 genes, including many encoding degradative enzymes or proteins involved in the synthesis of a nonribosomal peptide named kurstakin. These degradative enzymes are essential in vitro to degrade several substrates and are specifically expressed after host death suggesting that Bt has an active necrotrophic lifestyle in the cadaver. We show that kurstakin is essential for Bt survival during necrotrophic development. It is required for swarming mobility and biofilm formation, presumably through a pore forming activity. A nprR deficient mutant does not develop necrotrophically and does not sporulate efficiently in the cadaver. We report that necrotrophism is a highly regulated mechanism essential for the Bt infectious cycle, contributing to spore spreading.
Introduction
Saprophytism, probably one of the most common lifestyle for micro-organisms, involves living in dead or decaying organic matter. For most pathogens, saprophytism is limited to necrotrophism (the use of the host cadaver). This step of the infection process is essential for the proliferation and horizontal transmission of these microorganisms (transfer of infection within a single generation) [1]. However, there have been very few studies addressing this major issue. The transition from a pathogenic to a necrotrophic lifestyle implies substantial metabolic changes for microorganisms [2]. The death of the host is a critical event which compels the micro-organisms to cope with a new series of challenges: competition with the commensal organisms and opportunistic incomers, stress, and nutrient deficiencies. Therefore, necrotrophism is likely to be highly regulated.
The insect pathogen Bacillus thuringiensis (Bt) is a suitable model for studying the time course of the infection process, including necrotrophism in the insect cadaver. Bt is an ubiquitous spore-forming bacterium belonging to the Bacillus cereus (Bc) group [3]. Its spores are found in a large variety of environments, such as soils, dead and living insects and plant phylloplane [4]. However, Bt probably does not grow in soil and reports of natural epizootic episodes are very rare [5], [6]. Unlike soil bacteria, such as Streptomyces spp and B. subtilis, Bc group genomes contain a large number of genes involved in nitrogen metabolism [3]. It is therefore likely that Bt multiplies in the host cadaver [1], [6].
Bt carries plasmids encoding specific insecticidal toxins responsible for their insecticidal properties [7]. Bt spores and toxins are ingested by larvae, and the toxins bind to specific receptors on the midgut epithelial cells, inducing cell lysis and creating favorable conditions for the development of the bacteria [8]. The vegetative bacteria multiply in the insect hemocoel and cause septicemia [1], [9]. Bt also harbors genes encoding exported virulence factors including enterotoxins, hemolysins, phospholipases and proteases [10]. The transcription of most of these virulence genes in bacteria growing in a rich medium is activated at the onset of stationary phase by the quorum-sensing system PlcR-PapR [11], [12]. PlcR-regulated factors account for about 80% of the extracellular proteome of Bt during early stationary phase in rich medium [13]. In sharp contrast, the expression of the PlcR-regulated genes is repressed when the bacteria enter sporulation [14] and the stationary phase secretome of Bt and B. anthracis (Ba) growing in a sporulation medium is mainly composed of the metalloprotease NprA [15], [16]. NprA (also designated NprB and Npr599 in Ba) cleaves tissue components such as fibronectin, laminin and collagen, thus displaying characteristics of pathogenic factors [17]. Transcription of nprA is activated during the late stationary phase by the regulator NprR [16]. NprR is a quorum sensor activated by its cognate signaling peptide, NprX. NprR-NprX functions as a typical Gram-positive quorum-sensing system: the pro-signaling peptide NprX is exported from the cell, and after being processed to its active form is reimported, and binds to NprR allowing the recognition of its DNA target and the activation of nprA transcription [16].
The first stages of Bt infection are relatively well documented, but the fate of the bacteria after death of the host remains unclear. Here, we report evidence that the necrotrophic lifestyle of Bt is a specific and highly regulated process. The quorum-sensing system NprR-NprX controls at least 41 genes some of which are required for Bt to survive in the insect cadaver and to complete its development in vivo ending with the production of spores.
Results
NprR is activated after the host death
We tested whether NprR, the activator of nprA transcription [16], is involved in the pathogenicity of Bt. The LD50 s of the Bt 407 Cry− (wt) strain and of the nprR-deficient (ΔRX) strain in the insect model Galleria mellonella were measured in two ways: by feeding larvae with spores mixed with the insecticidal toxin Cry1C and by injection of vegetative bacteria into the insect hemocoel (Table S1). The LD50 s of two strains did not differ significantly in either of the two conditions indicating that NprR was not required for pathogenicity. Consistently, an nprA-deficient strain was similarly found not to be affected in pathogenicity (not shown).
We investigated the involvement of NprR in the infection process by comparing, in vivo, the expression kinetics of nprA with that of the protease gene mpbE, reflecting the transcriptional activities of NprR and PlcR, respectively [16], [18]. The reporter strains grew similarly in insect larvae and a constitutively expressed PaphA3-lacZ fusion was used as the reference standard (Figure 1A and Figure S1A). Transcription of mpbE increased between 0 h and 24 h after injection and gradually decreased thereafter. In contrast, nprA transcription was low between 0 h and 24 h, increased between 24 h and 48 h and then decreased sharply (Figure 1B). Thus, NprR is active later in the infection process than PlcR, and after the death of the host.
Fig. 1. mpbE and nprA are sequentially activated during infection. 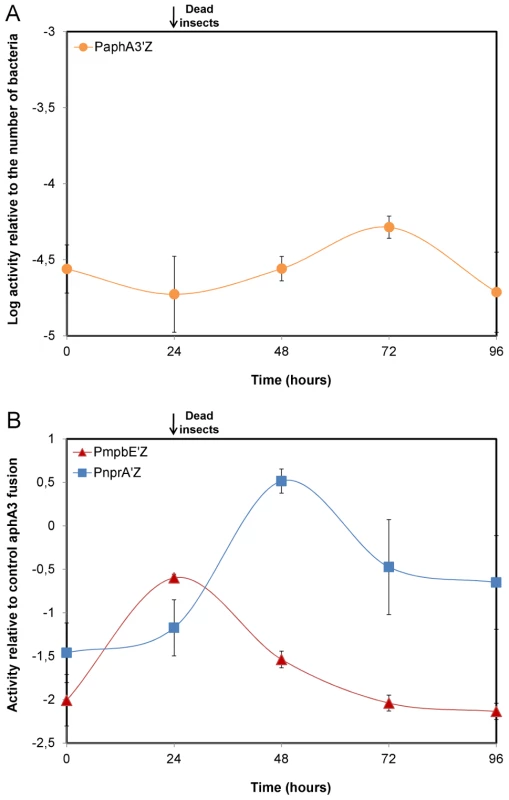
(A) Correlation between the β-galactosidase activity obtained with the PaphA3-lacZ fusion and the number of bacteria of the reporter strain 407 papha3′Z in infected insects. The activity of the aphA3 promoter remains constant over time in insects such that the PaphA3-lacZ fusion can be used as an in vivo reference standard. (B) Expression of the mpbE and nprA genes in vivo. Each point is the log-transformed ratio of the β-galactosidase activity obtained with the 407 pnprA′Z and 407 pmpbE′Z strains to that with the control strain 407 paphA3′Z. The black arrow indicates that assays were carried out with dead insects from 24 h post infection until the end of the experiment. Data are averages of at least three independent experiments (error bars are SEM from mean values). NprR allows Bt to survive in insect cadavers by a process independent of sporulation
To investigate the role of NprR during the late stage of infection, we compared the growth of the wt and ΔRX strains in insect larvae (Figure 2A). The total population of the two strains increased between 0 h and 24 h to reach about 1×108 cfu/mL. From 24 h to 96 h, the population of the wt strain remained stable, whereas the population of the ΔRX strain decreased sharply: 96 h post infection, the total population of the ΔRX strain was 6-log lower than that of the wt strain. Complementation of the ΔRX strain by pHT304-RX restored the wt phenotype. These findings indicate that NprR substantially improves the survival of Bt in insect cadavers.
Fig. 2. NprR allows Bt to survive in insect cadavers by a process independent of sporulation. 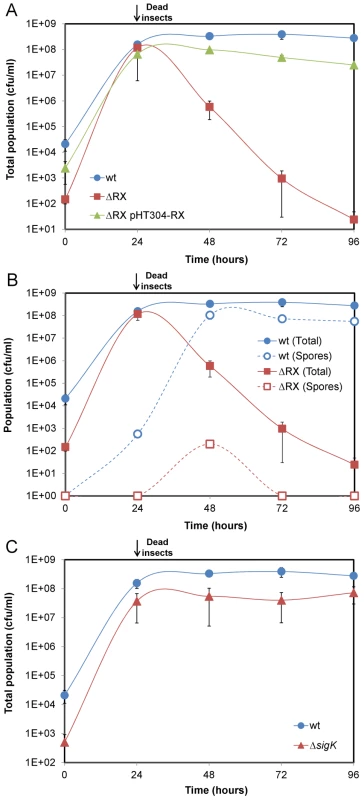
(A) Comparison of the wt and ΔRX strain survival in the insect host. The survival defect of the mutant is genetically complemented by pHT304-RX. (B) Effect of the nprR-nprX deletion on sporulation. (C) Comparison of the wt and ΔsigK strain survival in the insect host. Data are averages of at least four independent experiments (error bars are SEM from mean values). In sporulating microorganisms, sporulation is generally regarded as the key process ensuring survival in unfavorable conditions. We therefore investigated i) whether NprR was involved in the sporulation process of Bt in the insect cadaver, and ii) whether sporulation is responsible for the survival of the bacteria in the insect cadaver. We compared the sporulation efficiencies of the wt and ΔRX strains in both LB and sporulation-specific medium (HCT) (Table S2). In HCT, the sporulation efficiencies of the two strains were similar. However, in LB medium, the total number of viable spores of the ΔRX strain was half that for the wt strain (8.30×107 vs 1.58×108), suggesting that NprR is involved in the sporulation of Bt in rich medium. Next, we monitored the counts of wt and ΔRX strain spores in insect larvae over 96 h (Figure 2B and Table S2). For the wt strain, heat-resistant spores were detected 24 h after injection and their number increased until 48 h. From 48 h to 96 h, the number of spores remained stable and represented one third of the total bacterial population. The large number of non sporulated bacteria 96 h after the death of the insect suggests that sporulation was not the main mechanism allowing Bt to survive. For the ΔRX strain, less than one percent of the bacterial population was heat-resistant spores throughout the infection process. The decrease in the number of heat-resistant spores from 48 h to 72 h is likely due to the germination of the spores. We suspected that the low number of spores is not a cause but a consequence of the inability of the ΔRX strain to survive in the insect cadaver. To test this idea, we tested the survival of a sigK-deficient strain (Figure 2C): SigK is a sigma factor involved in the transcription of late sporulation genes in the mother cell, and sigK-deficient strains are not able to form viable spores [19], [20]. The total population of the sigK strain in the insect cadaver was similar to the total population of the wt strain, indicating that NprR ensures the survival of Bt by a process independent of sporulation.
NprR is a pleiotropic regulator involved in the necrotrophic development of Bt in the insect cadaver
The only gene described as being controlled by NprR was nprA. Therefore, we monitored the survival of a ΔnprA strain in infected larvae (Figure S2). The survival of the wt and ΔnprA strains was similar throughout the experiment, suggesting that other NprR-regulated genes are involved in bacterial survival. Microarray analysis was used to identify other NprR-regulated genes. Gene expression ratios between the wt and the ΔRX strains were determined 3 h after the onset of stationary phase (t3), when nprA transcription increases sharply [16]. For 107 genes, this expression ratio was greater than 2 (p<0.05) (http://www.ebi.ac.uk/arrayexpress/experiments/E-TABM-790), suggesting that NprR has a direct or indirect effect on their transcription. Thirty-nine genes, with a relative expression ratio greater than 4, and a significance value (p) smaller than 0.01, were considered for subsequent analysis. The genes matching probes for BC2622, a macrolide glycosyltransferase, and BC3725, an exochitinase, were also investigated due to their functional similarity to the genes fulfilling these criteria. Quantitative RT-PCR confirmed that these 41 genes were at least four times up - or downregulated. Fusions to lacZ were constructed for nine of these genes and used to confirm that they are differentially regulated in the ΔRX mutant and wt strains (Figure S3). The expression kinetics of these genes were similar, with a sharp increase of expression after the onset of stationary phase. The final list of NprR-regulated genes is presented in Table S3. Of the 41 genes directly or indirectly regulated by NprR, 37 were down-regulated in the ΔRX strain, suggesting that NprR primarily acts as a transcriptional activator. The subcellular localizations of the products of these genes were assessed: 46% were cytoplasmic, 20% were associated with the membrane and 34% were extracellular or associated with the cell wall. The NprR-regulated genes can be distributed into four functional groups. The first group is composed of genes encoding proteins potentially involved in stress resistance: they include the genes for cytochrome P450 (BC2613), cysteine dioxygenase (BC2617), and Transporter Drug/Metabolite exporter family members (BC1063). The second group is a four-gene locus encoding the oligopeptide permease system Opp required for the import of small peptides into the cell. The third group is a five-gene locus encoding a nonribosomal peptide synthesis (NRPS) system showing similarities with the systems involved in the synthesis of secreted factors like toxins and antibiotics. The last group codes for degradative enzymes (metalloproteases, esterases and chitinases) and for proteins which can bind organic material (chitin-binding protein and collagen adhesion protein). The role of NprR in the degradation of lipids, proteins and chitin was analyzed by growing the ΔRX and wt strains on specific culture media (Figure 3A). The lipolytic, proteolytic and chitinolytic activities of the ΔRX strain were significantly lower than those of the wt strain. We monitored the expression kinetics in infected insect larvae of two NprR-regulated genes encoding degradative enzymes (BC0429 and BC2167) (Figure 3B). The two genes were specifically expressed after the insect death, from 24 h to 96 h, suggesting that Bt displays a necrotrophic lifestyle in the insect cadaver.
Fig. 3. NprR is involved in the necrotrophic development of Bt in the insect cadaver. 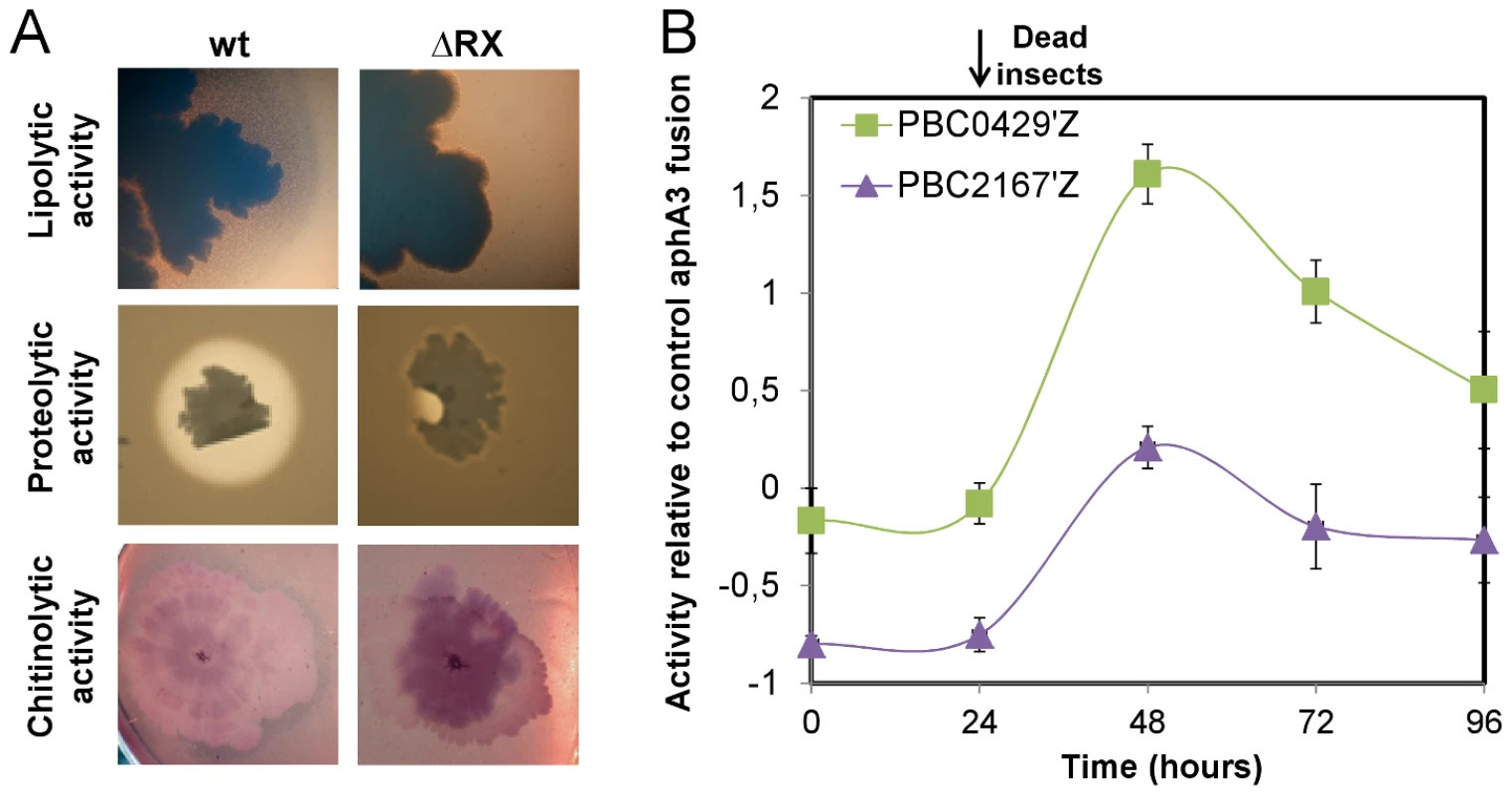
(A) Growth of the wt and ΔRX strains on specific culture media. The lipolytic activity was assayed on TS medium supplemented with Tween 80. Wild-type strain colonies were surrounded by a precipitate of oleic acid (a Tween 80 degradation product) whereas no precipitate was detected around the mutant strain colonies. The proteolytic and chitinolytic activities were assayed on HCT medium supplemented with cow's milk and on chitin medium, respectively. In both conditions, the wt strain was surrounded by a degradation ring whereas no such rings were detected for the mutant. (B) The genes BC0429 and BC2167 were specifically expressed after the insect death. Each point is the log-transformed ratio of the β-galactosidase activity obtained with the 407 pBC0429'Z and 407 pBC2167'Z strains to that with the control strain 407 paphaA3'Z. The growth kinetics of both reporter strains are presented in Fig. S1B. Data are averages of at least three independent experiments (error bars are SEM from mean values). A secreted factor regulated by NprR allows Bt to survive in insect cadavers
To identify the NprR-dependent survival factor we first tested whether this putative factor was secreted. Insect larvae were co-infected with two different ratios of wt and ΔRX strains: 90% of wt bacteria with 10% of ΔRX bacteria (90∶10), and 10% of wt bacteria with 90% of ΔRX bacteria (10∶90) (Figure 4A and Figure 4B). In insects infected with the ratio 10∶90, the total population of the wt and the ΔRX strains decreased after 24 h. This may result from the ΔRX strain capturing NprX without being able to express NprR-regulated genes, but nevertheless removing the peptide from the environment. Consequently, the amount of signaling peptide was insufficient to activate NprR-regulated genes in the wt, resulting in clearance of both populations. Co-infection with the ratio 90∶10 led to the survival of the two subpopulations during the 96 h of the experiment. In this condition, the concentration of NprX in the host was presumably sufficient to maintain the expression of the NprR-regulated genes in the wt subpopulation, and this expression allowed the survival of the ΔRX population. Therefore, the wt strain may produce a secreted factor that enables the ΔRX strain to survive in the insect cadaver.
Fig. 4. A NprR-regulated secreted factor allows Bt to survive in the insect host. 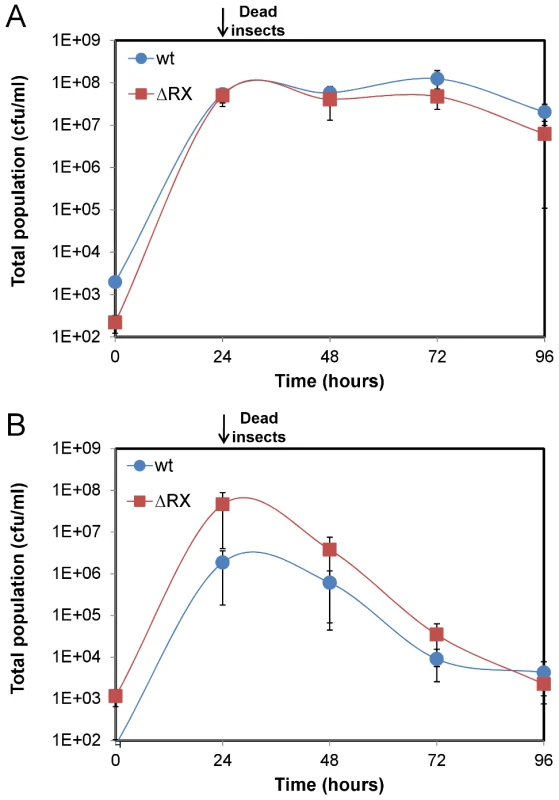
Insect larvae were co-infected with both wt and ΔRX strains mixed at two different ratios: (A) 90% wt bacteria with 10% ΔRX bacteria (90∶10) and (B) 10% wt bacteria with 90% ΔRX bacteria (10∶90). Data are averages of at least four independent experiments (error bars are SEM from mean values). The survival factor is a lipopeptide named kurstakin
NprR-dependent extracellular factors are degradative enzymes and the factor synthesized by the NRPS system. NprA, the major degradative enzyme produced during late stationary phase, is not required for bacterial survival in insect cadaver (Figure S2). The NRPS locus consists of seven open reading frames annotated BC2450 to BC2456 in the genome of the strain Bc ATCC 14579 used for designing the microarrays [3]. In silico analysis of all available sequenced Bt and Bc genomes, including that of strain Bt 407 used in this study, reveals that in all cases, this locus includes only four genes (http://www.ncbi.nlm.nih.gov/bioproject/29717). Several studies suggest that these four genes (designated krsA,B,C,E; Figure 5A) are involved in the production of the lipopeptide kurstakin [21], [22], [23]. KrsE is a presumed efflux protein and KrsA, B, C are the peptide synthetase subunits. The genes krsA, krsB and krsC were deleted from a wt strain and the survival of the mutant (ΔkrsABC) in insects was monitored for 96 h (Figure 5B). The total population of the ΔkrsABC strain declined from 2.107 cfu/ml at 24 h falling to 1.102 cfu/ml at 96 h. To test whether this effect was specifically dependent on the krsABC genes, we introduced a constitutive promoter upstream from these genes in the ΔRX strain. This NprR-independent expression of krsABC partially and significantly restored the survival of the ΔRX strain in the insect cadaver. These observations implicate the krsABC genes in the necrotrophic properties of Bt. We used MALDI-ToF-MS analysis to determine whether the krsABC genes are responsible for the production of kurstakin. Peaks characteristic of kurstakin were found for whole cells of the wt strain and not for those of the ΔkrsABC mutant (Figure 5C). This confirms that the krsABC genes are involved in kurstakin synthesis.
Fig. 5. The krsABC genes are involved in the production of kurstakin, a lipopeptide essential for the necrotrophic lifestyle of Bt. 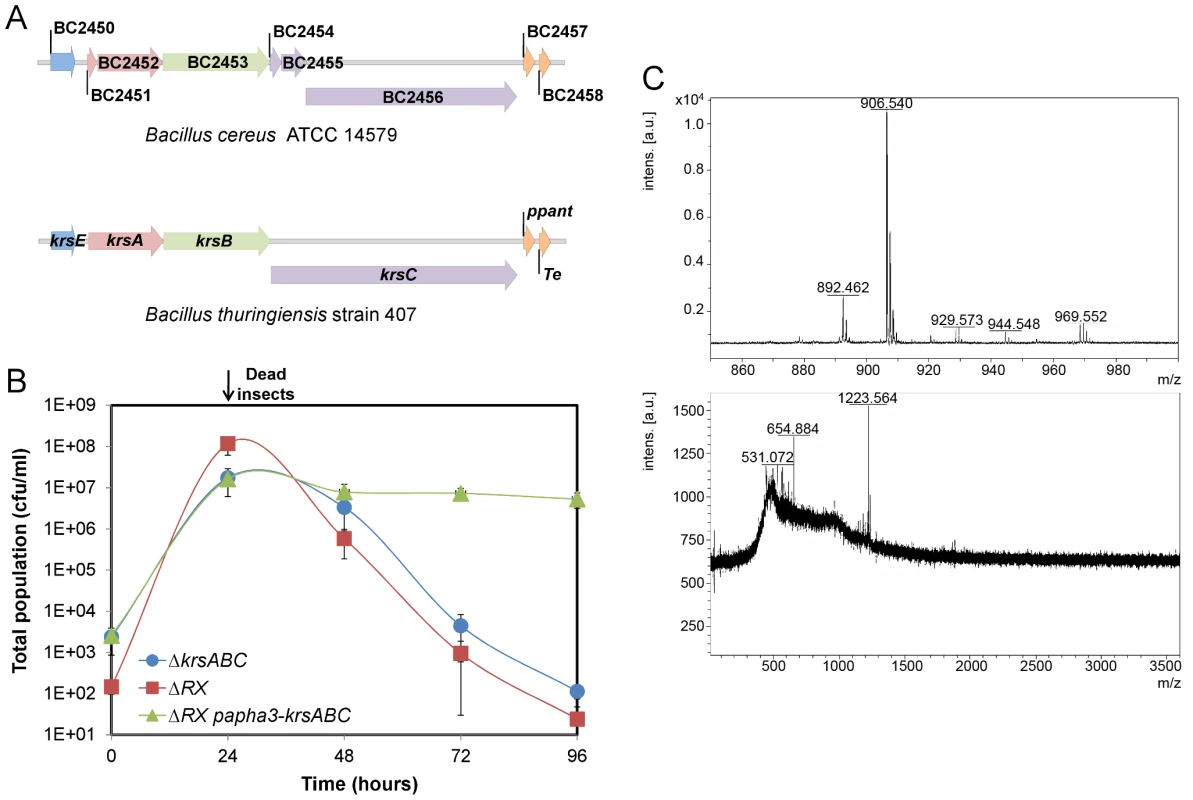
(A) Genetic organization of the krsEABC locus in Bc ATCC14579 and Bt strain 407. krsE encodes a putative efflux protein. krsA, krsB and krsC encode non ribosomal peptide synthetases KrsA, KrsB and KrsC, respectively. Immediately downstream is a gene encoding a phosphopanthetheinyl transferase (ppant) and a gene encoding a protein with a thioesterase domain; both these genes are involved in kurstakin synthesis. (B) The total population of the ΔkrsABC strain throughout the experiment was similar to the total population of the ΔRX strain. The constitutive expression of the krsABC genes in the ΔRX strain restored the survival of this mutant. (C) MALDI-ToF mass spectra of the wt (upper panel) and ΔkrsABC (lower panel) strains. Four peaks corresponding to kurstakin were identified for the wt strain and none of these peaks were detected in the ΔkrsABC strain. Molecular ions are apparent for kurstakins C12 [M+H+] at 892 and C13 [M+H+] at 906, [M+Na+] at 929, and [M+K+] at 944 identified by Price et al. (2007). Kurstakin is involved in swarming and biofilm formation through a pore-forming activity
We compared properties of the wt and ΔkrsABC strains on swimming plates (LB Agar 0.3%) and on swarming plates (LB Agar 0.7% and EPS Agar 0.7%) (Figure 6A). The wt and the ΔkrsABC strains grown on LB 0.3% agar covered the plates, indicating that both strains were swimming proficient. However, unlike the wt, the ΔkrsABC strain was unable to swarm or to form dendrites indicative of swarming mobility [24], [25]. Lipopeptides are known to enhance biofilm formation [26] and it has been shown that the Bt 407 strain forms a thick biofilm at the air / liquid interface in glass tubes [27]. We tested the ability of the two strains to form biofilm in glass tubes (Figure 6A). The wt strain produced a significant ring at the air / liquid interface, whereas biofilm formation was abolished for the ΔkrsABC strain. Kurstakin is therefore necessary for swarming and biofilm formation.
Fig. 6. Kurstakin is required for swarming mobility and biofilm formation. 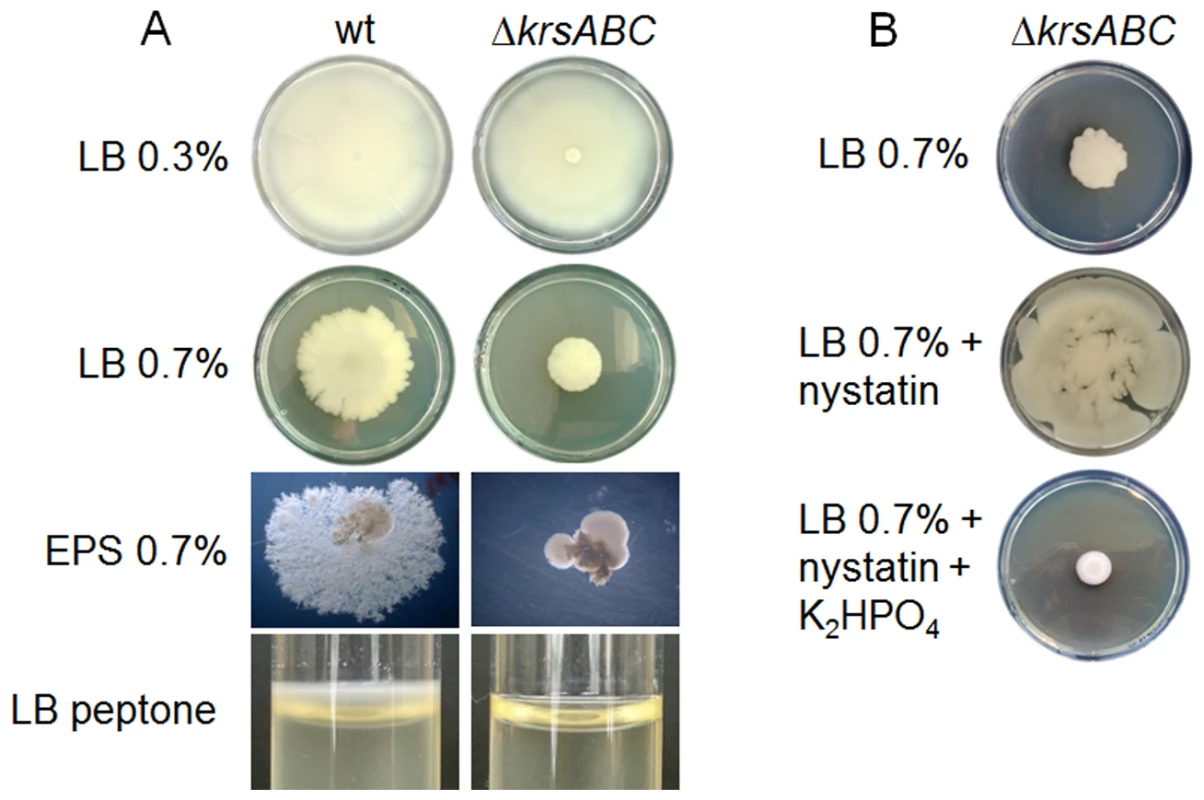
(A) Mobility and morphology of the wt and ΔkrsABC strains were compared on LB 0.3% agar, LB 0.7% agar and EPS 0.7% agar. Biofilm formation by the two strains was also assessed in glass tubes containing 2 ml of LB peptone. A ΔkrsABC strain is unable to swarm or to form biofilm at the air/liquid interface. (B) The swarming mobility of the ΔkrsABC strain is restored by the addition of nystatin, and this phenotype is reversed by the addition of K2HPO4. Lopez and coll. have shown that swarming mobility in B. subtilis is triggered by surfactin, a lipopeptide, which acts as a pore-forming molecule causing potassium leakage across the cytoplasmic membrane [26]. We tested the swarming mobility of the ΔkrsABC strain on swarming plates with nystatin (a pore-forming molecule) and with nystatin plus K2HPO4 (Figure 6B). Nystatin restored the swarming mobility of the ΔkrsABC strain and the addition of K2HPO4 reversed this phenotype. These results suggest that kurstakin is a pore forming molecule causing a potassium leakage across the cytoplasmic membrane of Bt.
Discussion
PlcR is the main virulence regulator in Bt and Bc [10], [12] and is required for the early steps of the infection process [9], [28]. We show here that another quorum sensor, NprR, is active after host death and is necessary for Bt to survive in the insect cadaver. NprR is a pleiotropic regulator directly or indirectly affecting the expression of at least 41 genes during the stationary phase. About 30% of the NprR-regulated genes encode extracellular or cell wall-associated proteins involved in the degradation of proteins, lipids and chitin. We report that nprA and two other NprR-regulated genes encoding degradative enzymes were expressed after death of the host. Therefore, it is likely that these enzymes allow Bt to use the content of the host, indicating that Bt displays a necrotrophic lifestyle in the insect cadaver. This nutrient acquisition may support the developmental program of Bt until sporulation. The degradative enzymes may also have other functions. Insect cuticles are made of chitin filaments arranged within a protein matrix which constitutes a physical barrier to the outside environment. Degradative enzymes may degrade this barrier and facilitate spore and toxin release into the environment. Degradative enzymes may also participate in cell protection against competitors. For example, the endochitinase ChiCW was reported as having antifungal properties [29], and InhA3 (BC2984) is a member of the Immune Inhibitor A metalloprotease family, which plays a key role in the resistance to the host immune defenses by degrading antimicrobial peptides [30], [31], [32]. In addition, two substrate-binding proteins (BC2827 and the BC3526) may increase the efficiency of these enzymes.
A large locus of three NprR-regulated genes (krsABC) codes for a NRPS system involved in the synthesis of a secreted lipopeptide called kurstakin. At least three suggestions could explain the important and surprising function of kurstakin:
-
We show that this lipopeptide is essential for Bt to survive in the cadaver. Lipopeptides have biosurfactant activity, and we show that kurstakin is necessary for swarming mobility and biofilm formation. Thus, the kurstakin may possibly allow Bt to spread across the cadaver, facilitating access to new substrates.
-
Lipopeptides are potent antimicrobials, and kurstakin has an antifungal activity [21], [33], [34]. Kurstakin may thus act as an antimicrobial molecule and prevent colonization and growth by competing microorganisms.
-
Lopez and coll. [26], [35] demonstrated that surfactin is a pore-forming molecule causing a potassium leakage across the membrane of B. subtilis. This molecule acts as a signal triggering multicellularity: one subpopulation of bacterial cells produces surfactin and another responds to it by producing extracellular matrix. Here, we show that kurstakin may similarly induce potassium leakage across the cytoplasmic membrane of Bt. In addition, bioassays in G. mellonella indicate that at least two bacterial subpopulations coexist after host death: one subpopulation enters into sporulation, while the other remains in a vegetative form (Figure 2B) and expresses the NprR-regulated genes (Figure 3B). By analogy, kurstakin may also be a signaling molecule allowing Bt cells to differentiate into subpopulations. Bacterial heterogeneity could provide an advantage to the bacteria for the survival to sudden changes in the insect cadaver environment.
Some NprR-regulated genes encoding cytoplasmic or membrane-associated proteins may participate in the necrotrophic development of Bt. A putative efflux system (BC1063), two macrolide glycosyl transferases (BC2066 and BC2622) and a N-hydroxyarylamine O-acetyltransferase could be involved in resistance to antimicrobial molecules, and cytochrome P450 (BC2613) may be involved in resistance to reactive oxygen species. The membrane-associated proteins are mainly components of an oligopeptide permease system (Opp) involved in the uptake of PapR, the signaling peptide required for PlcR activation [36]. The operon encoding this Opp system is downregulated by NprR suggesting that NprR controls PlcR expression negatively through the Opp system. Interestingly, nprA expression was only slightly reduced in a oppB mutant strain suggesting that another oligopeptide permease system is involved for the uptake of NprX (Figure S4). Consequently, a down-regulation of the opp genes could not have a significant effect on the expression of the NprR-regulated genes. Possibly, the extracellular concentration of NprX is a signal triggering the transition from a pathogenic to a necrotrophic lifestyle. During the early stage of infection, the PlcR regulon is expressed and the extracellular concentration of NprX might be low. Indeed, recent results indicate that nprX transcription starts in late stationary phase (TD, SP, DL, unpublished data). In view of these data, we hypothesize that after the host death, the extracellular concentration of NprX increases, leading to the expression of NprR-regulated genes and repression of the PlcR regulon via the Opp transporter. These various observations indicate that the necrotrophic lifestyle of Bt is a complex developmental stage, not limited to simple feeding on the host contents. They also imply that the transition from a pathogenic to a necrotrophic lifestyle is associated with significant metabolic changes.
It is becoming clear that the infectious cycle of Bt can be divided into four distinct and sequential phases starting with toxemia caused by the Cry proteins, followed by the action of PlcR in virulence, necrotrophism and the completion of the sporulation process involving NprR, and finally the dissemination of spores (Figure 7). We describe a nprR-nprX mutant that does not develop necrotrophically and is unable to sporulate efficiently, demonstrating that the necrotrophic properties of Bt are essential for horizontal transmission. It is remarkable that the developmental process through the complete infection cycle in the insect host is coordinated by three regulator-signaling peptide cell-cell communication systems: PlcR-PapR, NprR-NprX and the Rap-Phr complexes which control the phosphorylation of Spo0A [37], [38], [39].
Fig. 7. Sequential expression of master regulators during the infectious cycle of Bt. 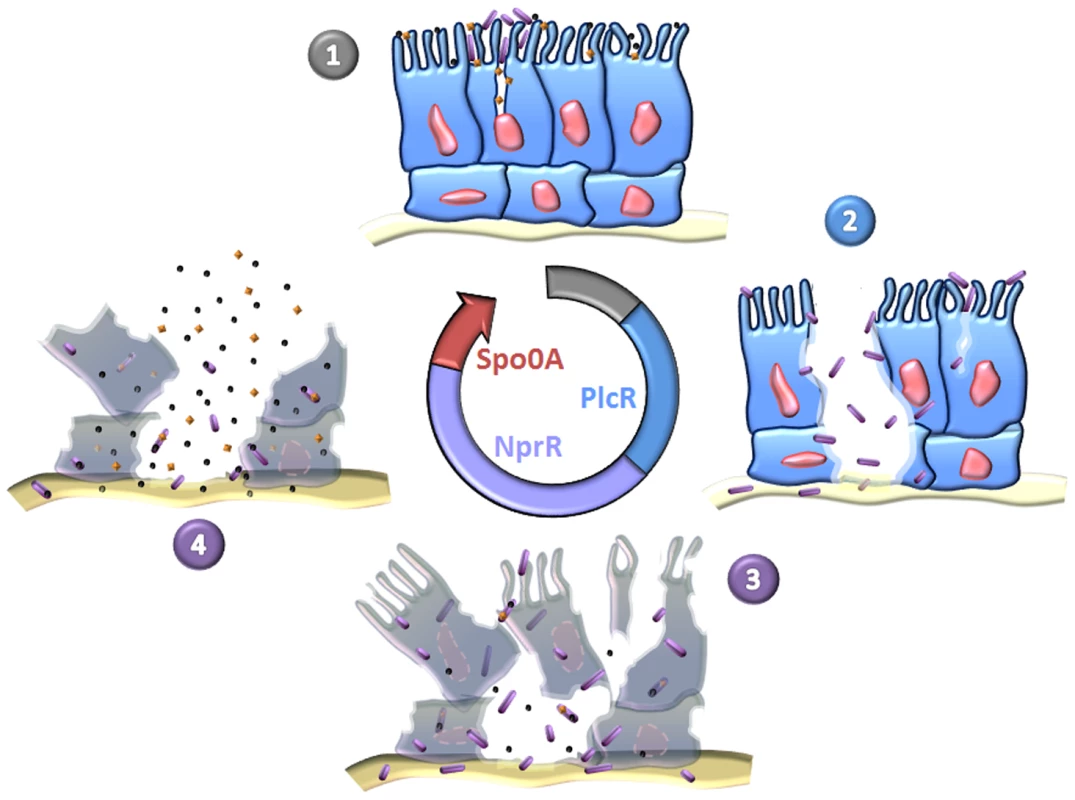
(1) Toxins and spores reach the intestinal epithelium. The toxins induce cell lysis and create favorable conditions for spore germination. The resulting vegetative bacteria adhere to cells weakened by toxins and can grow. (2) The bacteria produce PlcR-regulated virulence factors which attack the intestinal barrier. This event allows the bacteria to penetrate into deeper tissues, such that they can multiply in body fluids and cause host death. (3) The bacteria shift from a virulent lifestyle to a necrotrophic lifestyle and change their gene expression pattern. The pleiotropic regulator NprR activates the transcription of genes encoding proteins involved in food supply, stress resistance, protection against competitors and kurstakin synthesis. These factors increase bacterial fitness in the host cadaver, allowing the bacteria to produce Cry toxins (orange diamonds) and to pursue their developmental program until sporulation and production of resistant spores. (4) Spores and toxins disseminate in the environment through various abiotic (wind, rain) and biotic (parasites, predators) mechanisms. Members of the Bc group may be able to follow two different life cycles: an infectious cycle (described above) and an endosymbiotic cycle in which the bacteria live in a symbiotic relation with their invertebrate hosts [6], [40]. Here, we report that Bt multiplies efficiently in the insect cadaver and has a genetic developmental program to live in this biotope. Consequently, we propose the existence of a strictly necrotrophic life cycle in which Bt colonizes a wide variety of dead insects, and uses the cadaver as a bioreactor to multiply and to produce spores and toxins.
In silico analysis reveals that nprR is found in all strains of the Bc group, except that in Bc ATCC14579 nprR is disrupted by a transposon [3]. The published genome sequences of Ba and Bc strains provide no evidence for any loss of genetic determinants, which might be crucial for saprophytic survival [41]. Therefore, the function of NprR is probably conserved in the Bc group, and it would be interesting to determine whether the necrotrophic development of Bc and Ba in their mammalian hosts requires the same quorum-sensing regulated network. It is also important to characterize kurstakin to determine how it promotes survival in insect cadavers. Moreover, the properties of this molecule may indicate possibilities for the development of phytosanitary products or adjuvants to improve both the ecological fitness and the efficacy of biopesticides.
Materials and Methods
Bacterial strains and growth conditions
The Bt strain 407 Cry− is an acrystalliferous strain cured of its cry plasmid [42]. This strain shares high phylogenic similarity with Bc [43]. Bacillus 407 oppB::tet, Bacillus 407 sigK::aphA3, Bacillus 407 nprRX::tet (ΔRX), Bacillus 407 nprA::lacZ and Bacillus 407 nprRX::tet nprA::lacZ mutant strains were described previously [16], [20], [36]. Escherichia coli K-12 strain TG1 was used as host for the construction of plasmids and cloning experiments. Plasmid DNA for Bacillus electroporation was prepared from the Dam− Dcm− E. coli strain ET12567 (Stratagene, La Jolla, CA, USA). E. coli and Bt cells were transformed by electroporation as described previously [42], [44]. E. coli strains were grown at 37°C in Luria Broth (LB). Bacillus strains were grown at 30 or 37°C in LB or in HCT, a sporulation-specific medium [45]. The following concentrations of antibiotic were used for bacterial selection: 100 µg/ml ampicillin for E. coli; 200 µg/ml kanamycin, 10 µg/ml tetracycline, 200 µg/ml spectinomycin and 10 µg/ml erythromycin for Bacillus. Numbers of viable cells were counted as total colony-forming units (cfu) on LB plates. Numbers of spores were determined as heat-resistant (80°C for 12 min) cfu on LB plates.
In vivo experiments
Force-feeding and intrahemocelic injection experiments with G. mellonella were carried out as described previously [9]. LD50 data were analyzed using the program StatPlus 2007 of Analysoft. Bt cells in living and dead insects were counted as follows. For each strain, each larva was injected with 2.104 bacteria and kept at 30°C for 96 h; 24 h after injection, surviving insects were eliminated. At the injection time and every 24 h for the 96 h of the experiment, two larvae were crushed and homogenized in 10 ml of physiological water and dilutions were plated onto LB agar plates containing appropriate antibiotics. To follow the spore population, bacterial colony-forming units were determined before and after treatment of the insect homogenate for 12 min at 80°C. At least four independent replicates were performed for each time and for each strain tested. In vivo ß-galactosidase activity was assayed from 2 ml aliquots of the insect homogenate as described previously [16]. At least three independent measurements were performed for each time and for each transcriptional fusion tested.
DNA manipulations
Chromosomal DNA was extracted from Bt cells using the Puregene Yeast/Bact. Kit B (QIAgen, France). Plasmid DNA was extracted from E. coli using QIAprep spin columns (QIAgen, France). Restriction enzymes (New England Biolabs, USA) and T4 DNA ligase (New England Biolabs, USA) were used in accordance with the manufacturer's recommendations. Oligonucleotide primers (Table S4) were synthesized by Sigma-Proligo (Paris, France). PCRs were performed in a Applied Biosystem 2720 Thermak cycler (Applied Biosystem, USA). Amplified fragments were purified using the QIAquick PCR purification Kit (QIAgen, France). Digested DNA fragments were separated on 1% (w/V) agarose gels after digestion and extracted from gels using the QIAquick gel extraction Kit (QIAgen, France). Nucleotide sequences were determined by Beckman Coulter Genomics (Takeley, UK)
Plasmid constructions
The plasmid pRN5101 [46] was used for homologous recombination. The low-copy-number plasmid pHT304 was used for complementation experiments with wild-type nprR-nprX genes under their own promoters [16]. Transcriptional fusions were constructed in pHT304-18Z [47]. All the plasmids used in this study are described in Table S5.
Construction of the B. thuringiensis recombinant strains
The krsABC genes were disrupted by inserting a spectinomycin resistance gene into the coding sequence. The thermosensitive plasmid pRN5101ΩkrsABC::spc was used to disrupt the chromosomal wild-type copy of the krsABC genes in the Bacillus 407 wt strain by homologous recombination as described previously [46]. The recombinant strain, designated Bacillus 407 ΔkrsABC, was resistant to spectinomycin and sensitive to erythromycin. The thermosensitive plasmid pRN5101ΩPkrsABC::aphA3 was used to replace the natural promoter region of the krsABC genes in the Bacillus 407 ΔRX strain by aphA3 and its constitutive promoter. In the resulting Bacillus recombinant strain, the krsABC genes were transcribed from the aphA3 promoter; it was designated Bacillus 407 ΔRX PaphA3-krsABC, and was resistant to kanamycin and sensitive to erythromycin.
Phenotype analysis
The methods used to study the proteolitic activity, the chitinolytic activity and the lipolytic activity have been described previously [32], [48]. Swimming and swarming were evaluated using LB 0.3% agar plates and LB 0.7% agar plates, respectively. Biofilm formation was assayed in LB medium and in glass tubes as described previously [27]. Dendrite formation was evaluated on EPS 0.7% agar. Strains were cultured in LB medium at 37°C until the beginning of stationary phase and 2.106 bacteria were spotted onto the center of the agar plate. Plates were incubated at 37°C for 24 h to 96 h.
ß-Galactosidase assay
For in vitro ß-galactosidase activity measurements, Bt cells containing lacZ transcriptional fusions were cultured in LB medium at 37°C. In vivo ß-galactosidase activity was assayed from 2 ml aliquots of insect homogenate (see in vivo experiments). ß-Galactosidase activities were measured as described previously [49]. The specific activities are expressed in units of ß-galactosidase per milligram of protein (Miller units).
Samples for microarrays and quantitative RT-PCR
Prewarmed 500 ml baffled erlenmeyer flasks with 50 ml LB medium were inoculated with 1 ml overnight cultures of Bacillus 407 nprA::lacZ or Bacillus 407 ΔRX nprA::lacZ, and incubated at 37°C and 250 rpm. Samples for microarray analysis were taken three (t3) hours after the onset of the stationary phase. Samples were harvested as described previously [12] mixed with RLT buffer from the RNeasy midi kit (Qiagen, France) and frozen at −70°C. After thawing samples at 37°C for 15 min, RNA isolation, cDNA synthesis, labeling and purification were performed as described [12].
Microarray comparisons
The microarray slides were printed, prehybridized and hybridized as described previously [12], except that hybridization was extended to 17 hours. The slides were scanned on an Axon 4000B scanner (Molecular Devices). Gridding, spot annotation and calculation of raw spot intensities was done with the GenePix Pro 6.1 software (Molecular Devices). The LIMMA package [50], [51], [52] on the R 2.7.1 platform [53] was used for filtering, normalization and further analysis. The raw data were filtered and weighted by quality [54], and the four technical replicates on each slide were averaged to increase robustness. P-values were computed using a false discovery rate of 0.05. The analysis was based on hybridization to three slides, all employing biological replicates.
Quantitative RT-PCR
Gene expression was investigated in Bacillus 407 nprA::lacZ and Bacillus 407 ΔRX nprA::lacZ. Reverse transcription was performed according to the SuperScript III reverse transcriptase protocol from Invitrogen, but RNaseOUT was replaced with 0.1 µl SUPERase-In (Ambion). A negative control without reverse transcriptase was included. In all samples, the reaction volume was adjusted to 20 µl with DEPC-treated water (Ambion) before reverse transcription. The reaction product was diluted (1 µl in 39 µl) with water, and 8 µl applied to each well (2 µl for 5 s rRNA samples). Primers were added to a final concentration of 0.56 µM. A volume of 9 µl LightCycler 480 DNA SYBR Green I Master (Roche) was added, and the volume was adjusted to 18 µl. Primers (available on request) were designed to give PCR products of around 100 bp. The reference genes, gatB (BC4306) and 5 s rRNA, were included on every plate. The samples were analyzed on a Roche Lightcycler 480 (Roche Diagnostics GmbH, Mannheim, Germany). Cycling conditions were 95°C for 5 minutes followed by 45 cycles at 95°C for 10 seconds, 58°C for 10 seconds, and 72°C for 8 seconds. Cp values were determined using 2nd derivative max, and are averages of two technical replicates. The results were calculated by the delta-delta Ct approximation. The log2 expression ratios of Bacillus 407 ΔRX nprA::lacZ over Bacillus 407 nprA::lacZ in Table S3 are averages for three biological replicates.
Mass spectrometry analysis
Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry (MALDI-ToF MS) was used to screen kurstakin production from whole bacterial cells on solid media. Cultures were performed on AK agar plates incubated at 30°C for 24 h or 48 h. A saturated solution of α-cyano-4-hydroxy-cinnamic acid was prepared in 1∶2 (v/v) solution of CH3CN and H2O containing 0.1% TFA. Measurement was performed using UV laser MALDI-ToF spectrometer (Bruker UltraFlex TOF; Bruker Daltonics) equipped with a pulsed nitrogen laser (λ = 337 nm). The ions were extracted from the ionization source with an acceleration voltage of 20 kV. Samples were measured in the reflector mode, positive mode. The equivalent of about 1 µl of cell material was picked from agar plates with an automatic pipette. The tip with the culture was deposited in an eppendorf tube. 20 µl of matrix solution (saturated solution of α-cyano-4-hydroxycinnamic acid in a 1∶2 v/v solution of CH3CN/H2O with 0.1% TFA) are added. The eppendorf with the tip and the matrix solution was vortexed for 30 seconds. 1 µl of this sample solution was deposited on the MALDI target and let dry at room temperature. The spectrum was obtained with 5×30 shots on the sample. Analyses were performed on two different samples.
Supporting Information
Zdroje
1. RaymondBJohnstonPRNielsen-LeRouxCLereclusDCrickmoreN 2010 Bacillus thuringiensis: an impotent pathogen? Trends Microbiol 18 189 194
2. Toledo-AranaADussurgetONikitasGSestoNGuet-RevilletH 2009 The Listeria transcriptional landscape from saprophytism to virulence. Nature 459 950 956
3. IvanovaNSorokinAAndersonIGalleronNCandelonB 2003 Genome sequence of Bacillus cereus and comparative analysis with Bacillus anthracis. Nature 423 87 91
4. MartinPAWTraversRS 1989 Worldwide abundance and distribution of Bacillus thuringiensis isolates. Appl Environ Microbiol 55 2437 2442
5. Vilas-BoasLAVilas-BoasGFSaridakisHOLemosMVLereclusD 2000 Survival and conjugation of Bacillus thuringiensis in a soil microcosm. FEMS Microbiol Ecol 31 255 259
6. JensenGBHansenBMEilenbergJMahillonJ 2003 The hidden lifestyles of Bacillus cereus and relatives. Environ Microbiol 5 631 640
7. SchnepfECrickmoreNVan RieJLereclusDBaumJ 1998 Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol Mol Biol Rev 62 775 806
8. DuCNickersonKW 1996 Bacillus thuringiensis HD-73 Spores Have Surface-Localized Cry1Ac Toxin: Physiological and Pathogenic Consequences. Appl Environ Microbiol 62 3722 3726
9. SalamitouSRamisseFBrehélinMBourguetDGiloisN 2000 The plcR regulon is involved in the opportunistic properties of Bacillus thuringiensis and Bacillus cereus in mice and insects. Microbiology 146 2825 2832
10. AgaisseHGominetMØkstadOAKolstøABLereclusD 1999 PlcR is a pleiotropic regulator of extracellular virulence factor gene expression in Bacillus thuringiensis. Mol Microbiol 32 1043 1053
11. SlamtiLLereclusD 2002 A cell-cell signaling peptide activates the PlcR virulence regulon in bacteria of the Bacillus cereus group. EMBO J 21 4550 4559
12. GoharMFaegriKPerchatSRavnumSØkstadOA 2008 The PlcR virulence regulon of Bacillus cereus. PLoS ONE 3 e2793
13. GoharMØkstadOAGiloisNSanchisVKolstøA-B 2002 Two-dimensional electrophoresis analysis of the extracellular proteome of Bacillus cereus reveals the importance of the PlcR regulon. Proteomics 2 784 791
14. LereclusDAgaisseHGrandvaletCSalamitouSGominetM 2000 Regulation of toxin and virulence gene transcription in Bacillus thuringiensis. Int J Med Microbiol 290 295 299
15. ChitlaruTGatOGozlanYArielNShaffermanA 2006 Differential proteomic analysis of the Bacillus anthracis secretome: distinct plasmid and chromosome CO2-dependent cross talk mechanisms modulate extracellular proteolytic activities. J Bacteriol 188 3551 3571
16. PerchatSDuboisTZouhirSGominetMPoncetS 2011 A cell-cell communication system regulates protease production during sporulation in bacteria of the Bacillus cereus group. Mol Microbiol 82 619 633
17. ChungMCPopovaTGMillisBAMukherjeeDVZhouW 2006 Secreted neutral metalloproteases of Bacillus anthracis as candidate pathogenic factors. J Biol Chem 281 31408 31418
18. Hajaij-EllouzeMFedhilaSLereclusDNielsen-LeRouxC 2006 The enhancin-like metalloprotease from the Bacillus cereus group is regulated by the pleiotropic transcriptional activator PlcR but is not essential for larvicidal activity. FEMS Microbiol Lett 260 9 16
19. HilbertDWPiggotPJ 2004 Compartmentalization of gene expression during Bacillus subtilis spore formation. Microbiol Mol Biol Rev 68 234 262
20. BravoAAgaisseHSalamitouSLereclusD 1996 Analysis of cryIAa expression in sigE and sigK mutants of Bacillus thuringiensis. Mol Gen Genet 250 734 741
21. HathoutYHoYPRyzhovVDemirevPFenselauC 2000 Kurstakins: a new class of lipopeptides isolated from Bacillus thuringiensis. J Nat Prod 63 1492 1496
22. BumpusSBEvansBSThomasPMNtaiIKelleherNL 2009 A proteomics approach to discovering natural products and their biosynthetic pathways. Nat Biotechnol 27 951 956
23. AbderrahmaniATapiANatecheFCholletMLeclereV 2011 Bioinformatics and molecular approaches to detect NRPS genes involved in the biosynthesis of kurstakin from Bacillus thuringiensis. Appl Microbiol Biotechnol 92 571 81
24. KinsingerRFShirkMCFallR 2003 Rapid surface motility in Bacillus subtilis is dependent on extracellular surfactin and potassium ion. J Bacteriol 185 5627 5631
25. JulkowskaDObuchowskiMHollandIBSerorSJ 2004 Branched swarming patterns on a synthetic medium formed by wild-type Bacillus subtilis strain 3610: detection of different cellular morphologies and constellations of cells as the complex architecture develops. Microbiology 150 1839 1849
26. LopezDFischbachMAChuFLosickRKolterR 2009 Structurally diverse natural products that cause potassium leakage trigger multicellularity in Bacillus subtilis. Proc Natl Acad Sci U S A 106 280 285
27. HouryABriandetRAymerichSGoharM 2010 Involvement of motility and flagella in Bacillus cereus biofilm formation. Microbiology 156 1009 1018
28. FedhilaSGoharMSlamtiLNelPLereclusD 2003 The Bacillus thuringiensis PlcR-regulated gene inhA2 is necessary, but not sufficient, for virulence. J Bacteriol 185 2820 2825
29. HuangCJWangTKChungSCChenCY 2005 Identification of an antifungal chitinase from a potential biocontrol agent, Bacillus cereus 28-9. J Biochem Mol Biol 38 82 88
30. DalhammarGSteinerH 1984 Characterization of inhibitor A, a protease from Bacillus thuringiensis which degrades attacins and cecropins, two classes of antibacterial proteins in insects. Eur J Biochem 139 247 252
31. RamaraoNLereclusD 2005 The InhA1 metalloprotease allows spores of the B. cereus group to escape macrophages. Cell Microbiol 7 1357 1364
32. GuillemetECadotCTranSLGuinebretiereMHLereclusD 2010 The InhA metalloproteases of Bacillus cereus contribute concomitantly to virulence. J Bacteriol 192 286 294
33. CarrilloCTeruelJAArandaFJOrtizA 2003 Molecular mechanism of membrane permeabilization by the peptide antibiotic surfactin. Biochim Biophys Acta 1611 91 97
34. SteinT 2005 Bacillus subtilis antibiotics: structures, syntheses and specific functions. Mol Microbiol 56 845 857
35. LopezDVlamakisHLosickRKolterR 2009 Paracrine signaling in a bacterium. Genes Dev 23 1631 1638
36. GominetMSlamtiLGiloisNRoseMLereclusD 2001 Oligopeptide permease is required for expression of the Bacillus thuringiensis PlcR regulon and for virulence. Mol Microbiol 40 963 975
37. PeregoMHochJA 1996 Protein aspartate phosphatases control the output of two-component signal transduction systems. Trends Genet 12 97 101
38. LazazzeraBAGrossmanAD 1998 The ins and outs of peptide signaling. Trends Microbiol 6 288 294
39. BongiorniCStoesselRShoemakerDPeregoM 2006 Rap phosphatase of virulence plasmid pXO1 inhibits Bacillus anthracis sporulation. J Bacteriol 188 487 498
40. MargulisLJorgensenJZDolanSKolchinskyRRaineyFA 1998 The Arthromitus stage of Bacillus cereus: intestinal symbionts of animals. Proc Natl Acad Sci U S A 95 1236 1241
41. SaileEKoehlerTM 2006 Bacillus anthracis multiplication, persistence, and genetic exchange in the rhizosphere of grass plants. Appl Environ Microbiol 72 3168 3174
42. LereclusDArantesOChaufauxJLecadetM-M 1989 Transformation and expression of a cloned ∂-endotoxin gene in Bacillus thuringiensis. FEMS Microbiol Lett 60 211 218
43. KolstøABTourasseNJØkstadOA 2009 What sets Bacillus anthracis apart from other Bacillus species? Annu Rev Microbiol 63 451 476
44. DowerWJMillerJFRagsdaleCW 1988 High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res 16 6127 6145
45. LecadetMMBlondelMORibierJ 1980 Generalized transduction in Bacillus thuringiensis var. berliner 1715, using bacteriophage CP54 Ber. J Gen Microbiol 121 203 212
46. LereclusDValladeMChaufauxJArantesORambaudS 1992 Expansion of insecticidal host range of Bacillus thuringiensis by in vivo genetic recombination. Bio/Technology 10 418 421
47. AgaisseHLereclusD 1994 Structural and functional analysis of the promoter region involved in full expression of the cryIIIA toxin gene of Bacillus thuringiensis. Mol Microbiol 13 97 107
48. de BarjacHCosmao-DumanoirV 1975 Interest of some additional biochemical tests for the classification of “Bacillus” (author's transl). Ann Microbiol (Paris) 126 83 95
49. BouillautLRamaraoNBuissonCGiloisNGoharM 2005 FlhA influences Bacillus thuringiensis PlcR-regulated gene transcription, protein production, and virulence. Appl Environ Microbiol 71 8903 8910
50. SmythGK 2004 Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3 Article3
51. SmythGKSpeedT 2003 Normalization of cDNA microarray data. Methods 31 265 273
52. SmythGK 2005 Limma: Linear models for microarray data. GentlemanVCRDudoitSIrizarryRHuberW Bioinformatics and Computational Biology Solutions using R and Bioconductor New York Springer 397 420
53. R-Development-Core-Team 2005 R: A language and environment for statistical computing Vienna (Austria) R Foundation for Statistical Computing Available: http://www.R-project.org
54. BrulandTAnderssenEDosethBBergumHBeisvagV 2007 Optimization of cDNA microarrays procedures using criteria that do not rely on external standards. BMC Genomics 8 377
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Fungal Biofilms
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2012 Číslo 4- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Systematic Review of Mucosal Immunity Induced by Oral and Inactivated Poliovirus Vaccines against Virus Shedding following Oral Poliovirus Challenge
- The Arbuscular Mycorrhizal Symbiosis: Origin and Evolution of a Beneficial Plant Infection
- Modelling the Evolutionary Dynamics of Viruses within Their Hosts: A Case Study Using High-Throughput Sequencing
- Structural Basis of Cytotoxicity Mediated by the Type III Secretion Toxin ExoU from
- Fungal Biofilms
- The Problem of Auto-Correlation in Parasitology
- T Regulatory Cells Control Susceptibility to Invasive Pneumococcal Pneumonia in Mice
- Small-Molecule Inhibitors of Dengue-Virus Entry
- The Baculovirus Uses a Captured Host Phosphatase to Induce Enhanced Locomotory Activity in Host Caterpillars
- Matrix Metalloprotease 9 Mediates Neutrophil Migration into the Airways in Response to Influenza Virus-Induced Toll-Like Receptor Signaling
- Necrotrophism Is a Quorum-Sensing-Regulated Lifestyle in
- Modeling of the N-Glycosylated Transferrin Receptor Suggests How Transferrin Binding Can Occur within the Surface Coat of
- The Role of Cofactors in Prion Propagation and Infectivity
- The Role of Mast Cells in the Defence against Pathogens
- The Accessory Genome as a Cradle for Adaptive Evolution in Pathogens
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The Accessory Genome as a Cradle for Adaptive Evolution in Pathogens
- Systematic Review of Mucosal Immunity Induced by Oral and Inactivated Poliovirus Vaccines against Virus Shedding following Oral Poliovirus Challenge
- The Arbuscular Mycorrhizal Symbiosis: Origin and Evolution of a Beneficial Plant Infection
- Modelling the Evolutionary Dynamics of Viruses within Their Hosts: A Case Study Using High-Throughput Sequencing
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



