-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaDistinct Effects on Diversifying Selection by Two Mechanisms of Immunity against
Antigenic variation to evade host immunity has long been assumed to be a driving force of diversifying selection in pathogens. Colonization by Streptococcus pneumoniae, which is central to the organism's transmission and therefore evolution, is limited by two arms of the immune system: antibody - and T cell - mediated immunity. In particular, the effector activity of CD4+ TH17 cell mediated immunity has been shown to act in trans, clearing co-colonizing pneumococci that do not bear the relevant antigen. It is thus unclear whether TH17 cell immunity allows benefit of antigenic variation and contributes to diversifying selection. Here we show that antigen-specific CD4+ TH17 cell immunity almost equally reduces colonization by both an antigen-positive strain and a co-colonized, antigen-negative strain in a mouse model of pneumococcal carriage, thus potentially minimizing the advantage of escape from this type of immunity. Using a proteomic screening approach, we identified a list of candidate human CD4+ TH17 cell antigens. Using this list and a previously published list of pneumococcal Antibody antigens, we bioinformatically assessed the signals of diversifying selection among the identified antigens compared to non-antigens. We found that Antibody antigen genes were significantly more likely to be under diversifying selection than the TH17 cell antigen genes, which were indistinguishable from non-antigens. Within the Antibody antigens, epitopes recognized by human antibodies showed stronger evidence of diversifying selection. Taken together, the data suggest that TH17 cell-mediated immunity, one form of T cell immunity that is important to limit carriage of antigen-positive pneumococcus, favors little diversifying selection in the targeted antigen. The results could provide new insight into pneumococcal vaccine design.
Published in the journal: . PLoS Pathog 8(11): e32767. doi:10.1371/journal.ppat.1002989
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1002989Summary
Antigenic variation to evade host immunity has long been assumed to be a driving force of diversifying selection in pathogens. Colonization by Streptococcus pneumoniae, which is central to the organism's transmission and therefore evolution, is limited by two arms of the immune system: antibody - and T cell - mediated immunity. In particular, the effector activity of CD4+ TH17 cell mediated immunity has been shown to act in trans, clearing co-colonizing pneumococci that do not bear the relevant antigen. It is thus unclear whether TH17 cell immunity allows benefit of antigenic variation and contributes to diversifying selection. Here we show that antigen-specific CD4+ TH17 cell immunity almost equally reduces colonization by both an antigen-positive strain and a co-colonized, antigen-negative strain in a mouse model of pneumococcal carriage, thus potentially minimizing the advantage of escape from this type of immunity. Using a proteomic screening approach, we identified a list of candidate human CD4+ TH17 cell antigens. Using this list and a previously published list of pneumococcal Antibody antigens, we bioinformatically assessed the signals of diversifying selection among the identified antigens compared to non-antigens. We found that Antibody antigen genes were significantly more likely to be under diversifying selection than the TH17 cell antigen genes, which were indistinguishable from non-antigens. Within the Antibody antigens, epitopes recognized by human antibodies showed stronger evidence of diversifying selection. Taken together, the data suggest that TH17 cell-mediated immunity, one form of T cell immunity that is important to limit carriage of antigen-positive pneumococcus, favors little diversifying selection in the targeted antigen. The results could provide new insight into pneumococcal vaccine design.
Introduction
Diversifying selection on genes encoding pathogen antigens is a well known effect of host immunity [1], [2]. Diversifying selection can maintain multiple alleles of a gene at appreciable frequencies in a population [3]. Acquired immune responses provide a fitness advantage for antigenic variants that evade immune recognition, reducing the probability that the allele encoding the targeted antigen will fix with a single allele. In viruses such as HIV [4], [5], [6] and influenza [7], [8], neutralizing antibody and cytotoxic T-lymphocytes (CTLs) drive antigenic diversification. Strong diversifying selection was also identified in major antigen genes in the malaria parasite Plasmodium falciparum [9], [10]. In bacteria, diversity of surface structures (such as capsular polysaccharides) that are targeted by host antibodies is thought to result from such diversifying selection [1]. However, a few exceptions exist. Measles virus antigens show little variation, partially because exposure to the virus would generate polyclonal antibodies that efficiently neutralize a broad range of antigenic variants [11]. Human T cell epitopes of Mycobacterium tuberculosis show a substantially lower level of sequence variation than seen in other genomic regions, suggesting T cell immune responses might limit diversification in the antigen genes [12]. Therefore, we hypothesized that the effect of host immunity on diversifying selection depends on the specific mechanism involved.
Recent studies have indicated that acquired immunity elicited by natural exposure to Streptococcus pneumoniae includes three distinct arms [13]: (1) type-specific, antibody-mediated immunity to the highly variable polysaccharide capsule [14], [15], [16], [17], (2) antibody-mediated immunity to pneumococcal proteins, some of which are variable and some of which are more conserved [15], [18], [19], [20], [21], [22], [23], [24], and (3) CD4+ TH17 cell - mediated, antibody independent immunity to pneumococcal proteins and to the cell-wall polysaccharide [15], [25], [26], [27], [28]. The first two forms of immunity are thought to operate by the standard mechanisms of antibody binding to surface antigens, leading to opsonophagocytosis, reduced attachment and/or other mechanisms of reduced colonization [22], [29]. In the last form of immunity, antigen-specific CD4+ TH17 cells secrete interleukin (IL)-17A, leading to the activation and recruitment of effector cells (neutrophils and macrophages) that then kill pneumococci [25], [30], [31], [32]. TH17 cell-mediated immunity primarily accelerates the clearance of pneumococcus rather than preventing initiation of carriage [31]. Even in combination, these forms of immunity to S. pneumoniae are imperfect. Humans can be repeatedly colonized despite the immune responses from multiple arms.
While antibody binding is by definition specific to bacteria bearing the target antigen, we have previously shown that the CD4+ TH17-based effector activity may extend beyond antigen-expressing bacteria, accelerating the clearance of co-colonized pneumococci that even do not bear the relevant antigen [23]. It is unclear whether CD4+ TH17-mediated immunity would still create a fitness advantage for antigenic variants and thus promote diversifying selection on the genes encoding the targets of such immunity in S. pneumoniae.
Here we report the assessment of two hypotheses: first, a competition assay was performed to examine whether an antigen-negative strain shows a colonization advantage over the antigen-positive strain in mice with antigen-specific TH17 immunity. Second, pneumococcal genes that show signs of being under diversifying selection were systematically identified and their association with either Antibody antigens or TH17 antigens was examined. The results indicate little evidence of diversifying selection in the targets of CD4+ TH17 cell immunity, unlike the targets of antibody immunity.
Results
CD4+ TH17 cell-mediated immunity to pneumococcal carriage provides only weak selection for antigenic variation
Immunization with a pneumococcal whole cell vaccine displaying a peptide from ovalbumin (OVA323–339) delivered with cholera toxin (CT) adjuvant results in CD4+ TH17 cell-mediated and antibody-independent protection against subsequent pneumococcal colonization [23]. To examine whether the TH17 cell immunity against S. pneumoniae, given its in trans clearance effect [23], allows a competitive advantage for a non-recognizable (antigen-negative) strain, twenty BALB/c mice were immunized by either ovalbumin with adjuvant (OVA+CT) or adjuvant alone (CT). The mice were challenged with a 1∶1 mix of an antigen-negative strain (AVO) and an antigen-positive strain (OVA). The two strains were isogenic except that only the OVA strain displays OVA323–339 peptides that can be recognized by the ovalbumin-induced, TH17 immunity in mice [23]. The AVO strain can be viewed as an antigenic variant of the OVA strain and the AVO/OVA ratio would increase if there were a competitive advantage for the antigen-negative strain.
The mixture of pneumococci colonized the ovalbumin-immunized and control mice equally well on day 1. No significant difference in colonization density was observed (Figure 1A, p = 0.87, Mann-Whitney test). By day 4, the median colonization density in ovalbumin-immunized mice was about 7-fold lower than that in the control mice, although the difference was not statistically significant (Figure 1A, p = 0.48, Mann-Whitney test). By day 8, the median colonization density in the immunized mice was about 40-fold lower than that in the control mice and the difference was statistically significant (Figure 1A, p = 0.02, Mann-Whitney test). The effect was consistent with an accelerated clearance of colonization mediated by TH17 immunity [31].
Fig. 1. The benefit of antigenic variation in CD4+ TH17 epitope is limited. 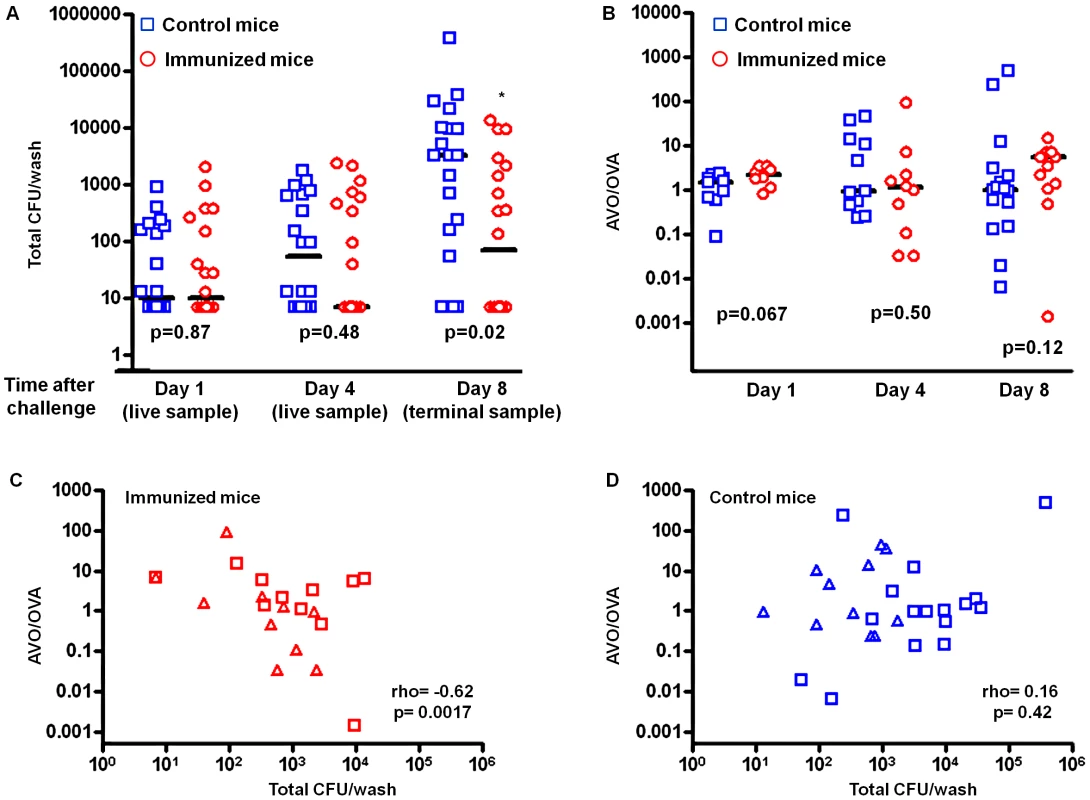
BALB/c mice were immunized by either CT alone (CT) or CT and ovalbumin (CT+OVA). All mice were challenged with a 1∶1 mixture of the antigen-negative (AVO) strain and the antigen-positive (OVA) strain. The density of intranasal colonization by pneumococcus in each mouse was determined on days 1, 4, and 8 after challenge as described in the Methods. Total CFU counts are shown in (A). The ratio between the two strains in each mouse was determined (B). The p values were derived from Mann-Whitney tests comparing the immunized with the control group on days 1, 4, and 8. Solid lines indicate group medians. The correlation between total CFU and the AVO/OVA ratio is shown for the immunized mice (C) and the control mice (D) that remained colonized on days 4 (triangle) and 8 (diamond). The AVO/OVA ratio remained approximately 1∶1 in the control mice during the course of the experiment (Figure 1B). The medians of log10 (AVO/OVA) were 0.185 (n = 10), −0.028 (n = 11), and 0.011 (n = 16) on days 1, 4 and 8, respectively (Table 1), indicating that the AVO strain was competitively neutral in the absence of antigen-specific immunity. In the ovalbumin-immunized mice, the medians of log10 (AVO/OVA) were 0.334 (n = 8), 0.042 (n = 10) and 0.730 (n = 13) on days 1, 4 and 8, respectively (Table 1). The median log10 (AVO/OVA) was not significantly different between the control and the immunized group on days 1, 4 or 8 (Figure 1B, p = 0.067, p = 0.50, and p = 0.12, respectively, Mann-Whitney test), although there was a trend toward an increase in AVO/OVA ratio in the immunized mice.
Tab. 1. Analysis of competitive advantage for the antigen-negative strain. 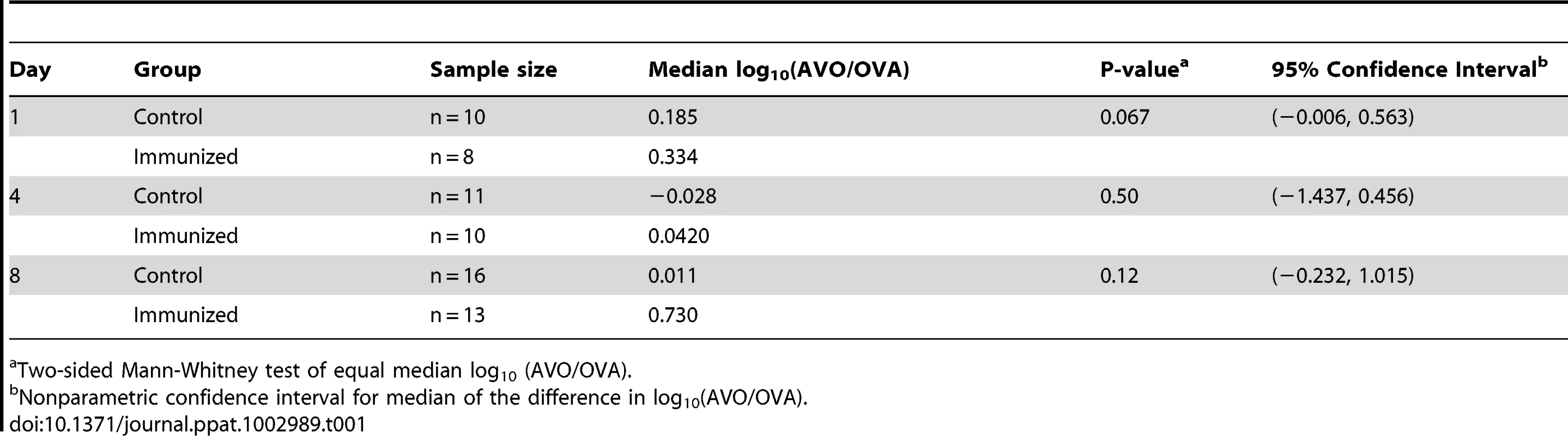
Two-sided Mann-Whitney test of equal median log10 (AVO/OVA). To better quantify the potential competitive advantage for the antigen-negative strain, we constructed nonparametric confidence intervals for the median of the difference in log10 (AVO/OVA) between the immunized group and the control group (Table 1). A median greater than 0 would indicate a competitive advantage for the AVO strain in the immunized group. The 95% confidence intervals for median difference in log10 (AVO/OVA) were (−0.006, 0.563), (−1.437, 0.456), and (−0.2319, 1.015) on days 1, 4, and 8, respectively (Table 1). Thus, the loss of an antigen was unlikely to provide a more than 10.4-fold (1.015 log10) median increase in competitive advantage for the AVO strain by day 8. We also note that the increased frequency of AVO strains was almost entirely found in mice who have nearly cleared colonization (Figure 1C). In absolute CFU numbers, therefore, the relative advantage is unlikely to be associated with much overall superiority.
In mice that remain colonized on days 4 and 8, a negative correlation between the AVO/OVA ratio and total CFU recovered was observed in the immunized group (Figure 1C) but not in the control group (Figure 1D). These results suggested that the antigen-negative strain gains a relative advantage only for the period where bacterial numbers are rather low.
Identification of human CD4+ TH17 antigens in pneumococcus
To determine whether CD4+ TH17 cell-mediated immunity to S. pneumoniae affects antigenic variation in the context of human colonization and disease, S. pneumoniae antigens recognized by human TH17 cells were identified. CD4+ TH 17 cells were enriched from peripheral blood cells and IL-17A secretion in response to pneumococcal protein pools was measured by ELISA (see Materials and Methods, Figure S1, and Figure S2). To identify the common antigens in the sample population of 36 healthy adults, a Mann-Whitney test was used to compare normalized values for each pool to the normalized values for E. coli expressing GFP. Each protein was then ranked by its antigenicity score, which was calculated by multiplying together the p-values resulting from the Mann-Whitney test for both pools containing the protein, lower antigenicity scores indicating more commonly recognized antigens. An N-terminal fragment of PtrA (SP0641.1) was the most strongly recognized antigen in the screen with an antigenicity score of 1.58×10−17 (Figure 2B). Clones with a score less than 0.05 were defined as the common antigens (Table 2).
Fig. 2. Identification of antigens recognized by human TH17 cells. 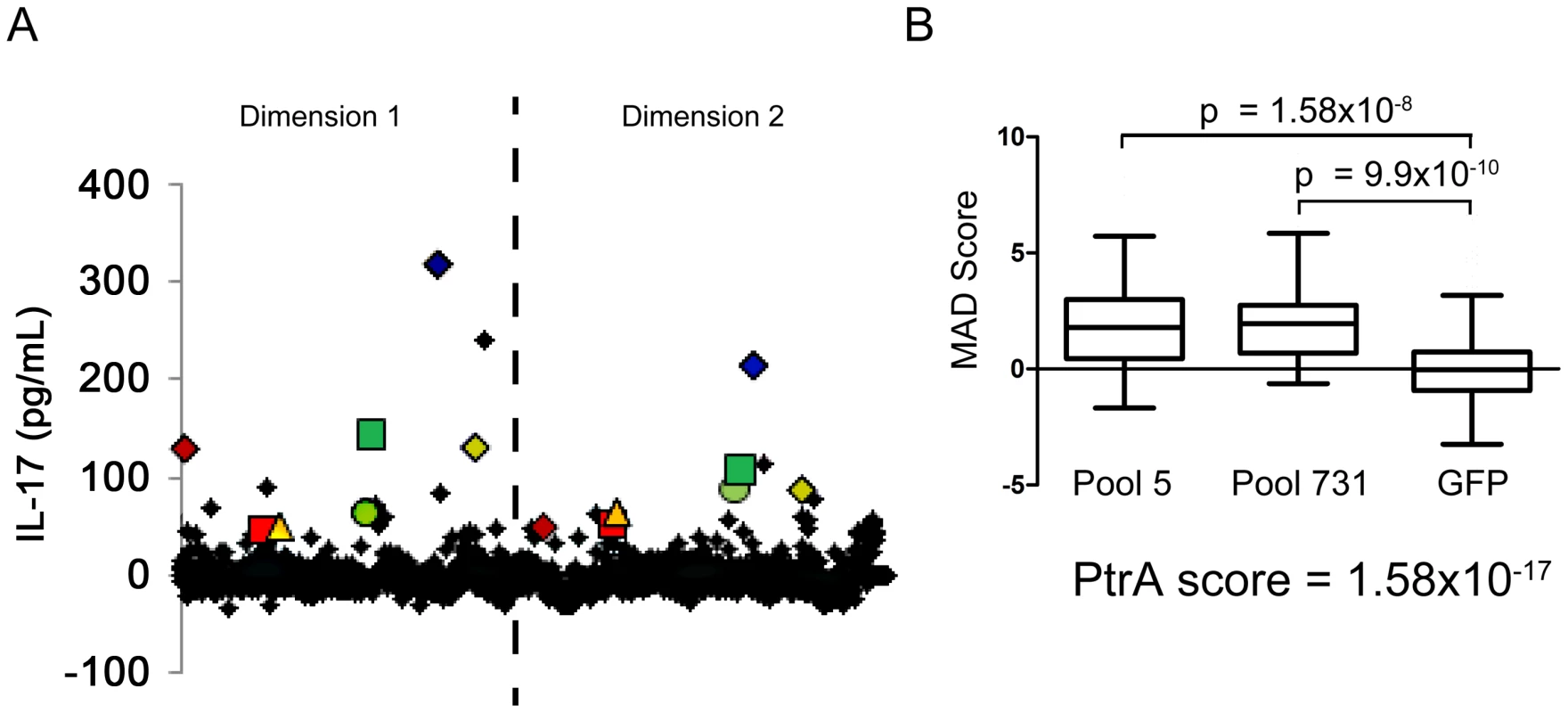
(A) The average of duplicate ELISA measurements of the IL-17A concentration in the supernatant for each protein pool is displayed. The dashed line separates data points from the two dimensions of the pooled library (see SI for details). Each pair of colored geometric data points marks the data from pools that contain the same clone. (B) The MAD score for each pool measured in screens of enriched TH17 cells from 36 subjects was compared to the MAD score for wells containing MoDCs pulsed with E. coli-expressing GFP using a two-tailed Mann-Whitney test. The antigenicity score for PtrA (SP0641.1), calculated by multiplying the p-values resulting from the Mann-Whitney test of the two pools containing the clone, is also displayed. Tab. 2. Identification of human T-cell antigens in pneumococcus. 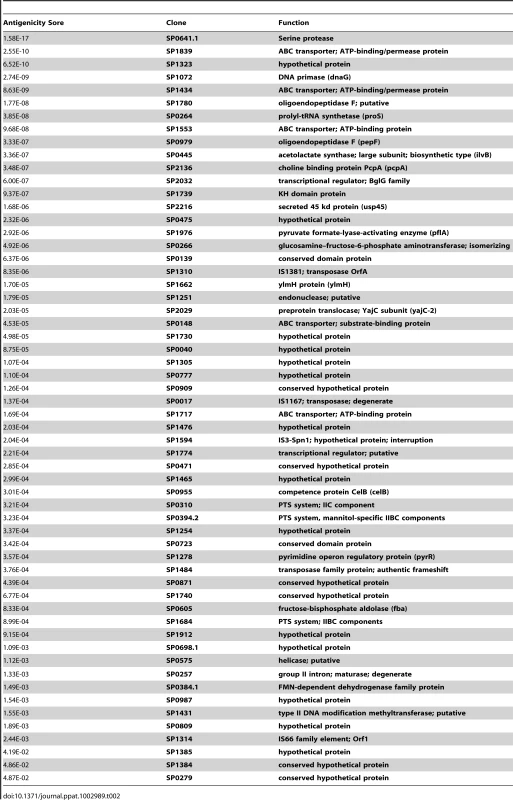
Detection of diversifying selection in pneumococcus
To evaluate genetic diversity and the underlying selection pressure on pneumococcal proteins, we systematically examined protein-encoding regions from the genome sequence data of 39 publicly-available pneumococcus strains for evidence of diversifying selection. Based on information accompanying the genome sequence data, the collection of strains covered 14 common serotypes (Table S1 in Text S1). Although the strains used in our study are not a random sample of any population and may overrepresent clinical (invasive) isolates, the distribution of serotype frequency in this study was reasonably consistent with distribution reported in human carriage [33] (Figure S3).
A flowchart of the analysis is shown in Figure 3A. Open reading frames (ORFs) that were inferred to represent the same gene in different strains were grouped together to form an orthologous group. A total of 2773 unique unambiguous groups were generated by the Proteinortho4 software [34]. Sequence alignment of genes within an orthologous group was performed using the PRANK software [35]. Extensive sequence variation was observed for many pneumococcal protein-encoding genes. The nucleotide diversity for a gene ranged from 0 to 0.23 with a median of 0.0091 (Figure 3B).
Fig. 3. Detection of diversifying selection in pneumococcus. 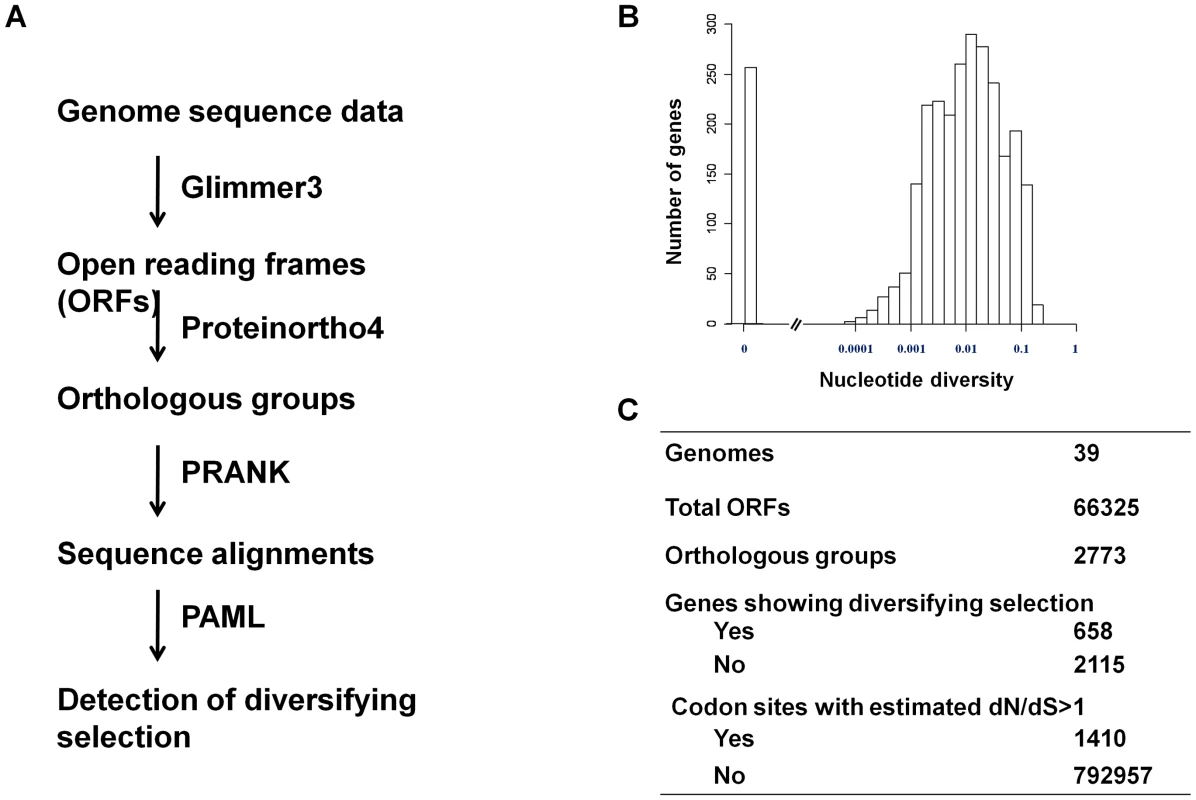
(A) Schematic of the workflow showing the procedures and software used to detect of diversifying selection in pneumococcus. (B) The distribution of nucleotide diversity (π) among pneumococcal genes. (C) A summary of number of genes and codon sites that show sign of being under positive selection. To identify pneumococcal genes that show signs of being under diversifying selection, we analyzed the non-synonymous to synonymous substitution (dN/dS) ratio for codon sites in each gene using the PAML package as described by Yang [36]. Signs of being under diversifying selection were detected by a likelihood ratio test in which a null model (dN/dS < = 1 for all codons) was compared with an alternative model (dN/dS>1for at least one codon), as described in the Materials and Methods. We concluded signs of diversifying selection for a gene if the null model was rejected at the significance level of 0.05. By this criterion, 658 genes (23.7%) showed signs of being under diversifying selection. The subsequent Bayes Empirical Bayes (BEB) analysis [37] identified 1410 codon sites, or 0.178% of total codon sites, to be under diversifying selection (Figure 3C). Codon sites under diversifying selection were enriched in cell envelope genes (Table S2 in Text S1), consistent with that interaction with antibodies might be a source of selection pressure on the pneumococcal protein antigens.
A link between immune recognition and diversifying selection
We hypothesized that if human immunity had promoted diversifying selection in pneumococcal antigens, the antigen genes would exhibit higher sequence diversity than non-antigen genes. Genes encoding CD4+ TH 17 antigens were identified as described above. Genes encoding Antibody antigens were obtained from the list published by Giefing et al [24]. TIGR4 genes belonging to an orthologous group of two or more genes were analyzed, including 1648 non-antigens, 48 TH17 antigens and 80 Antibody antigens. In addition, the regions of Antibody antigens genes that included epitopes were also noted by Giefing et al., facilitating our comparisons of non-antigens, Antibody antigen-encoding genes, and the epitope-containing and non-epitope-containing regions of these antigen-encoding genes.
The average non-synonymous substitution rate (dN) of Antibody antigens was significantly higher than that of non-antigens (Figure 4A; median 0.0032 vs. 0.0025; p = 0.022, Mann-Whitney test). However, there was no significant difference in dN between TH17 antigens and non-antigens. (Figure 4A; median 0.0026 vs. 0.0025; p = 0.65, Mann-Whitney test). Genes encoding Antibody antigens also showed a significantly higher proportion of genes with signs of being under diversifying selection (Figure 4B, OR = 1.95, p = 0.006, Fisher's Exact test). In contrast, TH17 antigen genes showed no evidence of being under diversifying selection (Figure 4B, OR = 0.77; p = 0.52; Fisher's Exact test).
Fig. 4. Antibody recognition is associated with stronger diversifying selection. 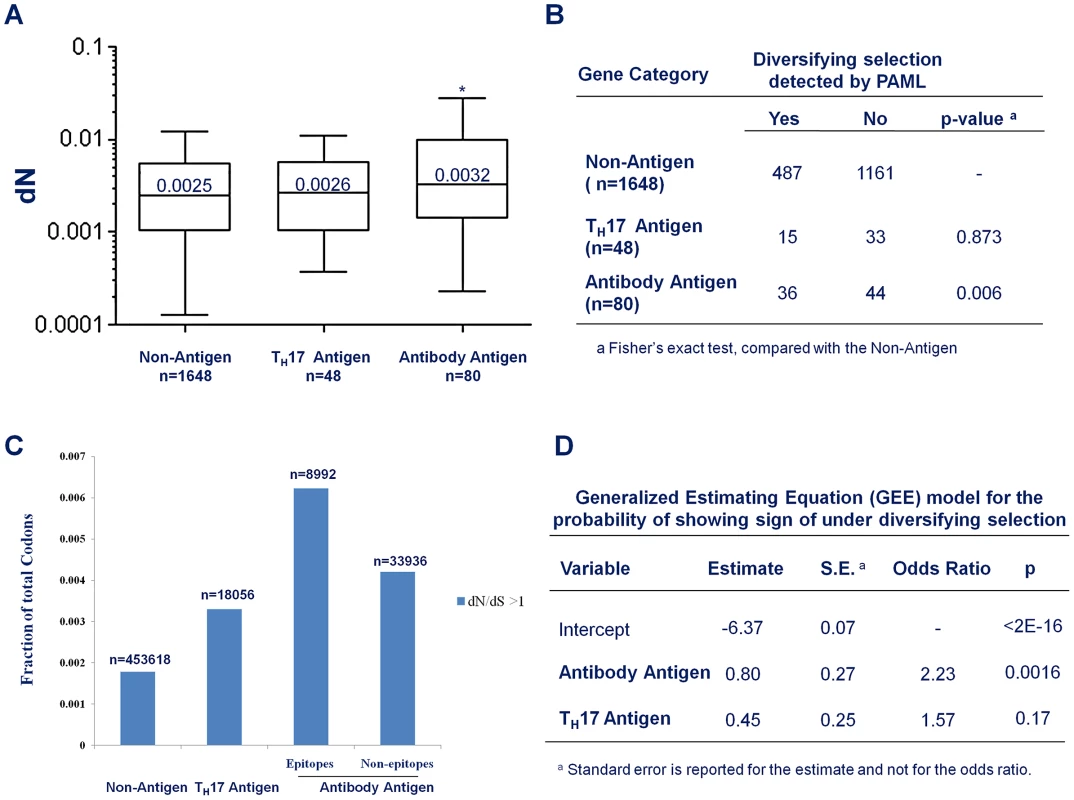
(A) Box plot to compare the average non-synonymous substitution rate (dN) of non-antigens, the Antibody antigens and CD4+ TH17 antigens. * p<0.05, Mann-Whitney test, compared with non-antigens (B) The fraction of genes that show evidence of being under diversifying selection in non-antigens, Antibody antigens and CD4+ TH17 antigens. (C) The fraction of codons with dN/dS>1 in non-antigen, the TH17 antigens, and the epitope and the non-epitope regions of the Antibody antigens. (D) Output of a generalized-estimating-equation (GEE) analysis for the effect of antibody-recognition (Antibody Antigen) and CD4+ TH17 cell- recognition (TH17 Antigen) on the probability that a gene shows signs of being under diversifying selection. Not all codon sites within a gene need be under the same selective force. To understand the contribution of host immunity to diversifying selection, we were particularly interested in whether the codon sites that did show an estimated dN/dS ratio greater than 1 were equally distributed among antigen categories. We found that 0.183% of the codon sites located in the non-antigen genes showed dN/dS ratio greater than 1 (Figure 4C). For codon sites in the CD4+ TH 17 antigen genes, a higher fraction (0.33%, Figure 4C) showed a dN/dS ratio greater than 1. An even higher fraction (0.46%, Figure 4C) of the Antibody antigen codon sites showed a dN/dS ratio greater than 1. Furthermore, within the Antibody antigens, the regions in antibody epitopes showed a higher density of codon sites with dN/dS greater than 1 than the non-epitope regions (0.62% vs. 0.42%, Figure 4C). Thus, the genomic regions that interact with antibody-mediated immunity appeared to be more enriched for codon sites with signs of being under diversifying selection, with a weaker signal of diversifying selection in the CD4+ TH17 antigens.
To account for correlations between different codon sites within a gene and for differences in gene length that would make longer genes more likely, by chance alone, to have sites with elevated dN/dS ratios, we employed a generalized-estimating-equation (GEE) model to examine the “population-averaged” effect of being recognized by human immunity on the probability that a gene is under diversifying selection [38]. Essentially, we treated the status of each individual codon in a gene (whether or not the codon showed sign of being under diversifying selection) as the outcome of a repeated measurement for the status of the gene (whether or not the gene showed sign of being under diversifying selection). During model fitting, the covariance structure across codon sites within a gene was treated as a nuisance parameter. The output of the model fitting showed that being an Antibody antigen is a highly significant predictor for being under diversifying selection (Figure 4D; OR = 2.23, p = 0.0016) and being a TH17 antigen is a weaker, and not statistically significant predictor (Fig. 4D, OR = 1.57, p = 0.17). Taken together, these results indicated that antibody immunity made a greater contribution than CD4+ TH 17 cell immunity to diversifying selection on antigen genes in S. pneumoniae.
To examine the robustness of our results, we carried out the analysis of diversifying selection using a different alignment algorithm [39], as well as another evolution model proposed by Wilson et al., which allows estimation of the dN/dS ratio in the presence of recombination [40]. All analyses yielded qualitatively similar results (Table S3 and Table S4 in Text S1).
Discussion
In this study, we investigated the contribution of host immunity to the diversifying selection in S. pneumoniae. We found that CD4+ TH17 cell-mediated immunity, elicited by exposure to pneumococci bearing a targeted antigen, cleared pneumococci that do not bear this antigen in trans almost as efficiently as it cleared the antigen-bearing cells. Thus, TH17 cell immunity limited the competitive benefit of antigenic variation within a colonized host, potentially reducing a driving force of diversifying selection. Consistent with this notion, we found a weak, and not statistically significant association between diversifying selection and recognition by human TH17 cell immunity. We hypothesize that this lack of selection is due to in trans killing of antigen-negative bacteria by innate cells recruited through TH17 cells recognition of antigen-expressing bacteria. However, the promiscuity of CD4+ T cell epitope recognition [41] could also play a role as it may be more difficult for bacteria to mutate the recognized antigens to avoid T cell recognition. In contrast to TH17 antigens, there was a significant association between recognition by human antibody and diversifying selection on the antigen. These data suggest that these two mechanisms of acquired immunity exert distinct selection forces on their respective antigens in S. pneumoniae.
We observed that an antigen-negative (AVO)/antigen-positive (OVA) ratio higher than 1 was associated with lower CFU in the ovalbumin-immunized mice but not in the control mice. This supported the antigen-specificity of the immunity recalled by the OVA strain. In principle, there are three stages of the pneumococcal life cycle in which escape from immunity might be beneficial: (1) an advantage for an escape variant by mutation or deletion of an antigen that is the target of an immune response during infection; (2) an advantage for a variant in colonizing a host already responding to a “wild-type” strain that is resident and targeted by the host's response; (3) an advantage for a variant in colonizing a host that is currently uncolonized with any pneumococcal strain, but has immunity to wild-type alleles of the antigen from previous exposure. Cis-acting immune effectors, such as antibodies, would be expected to provide an advantage for a variant at all three of these stages. Our animal experiments suggest that for CD4+ TH17 cells, the advantage of an immune-escape variant would be small at stages 1 and 2, because of in-trans killing; the first stage is particularly important because this is where a variant would likely first arise. Still, one would expect some advantage for CD4+ TH17 cell escape variants at the third stage – colonization of an uncolonized but partially immune host; this possibly may account for the weaker, less statistically convincing evidence of enrichment for diversifying selection in CD4+ TH17 antigens.
Escape from CD4+ TH 17 cell immunity in our in vivo model should be more favored than in natural settings, for two reasons. First, we constructed a model in which the TH17 epitope was completely deleted (and replaced with the reverse amino acid sequence), rather than creating a point mutation; given the promiscuity of T cell responses, many point mutations might make little or no difference to T cell recognition. Second, natural exposure to pneumococci would induce immunity to multiple T cell (and antibody) antigens, so that escape from a single response would not necessarily create a major advantage. The fact that we saw a modest benefit of losing the sole CD4+ T cell epitope against which the mice had been immunized argues that the benefit would be even weaker under natural conditions.
The high throughput screen was designed to pick up the antigens with the strongest TH17 responses in the studied sample. This strength includes both frequency of response in the studied population and the strength of the response within individuals. The Mann-Whitney analysis does not allow us to define whether an antigen was positive in any given subject. However, if we use a different analysis method of taking antigens that induce a response greater than 1.2 MAD above the median, we find that the most common antigen was recognized by 47% of the subjects, with most antigens present in 10–20% of subjects (data not shown), indicating a reasonably broad TH17 response. We acknowledge that there are weaker responses in these individuals that may have not been detected, but we posit that any selective pressure on TH17 antigens should be more robust in the strongly recognized antigens. Since no association between signs of diversifying selection and the human TH17 antigens we identified was found, the observation supports our hypothesis that CD4+ TH17 cell immunity in humans allows minimal competitive benefit for antigenic variation in S. pneumoniae. It is also important to note that only antigens recognized by IL-17A secreting T cells were identified. If the antigens recognized by different T cell lineages are distinct [42], [43], other T cells lineages may exert stronger selective pressure depending on their mechanism of action.
We found that genomic regions that showed signs of being under diversifying selection were enriched in the antibody antigen genes and further enriched in the epitopes targeted by antibodies. This finding was consistent with the conventional understanding that avoidance of antibody-recognition can provide a substantial competitive benefit. The magnitude of the enrichment was consistently modest among all analyses. It is possible that multiple ways to avoid antibody-recognition exist, reducing the dependence on non-synonymous substitutions in the antigens. For example, antigens can be temporarily down regulated at the expression level to escape from host immunity, as was seen in the malaria parasite Plasmodium falciparum [44] and suggested for meningococci under vaccine pressure [45]. Antigens are also proteins carrying out physiological functions for the pathogen at the same time. They might be subjected to diversifying, purifying or other selective forces in addition to those imposed by acquired immunity. However, the significant association between antibody-recognition and diversifying selection despite these putative competing mechanisms suggested that antibodies impose a strong fitness cost on the antigen-bearing pneumococcus. In addition, it would be interesting to understand whether the diversifying selection differs in selected genes according to the invasive potential and transformability of the strain. Appropriate comparison would require much larger samples, which we hope to investigate in future studies.
CD4+ T subsets other than the TH17 cells, such as the IFN-γ producing TH1 cells, have been proposed to play important roles in the control of pneumococcal invasive disease [42], [43] but not, to our knowledge, colonization. In fact, in our colonization model, the IFN-γ mediated mechanism appeared to be dispensable [31]. Our screen would not have picked up antigens that elicited CD4+ T responses unless they also stimulated IL-17A production. Further work might address the contribution of other forms of T cell mediated immunity to diversifying selection.
This study suggests that CD4+ TH17 cell immunity creates little selective pressure for antigenic variation while efficiently protecting against pneumococcal colonization, and suggest that the reason for this lack of selection may be due to efficient in trans killing of antigenic variants arising within a host. It is conceivable that a vaccine designed to induce TH17 cell immunity might limit the immune escape of antigenic variants and result in broader and longer protection. To this end, further research is ongoing to characterize the major TH17 cell antigens in pneumococcus and identify methods for eliciting this type of immunity through vaccination [27], [46].
Materials and Methods
Ethics statement
All human subjects enrolled in this study provided written informed consent. The protocols for this study were IRB-approved by Quorum Review, Inc.
All animal work has been conducted in compliance with the Animal Welfare Act and the guidelines of the U.S. Public Health Service Policy on Humane Care and Use of Laboratory Animals, and specifically approved by the Institutional Animal Care and Use Committee (IACUC) of Harvard Medical School. (Animal Welfare Assurance of Compliance A3431-01 and AAALAC Accreditation #000009, 6/19/09)
Strains and animals
The antigen-positive S. pneumoniae stain (OVA) was a serotype 6B strain 603 derivative that expressed the OVA323–339 peptide (ISQAVHAAHAEINEAGR) on the bacterial surface as fusion proteins with both pneumococcal surface protein A (PspA) and pneumolysin (Ply) [23]. To construct the antigen-negative S. pneumoniae (AVO), the OVA coding sequence in the pspA and ply loci of the OVA strain was replaced by a nucleotide sequence encoding the OVA323–339 peptide in reversed sequence (RGAENIEAHAAHVAQSI) by using a Janus-cassette mediated transformation protocol [47].
Wild-type, female BALB/c mice were obtained from the Jackson ImmunoResearch Laboratories, Barr Harbor, ME. All mice were 5 to 6 weeks old at the start of experiments and kept in a BL2 facility.
Immunization and challenge
Ovalbumin (Sigma-Aldrich, St. Louis, MO) and cholera toxin (CT) mucosal adjuvant (List Biological Laboratories, Compel, CA) were purchased and stored according to the manufacturer's protocols. Mice were intranasally immunized twice, one week apart, with10 µL of PBS containing 10 µg Ovalbumin plus 1 µg CT (OVA+CT) or 1 µg CT alone (CT).
Four weeks after the second immunization, mice were inoculated intranasally with a mix of the OVA and the AVO strains in 10 µl of PBS containing approximately 5×106 CFU of each strain. On days 1 and 4 after challenge, samples from live animals were collected by applying 10 µl of ice cold PBS to either nostril of a mouse and collecting droplets discharged by the animal. On day 8 after challenge, upper respiratory tract samples were collected post mortem from retrotracheal washes of sacrificed mice. Aliquots of sample were titered to determine the colonization density. The remaining samples were cultured on gentamicin plates overnight and the resulting colonies were harvested for genomic DNA extraction.
Quantitative PCR
Genomic DNA was purified from cultures of samples collected from animals using DNeasy Blood and Tissue kit (QIAGEN, Valencia, CA). The OVA strain - and the AVO strain-specific primer sets were designed based on the nucleotide sequence difference in the pspA locus between the two strains. The quantity of strain-specific genomic DNA in a sample was determined by absolute quantification protocol. A standard curve was built for each qPCR plate and was based on two replicates. All samples were measured based on averaged value of qPCR duplicate. The CFU ratio between the two strains was calculated by using the absolute amount of OVA DNA and AVO DNA in the same sample. The detection limit of AVO/OVA ratio was set as from (1×total CFU)−1 to (1×total CFU). The qPCR-derived ratios outside this range were rounded to the nearest detection limit.
Human CD4+ TH 17 antigen screen
Approval for blood collection was obtained from the Institutional Review Boards of each institution. IL-17A-secreting CD4+ T cells were first enriched from peripheral blood cells using negative magnetic selection of CD4+ T cells and a previously published IL-17A cytokine capture protocol [48]. S. pneumoniae-specific TH17 cells were further enriched by culturing the cells with autologous monocyte-derived dendritic cells (MoDCs) pulsed with inactivated S. pneumoniae. IL-17A secretion from the cells was measured after three days of co-culture with MoDCs pulsed with E. coli expressing a previously validated 2,547 clone ORFeome library of the S. pneumoniae TIGR4 genome [49] arrayed in pools of four clones. Enriched cells from 36 peripheral blood samples were screened with the pooled library (see SI for methods detail). The results of the IL-17A ELISA were first normalized by plate by averaging the duplicates for each well, subtracting the plate median from each average and then dividing the result by the median absolute deviation of the plate, yielding the MAD score for each well in the screen. The most common antigens recognized by the population were identified by comparing the population response to each pool in the library to the measured responses to the all the wells that received E. coli expressing GFP using a one-tailed Mann-Whitney test. Each individual antigen was then scored by multiplying the p-values from the Mann-Whitney test of the two wells in which it was present.
Genome sequences
Genome sequence data of 39 pneumococcal strains were retrieved from the NCBI FTP site, ftp://ftp.ncbi.nih.gov/genomes. The collection included 14 annotated genomes and 25 draft genomes. Accession numbers of genome sequence were listed in Table S1 in Text S1. For the annotated genomes, the annotation and nucleotide sequence of each gene were downloaded from the NCBI FTP site. For the draft genomes, putative protein-encoding genes were identified by using the Glimmer3 software [50]. Orthology analysis of pneumococcal proteins was carried out by using the Proteinortho4 software [34], which assigned orthologous proteins from different strains into a same orthologous group based on the reciprocal best alignment heuristic. Cellular roles of TIGR4 genes were categorized according to the JCVI Annotation Gene Attributes (http://cmr.jcvi.org).
Analysis of the non-synonymous to synonymous rate ratio (dN/dS ratio)
The gene sequences of each orthologous group were aligned based on the amino acid sequences they encode (codon alignment) and a gene tree was constructed using either the ClustalW software or the PRANK software [35], [39]. A likelihood ratio test was applied to compare a null model with an alternative model of the distribution of the dN/dS ratio parameter, ω, among codon sites, as described in [37]. In the null model (nearly-neutral model), each codon site within a gene is assumed to be either under purifying selection (ω0<1) or under neutral evolution (ω1 = 1). In the alternative model (positive selection model), a codon site can be under purifying selection (ω0<1), under neutral evolution (ω1 = 1) or under diversifying selection (ω2>1). For each model, the log likelihood value was calculated by the CodeML program from the package PAML [36]. If the null model was rejected by the likelihood ratio test at a significance level of 0.05, the gene represented by the orthologous group would be considered as being under diversifying selection. For such genes, a Bayes Empirical Bayes (BEB) analysis implemented in the CodeML program [36] was used to determine the particular codon sites that were under diversifying selection.
The output file of the CodeML program included non-synonymous substitution rate (dN) derived from pair wise sequence comparison. The average dN for each orthologous group was estimated by averaging over all pair wise dNs.
The dN/dS ratio for codon sites was also estimated by a method developed by Wilson et al., which applied a population genetics approximation to the coalescent to accommodate recombination events [40]. The codon alignment of each orthologous group was analyzed by Omegamap software with a prior exponential distribution of ω and a prior ω mean of 1. Each codon site was assumed to have independent ω and the posterior distributions of ω were obtains by 500,000 iterations. A codon site was defined to show evidence of being under diversifying selection if 95% of its posterior distribution of ω was above 1. A gene was considered to show evidence of being under diversifying selection if any codon site within the gene showed sign of being under diversifying selection. The analyses took 3–4 weeks on a Linux cluster comprised of 4708 processor cores.
Statistical analysis was performed by using the R package (http://www.r-project.org). Graphs were created in Graphpad Prism and in Microsoft Excel.
List of NCBI-Gene ID numbers for genes and proteins mentioned in the text: 929896 (PspA), 931915 (Pneumolysin), 930590 (PtrA).
Supporting Information
Zdroje
1. LipsitchM, O'HaganJJ (2007) Patterns of antigenic diversity and the mechanisms that maintain them. J R Soc Interface 4 : 787–802.
2. MaW, GuttmanDS (2008) Evolution of prokaryotic and eukaryotic virulence effectors. Curr Opin Plant Biol 11 : 412–419.
3. WeedallGD, ConwayDJ (2010) Detecting signatures of balancing selection to identify targets of anti-parasite immunity. Trends Parasitol 26 : 363–369.
4. FrostSD, WrinT, SmithDM, Kosakovsky PondSL, LiuY, et al. (2005) Neutralizing antibody responses drive the evolution of human immunodeficiency virus type 1 envelope during recent HIV infection. Proc Natl Acad Sci U S A 102 : 18514–18519.
5. LiuY, McNevinJ, CaoJ, ZhaoH, GenowatiI, et al. (2006) Selection on the human immunodeficiency virus type 1 proteome following primary infection. J Virol 80 : 9519–9529.
6. GoulderPJ, BranderC, TangY, TremblayC, ColbertRA, et al. (2001) Evolution and transmission of stable CTL escape mutations in HIV infection. Nature 412 : 334–338.
7. PlotkinJB, DushoffJ, LevinSA (2002) Hemagglutinin sequence clusters and the antigenic evolution of influenza A virus. Proc Natl Acad Sci U S A 99 : 6263–6268.
8. SimonsenL, ViboudC, GrenfellBT, DushoffJ, JenningsL, et al. (2007) The genesis and spread of reassortment human influenza A/H3N2 viruses conferring adamantane resistance. Mol Biol Evol 24 : 1811–1820.
9. PolleySD, ConwayDJ (2001) Strong diversifying selection on domains of the Plasmodium falciparum apical membrane antigen 1 gene. Genetics 158 : 1505–1512.
10. BaumJ, ThomasAW, ConwayDJ (2003) Evidence for diversifying selection on erythrocyte-binding antigens of Plasmodium falciparum and P. vivax. Genetics 163 : 1327–1336.
11. BirrerMJ, UdemS, NathensonS, BloomBR (1981) Antigenic variants of measles virus. Nature 293 : 67–69.
12. ComasI, ChakravarttiJ, SmallPM, GalaganJ, NiemannS, et al. (2010) Human T cell epitopes of Mycobacterium tuberculosis are evolutionarily hyperconserved. Nat Genet 42 : 498–503.
13. CobeyS, LipsitchM (2012) Niche and neutral effects of acquired immunity permit coexistence of pneumococcal serotypes. Science 335 : 1376–1380.
14. WeinbergerDM, DaganR, Givon-LaviN, Regev-YochayG, MalleyR, et al. (2008) Epidemiologic evidence for serotype-specific acquired immunity to pneumococcal carriage. J Infect Dis 197 : 1511–1518.
15. MalleyR (2010) Antibody and cell-mediated immunity to Streptococcus pneumoniae: implications for vaccine development. J Mol Med (Berl) 88 : 135–142.
16. GoldblattD, HussainM, AndrewsN, AshtonL, VirtaC, et al. (2005) Antibody responses to nasopharyngeal carriage of Streptococcus pneumoniae in adults: a longitudinal household study. J Infect Dis 192 : 387–393.
17. MalleyR, LipsitchM, BogaertD, ThompsonCM, HermansP, et al. (2007) Serum antipneumococcal antibodies and pneumococcal colonization in adults with chronic obstructive pulmonary disease. J Infect Dis 196 : 928–935.
18. TrzcinskiK, ThompsonC, MalleyR, LipsitchM (2005) Antibodies to conserved pneumococcal antigens correlate with, but are not required for, protection against pneumococcal colonization induced by prior exposure in a mouse model. Infect Immun 73 : 7043–7046.
19. RapolaS, JanttiV, HaikalaR, SyrjanenR, CarloneGM, et al. (2000) Natural development of antibodies to pneumococcal surface protein A, pneumococcal surface adhesin A, and pneumolysin in relation to pneumococcal carriage and acute otitis media. J Infect Dis 182 : 1146–1152.
20. SimellB, KorkeilaM, PursiainenH, KilpiTM, KayhtyH (2001) Pneumococcal carriage and otitis media induce salivary antibodies to pneumococcal surface adhesin a, pneumolysin, and pneumococcal surface protein a in children. J Infect Dis 183 : 887–896.
21. BrilesDE, HollingsheadSK, KingJ, SwiftA, BraunPA, et al. (2000) Immunization of humans with recombinant pneumococcal surface protein A (rPspA) elicits antibodies that passively protect mice from fatal infection with Streptococcus pneumoniae bearing heterologous PspA. J Infect Dis 182 : 1694–1701.
22. CuiY, ZhangX, GongY, NiuS, YinN, et al. (2011) Immunization with DnaJ (hsp40) could elicit protection against nasopharyngeal colonization and invasive infection caused by different strains of Streptococcus pneumoniae. Vaccine 29 : 1736–1744.
23. TrzcinskiK, ThompsonCM, SrivastavaA, BassetA, MalleyR, et al. (2008) Protection against nasopharyngeal colonization by Streptococcus pneumoniae is mediated by antigen-specific CD4+ T cells. Infect Immun 76 : 2678–2684.
24. GiefingC, MeinkeAL, HannerM, HenicsT, BuiMD, et al. (2008) Discovery of a novel class of highly conserved vaccine antigens using genomic scale antigenic fingerprinting of pneumococcus with human antibodies. J Exp Med 205 : 117–131.
25. MalleyR, TrzcinskiK, SrivastavaA, ThompsonCM, AndersonPW, et al. (2005) CD4+ T cells mediate antibody-independent acquired immunity to pneumococcal colonization. Proc Natl Acad Sci U S A 102 : 4848–4853.
26. BassetA, ThompsonCM, HollingsheadSK, BrilesDE, AdesEW, et al. (2007) Antibody-independent, CD4+ T-cell-dependent protection against pneumococcal colonization elicited by intranasal immunization with purified pneumococcal proteins. Infect Immun 75 : 5460–5464.
27. LuYJ, ForteS, ThompsonCM, AndersonPW, MalleyR (2009) Protection against Pneumococcal colonization and fatal pneumonia by a trivalent conjugate of a fusion protein with the cell wall polysaccharide. Infect Immun 77 : 2076–2083.
28. LuYJ, SkovstedIC, ThompsonCM, AndersonPW, MalleyR (2009) Mechanisms in the serotype-independent pneumococcal immunity induced in mice by intranasal vaccination with the cell wall polysaccharide. Microb Pathog 47 : 177–182.
29. HarfoucheC, FilippiniS, GianfaldoniC, RuggieroP, MoschioniM, et al. (2012) RrgB321, a fusion protein of the three variants of the pneumococcal pilus backbone RrgB, is protective in vivo and elicits opsonic antibodies. Infect Immun 80 : 451–460.
30. MalleyR, SrivastavaA, LipsitchM, ThompsonCM, WatkinsC, et al. (2006) Antibody-independent, interleukin-17A-mediated, cross-serotype immunity to pneumococci in mice immunized intranasally with the cell wall polysaccharide. Infect Immun 74 : 2187–2195.
31. LuYJ, GrossJ, BogaertD, FinnA, BagradeL, et al. (2008) Interleukin-17A mediates acquired immunity to pneumococcal colonization. PLoS Pathog 4: e1000159.
32. ZhangZ, ClarkeTB, WeiserJN (2009) Cellular effectors mediating Th17-dependent clearance of pneumococcal colonization in mice. J Clin Invest 119 : 1899–1909.
33. BogaertD, EngelenMN, Timmers-RekerAJ, ElzenaarKP, PeerboomsPG, et al. (2001) Pneumococcal carriage in children in The Netherlands: a molecular epidemiological study. J Clin Microbiol 39 : 3316–3320.
34. LechnerM, FindeissS, SteinerL, MarzM, StadlerPF, et al. (2011) Proteinortho: detection of (co−)orthologs in large-scale analysis. BMC Bioinformatics 12 : 124.
35. LoytynojaA, GoldmanN (2005) An algorithm for progressive multiple alignment of sequences with insertions. Proc Natl Acad Sci U S A 102 : 10557–10562.
36. YangZ (2007) PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24 : 1586–1591.
37. YangZ, WongWS, NielsenR (2005) Bayes empirical bayes inference of amino acid sites under positive selection. Mol Biol Evol 22 : 1107–1118.
38. ZegerSL, LiangKY (1986) Longitudinal data analysis for discrete and continuous outcomes. Biometrics 42 : 121–130.
39. ThompsonJD, GibsonTJ, HigginsDG (2002) Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics Chapter 2: Unit 2 3.
40. WilsonDJ, McVeanG (2006) Estimating diversifying selection and functional constraint in the presence of recombination. Genetics 172 : 1411–1425.
41. WucherpfennigKW (2004) T cell receptor crossreactivity as a general property of T cell recognition. Mol Immunol 40 : 1009–1017.
42. WeberSE, TianH, PirofskiLA (2011) CD8+ cells enhance resistance to pulmonary serotype 3 Streptococcus pneumoniae infection in mice. J Immunol 186 : 432–442.
43. SunK, SalmonSL, LotzSA, MetzgerDW (2007) Interleukin-12 promotes gamma interferon-dependent neutrophil recruitment in the lung and improves protection against respiratory Streptococcus pneumoniae infection. Infect Immun 75 : 1196–1202.
44. ScherfA, Lopez-RubioJJ, RiviereL (2008) Antigenic variation in Plasmodium falciparum. Annu Rev Microbiol 62 : 445–470.
45. MaidenMC, Ibarz-PavonAB, UrwinR, GraySJ, AndrewsNJ, et al. (2008) Impact of meningococcal serogroup C conjugate vaccines on carriage and herd immunity. J Infect Dis 197 : 737–743.
46. LuYJ, ZhangF, SayeedS, ThompsonCM, SzuS, et al. (2012) A bivalent vaccine to protect against Streptococcus pneumoniae and Salmonella typhi. Vaccine 30 : 3405–3412.
47. SungCK, LiH, ClaverysJP, MorrisonDA (2001) An rpsL cassette, janus, for gene replacement through negative selection in Streptococcus pneumoniae. Appl Environ Microbiol 67 : 5190–5196.
48. StreeckH, CohenKW, JolinJS, BrockmanMA, MeierA, et al. (2008) Rapid ex vivo isolation and long-term culture of human Th17 cells. J Immunol Methods 333 : 115–125.
49. MoffittKL, GierahnTM, LuYJ, GouveiaP, AldersonM, et al. (2011) T(H)17-based vaccine design for prevention of Streptococcus pneumoniae colonization. Cell Host Microbe 9 : 158–165.
50. DelcherAL, BratkeKA, PowersEC, SalzbergSL (2007) Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23 : 673–679.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2012 Číslo 11- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Egyptian H5N1 Influenza Viruses—Cause for Concern?
- Epigenetics of Host–Pathogen Interactions: The Road Ahead and the Road Behind
- Environmental Regulation of Prions in Yeast
- : Dissecting the Molecular Interface between Pathogen and Plant
- A Wolf in Sheep's Clothing: SV40 Co-opts Host Genome Maintenance Proteins to Replicate Viral DNA
- Exploring New Biological Functions of Amyloids: Bacteria Cell Agglutination Mediated by Host Protein Aggregation
- A Trade-off between the Fitness Cost of Functional Integrases and Long-term Stability of Integrons
- Intracellular Vesicle Acidification Promotes Maturation of Infectious Poliovirus Particles
- Distinct Effects on Diversifying Selection by Two Mechanisms of Immunity against
- Whole Genome Sequencing Reveals Local Transmission Patterns of in Sympatric Cattle and Badger Populations
- Early Mechanisms of Pathobiology Are Revealed by Transcriptional Temporal Dynamics in Hippocampal CA1 Neurons of Prion Infected Mice
- Revised Phylogeny and Novel Horizontally Acquired Virulence Determinants of the Model Soft Rot Phytopathogen SCC3193
- : Where Does It Live?
- The Role of Auxin-Cytokinin Antagonism in Plant-Pathogen Interactions
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- A Wolf in Sheep's Clothing: SV40 Co-opts Host Genome Maintenance Proteins to Replicate Viral DNA
- Intracellular Vesicle Acidification Promotes Maturation of Infectious Poliovirus Particles
- Revised Phylogeny and Novel Horizontally Acquired Virulence Determinants of the Model Soft Rot Phytopathogen SCC3193
- The Role of Auxin-Cytokinin Antagonism in Plant-Pathogen Interactions
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



