-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaDNA Watermarking of Infectious Agents: Progress and Prospects
article has not abstract
Published in the journal: . PLoS Pathog 6(6): e32767. doi:10.1371/journal.ppat.1000950
Category: Opinion
doi: https://doi.org/10.1371/journal.ppat.1000950Summary
article has not abstract
Following the 2001 anthrax attacks, infectious disease research laboratories and personnel were subjected to increased scrutiny amid concerns that the released agent originated from within such facilities. Since then, enhanced regulatory controls have been implemented to thwart the possibility of future releases. However, improved microbial forensics technologies have not been employed to facilitate fault attribution or to control and track agent inventories.
We believe that novel systems employing enhanced identity protection will instill new public confidence in scientists and avoid erroneous assignment of liability in the case of a release. We propose a DNA watermarking system that includes institution-, laboratory-, and/or investigator-specific watermarks in the genomes of organisms, especially Select Agents. The system will achieve five key goals critical to any watermarking system, phrased in general information theoretic terms: message fidelity, error tolerance, ease of interpretation, availability of signatures, and resistance to attack (Table 1).
Tab. 1. Goals and features of watermarking system. 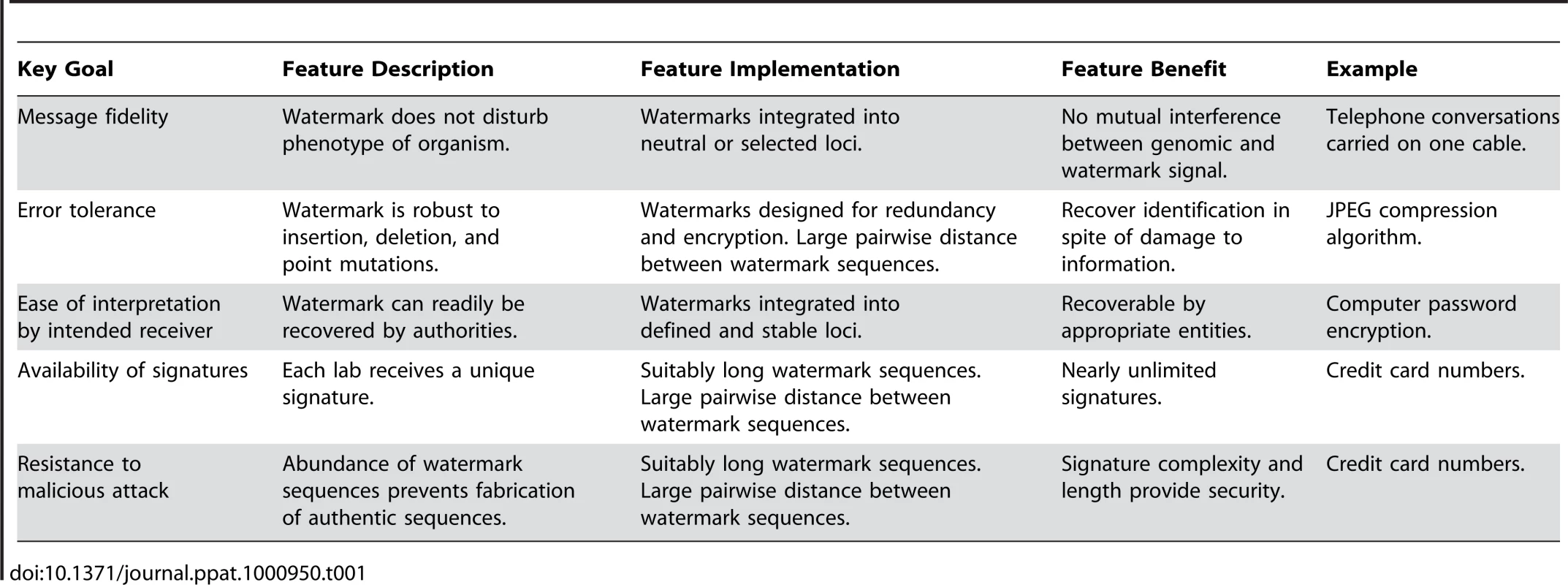
A DNA watermark is a unique synthetic DNA sequence embedded into the genome of a genetically tractable organism. The watermark provides a means for agent, isolate, or strain identification and tracking by PCR amplification and sequencing of the embedded tag. The power of watermarking for agent control emerges when the technology is linked to the activities of a trusted authorizing entity (Figure 1). This entity could, for example, be charged with distributing organisms containing unique watermark sequences to individual laboratories and/or investigators. These watermarks would distinguish their organisms from those of others in the research community. Laboratories would be encouraged, permitted, or required to use only strains that contain their approved, and confidential, watermark. In the event of release, the offending pathogen would be interrogated for the presence of an approved watermark. If such a watermark were present, then information about the possible source would become immediately available. Of course, pathogen-specific standard operating procedures (SOPs) that ensure the integrity of the watermarking system (to prevent cross-contamination, manage the sharing of strains, and prevent accidental or intentional misuse) would be a necessary component of any watermarking strategy.
Fig. 1. Proposed watermark implementation strategies. 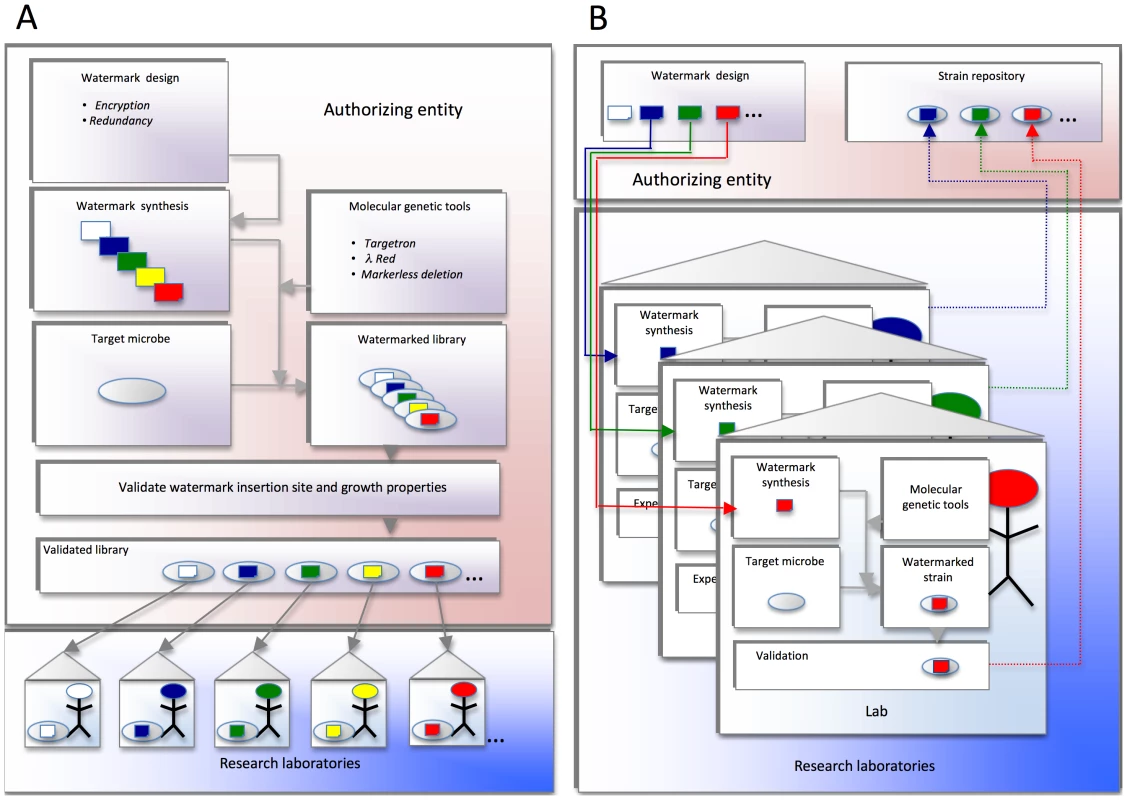
(A) Schema in which a central authorizing entity distributes watermarked strains to the research community. The authorizing entity designs and synthesizes confidential DNA watermark sequences. These sequences are introduced into genetically tractable target organisms using molecular genetic approaches that have proven efficacious in the agent. Proper insertion of watermarks at the desired loci is confirmed. The growth properties are also validated. The watermarked strains are finally distributed to individual labs, institutions, or investigators for experimental use. (B) Schema in which a central authorizing entity distributes to the research community DNA sequences that can be used for watermarking strains. The authorizing entity designs and distributes confidential DNA sequences for watermarking strains (solid lines). Individual institutions, laboratories, or investigators use this DNA to watermark their experimental strains. The individual research entities also confirm the proper integration of watermarked DNA sequences into the appropriate target genomic loci, and characterize the phenotypes of the resultant watermarked strains. The authorizing entity can also serve as a repository for watermarked strains (dotted lines). Previously developed watermarking technologies include approaches for embedding watermarks in microbial genomes [1]–[4] and strategies for encryption [2], [5], [6]. Each method seeks to develop a genetic cipher that is 1) robust to mutation, 2) easy for intended users to decipher, and 3) difficult for third parties to decipher or alter. While these strategies for manipulating watermarks have been successful at watermark encoding, placement in a genome, retrieval from a genome, and decoding, none of the techniques achieves all of the five goals (outlined in Table 1) that are necessary for a watermarking system for Select Agent tracking.
In our opinion, however, these techniques are worthy of further investigation, as regards their utility for the research and biosecurity communities. We propose investigation proceed on three discrete but interconnected fronts. First, the theoretical mathematical and information aspects of watermarking systems must be examined and rigorously tested in silico. Second, insertion and removal of watermarks from microbial genomes must be assessed, and the phenotypic invisibility of the watermarks tested. Finally, pathogen-specific SOPs must be developed, keeping in mind the need for transparency and collaboration in research, and tested in a “role playing” scenario. Our initial work indicates that the model is mathematically plausible. Previous work in the use of watermarks suggests that appropriately placed watermarks can be phenotypically neutral [1], [3]. The technologies to introduce watermarks into several of the highest risk Select Agent genomes are currently available, using site-specific insertion tools such as Targetron (intron-based homing) and Lambda red mutagenesis. Adaptation of these or comparable genetic tools for less tractable Select Agents would require technological advances that would also broadly benefit research of each agent.
Adoption of a watermarking strategy by research groups would need to be justified by a cost-benefit analysis, from an institutional liability perspective, and from the perspective of the research community. Several salient concerns can be readily identified. Our proposed system does not protect against covert usage of naturally occurring wild-type strains or remediate existing stocks, but instead provides a forward-looking strategy. To address cost concerns, funding agencies that require enhanced inventory control could be encouraged or required to support the cost of implementing watermarking systems. Similarly, these agencies could support or collaborate with private or public authorizing entities to develop SOPs for strain management. Finally, convincingly establishing phenotypic neutrality of genomic modifications will be non-trivial, and thus, will constitute an important area for future research. Despite these potential impediments, watermarking would nearly eliminate the potential for mistaken assignment of source for a suspected agent release. Moreover, the development and implementation costs may prove to be much less than other proposed measures for enhancing laboratory security, including around-the-clock security patrols.
We considered two variations on the operational infrastructure required (Figure 1). An authorizing entity could (Figure 1A) design, insert, and distribute, or (Figure 1B) simply distribute, the secured watermark to requesting laboratory. In the latter scenario, the requesting laboratory would be responsible for adapting genetic technology to deliver the watermark. We do not propose that previously generated modified strains (mutant collections, etc.) would be modified and restocked. The transition to marked strains would be incremental but stable. We speculate that an efficient approach to this scenario would be to provide funding opportunities to establish and validate agent-specific systems. While there are several potential impediments to implementing the proposed watermarking systems, the combination of positive impact on lay perception of responsible scientific activity and an increased confidence in control of liability by investigators and institutions provides a rationale to investigate the development of watermarking tools for Select Agent research.
Zdroje
1. HeiderD
BarnekowA
2008 DNA watermarks: a proof of concept. BMC Mol Biol 9 40
2. HeiderD
BarnekowA
2008 DNA-based watermarks using the DNA-Crypt algorithm. BMC Bioinformatics 8 176
3. AritaM
OhashiY
2004 Secret signatures inside genomic DNA. Biotechnol Prog 20 1605 1607
4. WongP
WongK
FooteH
2005 Organic data memory using the DNA approach. Ann N Y Acad Sci 1053 162 173
5. LeierA
RichterC
BanzhafW
RauheH
2000 Cryptography with DNA binary strands. Biosystems 57 13 22
6. SmithGC
FiddesCC
HawkinsJP
CoxJP
2003 Some possible codes for encrypting data in DNA. Biotechnol Lett 25 1125 1130
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Insight into the Mechanisms of Adenovirus Capsid Disassembly from Studies of Defensin NeutralizationČlánek Serum-Dependent Selective Expression of EhTMKB1-9, a Member of B1 Family of Transmembrane KinasesČlánek Host Cell Invasion and Virulence in Sepsis Is Facilitated by the Multiple Repeats within FnBPA
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2010 Číslo 6- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Cognitive Dysfunction Is Sustained after Rescue Therapy in Experimental Cerebral Malaria, and Is Reduced by Additive Antioxidant Therapy
- DNA Watermarking of Infectious Agents: Progress and Prospects
- Self-Protection against Gliotoxin—A Component of the Gliotoxin Biosynthetic Cluster, GliT, Completely Protects Against Exogenous Gliotoxin
- Modifies the Tsetse Salivary Composition, Altering the Fly Feeding Behavior That Favors Parasite Transmission
- Requirement of NOX2 and Reactive Oxygen Species for Efficient RIG-I-Mediated Antiviral Response through Regulation of MAVS Expression
- The Enteropathogenic Effector EspF Targets and Disrupts the Nucleolus by a Process Regulated by Mitochondrial Dysfunction
- Epithelial p38α Controls Immune Cell Recruitment in the Colonic Mucosa
- Coexpression of PD-1, 2B4, CD160 and KLRG1 on Exhausted HCV-Specific CD8+ T Cells Is Linked to Antigen Recognition and T Cell Differentiation
- Insight into the Mechanisms of Adenovirus Capsid Disassembly from Studies of Defensin Neutralization
- EspA Acts as a Critical Mediator of ESX1-Dependent Virulence in by Affecting Bacterial Cell Wall Integrity
- An RNA Element at the 5′-End of the Poliovirus Genome Functions as a General Promoter for RNA Synthesis
- Tetherin Restricts Productive HIV-1 Cell-to-Cell Transmission
- Epstein-Barr Virus-Encoded LMP2A Induces an Epithelial–Mesenchymal Transition and Increases the Number of Side Population Stem-like Cancer Cells in Nasopharyngeal Carcinoma
- Protein Kinase A Dependent Phosphorylation of Apical Membrane Antigen 1 Plays an Important Role in Erythrocyte Invasion by the Malaria Parasite
- Requirement for Ergosterol in V-ATPase Function Underlies Antifungal Activity of Azole Drugs
- The SOCS-Box of HIV-1 Vif Interacts with ElonginBC by Induced-Folding to Recruit Its Cul5-Containing Ubiquitin Ligase Complex
- A Crucial Role for Infected-Cell/Antibody Immune Complexes in the Enhancement of Endogenous Antiviral Immunity by Short Passive Immunotherapy
- Modulation of the Arginase Pathway in the Context of Microbial Pathogenesis: A Metabolic Enzyme Moonlighting as an Immune Modulator
- Role of Abl Kinase and the Wave2 Signaling Complex in HIV-1 Entry at a Post-Hemifusion Step
- Serum-Dependent Selective Expression of EhTMKB1-9, a Member of B1 Family of Transmembrane Kinases
- Epigenetic Repression of by Latent Epstein-Barr Virus Requires the Interaction of EBNA3A and EBNA3C with CtBP
- Host Cell Invasion and Virulence in Sepsis Is Facilitated by the Multiple Repeats within FnBPA
- Role of PKR and Type I IFNs in Viral Control during Primary and Secondary Infection
- A Kinome RNAi Screen Identified AMPK as Promoting Poxvirus Entry through the Control of Actin Dynamics
- Cryptococcal Cell Morphology Affects Host Cell Interactions and Pathogenicity
- NleG Type 3 Effectors from Enterohaemorrhagic Are U-Box E3 Ubiquitin Ligases
- Human Cytomegalovirus UL29/28 Protein Interacts with Components of the NuRD Complex Which Promote Accumulation of Immediate-Early RNA
- Complement Receptor 1 Is a Sialic Acid-Independent Erythrocyte Receptor of
- A Viral microRNA Down-Regulates Multiple Cell Cycle Genes through mRNA 5′UTRs
- Immunotoxin Complementation of HAART to Deplete Persisting HIV-Infected Cell Reservoirs
- Protein Expression Redirects Vesicular Stomatitis Virus RNA Synthesis to Cytoplasmic Inclusions
- Entry and Fusion of Emerging Paramyxoviruses
- Paramyxovirus Entry and Targeted Vectors for Cancer Therapy
- Suppressing Glucose Transporter Gene Expression in Schistosomes Impairs Parasite Feeding and Decreases Survival in the Mammalian Host
- Formation of Complexes at Plasmodesmata for Potyvirus Intercellular Movement Is Mediated by the Viral Protein P3N-PIPO
- Fungal Cell Gigantism during Mammalian Infection
- Two Novel Point Mutations in Clinical Reduce Linezolid Susceptibility and Switch on the Stringent Response to Promote Persistent Infection
- Rotavirus Structural Proteins and dsRNA Are Required for the Human Primary Plasmacytoid Dendritic Cell IFNα Response
- The Epigenetic Landscape of Latent Kaposi Sarcoma-Associated Herpesvirus Genomes
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Requirement of NOX2 and Reactive Oxygen Species for Efficient RIG-I-Mediated Antiviral Response through Regulation of MAVS Expression
- Formation of Complexes at Plasmodesmata for Potyvirus Intercellular Movement Is Mediated by the Viral Protein P3N-PIPO
- Insight into the Mechanisms of Adenovirus Capsid Disassembly from Studies of Defensin Neutralization
- Two Novel Point Mutations in Clinical Reduce Linezolid Susceptibility and Switch on the Stringent Response to Promote Persistent Infection
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



