-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaAn RNA Element at the 5′-End of the Poliovirus Genome Functions as a General Promoter for RNA Synthesis
RNA structures present throughout RNA virus genomes serve as scaffolds to organize multiple factors involved in the initiation of RNA synthesis. Several of these RNA elements play multiple roles in the RNA replication pathway. An RNA structure formed around the 5′ - end of the poliovirus genomic RNA has been implicated in the initiation of both negative - and positive-strand RNA synthesis. Dissecting the roles of these multifunctional elements is usually hindered by the interdependent nature of the viral replication processes and often pleiotropic effects of mutations. Here, we describe a novel approach to examine RNA elements with multiple roles. Our approach relies on the duplication of the RNA structure so that one copy is dedicated to the initiation of negative-strand RNA synthesis, while the other mediates positive-strand synthesis. This allows us to study the function of the element in promoting positive-strand RNA synthesis, independently of its function in negative-strand initiation. Using this approach, we demonstrate that the entire 5′-end RNA structure that forms on the positive-strand is required for initiation of new positive-strand RNAs. Also required to initiate positive-strand RNA synthesis are the binding sites for the viral polymerase precursor, 3CD, and the host factor, PCBP. Furthermore, we identify specific nucleotide sequences within “stem a” that are essential for the initiation of positive-strand RNA synthesis. These findings provide direct evidence for a trans-initiation model, in which binding of proteins to internal sequences of a pre-existing positive-strand RNA affects the synthesis of subsequent copies of that RNA, most likely by organizing replication factors around the initiation site.
Published in the journal: . PLoS Pathog 6(6): e32767. doi:10.1371/journal.ppat.1000936
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1000936Summary
RNA structures present throughout RNA virus genomes serve as scaffolds to organize multiple factors involved in the initiation of RNA synthesis. Several of these RNA elements play multiple roles in the RNA replication pathway. An RNA structure formed around the 5′ - end of the poliovirus genomic RNA has been implicated in the initiation of both negative - and positive-strand RNA synthesis. Dissecting the roles of these multifunctional elements is usually hindered by the interdependent nature of the viral replication processes and often pleiotropic effects of mutations. Here, we describe a novel approach to examine RNA elements with multiple roles. Our approach relies on the duplication of the RNA structure so that one copy is dedicated to the initiation of negative-strand RNA synthesis, while the other mediates positive-strand synthesis. This allows us to study the function of the element in promoting positive-strand RNA synthesis, independently of its function in negative-strand initiation. Using this approach, we demonstrate that the entire 5′-end RNA structure that forms on the positive-strand is required for initiation of new positive-strand RNAs. Also required to initiate positive-strand RNA synthesis are the binding sites for the viral polymerase precursor, 3CD, and the host factor, PCBP. Furthermore, we identify specific nucleotide sequences within “stem a” that are essential for the initiation of positive-strand RNA synthesis. These findings provide direct evidence for a trans-initiation model, in which binding of proteins to internal sequences of a pre-existing positive-strand RNA affects the synthesis of subsequent copies of that RNA, most likely by organizing replication factors around the initiation site.
Introduction
The genome of positive-strand RNA viruses has dual functions: as viral mRNA and as template for the synthesis of additional RNA genomes. Replication of the viral RNA occurs by a highly regulated, efficient mechanism, which produces tens of thousands of new RNA copies in only a few hours. Positive-strand RNA viruses follow a common strategy for replication: the viral genome is transcribed into a negative-strand intermediate, which, in turn, acts as a template for new positive-strand synthesis. The same RNA-dependent RNA polymerase synthesizes both RNA strands using viral and host factors. However, replication is a highly asymmetric process, resulting in the synthesis of many more positive - than negative-strands. Accordingly, a regulatory mechanism should exist to control levels of production of either strand, perhaps at the level of initiation.
Several viral RNA structures present within negative - and positive-strand RNA are important for initiation of RNA synthesis. RNA elements that organize RNA replication initiation complexes are believed to form around the 3′-termini of both positive and negative strands, but, surprisingly, they have also been found at the 5′ end of the viral RNA or within the coding region of the virus RNA. Furthermore, recent evidences suggest that interaction of the 5′ - and the 3′-ends of the viral genome is necessary to initiate negative-strand RNA synthesis [1], [2], [3], [4], [5]. These 5′-3′ interactions are mediated either by direct RNA-RNA interaction of complementary sequences in the genome [1], [2], [3], [4] or through an RNA-protein-protein-RNA bridge [5]. The global folding of the viral RNA genome could explain how RNA elements, dispersed throughout the genome, could assemble together into a complex that catalyzes initiation of negative-strand RNA synthesis. However, the relationship between structure and function of these complex elements remains poorly understood. A complication in dissecting the precise role of these RNA elements arises from the fact that some of these structures participate in multiple steps of the replication process. Such is the case for a 5′ - RNA element in the poliovirus genome that is proposed to contribute to the initiation of both negative - and positive-strand RNA synthesis. The overlapping functions of this element have limited our understanding of its structural and functional features.
Here, we examine the structure and function of the initiation complex of positive-strand RNA synthesis. We chose poliovirus, a member of the family Picornaviridae, as a model because both in vitro and in vivo systems are available to dissect the viral replication cycle [6], [7], [8]. Poliovirus contains a single positive-strand RNA genome of approximately 7500 nucleotides which is covalently linked to a small peptide, VPg, at the 5′-end and contains a poly(A) tail at its 3′-end [9], [10], [11], [12]. The viral RNA consists of an open reading frame flanked by two untranslated regions (UTR), at the 5 - and 3′-ends of the genome. The 5′-UTR contains two functional elements important for translation and replication: The internal ribosomal entry site (IRES) region spanning five stem loop structures within the 5′-UTR drives translation of the polyprotein via a cap-independent translation mechanism [13], [14]. The 5′-terminal 94 nucleotides fold into a cloverleaf-like structure, which plays a role in both translation and replication [15], [16], [17]. The cloverleaf structure is a key cis-acting element for initiation of negative-strand RNA synthesis [5], [18]. The cloverleaf structure forms a ternary complex with the cellular poly(rC) binding protein (PCBP; also known as hnRNP E or -αCP) [19], [20] and the uncleaved viral precursor of the polymerase, 3CD [15], [16], [17]. This complex can interact with the cellular factor, poly(A) binding protein (PABP), which binds to the poly(A)tail at the 3′-end of the genome. This leads to pseudo-circularization of the viral genome and initiation of negative-strand RNA synthesis [5], [18]. The ternary complex formed on the 5′-cloverleaf structure also functions in translation, thus providing a means to tune the balance between translation and RNA synthesis [21], [22]. The binding of the cellular protein, PCBP, to the cloverleaf RNA enhances viral translation (Gamarnik and Andino, unpublished). In contrast, binding of the viral polymerase precursor, 3CD, to the cloverleaf structure represses translation and promotes negative-strand synthesis of the viral RNA [22], [23].
Initial evidence also implicated the cloverleaf structure as a critical RNA element for positive-strand RNA synthesis. Certain mutations in this element resulted in reduced accumulation of positive-strand RNA without a significant effect on negative-strand levels [16]. This observation raised the question how the cloverleaf structure functions as a promoter for positive-strand RNA synthesis, which is initiated on the 3′-end of the negative-strand. Analyzing the precise role of the cloverleaf element in positive-strand RNA synthesis, however, is difficult because most mutations disrupting the structure and/or functions of the cloverleaf also inhibit negative-strand RNA synthesis.
We thus developed a novel approach to analyze RNA elements that play multiple roles during virus replication. Our approach relies on the duplication of the cloverleaf structure so that one cloverleaf is dedicated to the initiation of negative-strand RNA synthesis while the other can mediate positive-strand synthesis. This allows the study of the function of the cloverleaf RNA on positive-strand RNA synthesis. Our studies demonstrate that the cloverleaf structure formed at the 5′-end of the positive-strand is required for initiation of positive-strand RNA synthesis. Also required to initiate positive-strand RNA synthesis are the binding sites for the viral polymerase precursor, 3CD, and the host factor, PCBP. Furthermore, we identified specific nucleotide sequences within “stem a” that are essential for the initiation of positive-strand RNA synthesis
Results
Duplication of the 5′ cloverleaf RNA to examine its role in positive-strand RNA synthesis
To examine the role of the cloverleaf structure in positive-strand RNA synthesis we designed an artificial virus RNA genome with two independent RNA replication promoters dedicated to either positive - or negative-strand RNA synthesis (Fig. 1A to 1C). Previous results have shown that only the structure but not the specific sequences of the cloverleaf RNA stems are required for negative-strand synthesis [16]. In contrast, the specific nucleotide sequences of “stem a” are critical for efficient positive-strand initiation [24]. Furthermore, additional sequences at the 5′-end of the viral genome also lead to a defect in positive - but not negative-strand RNA synthesis [25]. We exploited these findings to construct a poliovirus luciferase replicon with tandem cloverleaf structures, in which the four A-U pairs in “stem a” of the downstream cloverleaf were replaced with G-C pairs (Fig. 1C, G/C-CL). In this construct, the downstream cloverleaf will only be able to participate in the initiation of negative-strand RNA synthesis, leaving the upstream, 5′-most cloverleaf open to the analysis of the elements required for positive-strand synthesis. Using enzymatic structural probing of the tandem cloverleaf structure in dCL-PLuc, we confirmed that the two cloverleaves fold as predicted (Fig. 1C), enabling them to function independently of each other (Fig. S1).
Fig. 1. Double cloverleaf replicons. 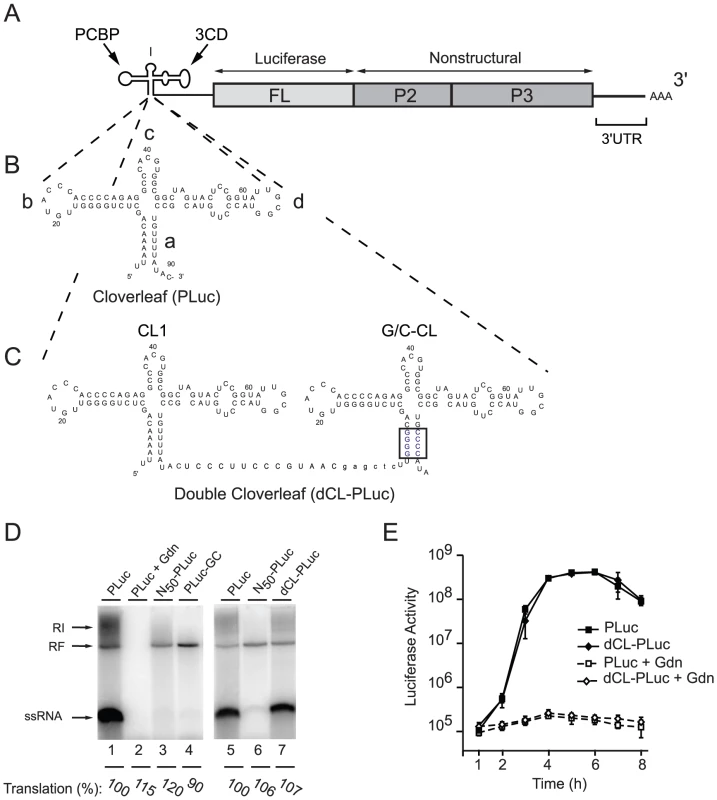
(A) Schematic representation of a poliovirus-luciferase replicon. (B and C) Secondary structure of the PLuc cloverleaf and the two tandem cloverleaves in dCL-PLuc with wild-type sequences in CL1 and four G-C pairs in “stem a” of the downstream cloverleaf G/C-CL (highlighted in blue). (D) RNA replication in a cell-free system. RNA transcripts either Pluc (lanes 1, 2+5), N50-Pluc (50 non-polio nucleotides at the 5′-end of the viral RNA) (lanes 3+6), Pluc-GC (lane 4), or dCL-Pluc (lane 7) were used to program HeLa cell S10 extract. After 4 hours of incubation at 30°C, translation levels were measured as luciferase activity (arbitrary units [AU]), and pre-initiation complexes were isolated by centrifugation in the presence of 2 mM guanidinium hydrochloride (Gdn). RNA synthesis was initiated by addition of NTP and monitored by [α32P]UTP incorporation for 2 hr. The RNA synthesized was analyzed using native agarose gels. (E) Replication of poliovirus replicon in intact HeLa S3 cells. RNA transcripts (PLuc or dCL-PLuc) were transfected into HeLa S3 cells, and luciferase activity [AU] corresponding to 2.5×105 cells was measured every hour for 8h. The cells were incubated either in the presence (dashed lines) or absence (solid lines) of 2 mM Gdn. Each measurement was carried out in triplicate; standard deviations are indicated by vertical bars. A cell-free system that supports complete poliovirus replication [8], was used to demonstrate that the cloverleaf RNA containing a GC “stem a” can only promote negative-strand RNA synthesis, resulting in accumulation of dsRNA replicative form (RF) (Fig. 1D, lane 4, PLuc-GC). The labeled RF RNA observed is composed of the input unlabeled positive-stranded and newly synthesized 32P-labeled negative-stranded RNA, thus RF can be taken as a direct measure of negative-strand RNA synthesis. As expected, a replicon containing a single wildtype cloverleaf at the 5′-end of the genome can support both negative - and positive-strand RNA synthesis and produced single stranded RNA (ssRNA) and replicative intermediate (RI), in addition to RF (Fig. 1D, lane 1, PLuc). Addition of guanidine hydrochloride (Gdn), which inhibits viral RNA replication [26], [27], [28], blocked formation of either species (lane 2) demonstrating that the bands observed correspond to bona fide poliovirus replication. Strikingly, when a wildtype cloverleaf structure was inserted 5′ from the G/C-CL both negative - and positive-strand RNA synthesis were observed (Fig. 1D, lane 7, double cloverleaf, dCL-PLuc). The level of translation (measured as luciferase activity, and normalized to the level of PLuc) was very similar for PLuc and PLuc-GC indicating that the stability of the virus RNA is not affected for PLuc-GC.
We next monitored replication in intact cells by transfecting PLuc and dCL-PLuc RNA into HeLa cells in the presence and in the absence of Gdn (Fig. 1E). Both constructs displayed identical replication kinetics. Monitoring luciferase activity in the presence of Gdn provides translation level of the input-RNA without replication. dCL-PLuc RNA translated with the same efficiency as PLuc RNA. These results indicate that dCL-PLuc RNA is able to support efficient replication in intact cells as well as in a cell-free system. Given that the downstream cloverleaf is dedicated only to negative-strand synthesis, this double-cloverleaf construct provides a system to study the effect of mutations within the cloverleaf that would affect positive-strand RNA synthesis.
Positive-strand RNA synthesis requires an intact 5′ end cloverleaf-structure
We next determined the specific 5′-terminal sequences critical for efficient initiation of positive-strand RNA synthesis. First, we determined the minimal 5′ sequences required for positive-strand synthesis by inserting fragments of increasing length corresponding to the wildtype poliovirus 5′-end genomic RNA upstream of the mutant G-C cloverleaf that only drives negative-strand synthesis (Fig. 2A). Given that the downstream G-C cloverleaf (G-C/CL) promotes negative-strand RNA synthesis at wildtype levels (Fig. 1D, lane 4), this experiment addressed the specific requirements for the initiation of positive-strand synthesis. We examined three constructs (Fig. 2A): a construct containing only the poliovirus 5′ most 9 nucleotides plus a linker that facilitated cloning (plus9), a construct in which the wildtype “stem a” was inserted (plus20), and a construct carrying “stem a” and “stem c” in front of the G-C cloverleaf (plus27). In the cell-free replication system these constructs were unable to produce positive-strand RNA (Fig. 2B, lanes 3 to 5). We observed relatively similar levels of negative-strand synthesis (within 2-fold from dCL-pLuc control) and translation (between 72 to 115% of dCL-PLuc translation control) for all partial cloverleaf constructs (Fig. 2B). Furthermore, a construct carrying the first 81 poliovirus wildtype nucleotides, in which the formation of “stem a” was disrupted, was also significantly defective in positive-strand RNA synthesis (Fig. 2B, lane 6).
Fig. 2. Efficient replication requires a full-length cloverleaf structure at the most 5′-end of the virus genome. 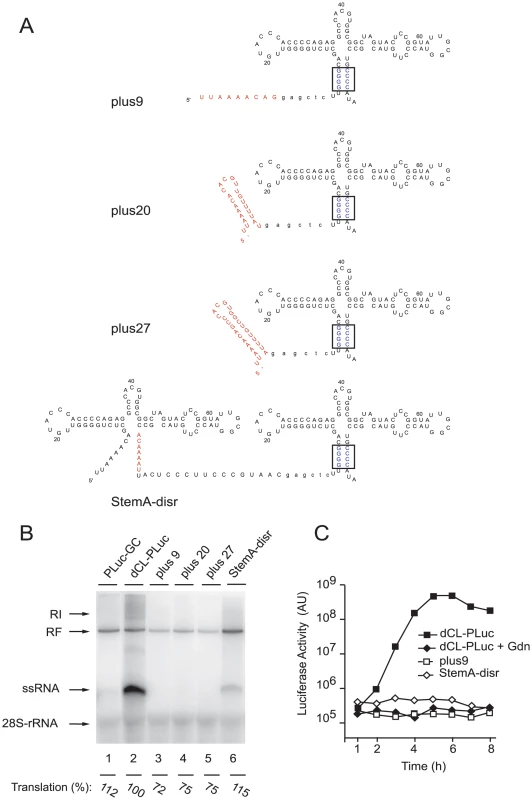
(A) Schematic representation of the secondary structure of the 5′- ends of plus9, plus20, plus27, and StemA-disr RNAs. A SacI site (in lower case letters) was introduced as a linker between the partial cloverleaf 5′-ends (in red) and a downstream G/C-CL cloverleaf. (B) RNA replication in a cell-free system. RNA transcripts of either PLuc-GC (lane 1), dCL-PLuc (lane 2), plus9 (lane 3), plus20 (lane 4), plus27 (lane 5), or StemA-disr (lane 6) were used to program a cell extract. (C) Replication of replicons bearing partial cloverleaf structure at the 5′-end. RNA transcripts (dCL-PLuc, plus9 or StemA-disr) were transfected into HeLa S3 cells, and luciferase activity [AU] was measured every hour for 8h. Incubations were carried out in the presence (+Gdn) or absence of 2 mM Gdn. The graphs are representative of three independent experiments. In vivo experiments were consistent with these results. Replication of plus9 and StemA-disr replicons were impaired in intact HeLa cells, while the efficiency of translation was comparable to dCL-PLuc in the presence of Gdn (Fig. 2C, dCL-PLuc + Gdn). These findings establish that not only specific 5′-sequences but also the entire structure of the 5′-cloverleaf is required for efficient positive-strand synthesis.
Features of the cloverleaf structure involved in positive-strand RNA synthesis
Bioinformatic analysis predicts the formation of a cloverleaf secondary structure either at the 5′-end of the positive-strand or at the complementary 3′-end of the negative-strand [16]. Since positive-strand RNA synthesis initiates at the 3′-end of the negative-strand template it has been proposed that the complementary cloverleaf structure in the negative-strand functions as a promoter of the initiation reaction [29], [30]. To examine this possibility, we used a particular property of G-U base pairs to selectively disrupt the structure in the positive - or the negative-strands. G-U pairs can replace A-U pairs in one strand while on the complementary strand the A-C base pairs cannot form, compromising the structure of the stem. We used this approach to incorporate G-U/A-C regions to selectively disrupt either the “stem b” or “stem d” regions of the 5′-cloverleaf of the positive - or negative strands (Fig. 3A). These were then tested in the context of the tandem cloverleaf replicons, to evaluate the positive-strand synthesis requirements for cloverleaf structure in either (+) or (−) strand.
Fig. 3. Elements within StemB or StemD required for RNA replication. 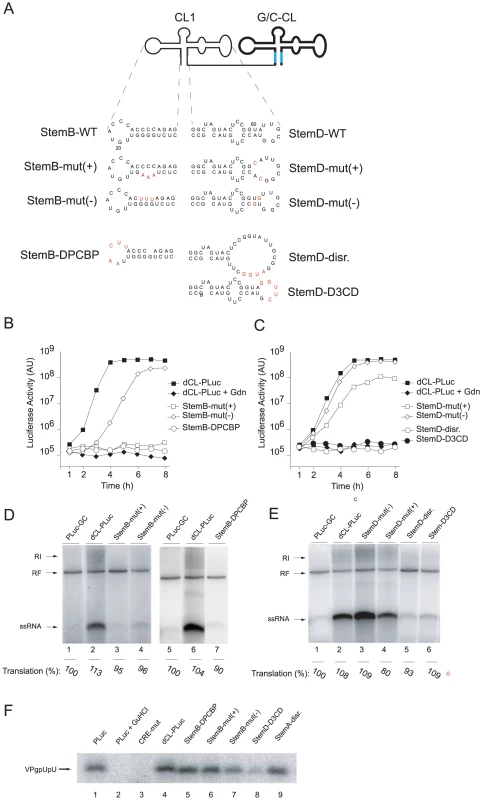
(A) Representation of the secondary structure of tandem cloverleaf replicons with either StemB or StemD mutations (highlighted in red) in the 5′-end cloverleaf. (B and C) Luciferase expression in replicon RNA-transfected HeLa S3 cells. RNA transcripts containing mutations either within StemB or StemD were transfected into HeLa S3 cells, and luciferase activity [AU] was determined every hour for 8h. Control experiment included the addition of 2 mM Gdn. The graphs are representative of three independent experiments. (D and E) RNA replication in a cell-free system. Replicon RNA transcripts with mutations in either StemB or StemD were used to program a cell extract. (F) VPg-uridylylation in a cell-free replication system. RNA transcripts corresponding to tandem cloverleaf replicons containing mutations in either StemB or StemD were employed to program cell-free replications systems. VPg-pU(pU) formation was monitored by incubating the extracts with [α32P]UTP for 1 hour. The radiolabeled RNA was immuno-precipitated using anti-VPg antibodies, separated on a Tris-Tricine SDS-Page gel and visualized by using autoradiography. First, we applied the asymmetric mutational analysis to “stem b” by mutating three consecutive base-pairs. In StemB-mut(+), the sequence GGG was replaced by AAA. This mutation should disrupt the stem in the positive-strand but should maintain the structure in the negative-strand. The second construct, StemB-mut(−) (CCC to UUU) disrupted the duplex structure in the negative-strand without modifying the structure in the positive-strand. While StemB-mut(+) did not produce any detectable positive-strand RNA, StemB-mut(−) was able to produce small, but detectable amounts of ssRNA. Strikingly, in intact cells, StemB-mut(+) was not able to replicate, whereas, StemB-mut(−), after an initial delay, replicated with almost wildtype kinetics, reaching similar maximum luciferase expression (Fig. 3B). The slight delay in StemB-mut(−) replication and the reduced level of positive-strand synthesis in the cell free system (Fig. 3D, lane 4) likely results from a weaker G-U base-pairing compared with the wildtype G-C base-pairs, implying that the function performed by the positive strand is sensitive to the stability of the stem. These results indicate that positive-strand RNA synthesis requires an intact “stem b” in the cloverleaf structure of the positive-strand RNA.
In “stem d” we first introduced mutations that disrupted base-pairing interactions (Fig. 3A, StemD-disr.). This mutant was unable to replicate in vivo or in vitro (Fig. 3C and 3E, lane 5). We then replaced two A-U pairs with A-C pairs, resulting in disruption of the “stem d” only on the positive-strand (StemD-mut(+)), or two G-U pairs (StemD-mut(−)), which should maintain the duplex structure on the positive-strand but should alter the structure on the negative-strand. In the cell-free replication system, StemD-mut(+) showed a reduced level of positive-strands as compared to dCL-PLuc (Fig. 3E, lane 2 and 4). In contrast, StemD-mut(−) was able to efficiently synthesize positive-strand RNA, resulting in even two-fold increase in positive-strand accumulation (Fig. 3E, lane 2 and 3). Our analysis in intact cells was consistent with the in vitro results. StemD-mut(−) replicated with wildtype kinetics and maximum luciferase expression, whereas, StemD-mut(+) showed a 10-fold decrease in replication (Fig. 3C).
Importantly, the defects in positive-strand synthesis were not due to decreased RNA stability because we observed no decrease in translation in any of the systems. The level of luciferase activity as an indirect measure for translation was approximately the same for all the mutants in comparison to wild-type in the cell-free replication system (see for example Fig. 1D, 2B, 3D and E). In addition, after transfection into cells, we monitored luciferase activity in the presence of Gdn for all mutants, which provided translation levels produced by the input RNA. Translation level for each mutant was within 10% of wildtype (data not shown). Therefore, we exclude RNA stability as an explanation for the decrease in positive-strand RNA accumulation. These results, as well as those obtained for “stem b”, indicate that compromising the cloverleaf structure on the positive-strand RNA leads to a defect in initiation of positive-strand synthesis, as originally suggested [16].
Binding-sites for PCBP and 3CD in the cloverleaf are also required for positive-strand RNA synthesis
The cloverleaf structure interacts with the host-cell factor, PCBP [19], [20], and the viral polymerase precursor, 3CD [15], [16], [17], to form a ternary complex that participates in initiation of negative-strand synthesis [5], [18]. Having defined the structural requirements of the cloverleaf RNA for positive-strand synthesis, we then examined whether binding of PCBP or 3CD is required for positive-strand RNA synthesis. A poly(C) stretch within the “stem b” of the cloverleaf has been identified as the binding-site for PCBP [19], [20]. We introduced a mutation that completely disrupts PCBP binding in the 5′-cloverleaf [19]. This mutant, StemB-ΔPCBP, showed a severe defect in positive-strand RNA synthesis in the cell-free replication system (Fig. 3D, lane 7) and in intact HeLa cells (Fig. 3B). We also engineered a deletion within the “stem d” region of the 5′-cloverleaf to disrupt 3CD binding [19]. This mutant, StemD-Δ3CD, was also severely impaired in its ability to replicate in the cell-free system (Fig. 3E, lane 6) and in intact cells (Fig. 3C). This result demonstrates that binding of PCBP and 3CD to the terminal cloverleaf structure is required for initiation of positive-strand synthesis.
The uridylated peptide VPg-pUpU functions as a primer for both negative - and positive-strand synthesis [31], [32], [33], [34]. An additional cis-acting replication element (CRE) within the 2C-coding region of the poliovirus genome functions as a template for the covalent linkage of two UMP nucleotides to the viral peptide, VPg, resulting in VPg-pUpU [35], [36], [37], [38]. Since evidence has been obtained for a role of the cloverleaf RNA in VPg-uridylylation [39], and given the severe defect in positive-strand synthesis exhibited by some of the mutants examined here, we analyzed whether the primary defect in our mutants impaired CRE(2C)-mediated VPg-uridylylation. HeLa S10 extract was programmed with replicon RNA and VPg-pUpU formation was analyzed by polyacrylamide gel electrophoresis. We examined the total amount of VPg uridylylation by specific immunoprecipitation using polyclonal anti-VPg-antibodies. Extracts programmed with PLuc RNA accumulated VPg-pUpU over the 2 hr incubation time, but Gdn prevented VPg uridylylation (Fig. 3F lane 1 and 2). In addition, a CRE-mutant virus RNA, which carries a mutation within the CRE(2C) region (A5 to C mutation) [31], was unable to catalyze VPg-pUpU formation (Fig. 3F, lane 3). These control experiments demonstrated that authentic VPg-pUpU was synthesized under our experimental conditions. As expected, dCL-PLuc also synthesized VPg-pUpU at wildtype levels (Fig. 3F, lane 4). VPg-pUpU formation was also observed in double cloverleaf replicons carrying mutations in the 5′ cloverleaf that either disrupted the “stem b” duplex structure on the positive - or the negative-strand (StemB-mut(+) and StemB-mut(−)), the “stem a” structure (StemA-disr) or the binding-site for PCBP (StemB-ΔPCBP) (Fig. 3F, lane 5–9). We observed a slight decrease of VPg-pUpU formation in the case of StemD-Δ3CD (Fig. 3F, lane 8). However, this reduction was not consistently observed from experiment to experiment (see Fig. S2). We concluded that the striking defect in positive-strand synthesis observed in cloverleaf mutants is not due to a defect in VPg-pUpU formation.
Specific sequence within “stem a” are essential for positive-strand synthesis
It was previously shown that “stem a” of the cloverleaf is needed for positive-strand synthesis [24]. We next defined the specific nucleotide sequence requirements at the “stem a” structure for positive-strand synthesis. We engineered a series of “stem a” mutations within the 5′ - cloverleaf of the double cloverleaf construct. Since disrupting the cloverleaf structure itself leads to a decrease in the level of positive-strand synthesis, sequence alterations were introduced together with compensatory mutations in the opposite side of the “stem a” to preserve the structure. The two 5′-terminal uridines were not mutated as they served as template for the 3′-terminal two A's on the negative-strand, which are believed to function as the binding site of the primer VPgpUpU. Instead we mutated a series of four A-U base-pairs (Fig. 4A). All mutated tandem cloverleaf RNAs were tested for positive-strand synthesis in the cell-free replication system and transfected into HeLa cells to examine their replication phenotype in vivo. We observed three types of replication phenotypes (Fig. 4B). One group of mutants had a mild effect on the replication rate ((+++) in Fig. 4B). These include mutants where the upper two A-U pairs were changed to U-A pairs, or either the first (starting from the lowest pair), third or fourth A-U pair replaced with G-C pairs (StemA-mut2, -mut4, -mut6, -mut7, respectively). In the cell free system, these mutants synthesized positive-strand RNA, although at a markedly reduced level (Fig. 4C, compare wildtype lane 2 with mutants in lane 4, 6, 8, 9). In intact cells, these mutants showed almost wildtype replication kinetics (Fig. 4D, black lines). The second group of mutants had a severe defect in positive-strand synthesis ((+) in Fig. 4B). These mutants include those where the lower two A-U pairs were replaced by U-A pairs, the second A-U pair was replaced by G-C, or the upper two A-U pairs were replaced with G-C pairs (StemA-mut1, StemA-mut5 and StemA-mut9, respectively). This group of mutants was unable to produce positive-strand RNA in the cell-free system (Fig. 4C, lanes 3, 7, 11), and showed significantly decreased, but still detectable, replication in intact cells (Fig. 4D, green lines). Lastly, some mutants had a dramatic loss-of-function phenotype in vitro and in vivo ((−) in Fig. 4B). These included the mutants with the most severe alterations in base-pairing. In StemA-mut3 all four A-U pairs were swapped into U-A pairs, in StemA-mut8 the lower two A-U pairs were replaced by G-C pairs, and in StemA-mut10 all four A-U pairs were replaced by G-C pairs. None of these mutants were able to synthesize detectable levels of positive-strands in cell extract (Fig. 4C, lanes 5, 10, 12) or in intact cells (Fig. 4D, red lines). These results establish that changes either in the upper and lower part of the “stem a” results in a defect in positive-strand synthesis. However, it seems that the bottom of the stem is more susceptible to sequence changes than the upper portion.
Fig. 4. Replication of Double Cloverleaf replicons bearing mutations within StemA. 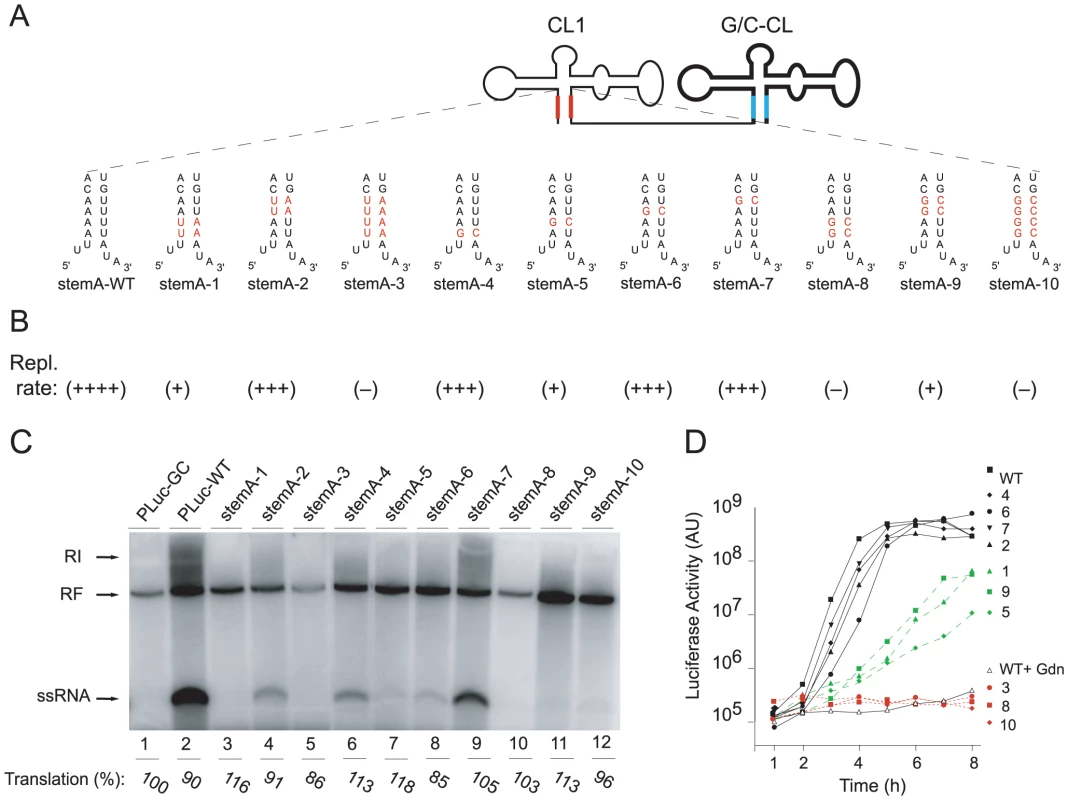
(A) Schematic representation of the secondary structure of Tandem cloverleaf structures replicons with either StemA mutations (highlighted in red) in the 5′-end cloverleaf. (B) Summary of replication rates of StemA mutants in a cell-free system and in HeLa S3 cells. (C) RNA replication in a cell-free system. RNA transcripts containing StemA mutations were used to program a cell extract. RNA products were analyzed on native agarose gels and detected by autoradiography. (D) Replication of virus RNA containing mutations within the StemA. Luciferase activity [AU] corresponding to 2.5×105 transfected HeLa-cells was measured every hour for 8h. The graphs are representative of three independent experiments. Reversion of “stem a” mutations reveals key role of nucleotide A4 of poliovirus genome
To further define the function of “stem a” in positive-strand RNA synthesis, we engineered the full-length poliovirus genome to carry two tandem cloverleaf structures (dCL-polio 1). A one-step growth curve demonstrated that dCL-polio 1 replicates with almost identical kinetics to wildtype poliovirus (Fig. 5A). Furthermore, the plaque-phenotype of dCL-polio 1 was identical to wildtype (not shown). We confirmed by RT-PCR and sequencing that the structure of the double cloverleaf of dCL-polio 1 was maintained during the entire course of the infection (data not shown). Thus, the full-length virus carrying the double cloverleaf replicates with wildtype characteristics and this construct provides a system to study the evolution of a virus carrying a mutated “stem a” during normal poliovirus replication in tissue culture.
Fig. 5. Analyzing the evolution of poliovirus carrying lesions within Stem A. 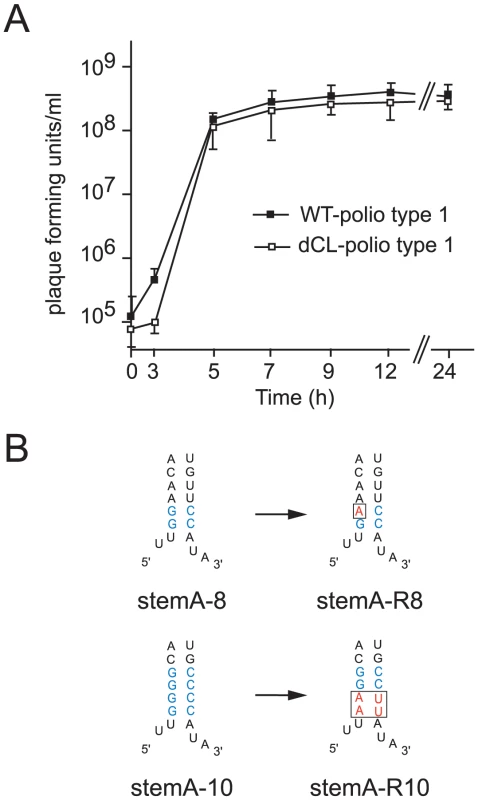
(A) One-step growth curve of poliovirus carrying either one cloverleaf (WT-polio type1) or tandem cloverleaf structures (dCL-polio type 1). HeLa S3 cells were infected at an MOI of 10 with either WT-polio type 1 or dCL-polio type 1 viruses. At indicated time-points viruses were harvested and their titers were determined according to standard plaque assays. The graphs are the mean of triplicate samples. Standard deviations are indicated by error bars. (B) Schematic representation of the cloverleaf StemA-8 and StemA-10 mutations (mutations highlighted in blue) and the changes in sequence observed in revertants, stemA-R8 or stemA-R10 (highlighted in red), with increased replication capacity. Multiple plaques from viruses with higher replication capacity were sequenced and they all contained the mutations highlighted in the figure. We examined two different mutations introduced in “stem a” of the 5′ terminal cloverleaf, StemA-mut8, and StemA-mut10. These mutations lead to a complete disruption of positive-strand synthesis (Fig. 4). However, poliovirus RNAs carrying these mutations were pseudo-infectious. Following high efficiency transfection with StemA-mut8 and StemA-mut10 viral RNA, we observed viral plaques, albeit at much lower efficiencies than for wildtype RNA. StemA-mut8, in which the lower two A-U pairs of “stem a” were replaced by G-C pairs, produced small plaques. StemA-mut10, in which all four A-U base pairs of “stem a” were replaced with G-C pairs, displayed a minute-plaque phenotype. We hypothesized that nucleotide changes accumulated after transfection of the mutants increased virus fitness allowing plaque formation. In order to identify the changes that allow these mutants to replicate, the complete genomes of several plaque-purified StemA-mut8-viruses and StemA-mut10-viruses were isolated and analyzed by sequencing. In StemA-mut8, a single point mutation was identified in all independently isolated revertants: a G to A transition in the second G-C base pair (Fig. 5B). Intriguingly, this sequence alteration, which allows the mutant to replicate, results in one mismatch within the “stem a” duplex structure (Fig. 5B), suggesting that the sequence is critical for initiation. Strikingly, the StemA-mut10 gain-of-function required a massive four point mutation, which was observed in all isolated viruses, whereby the lower two G-C base-pairs reverted back to wild-type A-U pairs (Fig. 5B). These results confirmed the significance of the specific sequences of “stem a” for positive-strand RNA synthesis. Furthermore, they imply a critical role for nucleotide A4 of the poliovirus genome in the initiation of positive-strand RNA synthesis.
Discussion
Viral RNA genomes often contain RNA elements that fulfill multiple regulatory roles. This is due in part to the compactness of the genome and the need to coordinate various functions of the viral RNA as both genome and mRNA. Dissecting the role of these multifunctional elements is usually hindered by the interdependent nature of the viral replication processes often associated with pleiotropic effects of mutations. This has been the case for the functional analysis of the enterovirus 5′-cloverleaf structure as well as for many other RNA elements in RNA viruses. Here we developed a broadly applicable experimental strategy to circumvent these problems and applied it to examining the role of the 5′-cloverleaf of poliovirus in positive-strand RNA synthesis. Our approach involved engineering of poliovirus replicons carrying tandem duplicated cloverleaf structures at their 5′-ends in which the downstream cloverleaf is defective for positive-strand synthesis but can initiate negative-strand synthesis. This leaves the 5′ most cloverleaf as the only RNA element that can participate in initiation of positive-strand RNA synthesis. This set-up enabled us to directly examine the effect of mutations in the 5′-end structure on positive-strand synthesis. A similar approach could be employed to examine the functional role of RNA elements of other virus families provided the structure can be dissected using mutations that disrupt specific functions.
Interestingly, bovine enteroviruses have two cloverleaf-like structures at the 5′-end of their genome [40]. Deletion of either one of them results in non-viable viruses. However, after exchange of the region spanning both cloverleaves with the coxsackievirus B3 (CBV3) cloverleaf a viable chimera was generated [40]. Thus, the two cloverleaf structures in bovine enteroviruses display the same roles as the single cloverleaf in CBV3, suggesting that one cloverleaf might function as a promoter for negative-strand and the other as a promoter for positive-strand RNA synthesis.
Functional dissection of cloverleaf regions implicated in positive-strand synthesis
Poliovirus genomes carrying duplicated cloverleaves are fully capable of initiating negative-strand RNA synthesis, and the stability of the positive-strand RNA is maintained by the downstream, intact cloverleaf even if the first structure is disrupted (Fig. 3 and 4). However, our study shows that mutations that disrupt either the structure of, or the binding of factors to the cloverleaf RNA result in reduced positive-strand accumulation (Fig. 4), as previously proposed [16].
Indeed, asymmetric mutations that disrupt “stem b” and “stem d” in either the positive-or the negative-strand RNA demonstrate that the cloverleaf structure formed in the positive-strand is directly involved in initiation of positive-strand synthesis (Fig. 3B–E). It has been proposed that the stability of viral RNAs carrying mutations within the cloverleaf structure could affect the positive-to-negative RNA ratio. If this were the case, we would expect a decrease in translation of the RNA. However, all mutants reached levels of translation similar to the wild-type RNA (data not shown). Therefore, we exclude RNA stability as an explanation for the decrease in positive-strand synthesis. Our results also show that the interaction of PCBP and 3CD with the cloverleaf is required for efficient positive-strand synthesis. We thus conclude that the same ternary complex used for negative-strand synthesis also plays a critical role in positive-strand synthesis.
Our analysis shows that the duplex structure and sequence of “stem a” in the cloverleaf is required for positive-strand synthesis. Disrupting “stem a” of the cloverleaf structure results in a virus lethal phenotype (pDNC-91) [16]. More recent studies demonstrated that “stem a” is also necessary for negative-strand synthesis [24]. Here, we extended those observations by showing that disruption of four base-pairs in “stem a” leads to a severe defect in positive-strand synthesis in the cell free system and completely abrogates replication in intact cells. Furthermore, the specific sequences of “stem a” are required for positive-strand synthesis (Fig. 4C), but not for negative-strand initiation. Examining the evolution of mutations that severely affect positive-strand RNA further defined the sequence requirements of “stem a”. One single change (A), at the second base-pair of “stem a” suffices to greatly increase the fitness of a mutant with a severe replication defect (Fig. 4, stemA-8). This result was consistent with the observation that mutating the second A-U base-pair to G-C resulted in dramatic defect in replication (compare stemA-4 and stemA-5). Interestingly, it appears that in this context a one base-pair disruption can be tolerated. Our results, however, cannot distinguish whether the sequence specificity of “stem a” is required in the positive-stranded cloverleaf or in the 3′-end of the negative-strand template. Given that the cloverleaf structure does not require specific sequences at “stem a” to facilitate initiation of the negative-strand synthesis, and assuming that the requirements for initiation complex formation are similar for negative-strand and positive-strand RNA synthesis, we propose that the specific sequence at “stem a” is required either during unwinding of positive - and negative-strand RNA or at the 3′-end of the negative-strand template initiation site.
An integrated model for enterovirus replication
A surprising corollary of our experiments is that the same RNA element at the 5′-end of the positive-strand functions as a promoter for both negative-strand and positive-strand synthesis, which is initiated at the 3′-end of the complementary strand. We propose a model that integrates all available data and explains how the same ternary complex formed around the cloverleaf structure can carry out a bifunctional role in two differently regulated steps during virus replication (Fig. 6). Negative-strand RNA synthesis is initiated by the circularization of the positive-strand genome via protein-protein bridge formed by the interaction of 3CD and PCPB, bound to the 5′-cloverleaf structure, with PABP associated with the 3′ poly-A tail. The cloverleaf recruits the polymerase as an uncleaved precursor, 3CD, via an RNA-protein interaction mediated by a domain within 3C. Active RdRp 3D is either locally produced by autoprocessing of 3CD or recruited by interactions with the 3D domain of 3CD. The peptide-nucleotide primer, VPg-pUpU, is produced with the assistance of CRE, which acts as template. VPg-pUpU primes the reaction and elongation proceeds resulting in a double-stranded intermediate (RF). In order to allow positive-strand synthesis, the cloverleaf RNA must fold at the 5′-end of the positive-strand RNA, thus the positive-negative duplex RNA intermediate must unwind. We hypothesize that a helicase should catalyze this step. It has been suggested that poliovirus protein 2C could carry out this role [41], [42]. 2C and its precursor 2BC interact with the 3′-end of the negative-strand [29], [43], and bioinformatic analyses demonstrated that the 2C nucleoside triphosphate binding domain belongs to the DEAD-box family of helicases. However, 2C has not been directly shown to have a helicase activity and the precise role of 2C during RNA synthesis is still ill-defined. On the other hand, it is also possible that the perfect double stranded RNA at the ‘left end” of the RF can spontaneously unwind to allow formation of the cloverleaf. Further investigation is necessary to define this critical step of enterovirus replication.
Fig. 6. An integrated model for enterovirus replication. 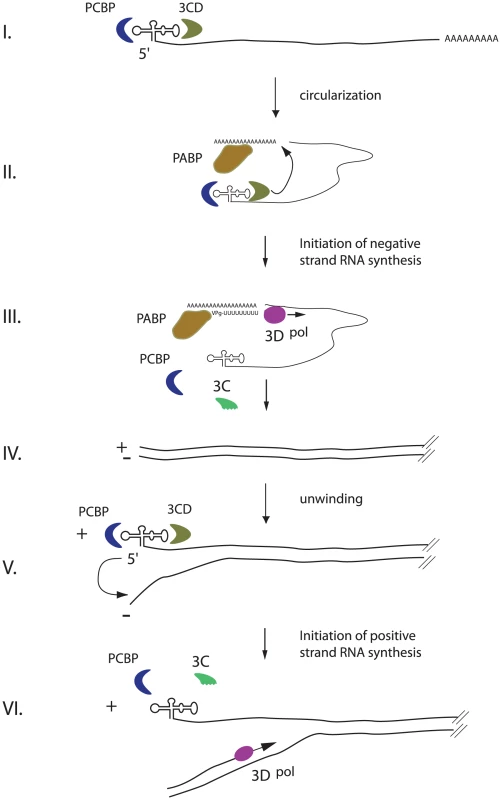
Negative-strand synthesis is initiated by circularization of the positive-strand genome via a protein-protein bridge through the interaction of the ternary complex at the 5′-end (3CD and PCBP bound to the cloverleaf structure) and PABP bound to the 3′-poly(A)tail (I. + II.). CRE-mediated VPg-pUpU acts as primer of the reaction and the polymerase 3D synthesizes the new negative-strand (III.), resulting in a double-stranded intermediate (RF) (IV.). The positive-negative duplex RNA intermediate unwinds, so that the cloverleaf structure at the 5′-end of the positive-strand can form. 3CD and PCBP bind to the cloverleaf to form a ternary complex, which, in turn, will initiate positive-strand synthesis on the 3′-end of the negative-strand (V.). The primer, VPg-pUpU, is recruited and binds to the 3′-terminal AA of the negative strand, and the new positive-strand is synthesized by the polymerase, 3D (VI.). Unwinding of the strands is a critical prerequisite for the formation of the cloverleaf structure on the 5′-end of the positive-strand. Following unwinding, PCBP and 3CD could then bind to the cloverleaf structure of the positive-strand. Our data suggest that this step is key for positive-strand synthesis. This should stabilize the cloverleaf structure as well as to keep the 3′-end of the negative strand single-stranded and available for the primer. The primer, VPg-pUpU, is recruited and binds to the 3′-terminal AA of the negative-strand. The simplest model for the initiation of positive-strand synthesis, is that the cloverleaf ternary complex is the organizing element that facilitates the delivery, in trans, of the RdRp polymerase, 3Dpol, from a preexisting positive-stranded cloverleaf to the initiation site, i.e. the 3′-end of the negative-strand template. Once positive-strand synthesis is initiated and the polymerase moves along the negative-strand, this nascent positive-strand will form double-stranded RNA until it is unwound. Unwinding the end is required to form the new cloverleaf ternary complex to initiate a new round of positive-strand synthesis. This mechanism may also determine the observed asymmetry of replication in which more positive - than negative-strands are made. One possible explanation is that the local concentration of cloverleaf at the positive-strand RNA initiation site results in a more efficient reaction compared to that of 7500 nucleotide downstream where negative-strand RNA initiates.
Why does a single RNA element function to initiate both negative and positive strand synthesis? Perhaps the underlining organizing and mechanistic principles are conserved for the initiation of negative and positive strand synthesis and thus the cis-acting elements involved in the processes, like the 5′-cloverleaf, are also conserved. However, because in infected cells positive strand RNA accumulates at much higher levels than negative strand, additional regulatory elements must exist to control the efficiency of each process. Accordingly, the cloverleaf RNA would act as a general promoter and additional elements would function as enhancers or regulators of the process. From an evolutionary point of view, it is possible to imagine that a single promoter element first evolved to facilitate initiation of RNA synthesis and then the process was optimized by the addition of regulatory elements. A mechanistic advantage of the proposed model is that by assembling the initiation complex in the positive strand, rather than in the 3′-end of the negative strand, RNA synthesis can initiate and proceed on a free negative strand template without interruptions. Considering these potential advantages of a trans-initiation model, it is possible that similar mechanisms may be at work on other positive-stranded RNA viruses. Further experiments in this area are warranted to establish the general characteristics of positive RNA virus replication.
Materials and Methods
Cells & viruses
HeLa S3 cells (ATCC CCL 2.2) were grown either (i) in tissue culture flasks in Dulbecco's modified Eagle medium-nutrient mixture/F-12 (Ham) (1∶1), supplemented with 2mM L-glutamine, 100 U of penicillin and streptomycin per ml, and 10% newborn calf serum or (ii) in suspension in suspension minimal essential medium (Joklik modified) supplemented with 2 mM L-glutamine, 100U of penicillin and streptomycin per ml, and 10% newborn calf serum. For virus production of rib(+)Xpa and double-Wt, in vitro RNA transcripts were electroporated into HeLa S3 cells under the same conditions as described in the section “RNA transfection for luciferase timecourses”. After electroporation 5 volumes of medium was added. Cells were then incubated at 37°C and 5% CO2 over night. After three freeze/thaw cycles viruses were further purified through centrifugation and stored at −80°C (P0 virus). The titers of the virus were determined according to standard plaque assays [44].
Plasmid design and RNA transcripts
PLuc-RNA is transcribed from prib(+)Luc-Wt, the luciferase-expressing, poliovirus-derived replicon; and WT-polio type 1-RNA is transcribed from prib(+)Xpa, containing the cDNA of the Mahoney strain of poliovirus, as previously described [25]. prib(+)Luc-Wt was used to introduce four G-C pairs in “stem a” of the cloverleaf (nucleotides A3-A6 of the poliovirus sequence were replaced by GGCC and nucleotides U91-U94 were replaced by GGCC) which resulted in pPLuc-GC. The complementary sequence in the hammerhead ribozyme was also altered to ensure efficient cleavage. A SacI-site was then introduced in front of the poliovirus sequence which resulted in pGC-SacI. The poliovirus Wt-sequence U1-C112 was then cloned into pGC-SacI, in front of the GC-pair cloverleaf, using the SacI-site, resulting in pdCL-PLuc. pGC-SacI was used as the parental construct for pPlus9, pPlus20, and pPlus27. The following sequences were inserted 5′ of the SacI-site: Poliovirus sequence U1-G9 in pPlus9; for pPlus20 poliovirus nucleotides U1-A8, followed by C38-U42, followed by U89-A95; and for pPlus27 poliovirus nucleotides U1-A8, followed by G35-C45, followed by U89-A95. prib(+)Luc-Wt was used as parental construct for pStemA-disr and all mutants depicted in Fig. 3–4. First, the respective mutations were cloned into prib(+)Luc-Wt resulting in prib(+)Luc-N (where N is the name of the mutation), respectively, then the mutated cloverleaf sequence nucleotide U1-C112 of prib(+)Luc-N was cloned into pGC-SacI, in front of the SacI-site, resulting in pStemB-N, pStemD-N or pStemA-N, respectively. For each mutation that was engineered at the 5′ end of the poliovirus sequence, the complimentary sequence in the hammerhead ribozyme was also altered to ensure efficient cleavage. The sequences of the two cloverleaves in pdCLuc-PLuc were cloned into prib(+)Xpa resulting in pdCL-polio type 1. For stemA-mut8-virus, and stemA-mut10-virus, the sequence of the two cloverleaves and the hammerhead ribozyme in the respective replicons (p-stemA-mut8 and p-stemA-mut10), were cloned into prib(+)Xpa. CREmut-RNA has been transcribed from pCB3-CREmut, the luciferase-expressing, Coxsackie virus B3-derived replicon with a mutation within the CRE-region (A5 in the CRE-loop has been changed to C) as previously described [31].
Poliovirus-specific plasmid DNAs were linearized with ApaI. pCB3-CREmut was linearized with SalI. RNAs were transcribed in vitro in reactions containing bacteriophage T7 RNA polymerase, 5×transcription buffer [400 mM Hepes-KOH (pH 7.5), 120 mM MgCl2, 10 mM spermidine, 200 mM DTT] and 7.5 mM NTP-mix. After incubation at 37°C for 3 h, DNaseI (Roche) was added and reactions incubated at 37°C for 15 min. RNA was precipitated by adding 50% (in volume) of LiCl2-solution [7.5mM LiCl2, 50 mM EDTA (pH 8.0)] and incubation over night at −20°C. After centrifugation the pellet was washed once with 70% ethanol and then resuspended in RNA storage solution (Ambion) and stored at −80°C.
Replication in cell-free extracts
Preparation of HeLa S10 cell extract and initiation factor has been described previously in detail [6]. Negative - and positive-strand RNA synthesis was analyzed as described before (Herold & Andino, 2000) with some minor modifications: 1 µg RNA transcripts was mixed with 25 µl HeLa S10 cell extract, 2 µl initiation factors, 5 µl 10×NTP/energy mix (Herold & Andino, 2000) and 1 µl 100mM guanidine hydrochloride in a total volume of 50 µl. After incubation at 30°C for 4h, 1 µl was removed and added to 50 µl cell culture lysis reagent (Promega) of which 10 µl was then used to measure luciferase activity to monitor translation. The rest of the original translation reaction was centrifuged and the pre-initiation complexes were resuspended in 25 µl labelling mix, containing 15 µl HeLa S10 cell extract, 2.5 µl 10×NTP/energy mix, 2.5 µl of puromycin (1mg/ml) and 30 µCi [α-32P]-UTP (3000Ci/mmol). After incubation at 30°C for 2 h (if not indicated otherwise), the samples were mixed with 175 µl TENSK buffer [50 mM Tris/HCl (pH7.5), 5 mM EDTA, 100 mM NaCl, 1% (v/v) SDS, 200 µg/ml proteinase K] to stop the reaction. After incubation at 37°C for 2 h, RNA was extracted with phenol/chloroform, and precipitated with ethanol. The pellet was resuspended in RNA-storage solution (Ambion) and gel-loading buffer was added prior to loading on a 0.8% agarose gel. The gel was run at 20 V constant current over night. After drying the gel, products were visualized by using autoradiography. Bands were quantified using a phosphorImager (Typhoon 9400; GE Healthcare)
RNA transfection for luciferase timecourse
HeLa S3 cells were trypsinized, washed three times with phosphate-buffered saline, and adjusted to 5×106 cells/ml. Then 800 µl aliquots were electroporated in 0.4 cm cuvettes with 20 µg of replicon RNA, using an Electro Cell Manipulator 600 (BTX Inc.) with the following settings: 300 V, 1000 µF, 24 Ω. Subsequently, 10 volumes of medium was added, the cells were divided in half, and guanidine hydrochloride (Sigma) was added to one half to a final concentration of 2 mM. 2×105 cells were plated per well in 12-well plates and incubated at 37°C in a 5% CO2 incubator.
Luciferase expression
Replicon-transfected cells were scraped off, washed once with phosphate-buffered saline, and then lysed in 100 µl cell culture lysis reagent (Promega). Luciferase activity in 10 µl of lysate was determined in a luminometer using the luciferase assay system (Promega).
VPg-uridylylation-assay
HeLa S10 cell extract was programmed with replicon RNA as described in the section “Replication in cell-free extract”. Pre-initiation complexes were resuspended as decribed above but incubated for 1 h rather than 2 at 30°C. The synthesis of VPg-pUpU was then analyzed by imunoprecipitation. Briefly, 500 µl Dynabeads-ProteinA (Invitrogen) were washed twice with 0.1 M Na PO4 buffer [pH8.0] and then resuspended in 500 µl of the same buffer. 125 µl of anti-VPg polyclonal antibodies were added to the Dynabeads-ProteinA and incubated rotating for 1 h at room temperature. The Dynabeads were washed twice again and resuspended in 500 µl 0.1 M NaPhosphate buffer [pH8.0]. After the 1 h incubation of the replication reaction as described above, 2.5 µl of 0.5 M NaPhosphate buffer and 25 µl of the Dynabeads-ProteinA coupled with anti-VPg antibodies were added. After incubation rotating at 4°C for 1 h, the Dynabeads were washed four times with phosphate-buffered saline. The Dynabeads were resuspended in 15 µl of tricine-sample buffer (Bio-Rad). The samples were heated at 94°C for 5 min and the supernatant was run on a 20% tris-tricine gel at 75 mA for 1 h and then for 72 h at 11 mA at 4°C. After drying the gel, products were visualized by using autoradiography using a phosphorImager (Typhoon 9400; GE Healthcare).
Growth curve
6-well plates were seeded with 106 HeLa S3 cells/well and incubated over night. Cells were washed with phosphate-buffered saline and infected with a multiplicity of infection of 10 of either WT-polio type 1 or dCL-polio type 1 viruses (P0-virus) in serum-free medium. After incubation at 37°C for 30 min, cells were washed twice with phosphate-buffered saline and fresh medium supplemented with 3% newborn calf serum was added to each well. The plates were incubated at 37°C and 5% CO2. At indicated time points viruses were harvested by three freeze/thaw cycles followed by centrifugation. The supernatant contained the virus and was stored at −80°C. The titers of the virus were determined according to standardplaque assays [44].
Virus production, plaque-purification and sequencing of poliovirion RNA
In vitro RNA transcript of viruses were transfected into HeLa S3 cells as described above in section “Cells and viruses”. Cells were incubated for 72 hours or until total cytopathic effect was reached. Viruses were harvested as described under section “growth curve.” Standard plaque-assays were performed [44]. For plaque-purification of viruses individual plaques were transferred to 6-well-plates (seeded with 106 cells/well the night before) before staining of the wells. The 6-well plates were incubated for 72 hours. Total RNA was purified by tryzol extraction (Invitrogen) and isopropanol precipitation. cDNA was synthesized using the Thermoscript RT-PCR system for First-Strand cDNA Synthesis (Invitrogen). Using standard PCR techniques and specific primers for the poliovirus genome, the viral genomes were amplified and the resulting PCR products were sequenced. For sequencing of the very 5′-end of the viral genome the 5′RACE system for rapid amplification of cDNA ends (Invitrogen) was used according to the manufacturer's instructions to amplify the 5′-ends and the resulting PCR products were sequenced.
Supporting Information
Zdroje
1. AlvarezDE
LodeiroMF
LuduenaSJ
PietrasantaLI
GamarnikAV
2005 Long-range RNA-RNA interactions circularize the dengue virus genome. J Virol 79 6631 6643
2. CorverJ
LenchesE
SmithK
RobisonRA
SandoT
2003 Fine mapping of a cis-acting sequence element in yellow fever virus RNA that is required for RNA replication and cyclization. J Virol 77 2265 2270
3. KhromykhAA
MekaH
GuyattKJ
WestawayEG
2001 Essential role of cyclization sequences in flavivirus RNA replication. J Virol 75 6719 6728
4. LoMK
TilgnerM
BernardKA
ShiPY
2003 Functional analysis of mosquito-borne flavivirus conserved sequence elements within 3′ untranslated region of West Nile virus by use of a reporting replicon that differentiates between viral translation and RNA replication. J Virol 77 10004 10014
5. HeroldJ
AndinoR
2001 Poliovirus RNA replication requires genome circularization through a protein-protein bridge. Mol Cell 7 581 591
6. BartonDJ
BlackEP
FlaneganJB
1995 Complete replication of poliovirus in vitro: preinitiation RNA replication complexes require soluble cellular factors for the synthesis of VPg-linked RNA. J Virol 69 5516 5527
7. GamarnikAV
AndinoR
1996 Replication of poliovirus in Xenopus oocytes requires two human factors. Embo J 15 5988 5998
8. MollaA
PaulAV
WimmerE
1991 Cell-free, de novo synthesis of poliovirus. Science 254 1647 1651
9. YogoY
WimmerE
1972 Polyadenylic acid at the 3′-ternegative of poliovirus RNA. Proc Natl Acad Sci U S A 69 1877 1882
10. FlaneganJB
PettersonRF
AmbrosV
HewlettNJ
BaltimoreD
1977 Covalent linkage of a protein to a defined nucleotide sequence at the 5′ ternegative of virion and replicative intermediate RNAs of poliovirus. Proc Natl Acad Sci U S A 74 961 965
11. LeeYF
NomotoA
DetjenBM
WimmerE
1977 A protein covalently linked to poliovirus genome RNA. Proc Natl Acad Sci U S A 74 59 63
12. RacanielloVR
BaltimoreD
1981 Molecular cloning of poliovirus cDNA and determination of the complete nucleotide sequence of the viral genome. Proc Natl Acad Sci U S A 78 4887 4891
13. PelletierJ
KaplanG
RacanielloVR
SonenbergN
1988 Cap-independent translation of poliovirus mRNA is conferred by sequence elements within the 5′ noncoding region. Mol Cell Biol 8 1103 1112
14. TronoD
PelletierJ
SonenbergN
BaltimoreD
1988 Translation in mammalian cells of a gene linked to the poliovirus 5′ noncoding region. Science 241 445 448
15. SilveraD
GamarnikAV
AndinoR
1999 The N-terminal K homology domain of the poly(rC)-binding protein is a major determinant for binding to the poliovirus 5′ untranslated region and acts as an inhibitor of viral translation. J Biol Chem 274 38163 38170
16. AndinoR
RieckhofGE
BaltimoreD
1990 A functional ribonucleoprotein complex forms around the 5′ end of poliovirus RNA. Cell 63 369 380
17. AndinoR
RieckhofGE
AchacosoPL
BaltimoreD
1993 Poliovirus RNA synthesis utilizes an RNP complex formed around the 5′ end of viral RNA. Embo J 12 3587 3598
18. BartonDJ
O'DonnellBJ
FlaneganJB
2001 5′ cloverleaf in poliovirus RNA is a cis-acting replication element required for negative-strand synthesis. Embo J 20 1439 1448
19. ParsleyTB
TownerJS
BlynLB
EhrenfeldE
SemlerBL
1997 Poly (rC) binding protein 2 forms a ternary complex with the 5′ terminal sequences of poliovirus RNA and the viral 3CD proteinase. Rna 3 1124 1134
20. GamarnikAV
AndinoR
1997 Two functional complexes formed by KH domain containing proteins with the 5′ noncoding region of poliovirus RNA. Rna 3 882 892
21. SimoesEA
SarnowP
1991 An RNA hairpin at the extreme 5′ end of the poliovirus RNA genome modulates viral translation in human cells. J Virol 65 913 921
22. GamarnikAV
AndinoR
1998 Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev 12 2293 2304
23. BartonDJ
MorascoBJ
FlaneganJB
1999 Translating ribosomes inhibit poliovirus negative-strand RNA synthesis. J Virol 73 10104 10112
24. SharmaN
O'DonnellBJ
FlaneganJB
2005 3′-Terminal sequence in poliovirus negative-strand templates is the primary cis-acting element required for VPg-pUpU-primed positive-strand initiation. J Virol 79 3565 3577
25. HeroldJ
AndinoR
2000 Poliovirus requires a precise 5′ end for efficient positive-strand RNA synthesis. J Virol 74 6394 6400
26. BartonDJ
FlaneganJB
1997 Synchronous replication of poliovirus RNA: initiation of negative-strand RNA synthesis requires the guanidine-inhibited activity of protein 2C. J Virol 71 8482 8489
27. BaltimoreD
EggersHJ
FranklinRM
TammI
1963 Poliovirus-induced RNA polymerase and the effects of virus-specific inhibitors on its production. Proc Natl Acad Sci U S A 49 843 849
28. RightselWA
DiceJR
McAR
TimmEA
McLIJr
1961 Antiviral effect of guanidine. Science 134 558 559
29. BanerjeeR
EcheverriA
DasguptaA
1997 Poliovirus-encoded 2C polypeptide specifically binds to the 3′-terminal sequences of viral negative-strand RNA. J Virol 71 9570 9578
30. RoehlHH
SemlerBL
1995 Poliovirus infection enhances the formation of two ribonucleoprotein complexes at the 3′ end of viral negative-strand RNA. J Virol 69 2954 2961
31. van OoijMJ
VogtDA
PaulA
CastroC
KuijpersJ
2006 Structural and functional characterization of the coxsackievirus B3 CRE(2C): role of CRE(2C) in negative - and positive-strand RNA synthesis. J Gen Virol 87 103 113
32. GoodfellowIG
PolacekC
AndinoR
EvansDJ
2003 The poliovirus 2C cis-acting replication element-mediated uridylylation of VPg is not required for synthesis of negative-sense genomes. J Gen Virol 84 2359 2363
33. MorascoBJ
SharmaN
ParillaJ
FlaneganJB
2003 Poliovirus cre(2C)-dependent synthesis of VPg-pUpU is required for positive - but not negative-strand RNA synthesis. J Virol 77 5136 5144
34. MurrayKE
BartonDJ
2003 Poliovirus CRE-dependent VPg uridylylation is required for positive-strand RNA synthesis but not for negative-strand RNA synthesis. J Virol 77 4739 4750
35. GerberS
LaneC
BrownDM
LordE
DiLorenzoM
2001 Human papillomavirus virus-like particles are efficient oral immunogens when coadministered with Escherichia coli heat-labile enterotoxin mutant R192G or CpG DNA. J Virol 75 4752 4760
36. PaulAV
RiederE
KimDW
van BoomJH
WimmerE
2000 Identification of an RNA hairpin in poliovirus RNA that serves as the primary template in the in vitro uridylylation of VPg. J Virol 74 10359 10370
37. PaulAV
PetersJ
MugaveroJ
YinJ
van BoomJH
2003 Biochemical and genetic studies of the VPg uridylylation reaction catalyzed by the RNA polymerase of poliovirus. J Virol 77 891 904
38. RiederE
PaulAV
KimDW
van BoomJH
WimmerE
2000 Genetic and biochemical studies of poliovirus cis-acting replication element cre in relation to VPg uridylylation. J Virol 74 10371 10380
39. LyonsT
MurrayKE
RobertsAW
BartonDJ
2001 Poliovirus 5′ terminal cloverleaf RNA is required in cis for VPg uridylylation and the initiation of negative-strand RNA synthesis. J Virol 75 10696 10708
40. ZellR
SidigiK
HenkeA
Schmidt-BraunsJ
HoeyE
1999 Functional features of the bovine enterovirus 5′ non-translated region. J Gen Virol 80(Pt 9) 2299 2309
41. TeterinaNL
KeanKM
GorbalenyaAE
AgolVI
GirardM
1992 Analysis of the functional significance of amino acid residues in the putative NTP-binding pattern of the poliovirus 2C protein. J Gen Virol 73(Pt 8) 1977 1986
42. DmitrievaTM
NorkinaKB
AgolVI
1991 Encephalomyocarditis virus RNA polymerase preparations, with and without RNA helicase activity. J Virol 65 2714 2717
43. BanerjeeR
TsaiW
KimW
DasguptaA
2001 Interaction of poliovirus-encoded 2C/2BC polypeptides with the 3′ ternegative negative-strand cloverleaf requires an intact stem-loop b. Virology 280 41 51
44. CrottyS
LohmanBL
LuFX
TangS
MillerCJ
1999 Mucosal immunization of cynomolgus macaques with two serotypes of live poliovirus vectors expressing simian immunodeficiency virus antigens: stimulation of humoral, mucosal, and cellular immunity. J Virol 73 9485 9495
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Insight into the Mechanisms of Adenovirus Capsid Disassembly from Studies of Defensin NeutralizationČlánek Serum-Dependent Selective Expression of EhTMKB1-9, a Member of B1 Family of Transmembrane KinasesČlánek Host Cell Invasion and Virulence in Sepsis Is Facilitated by the Multiple Repeats within FnBPA
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2010 Číslo 6- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Cognitive Dysfunction Is Sustained after Rescue Therapy in Experimental Cerebral Malaria, and Is Reduced by Additive Antioxidant Therapy
- DNA Watermarking of Infectious Agents: Progress and Prospects
- Self-Protection against Gliotoxin—A Component of the Gliotoxin Biosynthetic Cluster, GliT, Completely Protects Against Exogenous Gliotoxin
- Modifies the Tsetse Salivary Composition, Altering the Fly Feeding Behavior That Favors Parasite Transmission
- Requirement of NOX2 and Reactive Oxygen Species for Efficient RIG-I-Mediated Antiviral Response through Regulation of MAVS Expression
- The Enteropathogenic Effector EspF Targets and Disrupts the Nucleolus by a Process Regulated by Mitochondrial Dysfunction
- Epithelial p38α Controls Immune Cell Recruitment in the Colonic Mucosa
- Coexpression of PD-1, 2B4, CD160 and KLRG1 on Exhausted HCV-Specific CD8+ T Cells Is Linked to Antigen Recognition and T Cell Differentiation
- Insight into the Mechanisms of Adenovirus Capsid Disassembly from Studies of Defensin Neutralization
- EspA Acts as a Critical Mediator of ESX1-Dependent Virulence in by Affecting Bacterial Cell Wall Integrity
- An RNA Element at the 5′-End of the Poliovirus Genome Functions as a General Promoter for RNA Synthesis
- Tetherin Restricts Productive HIV-1 Cell-to-Cell Transmission
- Epstein-Barr Virus-Encoded LMP2A Induces an Epithelial–Mesenchymal Transition and Increases the Number of Side Population Stem-like Cancer Cells in Nasopharyngeal Carcinoma
- Protein Kinase A Dependent Phosphorylation of Apical Membrane Antigen 1 Plays an Important Role in Erythrocyte Invasion by the Malaria Parasite
- Requirement for Ergosterol in V-ATPase Function Underlies Antifungal Activity of Azole Drugs
- The SOCS-Box of HIV-1 Vif Interacts with ElonginBC by Induced-Folding to Recruit Its Cul5-Containing Ubiquitin Ligase Complex
- A Crucial Role for Infected-Cell/Antibody Immune Complexes in the Enhancement of Endogenous Antiviral Immunity by Short Passive Immunotherapy
- Modulation of the Arginase Pathway in the Context of Microbial Pathogenesis: A Metabolic Enzyme Moonlighting as an Immune Modulator
- Role of Abl Kinase and the Wave2 Signaling Complex in HIV-1 Entry at a Post-Hemifusion Step
- Serum-Dependent Selective Expression of EhTMKB1-9, a Member of B1 Family of Transmembrane Kinases
- Epigenetic Repression of by Latent Epstein-Barr Virus Requires the Interaction of EBNA3A and EBNA3C with CtBP
- Host Cell Invasion and Virulence in Sepsis Is Facilitated by the Multiple Repeats within FnBPA
- Role of PKR and Type I IFNs in Viral Control during Primary and Secondary Infection
- A Kinome RNAi Screen Identified AMPK as Promoting Poxvirus Entry through the Control of Actin Dynamics
- Cryptococcal Cell Morphology Affects Host Cell Interactions and Pathogenicity
- NleG Type 3 Effectors from Enterohaemorrhagic Are U-Box E3 Ubiquitin Ligases
- Human Cytomegalovirus UL29/28 Protein Interacts with Components of the NuRD Complex Which Promote Accumulation of Immediate-Early RNA
- Complement Receptor 1 Is a Sialic Acid-Independent Erythrocyte Receptor of
- A Viral microRNA Down-Regulates Multiple Cell Cycle Genes through mRNA 5′UTRs
- Immunotoxin Complementation of HAART to Deplete Persisting HIV-Infected Cell Reservoirs
- Protein Expression Redirects Vesicular Stomatitis Virus RNA Synthesis to Cytoplasmic Inclusions
- Entry and Fusion of Emerging Paramyxoviruses
- Paramyxovirus Entry and Targeted Vectors for Cancer Therapy
- Suppressing Glucose Transporter Gene Expression in Schistosomes Impairs Parasite Feeding and Decreases Survival in the Mammalian Host
- Formation of Complexes at Plasmodesmata for Potyvirus Intercellular Movement Is Mediated by the Viral Protein P3N-PIPO
- Fungal Cell Gigantism during Mammalian Infection
- Two Novel Point Mutations in Clinical Reduce Linezolid Susceptibility and Switch on the Stringent Response to Promote Persistent Infection
- Rotavirus Structural Proteins and dsRNA Are Required for the Human Primary Plasmacytoid Dendritic Cell IFNα Response
- The Epigenetic Landscape of Latent Kaposi Sarcoma-Associated Herpesvirus Genomes
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Requirement of NOX2 and Reactive Oxygen Species for Efficient RIG-I-Mediated Antiviral Response through Regulation of MAVS Expression
- Formation of Complexes at Plasmodesmata for Potyvirus Intercellular Movement Is Mediated by the Viral Protein P3N-PIPO
- Insight into the Mechanisms of Adenovirus Capsid Disassembly from Studies of Defensin Neutralization
- Two Novel Point Mutations in Clinical Reduce Linezolid Susceptibility and Switch on the Stringent Response to Promote Persistent Infection
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



