-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaHIV Capsid is a Tractable Target for Small Molecule Therapeutic Intervention
Despite a high current standard of care in antiretroviral therapy for HIV, multidrug-resistant strains continue to emerge, underscoring the need for additional novel mechanism inhibitors that will offer expanded therapeutic options in the clinic. We report a new class of small molecule antiretroviral compounds that directly target HIV-1 capsid (CA) via a novel mechanism of action. The compounds exhibit potent antiviral activity against HIV-1 laboratory strains, clinical isolates, and HIV-2, and inhibit both early and late events in the viral replication cycle. We present mechanistic studies indicating that these early and late activities result from the compound affecting viral uncoating and assembly, respectively. We show that amino acid substitutions in the N-terminal domain of HIV-1 CA are sufficient to confer resistance to this class of compounds, identifying CA as the target in infected cells. A high-resolution co-crystal structure of the compound bound to HIV-1 CA reveals a novel binding pocket in the N-terminal domain of the protein. Our data demonstrate that broad-spectrum antiviral activity can be achieved by targeting this new binding site and reveal HIV CA as a tractable drug target for HIV therapy.
Published in the journal: . PLoS Pathog 6(12): e32767. doi:10.1371/journal.ppat.1001220
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1001220Summary
Despite a high current standard of care in antiretroviral therapy for HIV, multidrug-resistant strains continue to emerge, underscoring the need for additional novel mechanism inhibitors that will offer expanded therapeutic options in the clinic. We report a new class of small molecule antiretroviral compounds that directly target HIV-1 capsid (CA) via a novel mechanism of action. The compounds exhibit potent antiviral activity against HIV-1 laboratory strains, clinical isolates, and HIV-2, and inhibit both early and late events in the viral replication cycle. We present mechanistic studies indicating that these early and late activities result from the compound affecting viral uncoating and assembly, respectively. We show that amino acid substitutions in the N-terminal domain of HIV-1 CA are sufficient to confer resistance to this class of compounds, identifying CA as the target in infected cells. A high-resolution co-crystal structure of the compound bound to HIV-1 CA reveals a novel binding pocket in the N-terminal domain of the protein. Our data demonstrate that broad-spectrum antiviral activity can be achieved by targeting this new binding site and reveal HIV CA as a tractable drug target for HIV therapy.
Introduction
Highly active antiretroviral therapies (HAART) against human immunodeficiency virus type 1 (HIV-1) have proven in recent years to be extremely effective at reducing viral load and significantly delaying disease progression [1]. However, there remains a pressing need to discover and develop new classes of HIV inhibitors. The virus continues to acquire resistance to currently administered antiretroviral drugs and the rate of transmitted resistance is increasing [2], [3]. The discovery of compounds that inhibit the replication of HIV-1 via new mechanisms offers the best hope of generating drugs that are active against all HIV-1 variants in the clinic. The potency of these compounds would not be affected by mutations that confer resistance to existing therapies [4].
The capsid protein (CA) of HIV-1 plays critical roles in both late and early stages of the viral replication cycle and is widely viewed as an important unexploited therapeutic target [4], [5], [6]. At the earliest stages of particle assembly, the interactions between CA domains of the Gag polyprotein help drive the formation of immature particles at the membrane of host cells [7]. After the release of immature particles from infected cells, proteolytic processing of the Gag polyprotein is completed, leading to capsid assembly and formation of the mature virus. During assembly, the viral RNA genome is packaged into a capsid particle composed of a lattice of CA protein hexamers that form a distinct fullerene cone shaped particle [8]. After virus fusion with a target cell, the core is released into the cytoplasm and CA is thought to undergo a controlled disassembly reaction in order for reverse transcription of the viral genome to occur properly [9].
The HIV-1 CA protein has attracted increased interest as a drug discovery target in recent years. A small molecule, CAP-1, and two versions of a peptide inhibitor, CAI and NYAD-1, have been described that target HIV-1 CA in vitro and appear to interfere with CA function in infected cells [10], [11], [12]. In addition, high resolution structural data on the hexameric lattice that forms the full core structure has been reported [13], [14]. These structures illustrate the distinct roles and importance of inter-subunit interfaces in the CA complex and have shed some light on the potential mechanisms of previously reported CA assembly inhibitors.
Here we describe a novel series of antiviral compounds that target HIV-1 CA in infected cells and appear to interfere with both the viral uncoating process and the formation of infectious particles. Mechanism-of-action and in vitro resistance studies of this series are described. A high resolution co-crystal structure has been determined and illustrates a novel binding pocket in the N-terminal domain (NTD) of HIV-1 CA that is distinct from any previously described. We demonstrate that targeting this new binding pocket with small molecules results in broad-spectrum antiviral activity. This study provides the starting point, where structure-based drug design is a viable option, for the development of a new class of HIV therapeutics.
Results
In vitro Antiviral Activity
PF-1385801 was identified as a hit in a high throughput screen for inhibitors of HIV replication [15]. Several analogs of this compound demonstrated activity in antiviral assays using the MT-2 T-cell line and HIV-1 NL4-3. PF-1385801 inhibited HIV-1 replication with a 50% effective concentration (EC50) of 4.5 µM and exhibited a 50% cytotoxic concentration (CC50) of 61 µM, resulting in a therapeutic index (TI, CC50/EC50) of 14 (Fig. 1). More potent analogs were subsequently designed and synthesized, as detailed in Fig. 1.
Fig. 1. Structures and antiviral activities of inhibitors that target HIV-1 capsid. 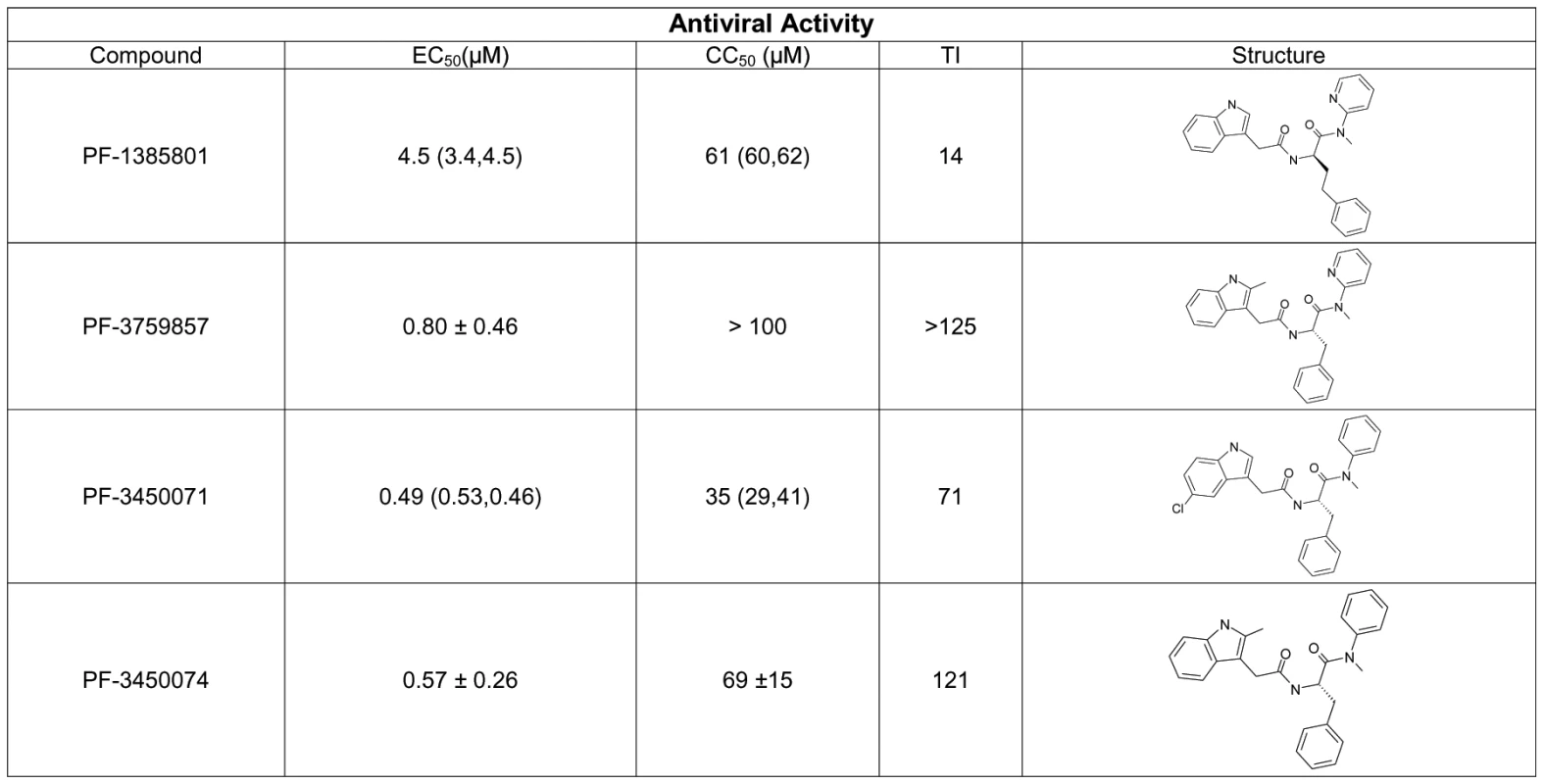
Antiviral activity was determined in CPE assays after infection of MT-2 cells with HIV-1 NL4-3. Results represent the mean ± standard deviations from 4–6 experiments, or the mean with individual values from 2 experiments. TI, therapeutic index is the ratio of mean CC50: mean EC50. A key issue in the development of novel HIV drugs is the potential therapeutic spectrum (consistency of activity across multiple strains of HIV). To address this, two of the compounds were evaluated in antiviral assays using peripheral blood mononuclear cells (PBMCs) infected with a diverse set of HIV-1 lab strains and clinical isolates covering 6 clades and both X4 and R5 tropic viruses (Fig. 2). PF-3450074 and PF-3759857 were active against all strains of HIV-1 tested with median EC50 values of 0.207 (range 0.113 to 0.362 µM) and 1.17 (range 0.51 to 3.17) µM, respectively (Fig. 2 and Tables S1 and S2). In addition, PF-3759857 was active against HIV-2 with an EC50 of 4.7 µM. This tight spectrum of activities against HIV-1 compared well with the marketed drugs, AZT, a nucleoside analog, and efavirenz (EFV), a non-nucleoside reverse transcriptase inhibitor (NNRTI) (Fig. 2 and Tables S3 and S4).
Fig. 2. Activity of inhibitors against diverse strains of HIV-1. 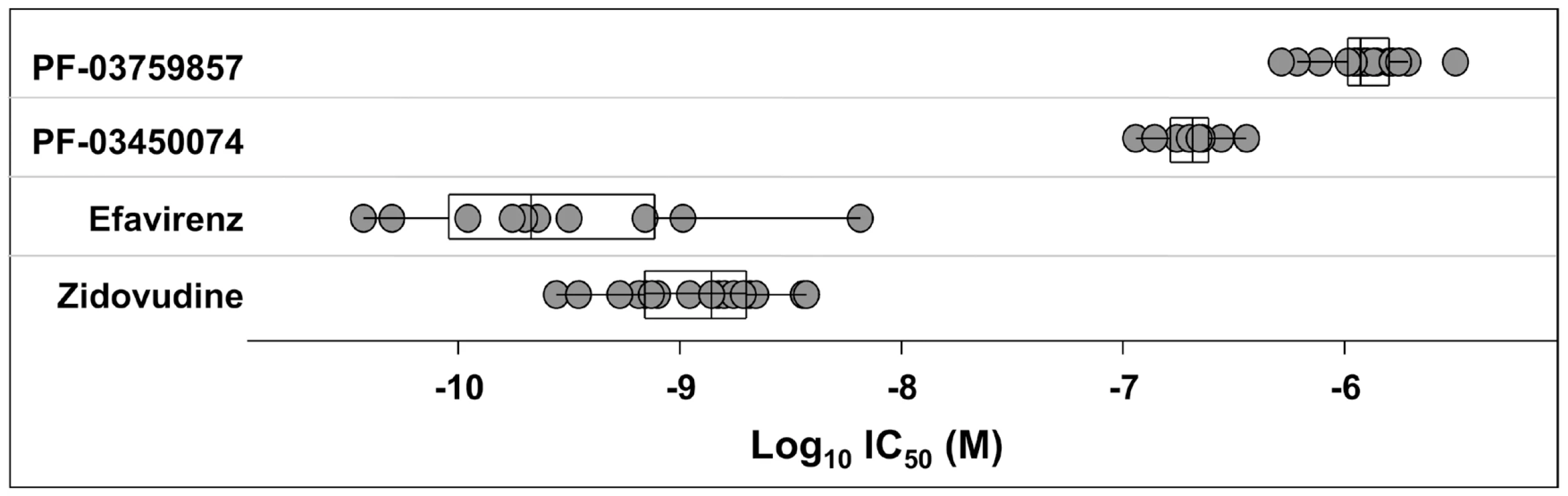
The spectrum of antiviral potencies was determined for two capsid inhibitors, efavirenz, and zidovudine against a panel of HIV isolates including clinical isolates and laboratory adapted strains. Each dot in the figure represents the geometric mean EC50 value for a single HIV isolate. The variability of the antiviral spectrum is further illustrated for each compound with a superimposed “box whisker” plot. The box represents the inter-quartile range and the whiskers highlight the 95% confidence interval. The vertical line in the box represents the geometric mean of the EC50 values. In vitro Resistance Studies to Identify the Antiviral Target
To positively identify the antiviral target of this class of inhibitors, PF-1385801 resistant viral variants were selected in in vitro serial passage experiments. Sequence analysis of cDNAs derived from resistant viral variants selected in the presence of PF-1385801 revealed a single mutation, T107N, located in the NTD of the CA protein. No mutations associated with resistance selection were identified in the integrase or reverse transcriptase coding sequences. Recombinant HIV-1 NL4-3 virus encoding the T107N substitution in HIV-1 CA exhibited an 11-fold reduction in susceptibility to PF-1385801 when compared to wild-type NL4-3 (Table 1). Similar reductions in susceptibility were observed for PF-3450071 (6-fold) and PF-3759857 (10-fold), while no shift was observed for EFV (NNRTI) or the integrase inhibitor AG-110079. In subsequent in vitro resistant virus selection experiments using a compound with a larger TI, PF-3759857, additional substitutions were identified in CA (T107N, H87P, Q67H, K70R and L111I), after 53 days in culture. Virus isolated from the final passage of this experiment showed a >60-fold reduction in susceptibility to PF-3759857 and recombinant NL4-3 virus encoding the five substitutions (5M) exhibited a >40-fold reduction in susceptibility to PF-3450074 (up to the cytotoxicity limit of the compound). Wild-type NL4-3, the T107N mutant, and the 5M mutant virus all exhibited similar levels of infectious virus production from transfected cells and comparable levels of viral replication in reporter gene-based infection assays (in the absence of compound), suggesting that the substitutions do not significantly impair replication capacity. However, more detailed studies, such as direct competition assays are required to accurately determine if the substitutions affect replication fitness.
Tab. 1. In vitro anti-viral activity against HIV-1 capsid wt and T107N mutant. 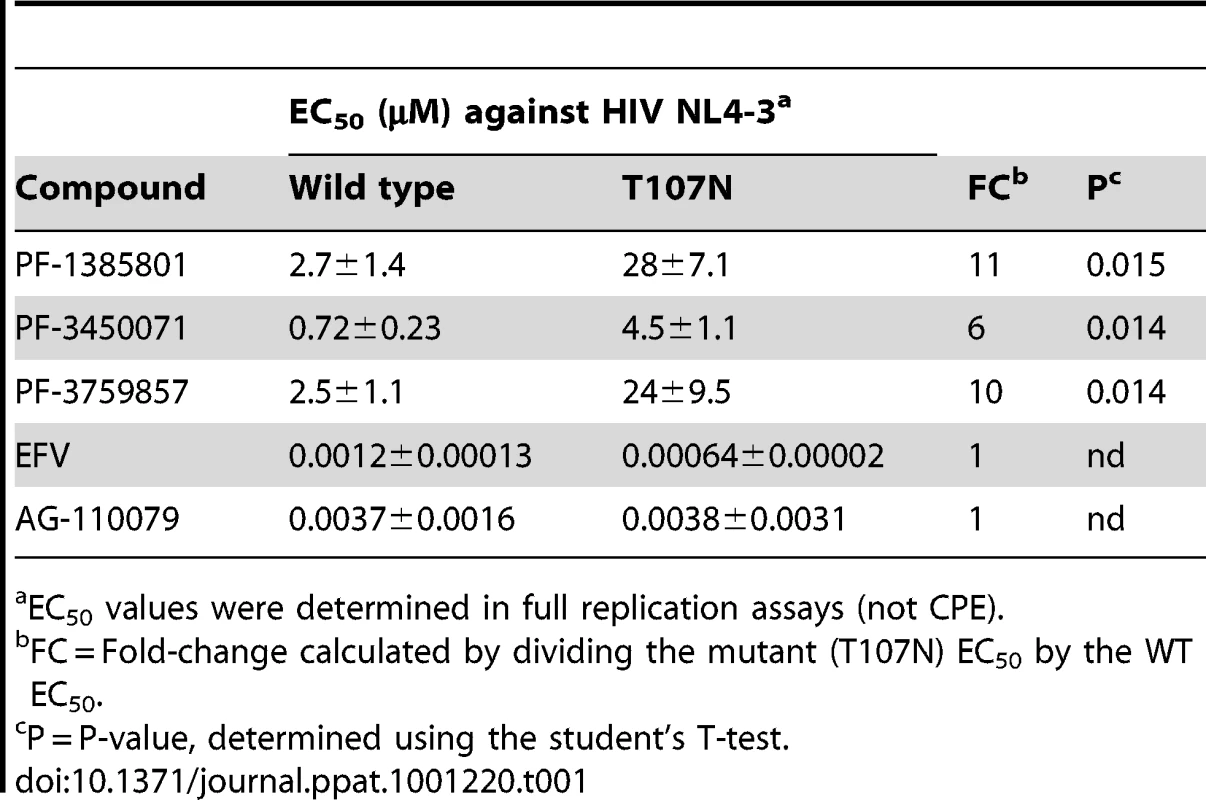
EC50 values were determined in full replication assays (not CPE). These results demonstrate that mutation(s) in HIV-1 CA (e.g. T107N) are sufficient to confer resistance to this new class of inhibitors and demonstrate that HIV-1 CA is the target in infected cells. Although additional work is needed to determine the individual contributions of the mutations identified in addition to T107N, it appears that more than one mutation is required to generate high levels of resistance to this inhibitor class. While it is a definitive tool for target identification, it should be noted that in vitro selection can be used to generate resistant mutants to any known antiviral and does not accurately predict the clinical barrier to resistance, which is a function of several in vivo parameters, including the pharmacokinetics of the compound.
Inhibition of Early Events in the HIV-1 Replication Cycle
To determine the stage of the replication cycle targeted by this new class of compounds, PF-3450071 and PF-3450074 were evaluated in single cycle infection assays. Such assays monitor the early steps of infection up to the integration of the viral cDNA into the host cell chromosome and expression of that sequence. PF-3450071 and PF-3450074 were tested in parallel infections with single-cycle HIV-1 virus packaged with either wild type HIV-1 envelope (NL4-3 pseudovirus) or vesicular stomatitis virus glycoprotein (VSVG), an envelope that allows viral entry by an alternative mechanism (VSV pseudovirus). In addition, three compounds with known mechanisms, AMD3100 (HIV CXCR4/entry inhibitor), EFV (NNRTI/early mechanism) and NFV (protease inhibitor (PI)/late mechanism) were evaluated in the assays. PF-3450071 and PF-3450074 were active against both the NL4-3 and VSV pseudoviruses (Table 2). This inhibition profile was similar to EFV. The profile was distinct from that of AMD3100, which inhibited the HIV-1 envelope NL4-3 pseudovirus, but not the VSVG pseudovirus, and NFV which was not active in the assay (Table 2). These data indicate that PF-3450071 and PF-3450074 act early in the HIV-1 replication cycle at a step following HIV-1 envelope-mediated entry.
Tab. 2. Activity of antiviral compounds in selected assays. 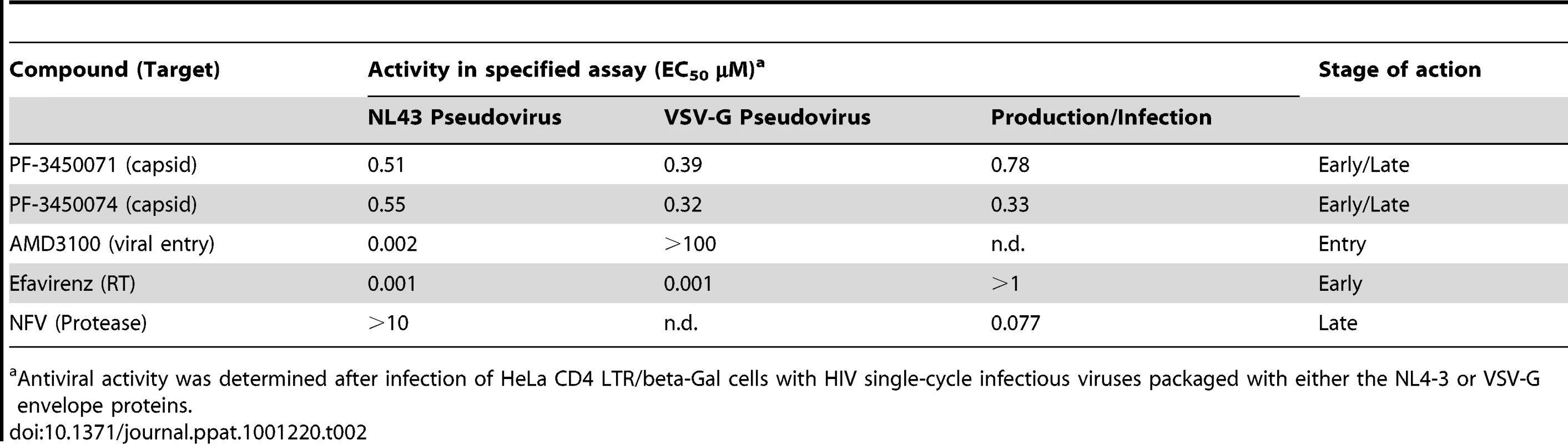
Antiviral activity was determined after infection of HeLa CD4 LTR/beta-Gal cells with HIV single-cycle infectious viruses packaged with either the NL4-3 or VSV-G envelope proteins. Elucidating the Early Stage Mechanism
To begin dissecting the early events of infection, DNA was isolated from single cycle infections of MT-2 cells in the presence or absence of inhibitors and analyzed by quantitative PCR. Primer sets were used that detect total viral cDNA produced, 2-LTR circles (a nuclear episomal form of the viral cDNA), or provirus (the viral genome integrated into host cell chromosomes) as described previously [16]. PF-3450074 inhibited the accumulation of total viral cDNAs (9% of control) and as result inhibited the levels of integrated provirus DNA measured (2% of control) (Fig. 3a). This profile was similar to that observed for the RT inhibitor EFV, indicating that PF-3450074 either inhibits reverse transcription or a step prior to it. In contrast, the integrase inhibitor, AG-110079, resulted in a 4-fold increase in 2-LTR circle accumulation and minimal reduction in total viral cDNA (78% of control) (Fig. 3a). As the profile for PF-3450074 did not match that of the integrase inhibitor control, integrase inhibition can be ruled out as a possible mechanism.
Fig. 3. Identification of mechanism of action. 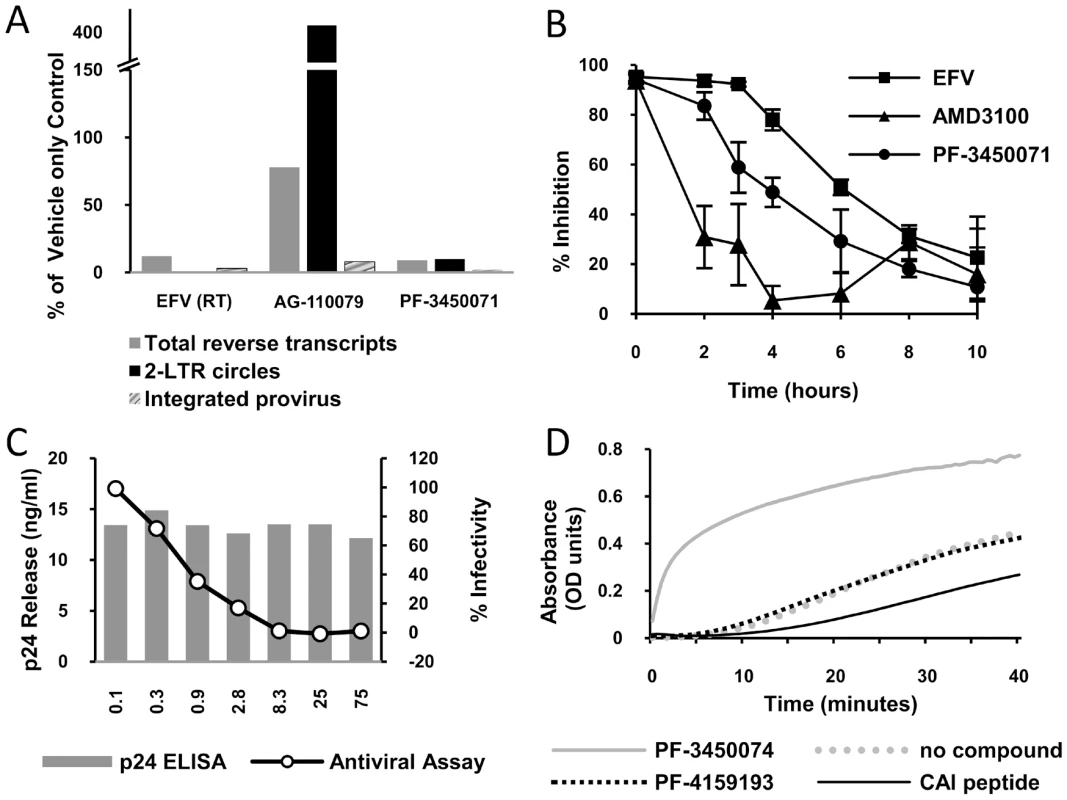
a) HIV DNA quantification. A Taqman quantitative PCR assay was performed to determine the effect of test compounds on the levels of total HIV DNA, 2-LTR circles and integrated provirus generated during infection. The data is presented as a percentage of amounts observed in an untreated control tested in parallel. b) Time-of-addition experiment. HeLa CD4 LTR/beta-Gal reporter cells were infected with HIV-1 (NL43) and test compounds (at 2×EC 90) were added at different times after infection. Reporter signal was monitored at 72 h post-infection as a measure of replication. c) Effect of PF-3450074 in a virus production assay. 293T cells transfected with pNL43 were incubated in varying concentrations of PF-3450074. After 48 hours supernatants were transferred to HeLa CD4 LTR/beta-Gal reporter cells. The line graph shows the percentage of β galactosidase production compared with a mock treated control. Solid bars indicate the concentration of HIV capsid protein (p24) secreted by the 293T cells at each concentration tested. d) Kinetics of CA multimerization. HIV CA spontaneously multimerizes in the presence of high salt, causing a change in optical density. Test compounds were incubated with CA protein and multimerization was initiated with the addition of concentrated salt solution. To further analyze the mechanism of action, the inhibition profile of PF-3450071 was compared to that of AMD3100 and EFV in synchronized time-of-addition experiments, where inhibitors are added a various time points following infection to monitor when the susceptible step has occurred. The viral entry inhibitor, AMD3100, showed a dramatic early loss in activity, even within the first two hours of infection. The NNRTI, EFV, retained the majority of inhibitory activity (>78%) when added up to 4 hours after infection and lost activity thereafter, with a mid-point around 6 hours after infection. PF-3450071 displayed a profile that was distinct from both comparators. The compound maintained the majority of its inhibitory activity (>84%) when added up to 2 hours after infection, and lost significant levels of activity when added 3 or more hours after infection (Fig. 3b). These results suggest that this new class of HIV-1 inhibitor targets an early step in the virus replication cycle, after viral entry but before reverse transcription, possibly viral uncoating. Consistent with these data, PF-3450074 did not inhibit recombinant HIV-1 RT in standard biochemical assays (IC50>100 µM).
Inhibition of Late Events in the HIV-1 Replication Cycle
Virus production-infectivity assays were conducted to determine if compounds affect the late stages of viral replication. Infectious virus was expressed from transfected producer cells in the presence or absence of compound and the infectivity of the resulting virus was tested by diluting the supernatant on to an indicator cell line. As infectious virus is produced from a transfected DNA, bypassing the early stages of the replication cycle, only inhibitors targeting the later stages of viral replication (i.e. post-integration) should show activity in the assay. We tested two compounds (PF-3450071 and PF-3450074) in the viral production-infectivity assay using NFV (PI) and EFV (NNRTI) as late stage and early stage inhibitor controls. Both PF-3450071 and PF-3450074 inhibited the production of infectious virus with EC50 values of 0.78 and 0.33 µM, respectively (Table 2). As expected the HIV-1 PI, NFV, inhibited infectious virus production while the RT inhibitor, EFV was not active in the assay (Table 2). Notably, the overall levels of Gag proteins released into the supernatant of transfected cells were not affected by PF-3450074, as measured by p24 ELISA (Fig. 3c). Thus, this series does not inhibit HIV-1 particle production but renders the nascent particles noninfectious.
Western blot analysis of viral supernatants with antibodies directed against the HIV-1 CA protein (p24) showed that PF-3450071 did not affect HIV-1 Gag proteolytic processing (Fig. S1), which demonstrates that the compounds do not inhibit HIV-1 protease or virion maturation in the manner described for inhibitors such as Bevirimat (PA-457) [17] and PF-46396 [18]. Collectively, these data suggest an effect of the series on proper assembly of fully processed CA protein in nascent viral particles.
Disruption of the Formation of Native-Like Particles
To observe the effects of this class of compound on the morphology of nascent viral particles, PBMCs were infected with HIV NL4-3 either in the absence or presence of PF-3450074 at 7 µM (∼10× EC50). The cells and resulting viral products were fixed and treated for transmission electron microscopy (TEM). In untreated infections, approximately 66% of particles (78/118 observed) were native-like, as defined by being ∼100nm in diameter and having a distinct central density representative of a mature capsid core). A close in view of a native-like particle from an untreated culture (Fig. 4a) clearly illustrates the conical capsid core in the center of the virion. A wider view shows the high level of uniformity among the particles produced in the untreated infections (Fig. 4b). Both native-like and apparent immature particles are present at consistent shape and size. In contrast, native-like particles were not observed in PF-3450074-treated infections. The particles produced in PF-3450074-treated infections lack a clear central density representative of a mature capsid (Fig. 4c). Consistent with the observation that these compounds do not affect p24 levels, the number of particles in PF-3450074-treated infections appears similar to untreated, however the morphology and size of particles is highly variable (Fig. 4d).
Fig. 4. EM analysis of the effects of PF-3450074 on nascent viral particles. 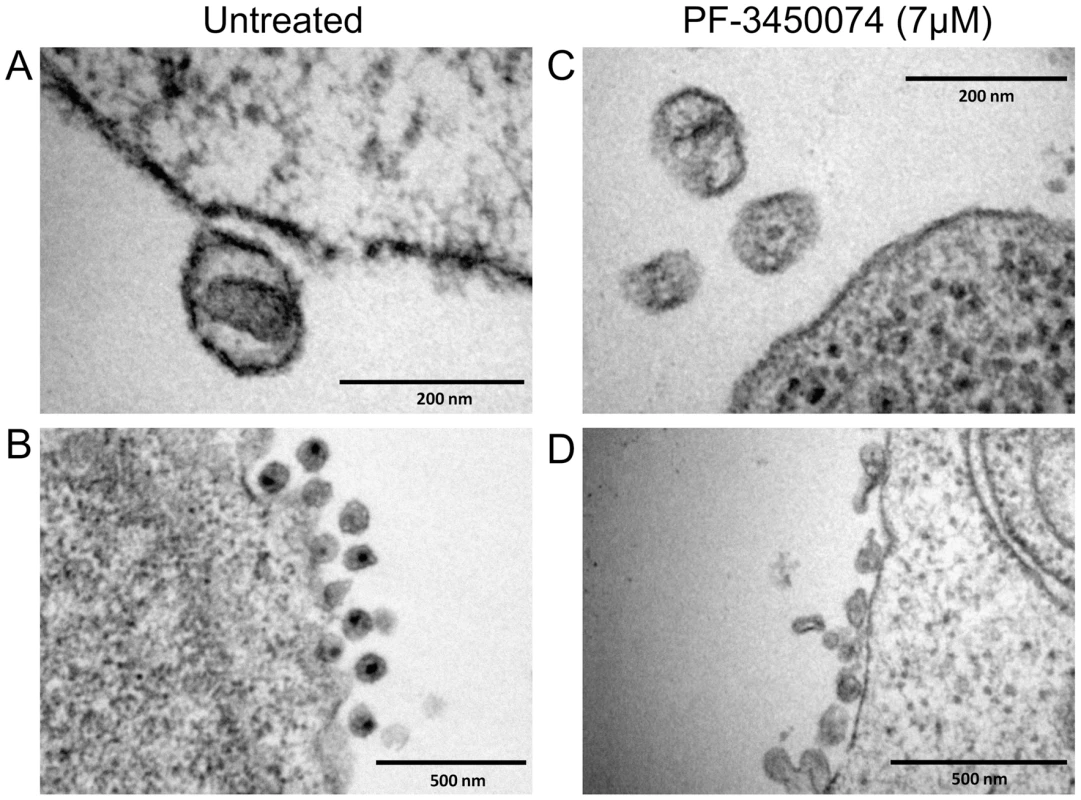
PBMCs were infected with HIV NL4-3 in medium containing either no compound or 7µM PF-3450074. The cells and resulting viral products were processed and observed by transition electron microscopy. Close in view of an untreated native-like particle (a) reveals the central conical capsid structure, while a wider view (b) shows the high level of uniformity in morphology and size among untreated particles. Close in view of PF-3450074-treated particles (c) shows consistent loss of the central capsid density, and a wider view (d) illustrates a much wider range in particle morphology and size. Compound Effects on CA Multimerization
The simplest explanation to reconcile the data above involves a model where this new class of inhibitors targets both early (viral core uncoating) and late (viral core assembly) events in the replication cycle by affecting CA – CA interactions and thus core stability. To study the effects on capsid assembly, we evaluated the inhibitors in in vitro CA multimerization assays [10]. Such assays can be used to measure the effect of compounds on the rate of formation of higher order CA multimers or tubes that are widely thought to represent many aspects of native core structure. Addition of PF-3450074 results in a significant increase in the rate of CA multimerization (Fig. 3d). In contrast, a structural analogue with no antiviral activity, PF-4159193 (Fig. S2), did not affect the kinetics of CA multimerization, indicating that this profound effect is correlated with antiviral activity and not a general physical effect of this series. As we have reproduced with the CAI peptide (Fig. 3d), all of the previously reported HIV CA assembly inhibitors decreased the rate of multimerization in this assay [10], [11], [12]. These data demonstrate indirectly that PF-3450074 interacts with HIV-1 CA and further suggest a mechanism that is fundamentally distinct from previously reported HIV CA inhibitors.
High Resolution Co-Crystal Structure Reveals a Novel Binding Pocket
To further understand the mode of action of this novel class of compounds, we determined the crystal structure of HIV-1 CA NTD protein in complex with PF-3450074 using a CA protein construct that contained a single glycine residue in place of the cyclophilin binding loop (residues 87–99). Although the cyclophilin binding loop is important for viral infection, it is not required for proper HIV CA protein folding and in vitro multimerization function [8]. Binding affinity of PF-3450074 to the crystallographic construct (Kd = 3.42 µM) is similar to that observed for both full length wild type CA (Kd = 2.79 µM) and isolated wild type NTD (Kd = 2.24 µM), as measured by isothermal titration calorimetry. Using this construct, the co-crystal structure of PF-3450074 was solved with the NTD of HIV-1 CA to 1.8 Å resolution. The structure of the complex showed that the overall fold of the CA protein is the same as previously described CA structures [19], [20], [21] and illustrates that neither the compound, PF-3450074, nor the loop deletion caused any significant shifts in the protein structure (Fig. 5a).
Fig. 5. Structure of the novel inhibitor binding site and context in the NTD. 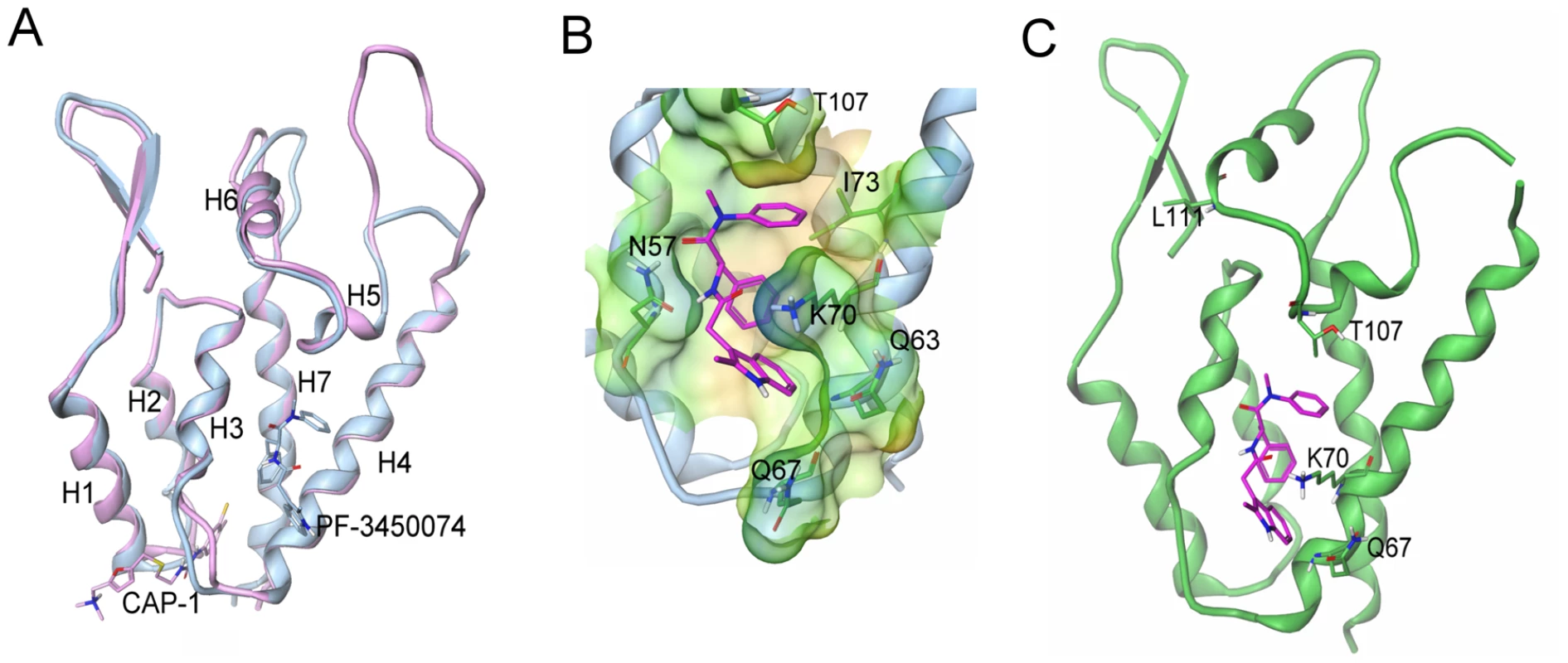
a) Overlays of capsid structures with PF-3450074 in blue and CAP-1 in pink bound to capsid N-terminal domain; b) Close up view of PF-3450074 site (binding site residues labelled in black, R1-3 sub-pockets labelled in purple). In the R1 sub-pocket PF-3450074 makes hydrophobic interactions with Ile-73, Ala-105, Thr-107, Tyr-130 and a stacking interaction with the side chain of Asn-53. The benzyl group R2 makes a number of hydrophobic interactions in a sub-pocket formed by the side chains of Met66, Leu69, Val59, Ile-73 and Leu-56. The R3 sub-pocket is only partially occupied by the indole group, forming interactions with the side chains of Met-66, Gln-67, Lys-70 and Gln-63 amide. The indole NH forms a hydrogen bond interaction with the side chain amide of Gln-67 via a water molecule, while an Asn-57 forms a key hydrogen bond with the cis amide bond of PF-3450074. c) Location of resistant mutations (purple) in relation to PF-3450074 capsid binding site. (PDB ID code 2XDE). PF-3450074 occupies a preformed pocket in the HIV-1 CA NTD bounded by helices 3, 4, 5 and 7 (Fig. 5a). The R1 and R2 aromatic moieties of the compound occupy two hydrophobic sub-pockets and provide most of the key interactions which anchor the compound to the NTD (Fig. 5b). The indole substituent protrudes from the NTD R3 sub-pocket close to Lys70. The binding site for PF-3450074 is distinct from the sites targeted by CAP-1 and CAI/NYAD-1 [10], [11], [22], [23], [24]. These results confirm that PF-3450074 directly binds HIV CA and are consistent with this class of inhibitors acting via a unique mechanism. Coordinates are stored in the Protein Data Bank (PDB ID code 2XDE).
Discussion
We describe a novel class of inhibitors that target HIV CA by a unique mechanism that interferes with both early and late events in the viral replication cycle. HIV CA plays an essential role in several stages of viral replication and is viewed as an important, yet unexploited target for therapeutic intervention [4], [5], [6]. This new series demonstrates that small molecules targeting HIV CA can have potent broad-spectrum antiviral activity. We demonstrate directly that HIV CA is the antiviral target of these inhibitors in infected cells by showing that mutations in HIV CA confer resistance to several members of the series. EM analysis shows that the series profoundly affects the morphology of nascent HIV particles. We demonstrate that the compounds affect CA protein multimerization in vitro and we have elucidated details of the novel compound binding site on HIV CA by solving a co-crystal structure of a compound from the series bound to the NTD of the protein. Our data strongly suggest that this new series of inhibitors targets HIV CA function during both the virion uncoating and viral core assembly processes.
Previous studies have described two molecules that target HIV-1 CA assembly in vitro, CAP-1 (a small molecule) and CAI (a dodecapeptide) [10], [11], [12]. CAP-1 acts in the late stage of viral replication and does not inhibit HIV-1 infection when added to pre-formed HIV-1 particles. A cell-permeable derivative of CAI, NYAD-1, inhibits formation of both immature and mature HIV-1 virus particles as well as early events in the replication cycle at low micromolar concentrations. The properties of the new class of CA inhibitors described in this study are clearly distinguished from those of other HIV-1 inhibitors, including previously described CA inhibitors. Unlike CAP-1, the small molecules described here inhibit both early and late events in the HIV replication cycle. In addition, PF-3450074 did not inhibit Gag particle production from HIV-1 transfected cells, suggesting that the compound series does not affect immature particle assembly. This is in contrast to the effects on immature particle assembly reported for NYAD-1. A co-crystal structure of a representative compound (PF-3450074) demonstrated a new binding site on HIV-1 CA distinct from those described for CAP-1 or CAI. Furthermore, PF-3450074 increased the rate of HIV-1 CA multimerization in vitro, while CAI and CAP-1 decreased the rate of CA multimerization in the same assay. While this does not necessarily define the action of these compounds on replicating virus, it does suggest a fundamentally different mechanism of inhibition from that of previously described CA inhibitors.
The proposed mechanism of action for both the early and late stage activities of this new class of inhibitors involves a direct effect on higher-order structures of HIV CA, in assembly and uncoating. Although the present data do not indicate whether the compounds enhance or inhibit the uncoating process, either effect is likely to interfere with proper reverse transcription [25]. HIV-1 capsid mutations proximal to the PF-3450074 binding pocket have been described that either destabilize or enhance the stability of viral cores and result in specific postentry defects in virus replication [24]. It is possible that such mutations and the compounds described in this study have analogous effects on inter-subunit capsid interactions. To gain further insights into the mode of action of PF-3450074, we generated a model of an assembled capsid hexamer in complex with PF-3450074 (Fig. 6a) based on superpositioning of published assembled capsid structures [13], [14] with the structure of the PF-3450074/CA complex. In the model, the R3 indole group which protrudes from the NTD in our structure localizes to the interface between capsid monomers in an assembled capsid and sits directly between the NTD of one capsid monomer and the C-terminal domain of another, making contacts to Tyr-169, Leu-172, Arg-173, Gln-179, and Lys-182 (Fig. 6b). This suggests the R3 indole group of PF-3450074 could play a critical role in modulating inter-subunit interactions. Both the CA NTD contact residues described by the co-crystal structure and these putative C-terminal contacts are well conserved across viral strains (Tables S5 and S6). This is consistent with the broad-spectrum antiviral activity observed for this series.
Fig. 6. Model of inhibitor effects at the NTD-CTD interface. 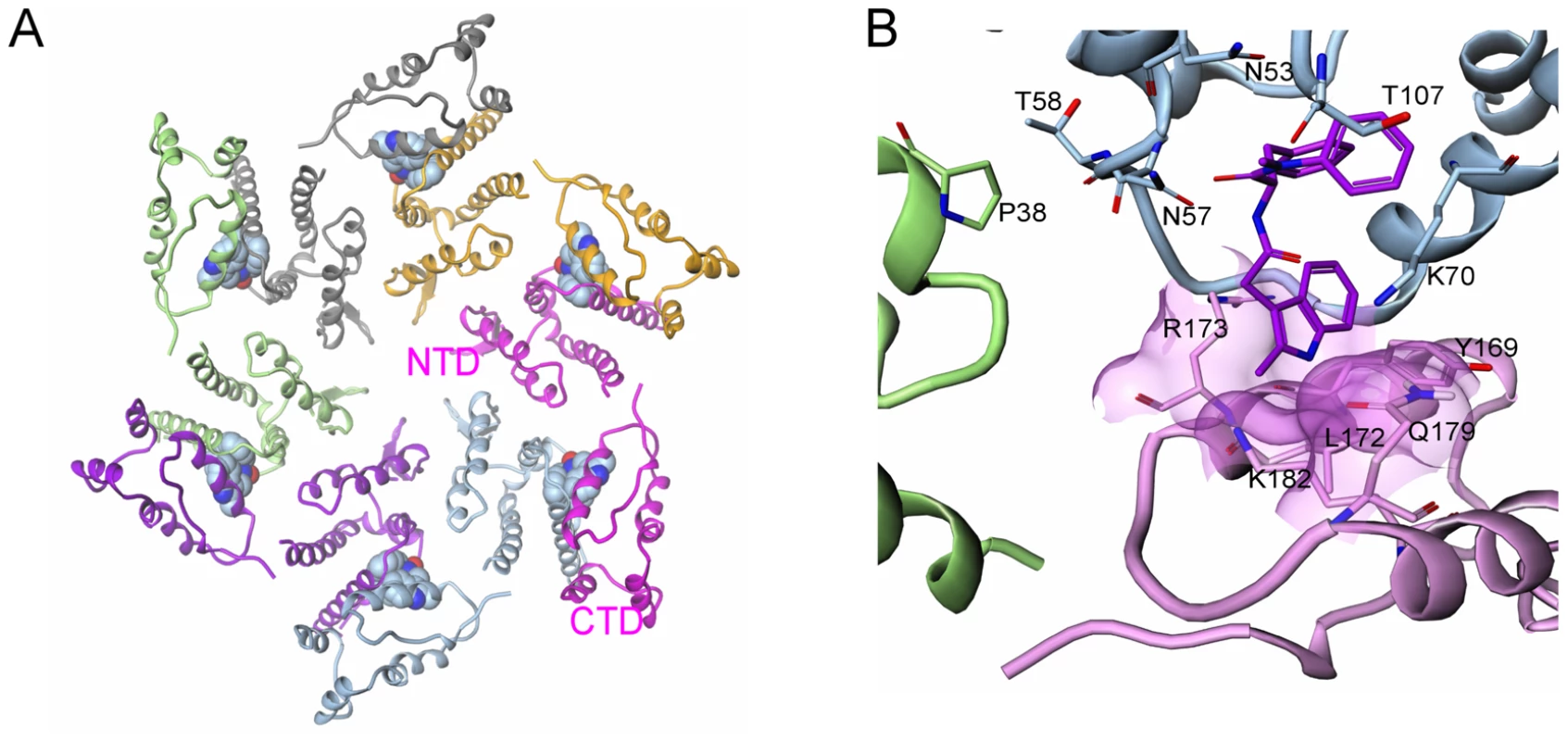
a) Model of HIV-1 capsid in hexameric complex with PF-3450074 bound in each of the binding pockets (each of the six full-length CA monomers are coloured differently); b) Close up view of model of interface between adjacent N-terminal subunits (green and blue) and the C-terminal domain of an adjacent monomer (pink) in assembled capsid. PF-3450074 highlighted in purple. Residues highlighted Pro38 (adjacent N-terminal domain green), Thr-107, Thr-58, Asn-57, Asn-53, Lys-70 (N-terminal domain blue), Tyr-169, Leu-172, Arg-173, Gln-179, Lys-182 (C-terminal domain pink). Although the sum of our results suggests a mechanism that affects interactions between capsid monomers, the early stage activity is consistent with other models. Cyclophilin A plays a critical role in the early stages of HIV-1 replication through interactions with the viral capsid [26]. Also, capsid-binding restriction factors such as the tripartite motif containing (TRIM) proteins prevent the infection of many primate cells with HIV or SIVs from other species [5], [9]. Thus, based on our data, we cannot dismiss the possibility that, during early infection, this new series might affect specific capsid-host protein interactions that mediate the viral uncoating process. A detailed study of the early stage activity of PF-3450074 has demonstrated direct destabilization of the HIV-1 capsid and a dependence on cyclophilin A, indicating that the compound induces premature uncoating of the virus, potentially through a mechanism similar to that of TRIM restriction [27].
In this study, we identify a new binding site on HIV-1 CA that can be targeted by small molecule inhibitors resulting in broad-spectrum antiviral activity. In addition, we describe the discovery and characterization of a novel series of compounds that act at this site and inhibit the virus at two points in the replication cycle. This series should serve as a good starting point for the development of a new class of HIV therapeutics through structure-based drug design or other approaches. The broad spectrum activity of this series is particularly exciting and highlights this novel mechanism as a significant therapeutic opportunity.
Material and Methods
Cells, Virus, and Compounds
HeLa CD4 LTR/beta-Gal, MT-2, PM1, CEM-SS and HEK 293 cells as well as pNL4-3 HIV-1 infectious clone, HIV-1 IIIB, HIV-1 RF, HIV-1 BaL, and all primary isolates were obtained through the National Institutes of Health (NIH) AIDS Research and Reference Reagent Program, Bethesda, MD. JC53BL cells were sourced from Tranzyme. Efavirenz (EFV) was kindly provided by DuPont Merck (Wilmington, DE). Nelfinavir (NFV), 135137, PF-3450074, and PF-3759857 were synthesized by Pfizer Inc.
Cytopathic Effect Assays (CPE)
As described [18], host cells were infected with HIV-1 NL4-3, HIV-1 IIIB, HIV-1 RF. The cytopathic effect was measured using XTT reagent and the therapeutic index (TI) calculated by dividing the CC50 (mock infected cells) value by the EC50.
Clinical Isolate Antiviral Assays
PHA-stimulated PBMC's were incubated for 1 hour with virus at an moi of 0.001–0.01. They were plated in 96-well plates at 5×105 cells/ml and incubated for 5 days at 37°C.10 µL of the supernatant was then transferred to a plate containing 40 µL of JC53BL cells at 0.5×105 cells/ml. After 2 days, the cells were processed for β-galactosidase activity using the FluorAce kit (Bio-Rad). PF-3794231 was tested against clinical isolates at the Southern Research Institute (Frederick, MD) as previously described [18].
Single-Cycle Infection Assays
The single-cycle infectious HIV-1 reporter viruses were generated as previously described [28] by co-transfecting HEK293 cells with an HIV-1 NL4-3 single-cycle infectious cDNA (pNL4-3deltaEnv) and an NL4-3 or VSV-G envelope expression vector. Half-log dilutions of test compounds were added to HeLa CD4 LTR/beta-Gal target cells, seeded in 96-well plates at a cell density of 1×104 cells per well in DMEM containing 10% FBS. Compound-treated or compound-free target cells were then infected with the HIV-1 single-cycle infectious virus and after 72 hours measured for the induction of beta-galactosidase Data were expressed as the percent of reporter gene activity in infected compound-treated cells relative to that of infected, compound-free cells.
Virus Production-Infectivity Assays
Envelope-deleted NL4-3 cDNA (pNL4-3deltaEnv) was co-transfected into HEK 293 cells with an HIV envelope expression vector as previously described [29]. Half-log dilutions of test compounds were then added to transfected cell cultures 3 hrs after transfection. The supernatants were harvested 72 hrs after transfection and infectious virus production was subsequently quantified by measuring the induction of the beta-galactosidase reporter gene after 100-fold dilution into fresh medium and incubation in the presence of HeLa CD4 LTR/beta-Gal target cells for 72 hours.
Selection and Characterization of Resistant Virus
Viral variants resistant to PF-1385801 or PF-3759857 were selected as described previously [18]. To construct NL4-3 recombinant virus containing the Q67H, K70R, Q87P, T107N and L111I amino acid substitutions identified in the serial passage studies, viral cDNAs that had been TOPO cloned for sequence analysis were digested with BssHII and ApaI and ligated back into a wild type pNL4-3 background. The effect of amino acid substitutions in HIV-1 Gag sequences on PF-3759857 susceptibility was measured in HIV Replication assays as described previously [15].
Protein Expression and Purification
Proteins were expressed in E. Coli BL21(DE3) in 2YT containing kanamycin. Cells were harvested following induction with IPTG and growth overnight at 20°C. CA1-231 was purified as described [30]. For CA1-146/Δ87-99G, cells were resuspended and lysed in buffer A (50 mM TrisHCl pH 7.4, 150 mM NaCl, 10 mM imidazole, and 2 mM β-mercaptoethanol). The lysate was clarified by centrifugation, filtered then applied to a His trap nickel affinity column (Sigma), eluting with buffer A supplemented with 300 mM imidazole. The eluted material was further purified on a Sephacryl 100HR column in 50 mM TrisHCl pH 7.4, 150 mM NaCl and 2 mM β-mercaptoethanol.
HIV Capsid Multimerisation Assay
Multimerisation assays were performed as previously described [10]. Compound was added to 30 µg of full-length CA protein in 50 mM sodium phosphate buffer, pH 8.0 in a volume of 30 µl. Capsid assembly was initiated by addition of a concentrated NaCl solution (50 µl 5 M NaCl in 50 mM sodium phosphate, pH 8.0). Optical density was monitored on a Molecular Devices SpectraMax spectrophotometer at 350 nm every 20 s for 1 h.
Transmission Electron Microscopy (TEM)
PBMCs were incubated with 7 µM PF-3450074 (approx. 10× IC50), for 30 minutes before being infected with mock virus (medium alone), or with NL4-3 at an MOI of 1. At 72 hours post-infection the supernatants were removed and the cells were fixed for 60 min in MacDowell's fixative (4% (v/v) paraformaldehyde, 1% glutaraldehyde in 0.1M phosphate Milliong buffer, pH 7.3) at 4 C. The cells were rinsed in 0.15M Phosphate Sörensen buffer with 0.2% (v/v) NaCl (pH 7.3) and suspended in warm 3.5% agar in water. The agar blocks were cooled to 4°C and washed three times for 10 minutes in 0.15M Phosphate Sörensen buffer with 0.2% (v/v) NaCl (pH 7.3). The agar blocks with the cells were stained using 2% Osmium tetroxide in 0.3M phosphate Sörensen buffer (pH 7.4) for 1 h at 4°C. The blocks were washed 4 times for 10 minutes in sterilized water at room temperature before they were dehydrated in graded ethanol (50%, 70%, 90%, 100%, 100%) for 15 minutes at each concentration at room temperature. The blocks were treated with propylene oxide three times for 15 minutes in room temperature before the resin was infiltrated for 1 hour at room temperature with a 1∶1 propylene oxide∶Epon mix (Epon contains epoxy embedding medium 20 mL (Epon 812 resin)) and 12 ml MNA (methyl nadic anhydride) and 9 ml DDSA (dodecenyl succinic anhydride)) before the resin was treated in epon overnight at room temperature. The resin was then embedded with Epon and BDMA 3% for 1 hour at room temperature. The resin was polymerised for 48h at 58–60°C. Approximately 1 micron thick semi-thin sections were cut and stained in toluidine blue and observed in light microscopy. Subsequently, ultra-thin sections of 70–90 nm were cut on an Ultracut E Reichert microtone, and stained with uranyl acetate and lead citrate. Observations were made using a Jeol 1200 EX II electron microscope.
Isothermal Calorimetry
Isothermal titration calorimetry (ITC) was performed using the VP-ITC calorimetric system (GE Healthcare). Protein solutions for ITC were dialyzed against buffer A (50 mM TrisHCl pH 7.5/150 mM NaCl/2 mM β-mercaptoethanol). The dialyzed protein solution (15 µM) in the calorimetric cell (1.4274 ml) was titrated at 25°C with ligand (200 µM) in the buffer A using 1×2 µL, followed by 25×10 µL injections. Heat evolved was obtained from the integral of the calorimetric signal and heat of dilution was negligible in titrations of the ligand into buffer only. Analysis was carried out with Origin 5.0 software (GE Healthcare). Binding parameters such as the number of binding sites (n), the binding constant (Ka, M−1), and the binding enthalpy (ΔHa, kcal/mol of bound ligand) were determined by fitting the experimental binding isotherms.
Protein Crystallization, Data Collection, and Structure Refinement
Protein was concentrated to 30 mg/ml and inhibitor added to 5 mM from a 100 mM DMSO stock solution. After standing for 2 hours, hanging drop crystallizations were set up consisting of 2 µl of protein and 1 µl of well solution (20% PEG 8000, 100 mM phosphate-citrate pH 4.2 and 200 mM sodium chloride). Crystals grew overnight at room temperature and were frozen for X-ray data collection following addition of 60% NDSB containing 40% ethylene glycol (4 µl). Data was collected at the ESRF (ID14-4) on an ADSC Q315 detector, integrated with mosflm [31] and scaled with CCP4 package SCALA [32]. The structure was solved by molecular replacement using MOLREP [32] with a truncated model of the N-terminal CA. The structure was refitted using QUANTA version 2000.1 (Accelrys Inc., San Diego, CA) and refinement was carried out using REFMAC [33]. Data collection and refinement statistics are shown in Table S7. Coordinates are stored in the Protein Data Bank (PDB ID code 2XDE).
Supporting Information
Zdroje
1. HammerSM
EronJJJr
ReissP
SchooleyRT
ThompsonMA
2008 Antiretroviral Treatment of Adult HIV Infection: 2008 Recommendations of the International AIDS Society-USA Panel. JAMA 300 555 570
2. TaiwoB
2009 Understanding transmitted HIV resistance through the experience in the USA. International Journal of Infectious Diseases 13 552 559
3. SmithRJ
OkanoJT
KahnJS
BodineEN
BlowerS
2010 Evolutionary dynamics of complex networks of HIV drug-resistant strains: the case of San Francisco. Science 327 697 701
4. AdamsonCS
FreedEO
2010 Novel approaches to inhibiting HIV-1 replication. Antiviral Research 85 119 141
5. MascarenhasAP
Musier-ForsythK
2009 The capsid protein of human immunodeficiency virus: interactions of HIV-1 capsid with host protein factors. FEBS Journal 276 6118 6127
6. NeiraJ
2009 The capsid protein of human immunodeficiency virus: designing inhibitors of capsid assembly. FEBS Journal 276 6110 6117
7. Ako-AdjeiD
JohnsonMC
VogtVM
2005 The Retroviral Capsid Domain Dictates Virion Size, Morphology, and Coassembly of Gag into Virus-Like Particles. J Virol 79 13463 13472
8. Ganser-PornillosBK
von SchwedlerUK
StrayKM
AikenC
SundquistWI
2004 Assembly properties of the human immunodeficiency virus type 1 CA protein. J Virol 78 2545 2552
9. AikenC
2006 Viral and cellular factors that regulate HIV-1 uncoating. Curr Opin HIV AIDS 1 194 199
10. TangC
LoeligerE
KindeI
KyereS
MayoK
2003 Antiviral inhibition of the HIV-1 capsid protein. J Mol Biol 327 1013 1020
11. StichtJ
HumbertM
FindlowS
BodemJ
MullerB
2005 A peptide inhibitor of HIV-1 assembly in vitro. Nat Struct Mol Biol 12 671 677
12. ZhangH
ZhaoQ
BhattacharyaS
WaheedAA
TongX
2008 A Cell-penetrating Helical Peptide as a Potential HIV-1 Inhibitor. Journal of Molecular Biology 378 565 580
13. Ganser-PornillosBK
ChengA
YeagerM
2007 Structure of Full-Length HIV-1 CA: A Model for the Mature Capsid Lattice. Cell 131 70 79
14. PornillosO
Ganser-PornillosBK
KellyBN
HuaY
WhitbyFG
2009 X-ray structures of the hexameric building block of the HIV capsid. Cell 137 1282 1292
15. CaoJ
IsaacsonJ
PatickAK
BlairWS
2005 High-throughput human immunodeficiency virus type 1 (HIV-1) full replication assay that includes HIV-1 Vif as an antiviral target. Antimicrob Agents Chemother 49 3833 3841
16. ButlerSL
HansenMS
BushmanFD
2001 A quantitative assay for HIV DNA integration in vivo. Nat Med 7 631 634
17. LiF
Goila-GaurR
SalzwedelK
KilgoreNR
ReddickM
2003 PA-457: A potent HIV inhibitor that disrupts core condensation by targeting a late step in Gag processing. Proceedings of the National Academy of Sciences of the United States of America 100 13555 13560
18. BlairWS
CaoJ
Fok-SeangJ
GriffinP
IsaacsonJ
2009 New Small-Molecule Inhibitor Class Targeting Human Immunodeficiency Virus Type 1 Virion Maturation. Antimicrob Agents Chemother 53 5080 5087
19. KellyBN
HowardBR
WangH
RobinsonH
SundquistWI
2006 Implications for Viral Capsid Assembly from Crystal Structures of HIV-1 Gag(1–278) and CA(N)(133–278). Biochemistry 45 11257 11266
20. GambleTR
VajdosFF
YooS
WorthylakeDK
HouseweartM
1996 Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell 87 1285 1294
21. Monaco-MalbetS
Berthet-ColominasC
NovelliA
BattaiN
PigaN
2000 Mutual conformational adaptations in antigen and antibody upon complex formation between an Fab and HIV-1 capsid protein p24. Structure 8 1069 1077
22. KellyBN
KyereS
KindeI
TangC
HowardBR
2007 Structure of the Antiviral Assembly Inhibitor CAP-1 Complex with the HIV-1 CA Protein. Journal of Molecular Biology 373 355 366
23. TernoisF
StichtJ
DuquerroyS
KrausslichHG
ReyFA
2005 The HIV-1 capsid protein C-terminal domain in complex with a virus assembly inhibitor. Nat Struct Mol Biol 12 678 682
24. BhattacharyaS
ZhangH
DebnathAK
CowburnD
2008 Solution structure of a hydrocarbon stapled peptide inhibitor in complex with monomeric C-terminal domain of HIV-1 capsid. J Biol Chem 283 16274 16278
25. ForsheyBM
von SchwedlerU
SundquistWI
AikenC
2002 Formation of a Human Immunodeficiency Virus Type 1 Core of Optimal Stability Is Crucial for Viral Replication. J Virol 76 5667 5677
26. LubanJ
2007 Cyclophilin A, TRIM5, and Resistance to Human Immunodeficiency Virus Type 1 Infection. J Virol 81 1054 1061
27. ShiJ
ZhouJ
ShahVB
AikenC
WhitbyK
2010 Small Molecule Inhibition of Human Immunodeficiency Virus Type 1 Infection by Virus Capsid Destabilization. J Virol epub ahead of print
28. BlairWS
IsaacsonJ
LiX
CaoJ
PengQ
2005 A novel HIV-1 antiviral high throughput screening approach for the discovery of HIV-1 inhibitors. Antiviral Research 65 107 116
29. BlairWS
CaoJ
JacksonL
JimenezJ
PengQ
2007 Identification and Characterization of UK-201844, a Novel Inhibitor That Interferes with Human Immunodeficiency Virus Type 1 gp160 Processing. Antimicrob Agents Chemother 51 3554 3561
30. LanmanJ
SextonJ
SakalianM
PreveligePEJr
2002 Kinetic analysis of the role of intersubunit interactions in human immunodeficiency virus type 1 capsid protein assembly in vitro. J Virol 76 6900 6908
31. LeslieAGW
1992 mosflm. Joint CCP4+ESF-EAMCB Newsletter on Protein Crystallography, No 26
32. Collaborative Computational Project N 1994 The CCP4 Suite: Programs for protein crystallography. Acta Crystallographica Section D Biological Crystallography 50 760 763
33. MurshudovGN
VaginAA
DodsonEJ
1997 Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53 240 255
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2010 Číslo 12- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Is Adherence to Erythrocytes a Factor in Extrapulmonary Dissemination?
- Identifying the Age Cohort Responsible for Transmission in a Natural Outbreak of
- Blockade of Immunosuppressive Cytokines Restores NK Cell Antiviral Function in Chronic Hepatitis B Virus Infection
- Metaeffector Exploits Host Proteasome to Temporally Regulate Cognate Effector
- The p53-Target Gene Drives Neutrophil-Mediated Protection against Lethal Bacterial Sepsis
- Development of an RNAi Protocol to Investigate Gene Function in the Filarial Nematode,
- Lysine Acetyltransferase GCN5-A Functions in the Cellular Response to Alkaline Stress and Expression of Cyst Genes
- Structural Basis for Apoptosis Inhibition by Epstein-Barr Virus BHRF1
- Molecular Architectures of Trimeric SIV and HIV-1 Envelope Glycoproteins on Intact Viruses: Strain-Dependent Variation in Quaternary Structure
- Interaction of c-Cbl with Myosin IIA Regulates Bleb Associated Macropinocytosis of Kaposi's Sarcoma-Associated Herpesvirus
- Glacial Refugia in Pathogens: European Genetic Structure of Anther Smut Pathogens on and
- Role for Sumoylation in Systemic Inflammation and Immune Homeostasis in Larvae
- Inflammasome Sensor Nlrp1b-Dependent Resistance to Anthrax Is Mediated by Caspase-1, IL-1 Signaling and Neutrophil Recruitment
- Noise Cancellation: Viral Fine Tuning of the Cellular Environment for Its Own Genome Replication
- Infectious Speciation Revisited: Impact of Symbiont-Depletion on Female Fitness and Mating Behavior of
- Eis Regulates Autophagy, Inflammation, and Cell Death through Redox-dependent Signaling
- NleC, a Type III Secretion Protease, Compromises NF-κB Activation by Targeting p65/RelA
- Early Myeloid Dendritic Cell Dysregulation is Predictive of Disease Progression in Simian Immunodeficiency Virus Infection
- HIV Capsid is a Tractable Target for Small Molecule Therapeutic Intervention
- Structural and Functional Studies of Nonstructural Protein 2 of the Hepatitis C Virus Reveal Its Key Role as Organizer of Virion Assembly
- Coming of Age—Sexual Reproduction in Species
- HIV-1 Envelope Subregion Length Variation during Disease Progression
- Compartmentation of Redox Metabolism in Malaria Parasites
- Evidence That Intracellular Stages of Utilize Amino Sugars as a Major Carbon Source
- Rapid End-Point Quantitation of Prion Seeding Activity with Sensitivity Comparable to Bioassays
- Dimeric 2G12 as a Potent Protection against HIV-1
- Hypoxia Induces an Immunodominant Target of Tuberculosis Specific T Cells Absent from Common BCG Vaccines
- CD4 Natural Regulatory T Cells Prevent Experimental Cerebral Malaria via CTLA-4 When Expanded In Vivo
- The Killing of African Trypanosomes by Ethidium Bromide
- H2A.Z Demarcates Intergenic Regions of the Epigenome That Are Dynamically Marked by H3K9ac and H3K4me3
- Large-Scale Field Application of RNAi Technology Reducing Israeli Acute Paralysis Virus Disease in Honey Bees (, Hymenoptera: Apidae)
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- HIV-1 Envelope Subregion Length Variation during Disease Progression
- Coming of Age—Sexual Reproduction in Species
- Evidence That Intracellular Stages of Utilize Amino Sugars as a Major Carbon Source
- Compartmentation of Redox Metabolism in Malaria Parasites
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



