-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaHost Genetics and HIV-1: The Final Phase?
This is a crucial transition time for human genetics in general, and for HIV host genetics in particular. After years of equivocal results from candidate gene analyses, several genome-wide association studies have been published that looked at plasma viral load or disease progression. Results from other studies that used various large-scale approaches (siRNA screens, transcriptome or proteome analysis, comparative genomics) have also shed new light on retroviral pathogenesis. However, most of the inter-individual variability in response to HIV-1 infection remains to be explained: genome resequencing and systems biology approaches are now required to progress toward a better understanding of the complex interactions between HIV-1 and its human host.
Published in the journal: . PLoS Pathog 6(10): e32767. doi:10.1371/journal.ppat.1001033
Category: Review
doi: https://doi.org/10.1371/journal.ppat.1001033Summary
This is a crucial transition time for human genetics in general, and for HIV host genetics in particular. After years of equivocal results from candidate gene analyses, several genome-wide association studies have been published that looked at plasma viral load or disease progression. Results from other studies that used various large-scale approaches (siRNA screens, transcriptome or proteome analysis, comparative genomics) have also shed new light on retroviral pathogenesis. However, most of the inter-individual variability in response to HIV-1 infection remains to be explained: genome resequencing and systems biology approaches are now required to progress toward a better understanding of the complex interactions between HIV-1 and its human host.
Introduction
Many fundamental questions about how and why humans differ in their susceptibility to HIV-1 remain largely unanswered. For example, it has long been known that a fraction of the human population cannot be infected by HIV-1 [1], [2]. We still do not know, however, whether most of those who are resistant to infection are resistant due to innate or adaptive immunity, or to some other mechanism. Nor are the precise pathways that allow apparently permanent control of the virus amongst a subset of those that do become infected well understood. These questions are obviously central in the effort to develop effective strategies to combat HIV-1, and at their heart, they are genetic.
Until recently, our capacity to systematically address these issues was limited. But genomic analyses have advanced to the point that comprehensive or nearly comprehensive analyses of the role of genetic variation in viral control is now within reach. A series of genome-wide association studies has already provided a detailed description of how common variation influences control of HIV-1 [3]–[9]. More importantly, next-generation sequencing is now sufficiently advanced such that a dedicated effort to uncover the role of rare variation has become feasible. Coupling these new developments with rich cohorts being built under the auspices of international groupings such as the Center for HIV/AIDS Vaccine Immunology (CHAVI) and the International HIV Controllers Study, all ingredients are now available for drawing conclusive answers to these fundamental questions.
Here, we first review what is known about how genetic variation influences HIV-1 acquisition and control. We then describe new developments and argue that a concerted effort is now appropriate to bring these elements together to answer these outstanding questions decisively and draw the appropriate lessons for vaccine development as well as understanding of pathogenesis.
Cohorts
Cohorts for the Study of HIV-1 Acquisition
In various high-risk populations, there are reports of individuals that have been repeatedly exposed to HIV-1 and yet have not been infected. Exposure itself has been assessed in various ways, but in all cases it appears that portions of the population are protected. Perhaps the most striking example is the case of hemophilia: a vast majority of severe hemophilia A patients born before 1979 became HIV-1 seropositive, due to virtually universal exposure to contaminated batches of factor VIII concentrates. Still, about 5% of them remained seronegative [1]. Other examples of exceptional resistance to infection have been described in cohorts of men who have sex with men reporting high-risk behavior [10] and of female sex workers in Nairobi, Kenya [11]. Finally, virtually all infectious disease clinicians report individual patients with very high levels of exposure that did not become seropositive; a celebrity such as Sir Elton John also described himself as a lucky person that mysteriously avoided infection. The genetic analysis of well-characterized highly exposed, yet uninfected, individuals is thus essential. An alternative approach to study HIV-1 acquisition is to test for differences in allelic distribution between patients with HIV-1 and large cohorts of presumably uninfected controls, since the infected population will then be depleted in protective factors or enriched in alleles conferring enhanced susceptibility to infection. Many groups active in HIV-1 host genetics research have recently created the International HIV Acquisition Consortium to initiate such a study.
Cohorts for the Study of Viral Control
The existence of clinical cohorts/studies prospectively collecting data and samples from individuals with HIV makes it more straightforward to study post-infection outcomes. Various measures of HIV-1 control and disease progression have been used as phenotypes in host genetic studies (Box 1). Plasma viral load and CD4+ T cell count are routinely collected during clinical follow-up and are thus widely available for large numbers of patients. These markers have been shown to be independent predictors of progression to severe immunodeficiency [12]. Later measures of progression like AIDS-defining events, specific opportunistic infections, or AIDS-related death are more likely to result from complex interactions between multiple genetic and environmental factors: the power to detect true genetic effects is thus reduced. In addition, later outcomes can only be assessed in historical cohorts, thanks to the efficacy of current antiretroviral treatments. The study of the earliest stages of infection represents an especially challenging task, since identification and recruitment of acutely infected patients is hampered by significant scientific and sociologic limitations. Finally, it is important to note that, so far, the vast majority of HIV-1 host genetic studies focused on patients of Western European ancestry, in striking contrast with the global distribution of HIV-1 burden. The creation and analysis of cohorts in other ethnic groups is clearly a priority for ethical reasons, but also because population diversity increases the likelihood of genetic discovery.
Box 1. Phenotypes That Have Been Used in Genetic Studies of HIV-1 Infection
HIV resistance/acquisition:
-
Mucosal exposure
-
Intravenous exposure
-
Mother-to-child transmission
HIV viral load:
-
Set point viral load
-
Intracellular HIV-1 DNA level
-
Viral control (elite control/viremic control)
HIV disease progression:
-
Slope of CD4+ T cell decrease
-
Time to CD4+ T cell decrease below a certain threshold
-
Time to AIDS 1987/AIDS 1993
-
Time to death
-
Long-term non-progression
-
Rapid disease progression
Cohorts for the Study of Therapeutic Outcomes
Human genomic approaches can also be used to study therapeutic intervention outcomes. Although antiretroviral therapy is highly effective, critical questions remain about how patients respond to treatment: genetic variation among individuals and populations may cause considerable variability in drug pharmacokinetics and pharmacodynamics. In particular, since anti-HIV medicines are used for life, even modest differences in susceptibilities to adverse events can become important. Identification of risk profiles could allow the tailoring of the drugs to minimize the long-term toxicities associated with treatment, like metabolic disturbances and cardiovascular diseases. Consequently, an important effort should be made to obtain informed consent for genetic testing from participants in randomized clinical trials, which represent an ideal setting for pharmacogenetic discovery, as recently demonstrated, for example, in hepatitis C research [13], [14]. Genetic variation will also be responsible for differences in vaccine immunogenicity and tolerability, and as such needs to be considered in the design and evaluation of current and future HIV-1 vaccine trials.
Common Variation and the Control of HIV-1
Most recent HIV-1 host genetic studies have interrogated human genetic variants for their association with viral load (either as a continuous trait, or as a categorical variable in studies of controllers) and CD4+ T cell decrease. Only a few confirmed genetic associations are understood in terms of the responsible causal sites, and even if it is the case, fundamental questions remain about the exact mechanisms involved in viral control. Most prominent is the example of human leukocyte antigen (HLA) class I variation, and notably of HLA-B*5701, whose protective effect has been shown to be the largest contributor to inter-individual variability for both viral set point and CD4+ T cell decline in genome-wide association studies performed in populations of recent European ancestry [3]–[7]. Interestingly, a very similar result has also been observed in a genome-wide study performed in African-Americans, where HLA-B*5703 was the most important determinant of viral control [9]. HLA-A, -B and -C are extremely polymorphic and encode protein products that are fundamental in the immune recognition process: expressed at the cell surface, they present antigenic epitopes including processed viral peptides to CD8+ T lymphocytes, thereby initiating a cytotoxic T cell (CTL) response. Other HLA-B types have been shown to associate with differences in HIV-1 outcomes [15]: HIV-1 control is better in the presence of HLA-B*27, B*51, and B*5801, but poorer in the presence of HLA-B*5802 and of alleles from the HLA-B35Px group [16]–[19]. Homozygosity for class I alleles also leads to faster progression and higher viremia, presumably because it reduces the diversity of the epitope recognition machinery, thereby impairing antiretroviral CTL response [16].
Beyond the key role they play in the induction of CTL responses, HLA class I molecules are also ligands for the killer cell immunoglobulin-like receptors (KIRs), expressed at the surface of natural killer (NK) cells. KIRs regulate NK cell activation status through inhibitory or activating signaling and can thereby have a direct modulating effect on the innate immune response to HIV-1 infection. Certain combinations of KIR genes and HLA class I alleles have epistatic influences on the outcome of HIV-1 infection [20]: KIR3DL1 and KIR3DS1 have been associated with better control of HIV-1 when they are found in patients that have HLA-B alleles with a Bw4 specificity. Recently, we showed that copy number variation of the KIR3DL1/KIR3DS1 locus results in differences in HIV-1 control in the presence of HLA-Bw4 (K. Pelak, A. C. Need, J. Fellay, K. V. Shianna, S. Feng et al., unpublished data).
Other genetic associations, albeit statistically unequivocal, are still poorly understood. A polymorphism located in the upstream region of HLA-C (HLA-C -35) associates with both HIV-1 control and expression levels of the gene [3], [21], suggesting that the number of HLA-C molecules expressed at the cell surface might play a role in the efficacy of the immune response. Genome-wide studies also detected additional independent associations in the major histocompatibility complex (MHC) [3]–[5], [7], but the long-range linkage disequilibrium structure of the region makes it virtually impossible to pinpoint the real causal sites using genetic data alone.
Outside of the MHC, nearly all genetics findings reported to date resulted from candidate gene studies. As a consequence, variants were identified in genes implicated in HIV-1 life cycle or in immune-related genes. The problem, however, is that most results are equivocal or controversial, due to technical or methodological limitations. In particular, the quasi-systematic absence of correction for population stratification before the genome-wide era has been responsible for a high number of false positive results [7], [22]. In fact, other than HLA and KIR variation, only polymorphisms located in the chemokine receptor cluster on chromosome 3 have been repeatedly associated with HIV-1 control: specifically, heterozygosity for a 32–base pair deletion in CCR5 (CCR5Δ32), variants of the CCR5 promoter region, and a non-synonymous coding change in CCR2 (V64I) have been consistently shown to associate with differences in viral load and/or disease progression [7], [23]–[28].
Common Variation and Acquisition
Variation in CCR5 remains the only human genetic determinant that has been proven to significantly impact HIV-1 acquisition: both the CCR5Δ32 variant (present in homozygous form in about 1% of Europeans) and the m303T>A point mutation (much rarer) result in a defective CCR5 protein product that is not expressed at the cell surface [23], [29]–[31]. When present in homozygous or combined heterozygous form, they confer complete resistance to infection by HIV-1 viruses that use CCR5 as co-receptor. Of note, those individuals remain susceptible to infection by CXCR4-using viruses (including dual-tropic viruses) that associate with more rapid HIV disease progression [32].
Other gene variants were reported to protect against acquisition or to increase susceptibility to infection, but they are at best supported by weak evidence from candidate gene studies. In fact, none could yet be replicated using contemporary standards widely accepted in human genetics, notably a correction for population stratification: for example, we and others reported a lack of association between HIV-1 susceptibility and the number of copies of CCL3L1 [33]–[35], or the allelic distribution of a DARC promoter variant [36]–[40].
Given the importance of the natural model of resistance to HIV-1 infection, it comes as a surprise that no genome-wide study has been published that looks at correlates of protection. It is clearly a priority for the HIV genetic field to carry out such studies.
The Role of Genetic Variation: The Complete Picture
It seems reasonable to conclude that most of the common variants important in the control of HIV-1 have now been identified, at least in individuals of European ancestry. Despite this, it appears that most of the inter-individual differences in control remain to be explained. The confirmed host genetic determinants of HIV-1 control are only able to explain about 20% of the observed variation in viral load or disease progression [7]. It is noteworthy that such limited genetic knowledge can already be used to refine the prediction of disease progression, beyond the information provided by viral load only, as shown in Figure 1.
Fig. 1. An additive genetic score helps predict HIV-1 disease progression. 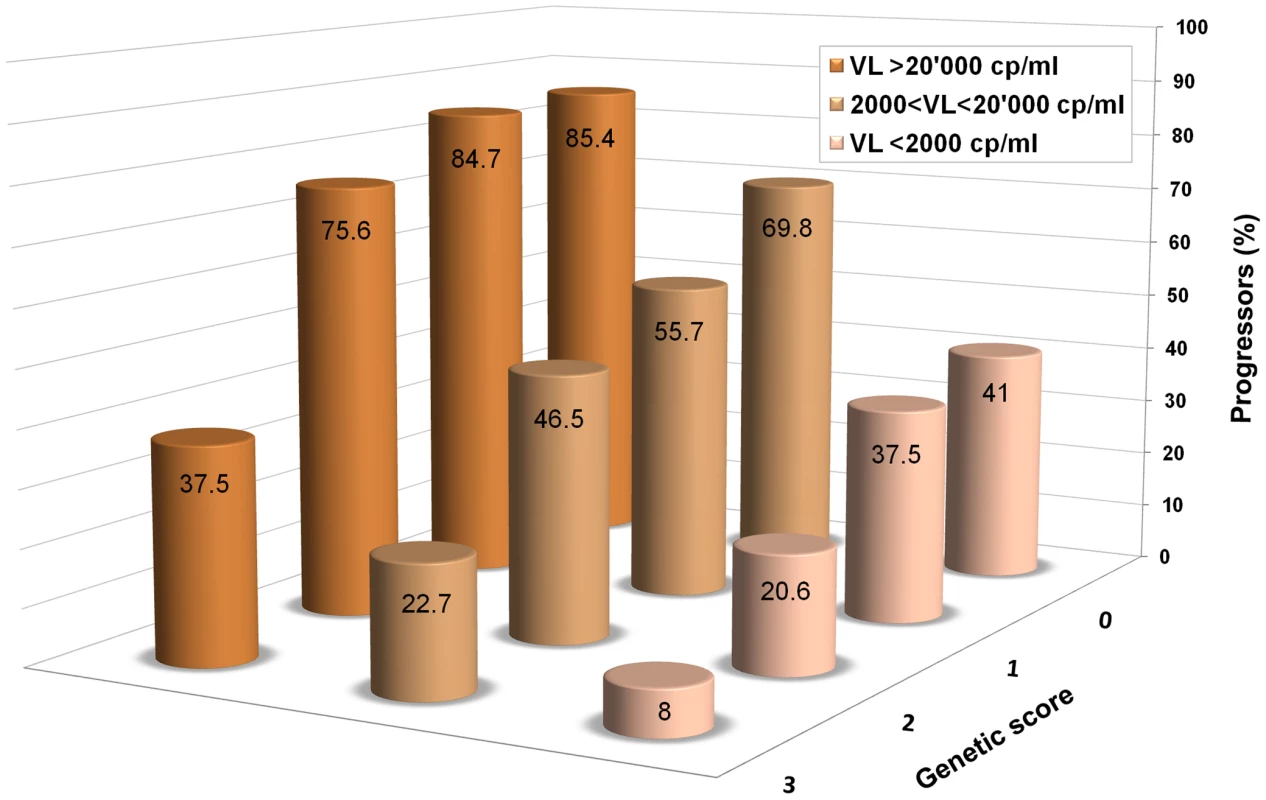
Data are from Fellay et al. [7]: 1,071 individuals of Caucasian ancestry with HIV-1 are included in the analysis. The columns show the proportions of individuals that reached a progression outcome (CD4+ T cells <350/ul or initiation of combined antiretroviral treatment with CD4+ T cells <500/ul) during the first 5 years after estimated date of seroconversion in categories defined by HIV-1 viral load and by a simple additive genetic score, in which one unit is counted for each “protective” allele. The minimum score is 0 for individuals that are homozygous for the major allele at rs2395029 (a proxy for HLA-B*5701), rs9264942 (HLA-C -35 variant), rs9261174 (ZNRD1), and CCR5-Δ32. The maximal observed score is 3 since no individual was heterozygous or homozygous for the minor allele at all four sites. Individuals were grouped in three categories to clearly show that the genetic score refines the prediction of progression, beyond the information provided by viral load only, throughout the range of set point values. So, what is responsible for the large variability in HIV-1 control that still remains unexplained? Clearly, part of it is attributable to the virus itself, as demonstrated by sudden changes in disease course and/or viral set point upon super-infection in chronically infected patients [41] and by sizeable differences in viral load set point that can be observed between the donor and the recipient in HIV-1 transmission pairs [42]. Environmental influences also play a role: for example, pro-inflammatory diseases are often associated with a significant increase in HIV-1 viral load, while co-infections with viruses like GB virus C, HTLV-1, and HIV-2 have been reported to have an inhibitory effect on HIV-1 replication [43].
However, as it is the case for many other human complex traits, it is not unreasonable to assume that rarer genetic variants are responsible for a sizeable fraction of the unexplained inter-individual differences [44]. For example, it is clear that a fraction of the population is highly resistant to infection by HIV-1. While homozygosity for CCR5Δ32 is responsible for some of these cases [23], [29], [30], it appears to explain only a minority of such observations. Several studies have shown a higher frequency of CCR5Δ32/Δ32 in HIV-uninfected hemophiliacs than in the general population (up to 25% compared to 1%, respectively), with the highest frequencies in those with severe hemophilia [45]–[47]. Homozygosity for CCR5Δ32 is also significantly enriched in highly exposed, yet seronegative homosexual men [23], [24]. While those numbers clearly illustrate the high degree of exposure in these populations, it also suggests that other protective mechanisms are responsible for most individual cases of resistance. So far, genome-wide studies in these groups also fail to reveal any strong common variants conferring further protection (D. Goldstein, unpublished data). The identification of the other variants responsible for protection therefore will require a deeper interrogation of the human genome than is possible using genome-wide association studies.
While still expensive and difficult to implement due to computational and bioinformatic challenges, it is feasible to carry out systematic discovery genetics using whole exome or whole genome sequencing [48]. Several recent reports demonstrated that the cause of Mendelian diseases can be identified using such resequencing strategies [49]–[51]. In the HIV field, one project already underway involves sequencing the complete genomes of 50 hemophilia patients (Figure 2). Discovery of variants in this framework will depend principally on three factors: 1) the initial population frequency, 2) the degree of enrichment in frequency in the exposed uninfected individuals, and 3) the defining characteristics of a rare causal variant, e.g., predicted functional consequence, clustering in a gene/pathway, conservation. Whatever the ease of recognition, it seems reasonable to expect that causal variants can be identified using a combination of sequencing in a discovery cohort and confirmation by genotyping in a much larger validation cohort.
Fig. 2. Project framework: human genome resequencing of hemophilia A individuals exposed to HIV-contaminated factor VIII in 1979–1984, yet uninfected. 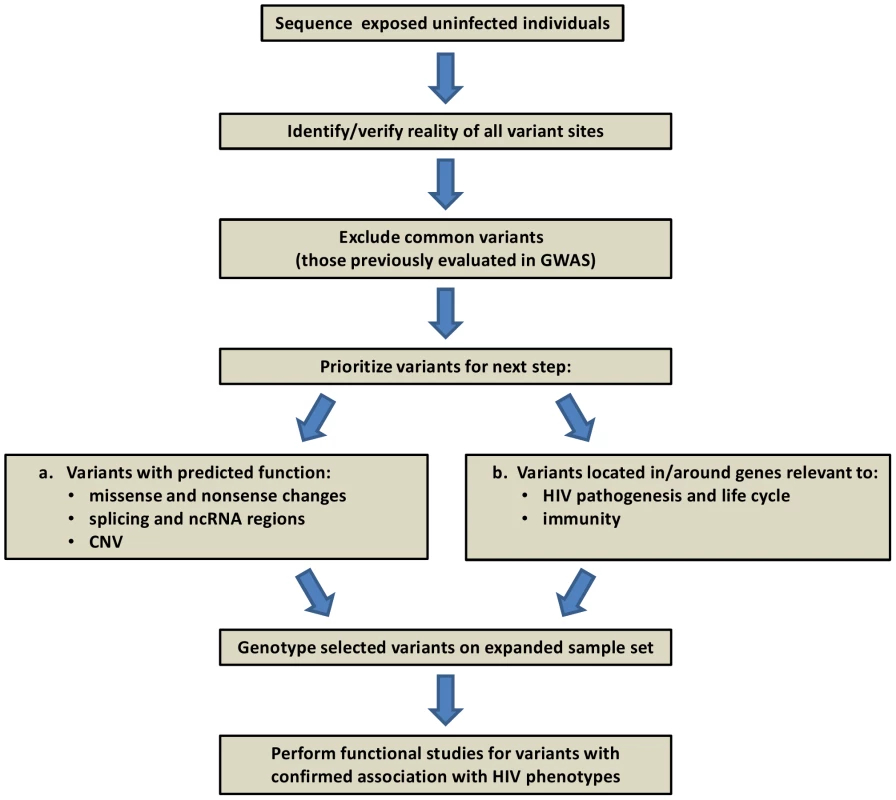
Other phenotypes related to HIV control that could be studied using a similar design include extremely rapid versus extremely slow progression, as well as resistance to infection through mucosal exposure. Other more complicated phenotypes may also be interesting targets for study, such as the occurrence of persistently high viral loads without progression to AIDS [52], extreme levels of microbial translocation during acute infection [53], or unusual immune activation patterns (Box 2).
Box 2. Examples of Target Phenotypes for Human Genome Resequencing Studies
-
Resistance to HIV-1 infection in highly exposed uninfected individuals
-
Very rapid disease progression
-
Elite viral control
-
Poor viral control in HLA-B*57 individuals
-
Persistently high viral loads without apparent disease progression
-
Degree of intestinal microbial translocation during acute infection
-
Unusual immune activation patterns
Systems Approach
Beyond the information generated by genome studies, the field can now press ahead with novel approaches that use a range of technologies. Prominent among these are the analyses of the transcriptome and proteome, and small interfering RNA (siRNA) screens (Figure 3). These genome-wide studies generate large data sets that can be analyzed in isolation, and, increasingly, in an integrated manner [54], [55]. Below we summarize key studies in the HIV field using these techniques, and the first efforts at feeding information across studies. The last section will address the prospects for a systems biology approach in the study of HIV-1 biology and pathogenesis.
Fig. 3. Genome-wide and large-scale studies published since 2007 in the HIV field. 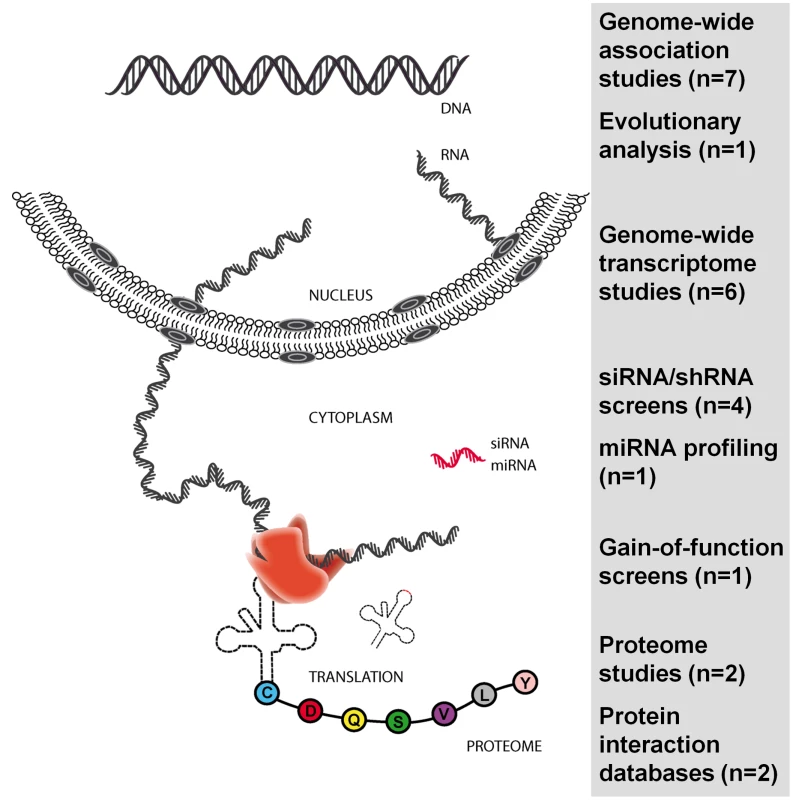
The number of studies is in parentheses. Diverse sets of results and data are compiled in an encyclopedia of overlaps between studies (http://www.hostpathogen.org/). This approach serves to identify networks used by HIV-1 to support its replication. Figure updated from reference [55] (http://F1000.com/Reports/Biology/content/1/71). Transcriptome Analyses
New microarray technologies have recently allowed the genome-wide analysis (>≈20,000 transcripts) of infected cells in vitro, and in vivo in individuals with HIV [56]–[59]. Dynamic analyses have been also completed in animal models. The cell types investigated varied from the collective study of peripheral blood mononuclear cells, to cell type–specific studies [60]. The overarching messages from these studies are (i) the massive modulation of the antiviral defense systems (the interferon response, including the antiretroviral intrinsic cellular defense apparatus), (ii) the prominent modulation of genes involved in the cell cycle and degradation/proteasome pathways, and (iii) the absence of a characteristic expression pattern of effective control of viral replication (e.g., in elite controllers). Evidence of a persistent deregulated interferon response upon infection is of particular interest in light of comparative studies of pathogenic and non-pathogenic animal models [61], [62]. Upon primary simian immunodeficiency virus infection of sooty mangabeys and of African green monkeys, these natural hosts display a strong interferon response at seroconversion followed by distinctive down-regulation despite persistence of ongoing active viral replication [63], [64]. The precision of transcriptome analyses will be greatly improved through the added resolution of RNA-Seq [65] and the capacity to look at the transcriptome in single cells [66].
Proteome Analyses
Large-scale studies are limited by the number of proteins that can be assessed in a quantitative fashion. Analyses of 2,000 to 3,200 proteins identified 15%–21% to be differentially expressed upon infection [67], [68], including changes in the abundance of proteins with known interactions with HIV-1 viral proteins. The NCBI HIV-1 Human Protein Interaction Database (http://www.ncbi.nlm.nih.gov/RefSeq/HIVInteractions/) summarizes over 3,000 interactions with almost 1,500 human genes [69]. Other datasets of interest include the human–pathogen protein–protein interactions (PPIs)/pathogen interaction gateway (PIG) [70] that reports that pathogens tend to interact with hubs (proteins with many interacting partners) and bottlenecks (proteins that are central to many paths in the network) in the human PPI network [71]. No integrated approaches have been used so far to analyze these data in the context of other genome-wide studies.
siRNA and Gain-of-Function Screens
Three siRNA transfection [72]–[74] and one short hairpin RNA (shRNA) [75] transduction studies have targeted the coding RNA for >20,000 human proteins. Approximately 1,000 proteins have been identified as potentially necessary for an optimal viral replication. However, there was minimal overlap across studies—possibly because of differences in cell types and in study design. None of the studies captured or were designed to identify genes that would restrict viral replication—i.e., their silencing would result in greater viral production. Overall, 34 genes were identified in two or more of the transfection screens. However, among those genes that were shared by one or more studies, a pattern emerged that involves the nuclear pore machinery, the mediator complex, a number of key kinases, and components of the NF-κB complex (Figure 4). One gain-of-function screen used a cDNA library representing 15,000 unique genes in an infectious HIV-1 system [76]. This led to the proposal of novel proviral host factors.
Fig. 4. Predicted interaction networks of genes identified as HIV dependency factors in siRNA/shRNA screens [72]–[75]. ![Predicted interaction networks of genes identified as HIV dependency factors in siRNA/shRNA screens <em class="ref">[72]</em>–<em class="ref">[75]</em>.](https://www.prolekarniky.cz/media/cache/resolve/media_object_image_small/media/image/02143e7599bccd493ebe923974160694.png)
Links have been predicted using STRING (http://string.embl.de/). Predicted interactions are depicted according to the type of available evidence. The interactions (see color labels) include direct (physical) and indirect (functional) associations; they are derived from four sources: genomic context, high-throughput experiments, conserved co-expression, and previous knowledge from literature. The nature of the supporting evidence is indicated by the color lines: yellow, text mining; purple, experimental; red, gene fusion; light blue, protein–protein interactions; blue, genomic co-occurrence evidence. Evolutionary Data
Genes involved in immunity and inflammation among those exhibiting the strongest signatures of positive selection both across species and within humans [77]–[84]. Increasingly, evolutionary and comparative sequence analyses across species or within human populations can be used to identify genes that have played a major role in host survival and are therefore likely to influence modern susceptibility to, or pathogenesis of, infectious diseases [78], [85]–[87]. Given the relevance of endogenous and exogenous retroviruses in primate evolution, the identification of genomic signatures can provide an additional layer of data for analysis of contemporary susceptibility to HIV-1. A number of targeted analyses of genes involved in cellular defense against retroviruses have been reported [88]–[92]. A systematic study of long-acting selective pressures on primate genomes (analysis of 140 genes proven or possibly involved in HIV-1 biology and pathogenesis) reached the following conclusions: (i) there are three general groups of genes presenting different evolutionary histories of their coding regions in primates, (ii) analyses allow a non a priori identification of candidate residues that affect host–pathogen interactions, and (iii) a subset of genes may remain under positive selective pressure in modern human populations [92].
Data Integration
Progressively, researchers aim at integrating different layers of data. Rotger et al. [59] examined the correspondence of results from genome-wide transcriptome analysis of differentially expressed mRNA in CD4 T cells from infected individuals with results from analysis of cis-acting genetic variants modulating gene expression in the same samples. In this work, 265 genes were differentially expressed in CD4 T cells across the range of viral set point, and 160 genes were shown to have cis-acting genetic variants associated with expression. However, the overlap between the two lists was minimal: only one gene was common to both lists: OAS1, an interferon-stimulated gene. However, SNPs in this gene are not associated with notable differences in viral set point or disease progression.
Bushman and colleagues [54] applied meta-analytical procedures to assess a wider range of genome-wide studies and public interaction databases. A higher level of signal would be obtained if the data were evaluated in the frame of specific networks and cellular systems. The approach led to the identification of at least 11 densely connected clusters. These clusters, which are enriched for proteins identified in multiple separate screens, specify cellular subsystems associated with HIV replication: the proteasome, subunits of RNA polymerase II and associated factors, the mediator complex, the Tat activation machinery, RNA binding and splicing proteins, and the BiP/GRP78/HSPA5 and CCT chaperones. The study went one additional step to organize data in an “encyclopedia” of host factors assisting HIV replication (http://www.hostpathogen.org/).
There is growing interest in applying non-reductionist approaches such as systems biology to the study of infectious diseases. The general premise of systems biology includes the high-throughput quantitative approach to a biological system that can be subjected to iterative cycles of perturbation, and the modeling of the collected data. HIV infection, which results in a perturbed environment that can be exogenously manipulated through treatment, or modulated by genetic determinants, should now be approached under this research paradigm.
Conclusion
The aim of HIV host genetic research is to comprehensively describe human genetic influences on HIV/AIDS. Some genetic factors have now been convincingly associated with viral control or resistance to infection, yet much effort is still needed to get the full picture. The field is now moving simultaneously towards greater depth in genome analysis and towards more breath and integration through systems biology. This ongoing transition brings renewed hopes that genetic analysis of the human host will contribute substantially to understanding HIV-1 pathogenesis and developing new strategies to stamp out the AIDS pandemic.
Zdroje
1. KronerBL
RosenbergPS
AledortLM
AlvordWG
GoedertJJ
1994 HIV-1 infection incidence among persons with hemophilia in the United States and western Europe, 1978–1990. Multicenter Hemophilia Cohort Study. J Acquir Immune Defic Syndr 7 279 286
2. KulkarniPS
ButeraST
DuerrAC
2003 Resistance to HIV-1 infection: lessons learned from studies of highly exposed persistently seronegative (HEPS) individuals. AIDS Rev 5 87 103
3. FellayJ
ShiannaKV
GeD
ColomboS
LedergerberB
2007 A whole-genome association study of major determinants for host control of HIV-1. Science 317 944 947
4. LimouS
Le ClercS
CoulongesC
CarpentierW
DinaC
2009 Genomewide association study of an AIDS-nonprogression cohort emphasizes the role played by HLA genes (ANRS Genomewide Association Study 02). J Infect Dis 199 419 426
5. DalmassoC
CarpentierW
MeyerL
RouziouxC
GoujardC
2008 Distinct genetic loci control plasma HIV-RNA and cellular HIV-DNA levels in HIV-1 infection: the ANRS Genome Wide Association 01 study. PLoS ONE 3 e3907 doi:10.1371/journal.pone.0003907
6. Le ClercS
LimouS
CoulongesCd
CarpentierW
DinaC
2009 Genomewide association study of a rapid progression cohort identifies new susceptibility alleles for AIDS (ANRS Genomewide Association Study 03). J Infect Dis 200 1194 1201
7. FellayJ
GeD
ShiannaKV
ColomboS
LedergerberB
2009 Common genetic variation and the control of HIV-1 in humans. PLoS Genet 5 e1000791 doi:10.1371/journal.pgen.1000791
8. HerbeckJT
GottliebGS
WinklerCA
NelsonGW
AnP
2010 Multistage Genomewide Association Study Identifies a Locus at 1q41 Associated with Rate of HIV-1 Disease Progression to Clinical AIDS. J Infect Dis 201 618 626
9. PelakK
GoldsteinDB
WalleyNM
FellayJ
GeD
2010 Host determinants of HIV-1 control in African Americans. J Infect Dis 201 1141 1149
10. DetelsR
LiuZ
HennesseyK
KanJ
VisscherBR
1994 Resistance to HIV-1 infection. Multicenter AIDS Cohort Study. J Acquir Immune Defic Syndr 7 1263 1269
11. FowkeKR
NagelkerkeNJ
KimaniJ
SimonsenJN
AnzalaAO
1996 Resistance to HIV-1 infection among persistently seronegative prostitutes in Nairobi, Kenya. Lancet 348 1347 1351
12. MellorsJW
RinaldoCRJr
GuptaP
WhiteRM
ToddJA
1996 Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science 272 1167 1170
13. GeD
FellayJ
ThompsonAJ
SimonJS
ShiannaKV
2009 Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature 461 399 401
14. FellayJ
ThompsonAJ
GeD
GumbsCE
UrbanTJ
2010 ITPA gene variants protect against anaemia in patients treated for chronic hepatitis C. Nature 464 405 408
15. GoulderPJ
WatkinsDI
2008 Impact of MHC class I diversity on immune control of immunodeficiency virus replication. Nat Rev Immunol 8 619 630
16. CarringtonM
NelsonGW
MartinMP
KissnerT
VlahovD
1999 HLA and HIV-1: heterozygote advantage and B*35-Cw*04 disadvantage. Science 283 1748 1752
17. GoulderPJ
BranderC
TangY
TremblayC
ColbertRA
2001 Evolution and transmission of stable CTL escape mutations in HIV infection. Nature 412 334 338
18. KiepielaP
LeslieAJ
HoneyborneI
RamduthD
ThobakgaleC
2004 Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature 432 769 775
19. KawashimaY
PfafferottK
FraterJ
MatthewsP
PayneR
2009 Adaptation of HIV-1 to human leukocyte antigen class I. Nature 458 641 645
20. CarringtonM
MartinMP
van BergenJ
2008 KIR-HLA intercourse in HIV disease. Trends Microbiol 16 620 627
21. ThomasR
AppsR
QiY
GaoX
MaleV
2009 HLA-C cell surface expression and control of HIV/AIDS correlate with a variant upstream of HLA-C. Nat Genet 41 1290 1294
22. FellayJ
2009 Host genetics influences on HIV type-1 disease. Antivir Ther 14 731 738
23. DeanM
CarringtonM
WinklerC
HuttleyGA
SmithMW
1996 Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Hemophilia Growth and Development Study, Multicenter AIDS Cohort Study, Multicenter Hemophilia Cohort Study, San Francisco City Cohort, ALIVE Study. Science 273 1856 1862
24. HuangY
PaxtonWA
WolinskySM
NeumannAU
ZhangL
1996 The role of a mutant CCR5 allele in HIV-1 transmission and disease progression. Nat Med 2 1240 1243
25. MartinMP
DeanM
SmithMW
WinklerC
GerrardB
1998 Genetic acceleration of AIDS progression by a promoter variant of CCR5. Science 282 1907 1911
26. MummidiS
AhujaSS
GonzalezE
AndersonSA
SantiagoEN
1998 Genealogy of the CCR5 locus and chemokine system gene variants associated with altered rates of HIV-1 disease progression. Nat Med 4 786 793
27. SmithMW
DeanM
CarringtonM
WinklerC
HuttleyGA
1997 Contrasting genetic influence of CCR2 and CCR5 variants on HIV-1 infection and disease progression. Hemophilia Growth and Development Study (HGDS), Multicenter AIDS Cohort Study (MACS), Multicenter Hemophilia Cohort Study (MHCS), San Francisco City Cohort (SFCC), ALIVE Study. Science 277 959 965
28. IoannidisJP
RosenbergPS
GoedertJJ
AshtonLJ
BenfieldTL
2001 Effects of CCR5-Delta32, CCR2-64I, and SDF-1 3'A alleles on HIV-1 disease progression: an international meta-analysis of individual-patient data. Ann Intern Med 135 782 795
29. LiuR
PaxtonWA
ChoeS
CeradiniD
MartinSR
1996 Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell 86 367 377
30. SamsonM
LibertF
DoranzBJ
RuckerJ
LiesnardC
1996 Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature 382 722 725
31. QuillentC
OberlinE
BraunJ
RoussetD
Gonzalez-CanaliG
1998 HIV-1-resistance phenotype conferred by combination of two separate inherited mutations of CCR5 gene. Lancet 351 14 18
32. SheppardHW
CelumC
MichaelNL
O'BrienS
DeanM
2002 HIV-1 infection in individuals with the CCR5-Delta32/Delta32 genotype: acquisition of syncytium-inducing virus at seroconversion. J Acquir Immune Defic Syndr 29 307 313
33. GonzalezE
KulkarniH
BolivarH
ManganoA
SanchezR
2005 The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science 307 1434 1440
34. UrbanTJ
WeintrobAC
FellayJ
ColomboS
ShiannaKV
2009 CCL3L1 and HIV/AIDS susceptibility. Nat Med 15 1110 1112
35. BhattacharyaT
StantonJ
KimEY
KunstmanKJ
PhairJP
2009 CCL3L1 and HIV/AIDS susceptibility. Nat Med 15 1112 1115
36. HeW
NeilS
KulkarniH
WrightE
AganBK
2008 Duffy antigen receptor for chemokines mediates trans-infection of HIV-1 from red blood cells to target cells and affects HIV-AIDS susceptibility. Cell Host Microbe 4 52 62
37. WalleyNM
JulgB
DicksonSP
FellayJ
GeD
2009 The Duffy antigen receptor for chemokines null promoter variant does not influence HIV-1 acquisition or disease progression. Cell Host Microbe 5 408 410; author reply 418–409
38. JulgB
ReddyS
van der StokM
KulkarniS
QiY
2009 Lack of Duffy antigen receptor for chemokines: no influence on HIV disease progression in an African treatment-naive population. Cell Host Microbe 5 413 415; author reply 418–419
39. HorneKC
LiX
JacobsonLP
PalellaF
JamiesonBD
2009 Duffy antigen polymorphisms do not alter progression of HIV in African Americans in the MACS cohort. Cell Host Microbe 5 415 417; author reply 418–419
40. WinklerCA
AnP
JohnsonR
NelsonGW
KirkG
2009 Expression of Duffy antigen receptor for chemokines (DARC) has no effect on HIV-1 acquisition or progression to AIDS in African Americans. Cell Host Microbe 5 411 413; author reply 418–419
41. SmithDM
RichmanDD
LittleSJ
2005 HIV superinfection. J Infect Dis 192 438 444
42. TangJ
TangS
LobashevskyE
ZuluI
AldrovandiG
2004 HLA allele sharing and HIV type 1 viremia in seroconverting Zambians with known transmitting partners. AIDS Res Hum Retroviruses 20 19 25
43. KannangaraS
DeSimoneJA
PomerantzRJ
2005 Attenuation of HIV-1 infection by other microbial agents. J Infect Dis 192 1003 1009
44. ManolioTA
CollinsFS
CoxNJ
GoldsteinDB
HindorffLA
2009 Finding the missing heritability of complex diseases. Nature 461 747 753
45. WilkinsonDA
OperskalskiEA
BuschMP
MosleyJW
KoupRA
1998 A 32-bp deletion within the CCR5 locus protects against transmission of parenterally acquired human immunodeficiency virus but does not affect progression to AIDS-defining illness. J Infect Dis 178 1163 1166
46. ZhangM
GoedertJJ
O'BrienTR
2003 High frequency of CCR5-delta32 homozygosity in HCV-infected, HIV-1-uninfected hemophiliacs results from resistance to HIV-1. Gastroenterology 124 867 868
47. KupferB
KaiserR
BrackmannHH
EffenbergerW
RockstrohJK
1999 Protection against parenteral HIV-1 infection by homozygous deletion in the C-C chemokine receptor 5 gene. AIDS 13 1025 1028
48. CirulliET
GoldsteinDB
2010 Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nat Rev Genet 11 415 425
49. ChoiM
SchollUI
JiW
LiuT
TikhonovaIR
2009 Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc Natl Acad Sci U S A 106 19096 19101
50. NgSB
TurnerEH
RobertsonPD
FlygareSD
BighamAW
2009 Targeted capture and massively parallel sequencing of 12 human exomes. Nature 461 272 276
51. NgSB
BuckinghamKJ
LeeC
BighamAW
TaborHK
2009 Exome sequencing identifies the cause of a mendelian disorder. Nat Genet 42 30 35
52. PaiardiniM
PandreaI
ApetreiC
SilvestriG
2009 Lessons learned from the natural hosts of HIV-related viruses. Annu Rev Med 60 485 495
53. BrenchleyJM
PriceDA
SchackerTW
AsherTE
SilvestriG
2006 Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat Med 12 1365 1371
54. BushmanFD
MalaniN
FernandesJ
D'OrsoI
CagneyG
2009 Host cell factors in HIV replication: meta-analysis of genome-wide studies. PLoS Pathog 5 e1000437 doi:10.1371/journal.ppat.1000437
55. TelentiA
2009 HIV-1 host interactions - integration of large scale datasets. F1000 Biology Reports 1 71
56. SedaghatAR
GermanJ
TeslovichTM
CofrancescoJJr
JieCC
2008 Chronic CD4+ T-cell activation and depletion in human immunodeficiency virus type 1 infection: type I interferon-mediated disruption of T-cell dynamics. J Virol 82 1870 1883
57. HyrczaMD
KovacsC
LoutfyM
HalpennyR
HeislerL
2007 Distinct transcriptional profiles in ex vivo CD4+ and CD8+ T cells are established early in human immunodeficiency virus type 1 infection and are characterized by a chronic interferon response as well as extensive transcriptional changes in CD8+ T cells. J Virol 81 3477 3486
58. GiriMS
NebozyhnM
RaymondA
GekongeB
HancockA
2009 Circulating monocytes in HIV-1-infected viremic subjects exhibit an antiapoptosis gene signature and virus - and host-mediated apoptosis resistance. J Immunol 182 4459 4470
59. RotgerM
DangKK
FellayJ
HeinzenEL
FengS
2010 Genome-wide mRNA expression correlates of viral control in CD4+ T-cells from HIV-1-infected individuals. PLoS Pathog 6 e1000781 doi:10.1371/journal.ppat.1000781
60. GiriMS
NebozhynM
ShoweL
MontanerLJ
2006 Microarray data on gene modulation by HIV-1 in immune cells: 2000–2006. J Leukoc Biol 80 1031 1043
61. MandlJN
BarryAP
VanderfordTH
KozyrN
ChavanR
2008 Divergent TLR7 and TLR9 signaling and type I interferon production distinguish pathogenic and nonpathogenic AIDS virus infections. Nat Med 14 1077 1087
62. LedererS
FavreD
WaltersKA
ProllS
KanwarB
2009 Transcriptional profiling in pathogenic and non-pathogenic SIV infections reveals significant distinctions in kinetics and tissue compartmentalization. PLoS Pathog 5 e1000296 doi:10.1371/journal.ppat.1000296
63. BosingerSE
LiQ
GordonSN
KlattNR
DuanL
2009 Global genomic analysis reveals rapid control of a robust innate response in SIV-infected sooty mangabeys. J Clin Invest 119 3556 3572
64. JacquelinB
MayauV
TargatB
LiovatAS
KunkelD
2009 Nonpathogenic SIV infection of African green monkeys induces a strong but rapidly controlled type I IFN response. J Clin Invest 119 3544 3555
65. WangZ
GersteinM
SnyderM
2009 RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10 57 63
66. TangF
BarbacioruC
WangY
NordmanE
LeeC
2009 mRNA-Seq whole-transcriptome analysis of a single cell. Nat Meth 6 377 382
67. RingroseJH
JeeningaRE
BerkhoutB
SpeijerD
2008 Proteomic studies reveal coordinated changes in T-cell expression patterns upon infection with human immunodeficiency virus type 1. J Virol 82 4320 4330
68. ChanEY
QianWJ
DiamondDL
LiuT
GritsenkoMA
2007 Quantitative analysis of human immunodeficiency virus type 1-infected CD4+ cell proteome: dysregulated cell cycle progression and nuclear transport coincide with robust virus production. J Virol 81 7571 7583
69. FuW
Sanders-BeerBE
KatzKS
MaglottDR
PruittKD
2009 Human immunodeficiency virus type 1, human protein interaction database at NCBI. Nucleic Acids Res 37 D417 D422
70. DriscollT
DyerMD
MuraliTM
SobralBW
2009 PIG–the pathogen interaction gateway. Nucl Acids Res 37 D647 D650
71. DyerMD
MuraliTM
SobralBW
2008 The landscape of human proteins interacting with viruses and other pathogens. PLoS Pathog 4 e32 doi:10.1371/journal.ppat.0040032
72. BrassAL
DykxhoornDM
BenitaY
YanN
EngelmanA
2008 Identification of host proteins required for HIV infection through a functional genomic screen. Science 319 921 926
73. KonigR
ZhouY
EllederD
DiamondTL
BonamyGM
2008 Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell 135 49 60
74. ZhouH
XuM
HuangQ
GatesAT
ZhangXD
2008 Genome-scale RNAi screen for host factors required for HIV replication. Cell Host Microbe 4 495 504
75. YeungML
HouzetL
YedavalliVS
JeangKT
2009 A genome-wide short hairpin RNA screening of jurkat T-cells for human proteins contributing to productive HIV-1 replication. J Biol Chem 284 19463 19473
76. NguyenDG
YinH
ZhouY
WolffKC
KuhenKL
2007 Identification of novel therapeutic targets for HIV infection through functional genomic cDNA screening. Virology 362 16 25
77. SabetiPC
SchaffnerSF
FryB
LohmuellerJ
VarillyP
2006 Positive natural selection in the human lineage. Science 312 1614 1620
78. SabetiPC
VarillyP
FryB
LohmuellerJ
HostetterE
2007 Genome-wide detection and characterization of positive selection in human populations. Nature 449 913 918
79. VoightBF
KudaravalliS
WenX
PritchardJK
2006 A map of recent positive selection in the human genome. PLoS Biol 4 e72 doi:10.1371/journal.pbio.0040072
80. BarreiroLB
LavalG
QuachH
PatinE
Quintana-MurciL
2008 Natural selection has driven population differentiation in modern humans. Nat Genet 40 340 345
81. BustamanteCD
Fledel-AlonA
WilliamsonS
NielsenR
HubiszMT
2005 Natural selection on protein-coding genes in the human genome. Nature 437 1153 1157
82. ClarkAG
GlanowskiS
NielsenR
ThomasPD
KejariwalA
2003 Inferring nonneutral evolution from human-chimp-mouse orthologous gene trios. Science 302 1960 1963
83. ConsortiumTCSaA
2005 Initial sequence of the chimpanzee genome and comparison with the human genome. Nature 437 69 87
84. KosiolC
VinarT
da FonsecaRR
HubiszMJ
BustamanteCD
2008 Patterns of positive selection in six mammalian genomes. PLoS Genet 4 e1000144 doi:10.1371/journal.pgen.1000144
85. SawyerSL
EmermanM
MalikHS
2004 Ancient adaptive evolution of the primate antiviral DNA-editing enzyme APOBEC3G. PLoS Biol 2 e275 doi:10.1371/journal.pbio.0020275
86. SawyerSL
WuLI
EmermanM
MalikHS
2005 Positive selection of primate TRIM5alpha identifies a critical species-specific retroviral restriction domain. Proc Natl Acad Sci U S A 102 2832 2837
87. Quintana-MurciL
AlcaisA
AbelL
CasanovaJL
2007 Immunology in natura: clinical, epidemiological and evolutionary genetics of infectious diseases. Nat Immunol 8 1165 1171
88. EsnaultC
HeidmannO
DelebecqueF
DewannieuxM
RibetD
2005 APOBEC3G cytidine deaminase inhibits retrotransposition of endogenous retroviruses. Nature 433 430 433
89. StremlauM
OwensCM
PerronMJ
KiesslingM
AutissierP
2004 The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature 427 848 853
90. SheehyAM
GaddisNC
ChoiJD
MalimMH
2002 Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418 646 650
91. NeilSJ
ZangT
BieniaszPD
2008 Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451 425 430
92. OrtizM
GuexN
PatinE
MartinO
XenariosI
2009 Evolutionary trajectories of primate genes involved in HIV pathogenesis. Mol Biol Evol 26 2865 2875
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek The Combined Effect of Environmental and Host Factors on the Emergence of Viral RNA RecombinantsČlánek Functional Interchangeability of Late Domains, Late Domain Cofactors and Ubiquitin in Viral BuddingČlánek Parvovirus Minute Virus of Mice Induces a DNA Damage Response That Facilitates Viral ReplicationČlánek Phenolglycolipid-1 Expressed by Engineered BCG Modulates Early Interaction with Human Phagocytes
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2010 Číslo 10- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Social Media and Microbiology Education
- Antimicrobial Peptides: Primeval Molecules or Future Drugs?
- Phylodynamics and Human-Mediated Dispersal of a Zoonotic Virus
- Retroviral RNA Dimerization and Packaging: The What, How, When, Where, and Why
- Distinct Clones of Caused the Black Death
- Strain-Specific Differences in the Genetic Control of Two Closely Related Mycobacteria
- The Combined Effect of Environmental and Host Factors on the Emergence of Viral RNA Recombinants
- MHC Class I Bound to an Immunodominant Epitope Demonstrates Unconventional Presentation to T Cell Receptors
- Stimulates Immune Gene Expression and Inhibits Development in
- Crystal Structure of DotD: Insights into the Relationship between Type IVB and Type II/III Secretion Systems
- Cytomegalovirus microRNAs Facilitate Persistent Virus Infection in Salivary Glands
- Strategies to Avoid Killing by Human Neutrophils
- Transforming Growth Factor-β: Activation by Neuraminidase and Role in Highly Pathogenic H5N1 Influenza Pathogenesis
- Autoimmunity in Arabidopsis Is Mediated by Epigenetic Regulation of an Immune Receptor
- Functional Interchangeability of Late Domains, Late Domain Cofactors and Ubiquitin in Viral Budding
- Dengue Virus Ensures Its Fusion in Late Endosomes Using Compartment-Specific Lipids
- Nucleocapsid Promotes Localization of HIV-1 Gag to Uropods That Participate in Virological Synapses between T Cells
- Host Genetics and HIV-1: The Final Phase?
- Variations in the Hemagglutinin of the 2009 H1N1 Pandemic Virus: Potential for Strains with Altered Virulence Phenotype?
- High-Resolution Functional Mapping of the Venezuelan Equine Encephalitis Virus Genome by Insertional Mutagenesis and Massively Parallel Sequencing
- Viral Replication Rate Regulates Clinical Outcome and CD8 T Cell Responses during Highly Pathogenic H5N1 Influenza Virus Infection in Mice
- Calcineurin Inhibition at the Clinical Phase of Prion Disease Reduces Neurodegeneration, Improves Behavioral Alterations and Increases Animal Survival
- Parvovirus Minute Virus of Mice Induces a DNA Damage Response That Facilitates Viral Replication
- -Induced Inactivation of the Macrophage Transcription Factor AP-1 Is Mediated by the Parasite Metalloprotease GP63
- Epstein Barr Virus-Encoded EBNA1 Interference with MHC Class I Antigen Presentation Reveals a Close Correlation between mRNA Translation Initiation and Antigen Presentation
- Fidelity Variants of RNA Dependent RNA Polymerases Uncover an Indirect, Mutagenic Activity of Amiloride Compounds
- The Spread of Tomato Yellow Leaf Curl Virus from the Middle East to the World
- Concerted Action of Two Formins in Gliding Motility and Host Cell Invasion by
- Requirements for Receptor Engagement during Infection by Adenovirus Complexed with Blood Coagulation Factor X
- Release of Intracellular Calcium Stores Facilitates Coxsackievirus Entry into Polarized Endothelial Cells
- Gene Annotation and Drug Target Discovery in with a Tagged Transposon Mutant Collection
- Nuclear Export and Import of Human Hepatitis B Virus Capsid Protein and Particles
- Retention and Loss of RNA Interference Pathways in Trypanosomatid Protozoans
- Identification and Genome-Wide Prediction of DNA Binding Specificities for the ApiAP2 Family of Regulators from the Malaria Parasite
- Controlling Cellular P-TEFb Activity by the HIV-1 Transcriptional Transactivator Tat
- Direct Visualization of Peptide/MHC Complexes at the Surface and in the Intracellular Compartments of Cells Infected by
- Phenolglycolipid-1 Expressed by Engineered BCG Modulates Early Interaction with Human Phagocytes
- In Vitro and In Vivo Studies Identify Important Features of Dengue Virus pr-E Protein Interactions
- Inhibition of Nipah Virus Infection In Vivo: Targeting an Early Stage of Paramyxovirus Fusion Activation during Viral Entry
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Retroviral RNA Dimerization and Packaging: The What, How, When, Where, and Why
- Viral Replication Rate Regulates Clinical Outcome and CD8 T Cell Responses during Highly Pathogenic H5N1 Influenza Virus Infection in Mice
- Antimicrobial Peptides: Primeval Molecules or Future Drugs?
- Crystal Structure of DotD: Insights into the Relationship between Type IVB and Type II/III Secretion Systems
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



