-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaGenetic Predictors of Response to Serotonergic and Noradrenergic Antidepressants in Major Depressive Disorder: A Genome-Wide Analysis of Individual-Level Data and a Meta-Analysis
Background:
It has been suggested that outcomes of antidepressant treatment for major depressive disorder could be significantly improved if treatment choice is informed by genetic data. This study aims to test the hypothesis that common genetic variants can predict response to antidepressants in a clinically meaningful way.Methods and Findings:
The NEWMEDS consortium, an academia–industry partnership, assembled a database of over 2,000 European-ancestry individuals with major depressive disorder, prospectively measured treatment outcomes with serotonin reuptake inhibiting or noradrenaline reuptake inhibiting antidepressants and available genetic samples from five studies (three randomized controlled trials, one part-randomized controlled trial, and one treatment cohort study). After quality control, a dataset of 1,790 individuals with high-quality genome-wide genotyping provided adequate power to test the hypotheses that antidepressant response or a clinically significant differential response to the two classes of antidepressants could be predicted from a single common genetic polymorphism. None of the more than half million genetic markers significantly predicted response to antidepressants overall, serotonin reuptake inhibitors, or noradrenaline reuptake inhibitors, or differential response to the two types of antidepressants (genome-wide significance p<5×10−8). No biological pathways were significantly overrepresented in the results. No significant associations (genome-wide significance p<5×10−8) were detected in a meta-analysis of NEWMEDS and another large sample (STAR*D), with 2,897 individuals in total. Polygenic scoring found no convergence among multiple associations in NEWMEDS and STAR*D.Conclusions:
No single common genetic variant was associated with antidepressant response at a clinically relevant level in a European-ancestry cohort. Effects specific to particular antidepressant drugs could not be investigated in the current study.
Please see later in the article for the Editors' Summary
Published in the journal: . PLoS Med 9(10): e32767. doi:10.1371/journal.pmed.1001326
Category: Research Article
doi: https://doi.org/10.1371/journal.pmed.1001326Summary
Background:
It has been suggested that outcomes of antidepressant treatment for major depressive disorder could be significantly improved if treatment choice is informed by genetic data. This study aims to test the hypothesis that common genetic variants can predict response to antidepressants in a clinically meaningful way.Methods and Findings:
The NEWMEDS consortium, an academia–industry partnership, assembled a database of over 2,000 European-ancestry individuals with major depressive disorder, prospectively measured treatment outcomes with serotonin reuptake inhibiting or noradrenaline reuptake inhibiting antidepressants and available genetic samples from five studies (three randomized controlled trials, one part-randomized controlled trial, and one treatment cohort study). After quality control, a dataset of 1,790 individuals with high-quality genome-wide genotyping provided adequate power to test the hypotheses that antidepressant response or a clinically significant differential response to the two classes of antidepressants could be predicted from a single common genetic polymorphism. None of the more than half million genetic markers significantly predicted response to antidepressants overall, serotonin reuptake inhibitors, or noradrenaline reuptake inhibitors, or differential response to the two types of antidepressants (genome-wide significance p<5×10−8). No biological pathways were significantly overrepresented in the results. No significant associations (genome-wide significance p<5×10−8) were detected in a meta-analysis of NEWMEDS and another large sample (STAR*D), with 2,897 individuals in total. Polygenic scoring found no convergence among multiple associations in NEWMEDS and STAR*D.Conclusions:
No single common genetic variant was associated with antidepressant response at a clinically relevant level in a European-ancestry cohort. Effects specific to particular antidepressant drugs could not be investigated in the current study.
Please see later in the article for the Editors' SummaryIntroduction
Major depressive disorder (MDD) is a disabling illness, affecting a high proportion of individuals at some point in their life [1]. Prescription of antidepressants is the most common initial step in treating MDD, but less than half of individuals achieve remission of symptoms with their first antidepressant [2],[3]. It has been proposed that common genetic variants could be used to personalize psychiatric treatment and significantly improve outcomes [4]–[7]. However, to date there has not been a robust, well-replicated finding of sufficient effect size to be worth translating into a clinical setting.
Identification of genetic determinants of antidepressant response has the potential to improve the treatment of MDD in two important ways. First, genetic and molecular predictors of poor treatment outcome with available antidepressants can provide targets for the development of novel therapeutic agents that may be effective for the type of depression that is resistant to current treatments. Second, for many individuals with MDD, delay in reaching recovery is avoidable, since they have the potential to respond to one of the currently available treatments. If a predictor of differential outcome with alternative treatments is identified, a clinician could use it to select the antidepressant that is most likely to alleviate depression in a given individual. For both applications, the clinical implications are predicated on the effect size of the prediction. A consensus criterion has been set for what size of difference in depressive symptoms is clinically meaningful: a panel of experts and service users has concluded that a difference in outcome equal or greater than three points on the Hamilton Rating Scale for Depression is noticeable to the patients and their relatives and can be considered as clinically significant [8],[9]. This criterion is equal to 6.33% of variance in outcome explained, which can be applied to assess whether a genetic biomarker provides clinically significant prediction [9]. Currently, no clinically significant predictor is available [10]. The aims of this NEWMEDS study address the two potential avenues for using genomic information to improve treatment of depression.
The first aim is to identify common genetic polymorphisms that predict unfavourable outcome of treatment with currently available antidepressants. Addressing this issue in the large combined NEWMEDS sample will substantially expand on the evidence from the first genome-wide studies on outcomes for single-drug treatment [11] or naturalistic inpatient treatment [12] of depression and could provide novel targets for the development of new treatments.
The second aim is to obtain predictors of differential outcomes of treatment with antidepressants with different modes of action in the largest comparative pharmacogenetic study to date. Specifically, we aim to identify common genetic variants that differentially predict outcome of treatment with antidepressants that act primarily through the inhibition of serotonin reuptake (serotonin reuptake inhibitors [SRIs]) or act primarily through the inhibition of norepinephrine reuptake (noradrenaline reuptake inhibitors [NRIs]). For the first time, to our knowledge, these two aims will be pursued in a sample that is large enough to provide sufficient power to ensure interpretable results.
Methods
Samples
As part of the NEWMEDS consortium (http://www.newmeds-europe.com) [13], three studies conducted by academic institutions (GENDEP, a part-randomized open study of two active antidepressants, n = 868; GENPOD, a randomized controlled trial of two active antidepressants, n = 601; and GODS, a treatment cohort of severe depression, n = 131) [14]–[16] and two studies by pharmaceutical industry members of the European Federation of Pharmaceutical Industries and Associations (active comparator arms from randomized controlled trials by Pfizer, n = 355, and GlaxoSmithKline, n = 191) were combined to obtain a sample of 2,146 adult individuals (1,205 women and 941 men; see Table 1 in Text S1 for description by contributing study) diagnosed with unipolar MDD according to the Diagnostic and Statistical Manual of Mental Disorders–IV and the International Classification of Diseases–10, with prospective data on outcome of treatment with SRI or NRI antidepressants. Diagnoses of schizophrenia, schizoaffective disorder, or bipolar disorder, or current alcohol or drug dependence constituted exclusion criteria. All individuals were treated for 6 to 12 wk with either an antidepressant that acts primarily through blocking the reuptake of serotonin (SRIs: escitalopram, citalopram, paroxetine, sertraline, fluoxetine) or an antidepressant that acts primarily through blocking the reuptake of norepinephrine (NRIs: nortriptyline, reboxetine). These groupings are based on previous evidence that at least some genetic predictors of treatment response are specific to antidepressants that block serotonin or noradrenaline reuptake [14]. The two largest studies (GENDEP and GENPOD) included an open-label randomized comparison between an SRI (escitalopram, citalopram) and an NRI (nortriptyline, reboxetine). Detailed information on individual studies, including inclusion and exclusion criteria for each study, can be found in Text S1 (section 1.1).
Genotyping
From each study, individuals with self-reported white European ancestry, available high-quality blood DNA samples, and valid information on treatment outcome with either an SRI or an NRI were selected for genotyping.
Samples were genotyped on Illumina Human610-Quad BeadChips (n = 727) or Illumina Human660W-Quad BeadChips (n = 1,166), which have identical tag SNP coverage. More specifically, 727 samples from the GENDEP study were genotyped at the Centre National de Génotypage (Evry Cedex, France) using the Illumina Human610-Quad BeadChip and were subjects of previous reports [17],[18]. The GENPOD, GlaxoSmithKline, Pfizer, GODS, and 93 additional GENDEP samples were genotyped at the University of Geneva Medical School (Geneva, Switzerland) using the Illumina Human660W-Quad BeadChip for the NEWMEDS consortium.
Quality Control
Quality control was implemented in PLINK [19], first on the level of the genetic marker and then on the level of the individual. Markers with minor allele frequency over 0.01 and at least 97% complete genotyping were retained. To avoid batch artefacts, markers that differed significantly (p<1×10−3) by genotyping centre were excluded. Overall, 520,978 (99.0%) of the 526,424 genotyped SNPs passed all stages of quality control and were included in pharmacogenetic analyses. Hardy–Weinberg equilibrium was tested, but was not used as an exclusion criterion for markers, since departures from Hardy–Weinberg equilibrium are expected in a case-only study [20].
Individuals were excluded for ambiguous sex (genotypic sex different from phenotypic sex) (n = 22), abnormal heterozygosity (n = 16), cryptic relatedness up to third-degree relatives by identity by descent (n = 20), genotyping completeness less than 97% (n = 9), and non-European ethnicity admixture detected as outliers in iterative EIGENSTRAT analyses of a linkage-disequilibrium-pruned dataset (n = 35). One additional individual was excluded because of invalid phenotypic information, leaving 1,790 (94.6%) of the 1,893 genotyped individuals for the pharmacogenetic analyses.
Figure 1 shows the flow of individuals through genotyping and quality control.
Fig. 1. Flow of samples through quality control. 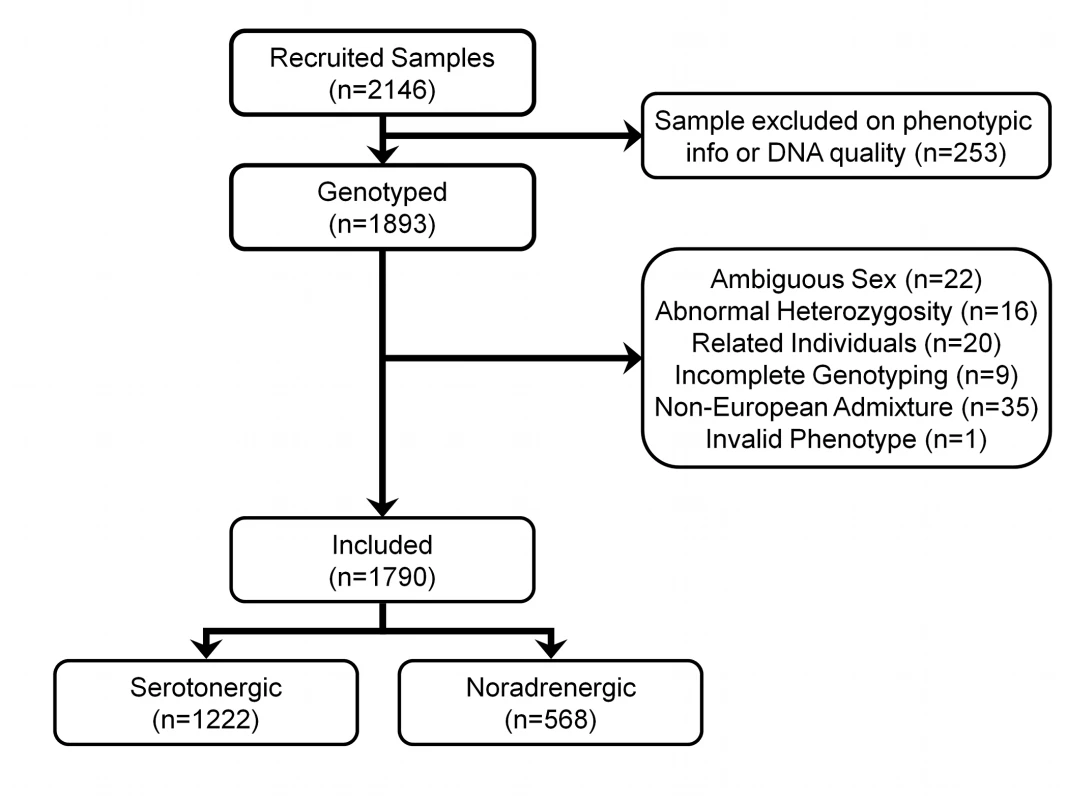
Definition of Antidepressant Response Phenotype
Response to antidepressants involves changes in depressive symptoms over a number of weeks and is more accurately captured by continuous than by dichotomous variables [17],[21]–[23]. We defined response as a continuous variable, reflecting proportional reduction in depression severity from baseline to end of treatment. This measure is uncorrelated with initial severity (−0.10<r<0.10 in all component studies), is independent of depression rating scale used, and is clinically relevant since it closely reflects clinician's impression of improvement [17],[24].
Studies included in NEWMEDS used several outcome measures. The Montgomery–Åsberg Depression Rating Scale [25] was the primary outcome measure in GENDEP and GODS, the 17-item Hamilton Rating Scale for Depression (HRSD-17) [26] was the primary outcome measure in the studies conducted by Pfizer and GlaxoSmithKline, and the Beck Depression Inventory [27] was the primary outcome measure in GENPOD. While the outcome measures used differ in details, here we are interested in generalizable effects related to depression as a whole rather than effects specific to a particular measure. Previous research has shown no difference between classes of antidepressants in response as measured by these three scales [28]. In addition, we took the following steps to minimize the effects of scale differences. To allow for an unbiased analysis of the combined dataset, we converted the outcome measures within each study to a single continuous metric: a standardized change score, adjusted for sex, age, and recruitment centre within each contributing study. The adjusted change score was z-transformed within each study to remove any correlation between data origin and outcome prior to the genetic analysis, and to remove effects that are specific to individual contributing studies. Detailed information about the definition of the phenotype is provided in Text S1 (section 1.3).
Statistical Analysis
Data analysis was carried out according to a protocol specified prior to data acquisition (Text S2). Joint analysis of individual-level data was conducted to allow for rigorous quality control and to retain maximum statistical power when combining studies that varied in size. The whole sample was analyzed jointly since this is a more efficient and powerful approach than discovery–replication design [29],[30]. Quality-controlled genotypes were tested for association with the adjusted percentage change in depression severity using linear regression under an additive genetic model in PLINK [19]. Four genome-wide analyses were performed. A first linear regression searched for common genetic markers that predict response to both types of antidepressants in the whole sample of 1,790 individuals. The second and third analyses tested predictors of response to SRIs (n = 1,222) and NRIs (n = 568). A fourth analysis, of primary interest to our second aim, searched the genome for common variants that differentially predict response to SRIs and NRIs. To avoid confounding by covariation between antidepressant drug and genetic background due to different studies contributing unequal numbers of individuals to each antidepressant group, we tested this hypothesis as a drug-by-genotype interaction in a sample restricted to individuals that were randomly allocated to treatment with either an SRI or an NRI (n = 949), thus ensuring full comparability of the two drug groups on measured and unmeasured confounders. In all analyses, the influence of genetic population structure was controlled by the inclusion of the four significant principal components from the final iteration of the EIGENSTRAT analysis of linkage-disequilibrium-pruned genetic data. We defined genome-wide statistical significance at the generally accepted threshold p<5×10−8 [31].
Associations reaching a less stringent threshold of p<5×10−6 are reported in Text S1 (section 2). Analyses performed by component study, summary data meta-analysis (Text S1, section 6), and pharmacogenetic associations within genes reported in previous candidate gene and genome-wide studies (Text S1, section 7) are also provided.
Power Analysis
Our aim was to determine whether any common genetic variant predicts a clinically significant difference in the outcome of treatment with antidepressants, taking a difference of at least three points in the reduction of depression symptom severity on HRSD-17 as the benchmark for clinical significance [8]. Specifically, we aimed to achieve 80% power to detect an additive genetic effect that explains 6.33% of variance in outcome, corresponding to an HRSD-17 three-point difference in a drug comparison study [9]. Since not all common genetic variants were directly genotyped, we factored in imperfect tagging (at R2 = 0.8) to estimate power for detecting effects of genotyped and ungenotyped variants. The quality-controlled sample provided a power well above 80% to detect a clinically significant effect at the genome-wide significance level for three of the four analyses (overall, SRI, and genotype–drug interaction). The meta-analysis of NEWMEDS and STAR*D samples had an adequate power to detect even an effect that was half of what would be considered clinically significant. Details of the power analysis can be found in Text S1 (section 1.4).
Pathway Analysis
Enrichment of genome-wide association signal in genes that belong to known biological pathways was tested using ALIGATOR [32]. This method takes a predefined list of significant SNPs, and tests whether these SNPs cluster in genes belonging to a particular pathway more than would be expected by chance, allowing for varying numbers of SNPs per gene and non-independence of SNPs within and between genes. ALIGATOR corrects the significance levels of pathway-specific enrichment for the testing of multiple non-independent pathways. Further details of the pathway analysis can be found in Text S1 (section 3).
Meta-Analysis with STAR*D
A meta-analysis was undertaken between NEWMEDS and data subsequently obtained from the first level of the STAR*D (Sequenced Treatment Alternatives to Relieve Depression) trial, when all participants with MDD were treated with citalopram (an SRI), in the hope of finding genome-wide significant associations. For further information about the STAR*D sample, genotyping, and quality control procedures see Text S1 (section 4.1).
To maximize the overlap between the two samples and genome coverage, both NEWMEDS and STAR*D were imputed to include over 1.4 million markers using BEAGLE 3.3 [33] and the HapMap phase 3 CEU population as the reference dataset. Meta-analysis was undertaken using the meta command in PLINK [19] in the entire samples and in a sample limited to individuals treated with a serotonergic antidepressant. More information about these methods used can be found in Text S1 (section 4).
Polygenic Scoring
While both our main analysis in NEWMEDS and the meta-analysis with STAR*D had sufficient power to detect clinically significant genetic associations, there could also exist an underlying weak signal from across the genome that could offer insight into the mechanism of antidepressant response. The methodology of polygene scoring allows for the detection of such weakly distributed signal [34]. Details on this method can be found in Text S1 (section 5). Polygenic scores were created based on the NEWMEDS results and used to predict outcomes in STAR*D using linear regression. Two polygenic tests, one based on the entire NEWMEDS sample and the other restricted to SRI-treated participants, were carried out.
Results
Response to Any Antidepressant
Linear regression assessed the influence of 520,978 SNPs on the adjusted percentage change in depression severity in the whole sample of 1,790 antidepressant-treated individuals. A quantile–quantile plot showed a uniform distribution of p-values, with no inflation of the test statistic (median lambda = 1.0034; Figure 2). No association reached the genome-wide level of significance (Figure 3).
Fig. 2. Quantile–quantile plots for the four genome-wide analyses. 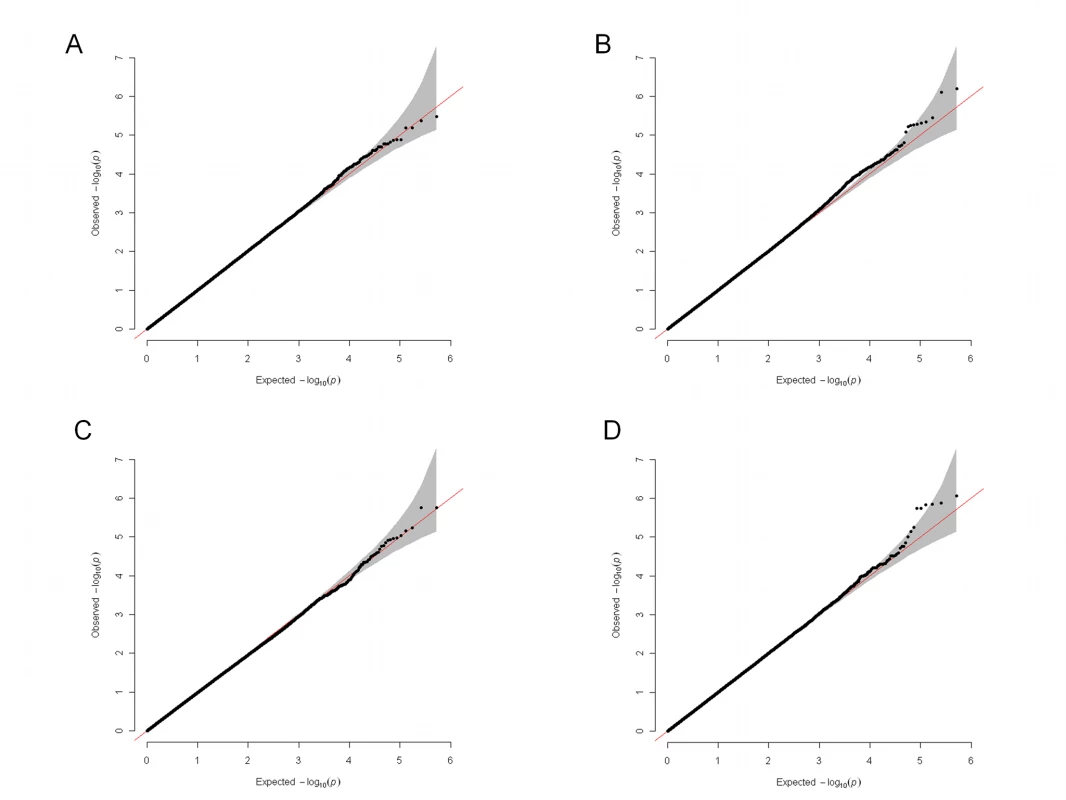
(A) Analysis of the whole sample (n = 1,790); (B) analysis of SRI-treated individuals (n = 1,222); (C) analysis of NRI-treated individuals (n = 568); (D) gene-by-drug interaction analysis in the randomly allocated individuals (n = 949). The shaded area is the 95% confidence interval of values expected under a uniform distribution. Fig. 3. Manhattan plots for the genome-wide pharmacogenetic analyses showing results by −log10 p-value and chromosome location. 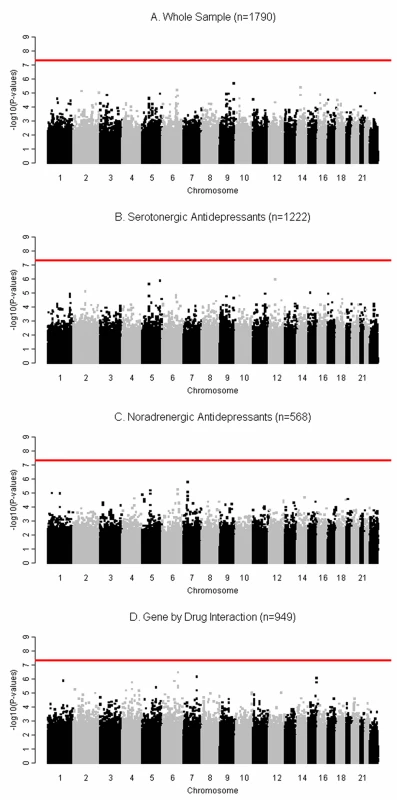
The red line indicates the genome-wide significance level (p<5.0×10−8), and the absence of data points above this line indicates the lack of significant associations in the four analyses: (A) analysis of the whole sample (n = 1,790); (B) analysis of SRI-treated individuals (n = 1,222); (C) analysis of NRI-treated individuals (n = 568); (D) gene-by-drug interaction analysis in the randomly allocated individuals (n = 949). Response to Serotonergic Antidepressants
A linear regression tested association between 520,978 SNPs and the adjusted percentage change in depression severity in 1,222 SRI-treated individuals. The quantile–quantile plot showed a uniform distribution of p-values, indicating no inflation of the test statistic (median lambda = 1.0094; Figure 2). No SNP was associated at the genome-wide level of significance (Figure 3).
Response to Noradrenergic Antidepressants
A linear regression tested association between 520,978 SNPs and adjusted percentage change in depression severity in 568 NRI-treated individuals. The quantile–quantile plot showed a uniform distribution of p-values, with no inflation of the test statistic (median lambda = 0.9875; Figure 2). There were no significant associations (Figure 3).
Differential Response to Serotonergic and Noradrenergic Antidepressants
A linear regression tested the interaction between 520,978 SNPs and antidepressant type (SRI versus NRI) in their effects on the adjusted percentage change in depression severity among the 949 individuals randomly allocated to SRI or NRI antidepressant. The quantile–quantile plot showed a uniform distribution of p-values, with no inflation of the test statistic (median lambda = 1.0015; Figure 2). No genotype–drug interactions were detected at the genome-wide level of significance (Figure 3).
For all four analyses, a meta-analysis of results from contributing studies also gave negative results (see Text S1, section 6).
Pathway Analysis
Pathway analysis tested whether any biological pathways had more genes in the top 5% of genes (ranked by their most significant SNP) than expected by chance. None of the four analyses (response to any antidepressant, serotonergic antidepressants, noradrenergic antidepressants, and differential response to serotonergic and noradrenergic antidepressants) showed a significant excess in the number of enriched pathways, and no single pathway was significantly enriched after correcting for multiple testing. Full results are given in Text S1 (section 3.2).
Meta-Analysis with STAR*D
A meta-analysis tested percentage improvement in 2,897 individuals from NEWMEDS and STAR*D using over 1.1 million genotyped and imputed SNPs and found no genome-wide significant results. A meta-analysis restricted to SRI-treated individuals (n = 2,329) found no genome-wide significant results. For more information see Text S1 (section 4.4).
Polygenic Scoring
Polygenic scores were calculated to test the combined effect of multiple weak associations across the genome. Scores were created using NEWMEDS and used to predict outcomes in STAR*D. In the analysis that included all individuals (NEWMEDS, n = 1,790; STAR*D, n = 1,107), there was no significant prediction across the 13 progressive p-value thresholds. In a sample restricted to SRI-treated individuals (NEWMEDS, n = 1,222; STAR*D, n = 1,107), there was no significant prediction from any of the 13 progressive p-value thresholds either. Further information about the results can be found in Text S1 (section 5.2).
Discussion
In a large pharmacogenetic analysis, including 1,790 antidepressant-treated individuals with MDD, none of the more than 500,000 genetic markers predicted treatment outcome after genome-wide correction. Since our study had adequate statistical power to detect common genetic variants with a clinically significant predictive effect, the results suggest that single marker prediction will not contribute to personalizing prescription of currently available antidepressants. Increasing sample size may aid in obtaining positive results in future studies, which may provide insight into the mechanism of the therapeutic action of antidepressants, even though the effect size will likely be too small to translate into clinical applications.
The lack of genome-wide significant or even moderately strong associations among a comprehensive list of candidate genes (details in Text S1, section 7) puts previously reported positive results from smaller candidate gene studies [4],[35] into a sobering perspective. The current study also fails to strengthen associations found previously in the genome-wide association study of the GENDEP sample [17], the largest sub-sample in the NEWMEDS consortium, or other genome-wide pharmacogenetic studies [11],[12],[17]. Furthermore, the current investigation fails to find a genome-wide significant association in the largest pharmacogenetic meta-analysis to date, which included 2,897 individuals. Pathway analysis has not shown any enrichment for a known biological pathway. Polygenic prediction has not found any evidence for a distributed convergent signal between the two largest pharmacogenetic samples collected to date. It is therefore possible that common polymorphisms will not help predict the outcome of treatment with commonly used antidepressants in a clinical meaningful way.
Limitations
The present study benefited from a large sample through the combination of participants from multiple studies. This limits the interpretation of the present results in several ways. One limitation is the use of multiple antidepressants, each differing slightly in its chemical structure, transporter affinity, and receptor binding profile. Based on previous pharmacogenetic data [14], we hypothesized the existence of common genetic predictors of response to antidepressants with broadly defined modes of action, such as serotonin and noradrenaline reuptake inhibition. While we found no predictors of response to SRI or NRI types of antidepressants, our results are compatible with the existence of pharmacogenetic effects that are specific to a particular antidepressant compound. Large groups treated with the same drug may uncover genetic predictors that were not detected in the present study. However, the largest existing sample treated with the same antidepressant has also failed to detect significant genetic predictors in a genome-wide analysis [11], and it is unlikely that even larger samples with homogeneous treatment will be collected in the near future. The studies included in NEWMEDS also differed in other aspects, e.g., in the depression rating scale used and in the way participants were recruited. The adjusted change score used as the phenotype in this study removed effects specific to each individual contributing study. This measure was intended to minimize the risk of spurious findings, but it could have reduced the impact of a genuinely larger treatment effect in a particular study. However, our aim was to detect pragmatic predictors that generalize to multiple settings rather than effects specific to a particular homogeneous group. It is unlikely that pharmacogenetic predictions limited to a particular depression rating scale or to a more homogeneous subgroup of patients would be clinically meaningful or commercially viable. Furthermore, a meta-analysis conducted across the individual studies provided similar results (see Text S1, section 6). A related limitation was the smaller size of the NRI-treated sample, meaning that only relatively strong predictors could be identified in this arm of the study. Additional limitations pertain to the scope of the present study. Our results are limited to the influence of common polymorphisms on the therapeutic effects of several monoaminergic antidepressants under study in individuals of European ancestry. Studies in other populations and studies of antidepressants with a non-monoaminergic mode of action are needed to extend the scope of pharmacogenetic exploration.
Conclusions and Future Directions
Our study adds to the growing literature of genome-wide pharmacogenetic studies, offering, to our knowledge, the largest body of pharmacogenetic data available to date. The absence of pharmacogenetic associations with clinically meaningful effect suggests that common genetic variation is not ready to inform personalization of treatment for depression. Future studies may need to combine clinical, genetic, epigenetic, transcriptomic, and proteomic information to obtain clinically meaningful prediction of how an individual with major depression will respond to antidepressant treatment.
Supporting Information
Zdroje
1. KesslerRC, ChiuWT, DemlerO, MerikangasKR, WaltersEE (2005) Prevalence, severity, and comorbidity of 12-month DSM-IV disorders in the National Comorbidity Survey Replication. Arch Gen Psychiatry 62 : 617–627.
2. RushAJ, TrivediMH, WisniewskiSR, NierenbergAA, StewartJW, et al. (2006) Acute and longer-term outcomes in depressed outpatients requiring one or several treatment steps: a STAR*D report. Am J Psychiatry 163 : 1905–1917.
3. TrivediMH, RushAJ, WisniewskiSR, NierenbergAA, WardenD, et al. (2006) Evaluation of outcomes with citalopram for depression using measurement-based care in STAR*D: implications for clinical practice. Am J Psychiatry 163 : 28–40.
4. KatoM, SerrettiA (2010) Review and meta-analysis of antidepressant pharmacogenetic findings in major depressive disorder. Mol Psychiatry 15 : 473–500.
5. KirchheinerJ, FuhrU, BrockmollerJ (2005) Pharmacogenetics-based therapeutic recommendations—ready for clinical practice? Nat Rev Drug Discov 4 : 639–647.
6. MrazekDA, LermanC (2011) Facilitating clinical implementation of pharmacogenomics. JAMA 306 : 304–305.
7. KimH, LimSW, KimS, KimJW, ChangYH, et al. (2006) Monoamine transporter gene polymorphisms and antidepressant response in koreans with late-life depression. JAMA 296 : 1609–1618.
8. National Institute for Health and Clinical Excellence (2004) Depression: management of depression in primary and secondary care. London: National Institute for Health and Clinical Excellence.
9. UherR, TanseyKE, MalkiK, PerlisRH (2012) Biomarkers predicting treatment outcome in depression: what is clinically significant? Pharmacogenomics 13 : 233–240.
10. SimonGE, PerlisRH (2010) Personalized medicine for depression: can we match patients with treatments? Am J Psychiatry 167 : 1445–1455.
11. GarriockHA, KraftJB, ShynSI, PetersEJ, YokoyamaJS, et al. (2010) A genomewide association study of citalopram response in major depressive disorder. Biol Psychiatry 67 : 133–138.
12. IsingM, LucaeS, BinderEB, BetteckenT, UhrM, et al. (2009) A genomewide association study points to multiple loci that predict antidepressant drug treatment outcome in depression. Arch Gen Psychiatry 66 : 966–975.
13. HughesB (2009) Novel consortium to address shortfall in innovative medicines for psychiatric disorders. Nat Rev Drug Discov 8 : 523–524.
14. UherR, Huezo-DiazP, PerroudN, SmithR, RietschelM, et al. (2009) Genetic predictors of response to antidepressants in the GENDEP project. Pharmacogenomics J 9 : 225–233.
15. ThomasL, MulliganJ, MasonV, TallonD, WilesN, et al. (2008) Genetic and clinical predictors of treatment response in depression: the GenPod randomised trial protocol. Trials 9 : 29.
16. PerroudN, BondolfiG, UherR, Gex-FabryM, AubryJM, et al. (2011) Clinical and genetic correlates of suicidal ideation during antidepressant treatment in a depressed outpatient sample. Pharmacogenomics 12 : 365–377.
17. UherR, PerroudN, NgMY, HauserJ, HenigsbergN, et al. (2010) Genome-wide pharmacogenetics of antidepressant response in the GENDEP project. Am J Psychiatry 167 : 555–564.
18. MalkiK, UherR, Paya-CanoJ, BinderE, RietschelM, et al. (2010) Convergent animal and human evidence suggests a role of PPM1A gene in response to antidepressants. Biol Psychiatry 69 : 360–365.
19. PurcellS, NealeB, Todd-BrownK, ThomasL, FerreiraMA, et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81 : 559–575.
20. Wittke-ThompsonJK, PluzhnikovA, CoxNJ (2005) Rational inferences about departures from Hardy-Weinberg equilibrium. Am J Hum Genet 76 : 967–986.
21. RoystonP, AltmanDG, SauerbreiW (2006) Dichotomizing continuous predictors in multiple regression: a bad idea. Stat Med 25 : 127–141.
22. StreinerDL (2002) Breaking up is hard to do: the heartbreak of dichotomizing continuous data. Can J Psychiatry 47 : 262–266.
23. UherR, MuthenB, SoueryD, MorsO, JaraczJ, et al. (2010) Trajectories of change in depression severity during treatment with antidepressants. Psychol Med 40 : 1367–1377.
24. LeuchtS, KaneJM, EtschelE, KisslingW, HamannJ, et al. (2006) Linking the PANSS, BPRS, and CGI: clinical implications. Neuropsychopharmacology 31 : 2318–2325.
25. MontgomerySA, AsbergM (1979) A new depression scale designed to be sensitive to change. Br J Psychiatry 134 : 382–389.
26. HamiltonM (1960) A rating scale for depression. J Neurol Neurosurg Psychiatry 23 : 56–62.
27. BeckAT, WardCH, MendelsonM, MockJ, ErbaughJ (1961) An inventory for measuring depression. Arch Gen Psychiatry 4 : 561–571.
28. UherR, MaierW, HauserJ, MarusicA, SchmaelC, et al. (2009) Differential efficacy of escitalopram and nortriptyline on dimensional measures of depression. Br J Psychiatry 194 : 252–259.
29. SkolAD, ScottLJ, AbecasisGR, BoehnkeM (2006) Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat Genet 38 : 209–213.
30. KallbergH, AlfredssonL, FeychtingM, AhlbomA (2010) Don't split your data. Eur J Epidemiol 25 : 283–284.
31. DudbridgeF, GusnantoA (2008) Estimation of significance thresholds for genomewide association scans. Genet Epidemiol 32 : 227–234.
32. HolmansP, GreenEK, PahwaJS, FerreiraMA, PurcellSM, et al. (2009) Gene ontology analysis of GWA study data sets provides insights into the biology of bipolar disorder. Am J Hum Genet 85 : 13–24.
33. BrowningSR, BrowningBL (2007) Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet 81 : 1084–1097.
34. PurcellSM, WrayNR, StoneJL, VisscherPM, O'DonovanMC, et al. (2009) Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 460 : 748–752.
35. HorstmannS, BinderEB (2009) Pharmacogenomics of antidepressant drugs. Pharmacol Ther 124 : 57–73.
Štítky
Interní lékařství
Článek vyšel v časopisePLOS Medicine
Nejčtenější tento týden
2012 Číslo 10- Berberin: přírodní hypolipidemikum se slibnými výsledky
- Léčba bolesti u seniorů
- Příznivý vliv Armolipidu Plus na hladinu cholesterolu a zánětlivé parametry u pacientů s chronickým subklinickým zánětem
- Červená fermentovaná rýže účinně snižuje hladinu LDL cholesterolu jako vhodná alternativa ke statinové terapii
- Jak postupovat při výběru betablokátoru − doporučení z kardiologické praxe
-
Všechny články tohoto čísla
- The Double Burden of Obesity and Malnutrition in a Protracted Emergency Setting: A Cross-Sectional Study of Western Sahara Refugees
- PRISMA-Equity 2012 Extension: Reporting Guidelines for Systematic Reviews with a Focus on Health Equity
- Human Rights Research and Ethics Review: Protecting Individuals or Protecting the State?
- Serious and Life-Threatening Pregnancy-Related Infections: Opportunities to Reduce the Global Burden
- Donor Funding for Newborn Survival: An Analysis of Donor-Reported Data, 2002–2010
- The Potential Impact of Pre-Exposure Prophylaxis for HIV Prevention among Men Who Have Sex with Men and Transwomen in Lima, Peru: A Mathematical Modelling Study
- Removing the Age Restrictions for Rotavirus Vaccination: A Benefit-Risk Modeling Analysis
- Where There Is No Paramedic: The Sachigo Lake Wilderness Emergency Response Education Initiative
- Associations between Mode of HIV Testing and Consent, Confidentiality, and Referral: A Comparative Analysis in Four African Countries
- Strengthening Medical Product Regulation in Low- and Middle-Income Countries
- Bringing Clarity to the Reporting of Health Equity
- Developing a National Mental Health Policy: A Case Study from Uganda
- Psychosocial Factors That Shape Patient and Carer Experiences of Dementia Diagnosis and Treatment: A Systematic Review of Qualitative Studies
- Research Conducted Using Data Obtained through Online Communities: Ethical Implications of Methodological Limitations
- Genetic Predictors of Response to Serotonergic and Noradrenergic Antidepressants in Major Depressive Disorder: A Genome-Wide Analysis of Individual-Level Data and a Meta-Analysis
- Risk of Cardiovascular Disease and Total Mortality in Adults with Type 1 Diabetes: Scottish Registry Linkage Study
- PLOS Medicine
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Human Rights Research and Ethics Review: Protecting Individuals or Protecting the State?
- The Potential Impact of Pre-Exposure Prophylaxis for HIV Prevention among Men Who Have Sex with Men and Transwomen in Lima, Peru: A Mathematical Modelling Study
- Psychosocial Factors That Shape Patient and Carer Experiences of Dementia Diagnosis and Treatment: A Systematic Review of Qualitative Studies
- Genetic Predictors of Response to Serotonergic and Noradrenergic Antidepressants in Major Depressive Disorder: A Genome-Wide Analysis of Individual-Level Data and a Meta-Analysis
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



