-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaRNA Processing Factors Swd2.2 and Sen1 Antagonize RNA Pol III-Dependent Transcription and the Localization of Condensin at Pol III Genes
Failure to condense chromosomes prior to anaphase onset can lead to genome instability. The evolutionary-conserved condensin complex drives chromosome condensation, probably by changing the topology of chromatin around its binding sites. Condensin localizes to regions of high transcription, suggesting that some transcription-associated feature(s) direct its association with chromatin. Here we considered that transcription-dependent DNA:RNA hybrids or topological stress could be involved in recruiting condensin. Our data show that condensin is indeed enriched at regions accumulating DNA:RNA hybrids but that they are not involved in its recruitment. Rather, we identify a mutant combination where increased transcription by RNA Pol III is associated locally with stronger topological stress. Strikingly the localization of condensin is dramatically enhanced at the same loci and we show that topological stress contributes to this enhanced association. Our data strengthen the idea that transcription creates the environment necessary to recruit condensin in mitosis.
Published in the journal: . PLoS Genet 10(11): e32767. doi:10.1371/journal.pgen.1004794
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004794Summary
Failure to condense chromosomes prior to anaphase onset can lead to genome instability. The evolutionary-conserved condensin complex drives chromosome condensation, probably by changing the topology of chromatin around its binding sites. Condensin localizes to regions of high transcription, suggesting that some transcription-associated feature(s) direct its association with chromatin. Here we considered that transcription-dependent DNA:RNA hybrids or topological stress could be involved in recruiting condensin. Our data show that condensin is indeed enriched at regions accumulating DNA:RNA hybrids but that they are not involved in its recruitment. Rather, we identify a mutant combination where increased transcription by RNA Pol III is associated locally with stronger topological stress. Strikingly the localization of condensin is dramatically enhanced at the same loci and we show that topological stress contributes to this enhanced association. Our data strengthen the idea that transcription creates the environment necessary to recruit condensin in mitosis.
Introduction
Mitotic chromosome condensation is essential for genome integrity. When defective, chromosomes often remain entangled and fail to segregate properly in anaphase. A key driver of chromosome condensation is the highly conserved condensin complex. Condensin is made of five sub-units (SMC2Cut14, SMC4Cut3, CAP-D2Cnd1, CAP-GCnd3 and CAP-HCnd2, name of the human protein followed by its name in fission yeast) and it is one of the main components of mitotic chromosomes [1]. In vitro, purified condensin can introduce positive supercoils into a relaxed plasmid in the presence of topoisomerase I [2], [3]. These observations support the idea that condensin shapes mitotic chromosomes by changing the topology of chromatin around its binding sites. However, the mechanisms underlying the association of condensin with chromatin remain poorly understood (reviewed in [4]).
Several studies have illustrated the paradoxical relationships linking gene transcription and the localization of condensin. From pro - to eukaryotes, condensin is preferentially enriched at highly transcribed genes [5], [6], [7], [8], suggesting that some highly conserved transcription-associated feature(s) that predate(s) the appearance of nucleosomes help to recruit condensin. However, experiments in yeast indicated that RNA polymerases must be silenced before condensin can bind, at least at repetitive sequences such as the rDNA or the sub-telomeres [9], [10]. These somewhat contradictory observations could potentially be reconciled if one hypothesizes that a by-product of the transcription process facilitates the recruitment of condensin. In this study, we have considered that such a by-product could be R-Loops or transcription-associated topological stress.
R-Loops result from the formation of stable DNA:RNA hybrids in the genome. As a consequence of the hybridization of the RNA to the template, the non-transcribed strand of the DNA remains single-stranded (reviewed in [11]). Interestingly, the hinge domain of the Smc2/Smc4 heterodimer in condensin shows high affinity in vitro for single-stranded DNA [12], [13]. Moreover, a recent study proposed that chromatin is less accessible to restriction enzymes in mutants where R-Loops accumulate, consistent with the idea that R-Loop formation favours chromatin compaction [14]. Interestingly, fission yeast condensin can disassemble DNA:RNA hybrids in vitro [15] and its chicken counterpart localizes to CpG islands [6], which constitute major R-Loop forming regions in the genome [16]. Taken together, these observations support the idea that R-Loops and condensin could interact functionally in vivo [14].
According to the twin supercoiled domain model, high rates of transcription induce positive supercoiling of the chromatin in front of the elongating polymerase, whilst negative supercoiling accumulate upstream of the polymerase [17]. As such, highly expressed genes represent regions of the genomes that accumulate topological stress. As confirmed in vivo recently, this stress is monitored by topoisomerase I and topoisomerase II [18], [19], [20]. Interestingly, in vitro assays have indicated that condensin binds preferentially to positively supercoiled plasmids in the presence of ATP [21]. Whether or not this transcription-associated topological stress contributes to the binding of condensin in vivo has not been addressed.
In order to clarify the functional relationships between transcription and chromosome condensation, we recently carried out a genetic screen in fission yeast to identify deletions of transcription-associated factors that would rescue a condensin deficiency [22]. For this, we isolated loss-of-function mutations that could rescue the thermo-sensitivity of the condensin mutant cut3-477 [23]. Two of the mutations we isolated were the deletions of swd2.2 (swd2.2Δ) and sen1 (sen1Δ) [22]. Swd2.2 is a non-essential component of the Cleavage and Polyadenylation Factor (CPF), the complex responsible for 3′end maturation of RNA Pol II transcripts in yeast (reviewed in [24]), where it acts to maintain the proper levels of CPF-associated phosphatases [22]. Fission yeast Sen1 is the homologue of human Senataxin and has been shown to unwind DNA:RNA hybrids in vitro [25]. Budding yeast Sen1 is involved in transcription termination [26] but its role in fission yeast has not been characterized. Here we show that both factors act directly at Pol III-transcribed genes to limit the association of condensin and the accumulation of topological stress. Furthermore, topological stress at Pol III-transcribed genes facilitates the association of condensin when Swd2.2 and Sen1 are missing.
Results
Swd2.2 and Sen1 negatively regulate the accumulation of condensin at Pol III-transcribed genes
On their own, the deletions of swd2.2 (swd2.2Δ) and sen1 (sen1Δ) partly restored growth of cut3-477 cells at the restrictive temperature (Figure 1A) and reduced the proportion of anaphase cells displaying chromosome segregation defects (Figure 1B). Combining both deletions (sen1Δswd2.2Δ) resulted in a stronger suppressor effect (Figure 1AB). The double mutant sen1Δswd2.2Δ also suppressed the other condensin mutant cut14-208 (Figure S1). Strikingly, Chromatin Immunoprecipitation (ChIP) analysis in cycling cell populations showed that the localization of condensin was altered at specific loci when Swd2.2 and Sen1 were both missing: its recruitment increased significantly at genes transcribed by RNA Pol III (Gln.04, Met.07, Ser.13, Pro.09, Tyr.04, Gly.05, 5S rRNA, Arg.04 on Figure 1C), whereas it was significantly reduced at the rDNA arrays (18S&Rfb2). The binding of condensin remained unaffected at kinetochores (cnt1) or at highly transcribed Pol II genes (Act1, Adh1, Fba1 and SPAC27E2.11c). The sequences of all the primers used in this study are available on Table S1. The mitotic indexes of both cell populations (swd2.2+sen1+ and swd2.2Δsen1Δ) were comparable (Figure 1D), ruling out that the changes in the association of condensin are due to indirect, cell-cycle defects. These data established that Sen1 and Swd2.2 act to limit the localization of condensin at Pol III-transcribed genes. The reasons why the association of condensin at the rDNA arrays is reduced in the absence of Swd2.2 and Sen1 will be explained elsewhere.
Fig. 1. The double deletion of Swd2.2 and Sen1 facilitates the localization of condensin at Pol III-transcribed genes. 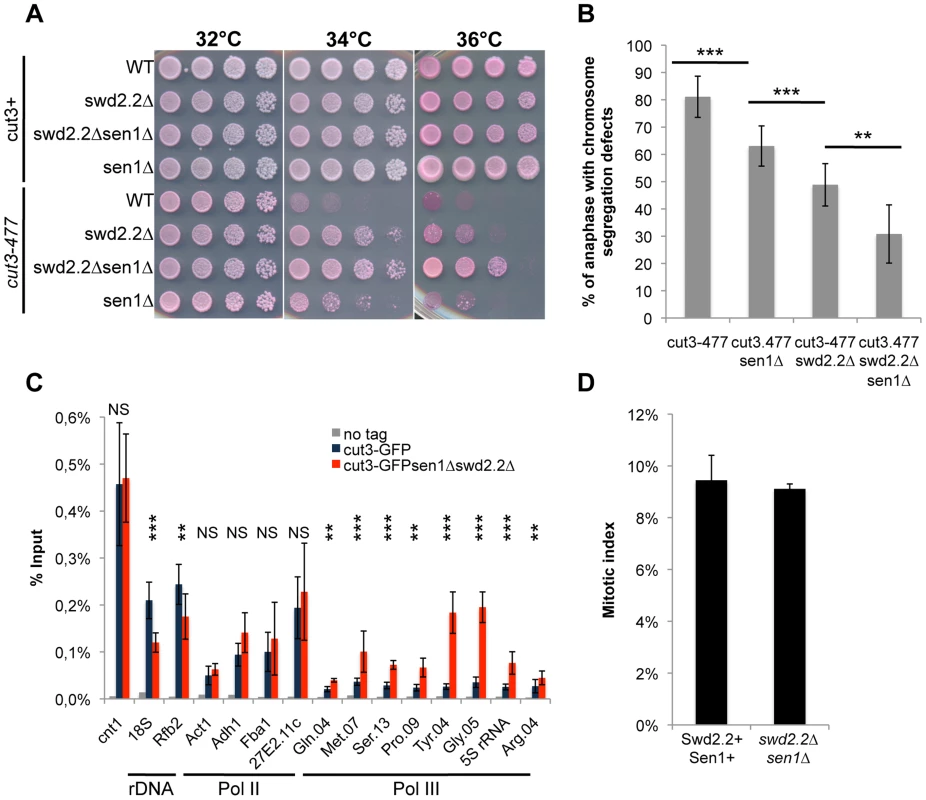
A. Serial dilutions of the indicated strains were plated on rich media at the indicated temperatures. B. Chromosome segregation in anaphase was monitored in the indicated strains after growing cells for one generation at 34°C. For each genotype, a minimum of 6 independent experiments was performed in which a minimum of 100 anaphase cells were scored (***<0.001; **<0.01 Wilcoxon - Mann Whitney). Anaphases were scored as defective when chromatin was detected lagging between the two main DNA masses C. ChIP-qPCR analysis of the amount of GFP-tagged Cut3 cross-linked to chromatin in cell populations of the indicated genotypes grown at 30°C (mean ± standard deviation from 6 biological replicates (NS: not significant, *P<0.05; **P<0.01; ***P<0.001 Wilcoxon - Mann Whitney). The primers used in this study are shown on Table S1. D. Mitotic indexes of the cell populations used in C. Cells were fixed with cold methanol and processed for immuno-fluorescence using an anti-tubulin antibody. Cells with a spindle were counted as mitotic. Swd2.2 and Sen1 localize at Pol III-transcribed genes and regulate the transcription cycle of RNA Pol III
We found previously that Swd2.2 associates with Pol III-transcribed genes and that lack of Swd2.2 restored the localization of condensin at Pol III-transcribed genes in the condensin-deficient mutant cut3-477 [22]. Here, we show that Sen1 is also significantly enriched at Pol III-transcribed genes and that its binding is independent of Swd2.2 (Figure 2A). Furthermore, affinity purification of Sen1 followed by mass-spectrometry analysis of its associated proteins identified most sub-units of the RNA Pol III complex as its most stable binding partners (Table S2). We confirmed this interaction by showing that the RNA Pol III sub-unit Rpc25 co-precipitates with Sen1 (Figure 2B). Note however that Sen1 did not co-precipitate with Sfc6, a sub-unit of TFIIIC (Figure S2), a complex required for the association of RNA Pol III with chromatin [27]. ChIP analysis showed that the association of Rpc25 with chromatin was significantly increased in the absence of Sen1 (Figure S3) or in swd2.2Δsen1Δ cells (Figure 2C&D). In swd2.2Δsen1Δ cells, the stabilization of RNA Pol III on chromatin was associated with an increase in the steady-state level of tRNAs, as detected by RT-qPCR analysis (Figure 2E). Taken together, these experiments concur to show that Swd2.2 and Sen1 play a direct role at Pol III-transcribed genes, where they limit the association of RNA Pol III and the accumulation of transcripts. These results show that the accumulation of condensin at Pol III-transcribed genes in swd2.2Δsen1Δ cells is concomitant with an enhanced transcriptional activity.
Fig. 2. Transcription is enhanced at Pol III-transcribed genes when Swd2.2 and Sen1 are missing. 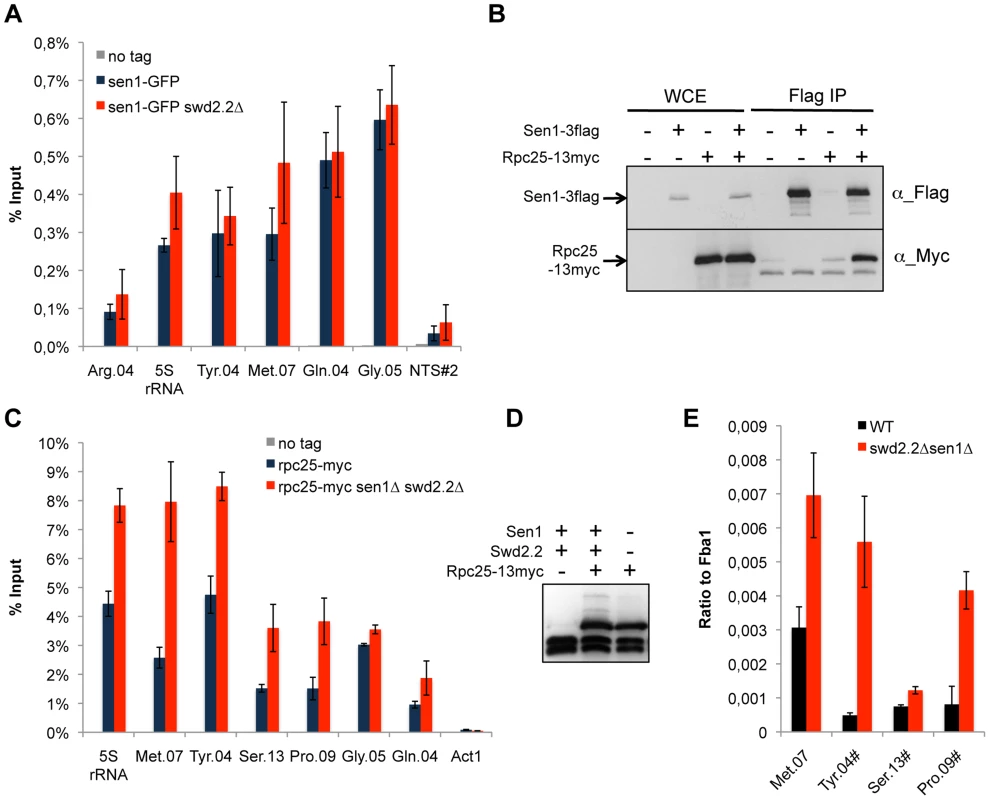
A. Sen1 is enriched at Pol III-transcribed genes. ChIP qPCR of the indicated strains grown in cycling conditions at the indicated loci (mean ± standard deviation from 3 biological replicates). NTS#2 is a site within the Replication Fork Barrier of the rDNA and is shown as a comparison. B. Flag-tagged Sen1 co-immunoprecipitates with Myc-tagged Rpc25. Whole cell extracts (WCE) and the immuno-precipitated material (Flag IP) of the indicated strains were analyzed by western blot. C. Rpc25 becomes more abundant at Pol III-transcribed genes when Swd2.2 and Sen1 are missing. ChIP qPCR of the indicated strains grown in cycling conditions at the indicated loci (mean ± standard deviation from 3 biological replicates). D. Western blot analysis of the stability of Rpc25-13myc. Tubulin is used as a loading control. E. Pol III transcripts are more abundant when Swd2.2 and Sen1 are missing. Total RNAs extracted from swd2.2+sen1+ or swd2.2Δsen1Δ cells grown in rich medium at 30°C were analyzed by RT-qPCR (3 biological replicates, 2 RT per replicate). R-Loops accumulate strongly at Pol III-transcribed genes
It was recently argued that budding yeast Sen1 limits the accumulation of DNA:RNA hybrids, including at Pol III-transcribed genes [28]. Fission yeast Sen1 similarly was shown to display a DNA:RNA helicase activity in vitro [25]. These observations and the additional arguments detailed in the introduction prompted us to test the possibility that R-Loops could represent a transcription by-product facilitating the association of condensin with chromatin. We speculated that lack of Sen1 and Swd2.2 could result in the accumulation of R-Loops at Pol III-transcribed genes where they might contribute to increase the association of condensin.
To establish whether or not R-Loops form at Pol III-transcribed genes in fission yeast, we first monitored by ChIP the chromatin association of RNase H1, one of the endogenous enzymes known to disassemble R-Loops. More specifically, we introduced at the endogenous locus a point mutation (D129N) in the fission yeast RNase H1 (Rnh1), because the same mutation was shown to weaken the catalytic activity of human RNase H1 [29]. Consistent with this, the D129N mutation did stabilize the interaction of Rnh1 with Pol III-transcribed genes (Figure 3A). Furthermore, the interaction of Rnh1D129N with Pol III-transcribed genes was lost upon over-expression in vivo of RnhA, the RNase H1 enzyme from E.coli (Figure 3B). Upon over-expression, RnhA itself did not stably associate with Pol III-transcribed genes (Figure S4), showing that the loss of Rnh1D129N from Pol III-transcribed genes upon over-expression of RnhA cannot be explained by its mere replacement by bacterial RnhA. Finally, Figure S5 shows that the association of Rnh1D129N with the rDNA repeats increased significantly in the absence of topoisomerase I (top1Δ), consistent with the observations reported previously that lack of Top1 triggers the accumulation of R-Loops at rDNA in budding yeast [30]. This confirmed that Rnh1D129N was able to detect significant changes in R-Loop accumulation. Taken together, these data show that ChIP with Rnh1D129N is a reliable way to identify R-Loop forming regions in fission yeast.
Fig. 3. R-Loops form in abundance at Pol III-transcribed genes but they do not significantly impact the association of condensin. 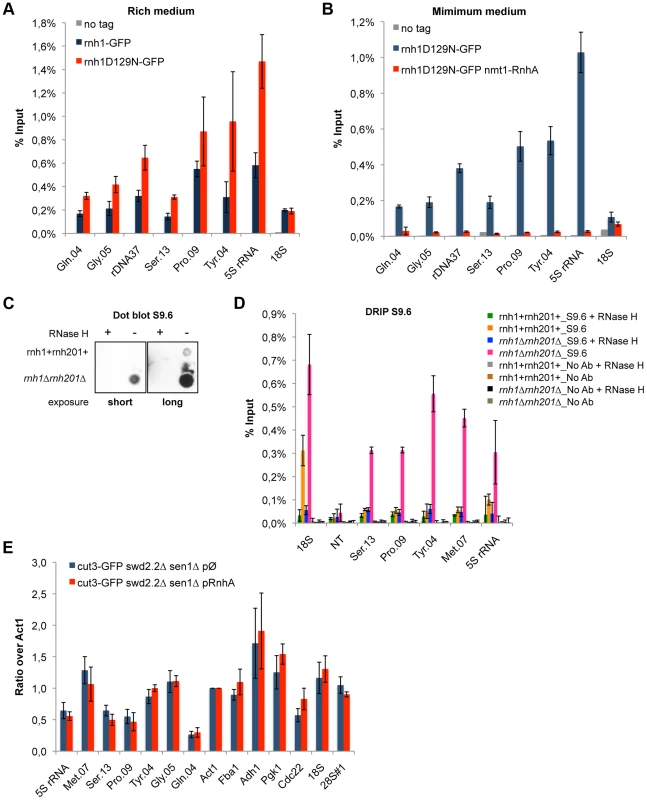
A. ChIP qPCR of the indicated strains grown in cycling conditions at the indicated loci (mean ± standard deviation from 3 biological replicates). B. As in A. Cells were grown in minimal medium for a minimum of 18 hours to promote the over-expression of RnhA driven by the nmt promoter. C. Genomic DNA was extracted from rnh1+rnh201+ and rnh1Δrnh201Δ cells in preparation for the DRIP procedure. Equal amount of genomic DNA were spotted on a nylon membrane and incubated with 2 µg/mL of purified S9.6 antibody. The amount of S9.6 bound to the DNA was revealed using chemiluminescence. D. DRIP-qPCR of the indicated strains grown in cycling conditions at the indicated loci (mean ± standard deviation from 3 biological replicates). E. Cells of the indicated genotypes were grown in minimum medium lacking thiamine for a minimum of 18 hours to drive the over-expression of RnhA. ChIP-qPCR was then performed (mean ± standard deviation from 3 biological replicates). We sought to confirm the formation of R-Loops at genes transcribed by RNA Pol III using another approach. A method that is commonly used to map R-Loop forming regions in yeast is to perform ChIP using the S9.6 antibody because of its high affinity for DNA:RNA hybrids [31]. ChIP requires formaldehyde cross-linking followed by sonication of the chromatin. We found that the ability of S9.6 to detect R-Loops generated after transcription in vitro was greatly diminished both by formaldehyde cross-linking and by sonication (Figure S6). We do not know at this stage whether this is because R-Loops are partly destroyed by these treatments or because these treatments reduce the affinity of the antibody for R-Loops. To circumvent these issues, we extracted genomic DNA from unfixed cells, digested soluble RNA using RNase A and sheared the DNA using a cocktail of restriction enzymes (see Methods). Dot blot analysis using the S9.6 antibody confirmed that our procedure largely preserved R-Loops (Figure 3C). We then performed DNA:RNA immuno-precipitation (DRIP) using the S9.6 antibody in stringent conditions, in the presence of 500 mM NaCl. As expected, the DRIP signal at 18S, the canonical R-Loop forming region within the rDNA repeats [30], increased significantly in the absence of RNase H1 and RNase H2 (rnh1Δrnh201Δ cells) and disappeared almost entirely upon treatment of the genomic DNA with commercial RNase H (Figure 3D). On the contrary, the DRIP signal detected at a non-transcribed region NT (chr I, 3009300-3009500, [32]) remained low both in rnh1Δrnh201Δ cells and upon treatment with RNase H. Those controls demonstrated that the signals we detected using DRIP were specific. In agreement with the results obtained using ChIP of Rnh1D129N as a reporter for the presence of R-Loops, we detected strong DRIP signals at Pol III-transcribed genes in the absence of RNase H1 and RNase H2 (Figure 3D). In conclusion, the two methods we have set up to map R-Loop forming regions establish that R-Loops are a prominent feature of Pol III-transcribed genes in fission yeast.
Using ChIP of Rnh1D129N, we established that R-Loops accumulate to similar levels at Pol III-transcribed genes in cycling cells (>90% of interphase cells) and in cells synchronized in early mitotis (Figure S7A). Consistent with this, ChIP established that the association of RNA Pol III with chromatin is largely maintained in mitosis (Figure S7B). Taken together these experiments support the idea that transcription at Pol III-transcribed genes is maintained in mitosis, at a time when condensin is loaded on chromosomes in fission yeast.
Finally, lack of Swd2.2 and Sen1 resulted in a small but significant increase in the formation of R-Loops at some but not all Pol III-transcribed genes (Figure S8). Note however that this increase could be due to the fact that Pol III transcription is stimulated in the absence of Swd2.2 and Sen1 (Figure 2C&D). As such, these observations therefore do not prove that Swd2.2 and Sen1 antagonize R-Loop formation at Pol III-transcribed genes directly.
Stable R-Loop formation is not necessary to recruit condensin
To establish whether R-Loops at Pol III-transcribed genes could contribute to the accumulation of condensin, we prevented the formation of stable R-Loops by over-expressing RnhA. ChIP analysis showed that over-expression of RnhA did not reduce the amount of condensin recruited at Pol III-transcribed genes in swd2.2Δsen1Δ cells (Figure 3E) or in wild-type mitotic cells (Figure S9). These data concur to demonstrate that stable, long-lived R-Loops play little or no part in recruiting condensin. Note that over-expression of RnhA did not interfere either with the association of RNA Pol III (Figure S10A) or Sen1 (Figure S10B).
Topological constraints accumulate in cells lacking Swd2.2 and Sen1
Because Xenopus condensin shows greater affinity in vitro for positively supercoiled DNA [21], we speculated that the cue facilitating the accumulation of condensin at Pol III-transcribed genes in the absence of Swd2.2 and Sen1 could be local topological constraints. Consistent with an increase in topological stress in swd2.2Δsen1Δ cells, ChIP analysis detected strong accumulation of topoisomerase I (Top1) at most loci (Figure 4A), although the protein levels of Top1 remained unaffected (Figure 4B). We also detected enhanced accumulation of topoisomerase II (Top2), mostly at Pol III-transcribed genes (Figure 4C), when the protein levels of Top2 remained unaffected (Figure 4D). Transcription-associated topological stress was recently shown to destabilize nucleosomes [19]. At some but not all Pol III-transcribed genes that we tested, we detected a significant reduction in the recruitment of histone H3 (Figure 4E) in swd2.2Δsen1Δ cells, which is consistent with the local depletion of nucleosomes. The concomitant accumulation of Top1 and Top2 and the depletion of nucleosomes suggest that topological stress is greater at Pol III-transcribed genes in swd2.2Δsen1Δ cells. We speculate that the increased transcription of Pol III-transcribed genes in swd2.2Δsen1Δ cells could contribute at least in part to this enhanced topological stress.
Fig. 4. Lack of Swd2.2 and Sen1 results in local topological stress at Pol III-transcribed genes. 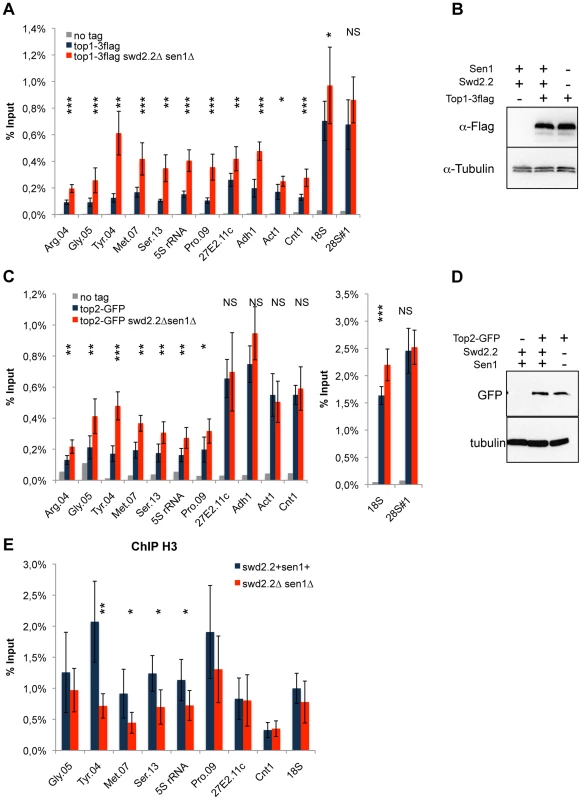
A. ChIP qPCR of the indicated strains grown in cycling conditions at the indicated loci (mean ± standard deviation from 6 biological replicates. NS: not significant *P<0.05; **P<0.01; ***P<0.001 Wilcoxon - Mann Whitney). B. Western blot analysis of the stability of Top1-3flag. Tubulin is used as a loading control. C. ChIP qPCR of the indicated strains grown in cycling conditions at the indicated loci (mean ± standard deviation from 6 biological replicates. NS: not significant *P<0.05; **P<0.01; ***P<0.001 Wilcoxon - Mann Whitney). D. Western blot analysis of the stability of Top2-GFP. Tubulin is used as a loading control. E. ChIP qPCR of histone H3 in the indicated strains grown in cycling conditions at the indicated loci (mean ± standard deviation from 6 biological replicates. NS: not significant *P<0.05; **P<0.01; Wilcoxon - Mann Whitney). As R-Loops unwind the DNA, it was possible that the abundance of R-Loops formed at Pol III-transcribed genes (Figure 3) could contribute to this topological stress. To test this possibility, we monitored by ChIP the localization of Top2 upon over-expression of RnhA. Surprisingly, the localization of Top2 was not altered at Pol III-transcribed genes upon over-expression of RnhA, whilst it was reduced at the Pol I-transcribed 18S (Figure S11). This suggested that the impact of R-Loop formation on the surrounding chromatin depends on where in the genome R-Loops form.
Topological stress contributes to the loading of condensin at Pol III-transcribed genes in the absence of Swd2.2 and Sen1
Based on these results, we envisaged two possible models to explain the increased localization of condensin at Pol III-transcribed genes in the absence of Swd2.2 and Sen1: either the accumulation of Top1 and/or Top2 helps to recruit and/or stabilize condensin, or topological stress facilitates the association of condensin at Pol III-transcribed genes. We previously identified the deletion of Top1 (top1Δ) as a suppressor of cut3-477 [22], suggesting that the accumulation of Top1 that results from lack of Swd2.2 and Sen1 is unlikely to facilitate the association of condensin with chromatin. Figures 5A&B show that the triple deletion swd2.2Δsen1Δtop1Δ was a better suppressor of cut3-477 than the double deletion swd2.2Δsen1Δ. This genetic evidence suggested that failure to monitor topological stress in top1Δ cells might facilitate the association/function of condensin. In support of this, ChIP analysis showed that there was a small but significant increase in the association of condensin at most Pol III-transcribed genes in cells deleted for Swd2.2, Sen1 and Top1 (swd2.2Δsen1Δtop1Δ cells) (Figure 5C). Taken together, these data support the following model: the absence of Swd2.2 and Sen1 increases the transcriptional activity at Pol III-transcribed genes and this might contribute to enhance local topological constraints. These constraints, either directly or indirectly, contribute to recruit or maintain condensin at Pol III-transcribed genes (Figure 5D).
Fig. 5. Lack of Top1 further increases the association of condensin with Pol III-transcribed genes when Swd2.2 and Sen1 are missing. 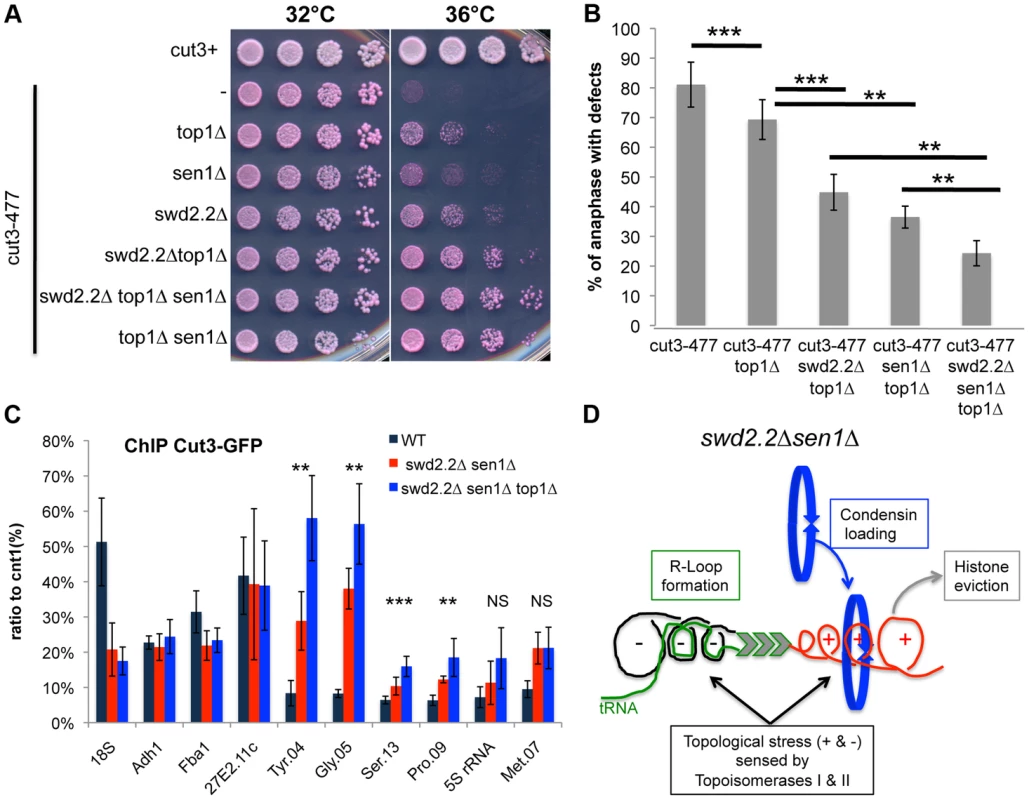
A. Serial dilutions of the indicated strains were plated on rich media at the indicated temperatures. B. Chromosome segregation in anaphase was monitored in the indicated strains after growing cells for one generation at 34°C. For each genotype, a minimum of 6 independent experiments was performed in which a minimum of 100 anaphase cells were scored (***<0.001; **<0.01 Wilcoxon - Mann Whitney). Anaphases were scored as defective when chromatin was detected lagging between the two main DNA masses C. ChIP qPCR of the indicated strains grown in cycling conditions at the indicated loci (mean ± standard deviation from 6 biological replicates. NS: not significant *P<0.05; **P<0.01; ***P<0.001 Wilcoxon - Mann Whitney). D. Model. Lack of Swd2.2 and Sen1 increases gene transcription at Pol III-transcribed genes. According to the twin supercoiled domain model, this results in more positive supercoils downstream of the polymerase and compensatory negative supercoils upstream of the polymerase. Negative supercoils favor the formation of R-Loops (reviewed in [41]). Positive supercoils result in nucleosome eviction. This topological stress also facilitates the recruitment of condensin, either directly or indirectly. Topological stress is not sufficient to recruit condensin
To establish whether topological stress was sufficient to stimulate the association of condensin with chromatin, we monitored the association of condensin in the temperature-sensitive Top2 mutant top2-191 ([33]) at the semi-restrictive temperature of 28°C. This analysis showed that the association of condensin was not significantly disrupted in these conditions (Figure S12A). Similarly, lack of Top1 on its own did not significantly impact the association of condensin (Figure S12B). Taken together, these observations suggest that topological stress on its own is not sufficient to stimulate the association of condensin with chromatin.
Discussion
Topological stress facilitates the recruitment of condensin at Pol III-transcribed genes in the absence of Swd2.2 and Sen1
In order to explain that condensin localizes to highly expressed genes from pro - to eukaryotes, whatever the RNA polymerase involved, we first hypothesized that a transcription by-product could facilitate the association of condensin with chromatin (see Introduction). We speculated that this mechanism could represent the ancestral way of recruiting condensin to chromatin. Complementary cis-acting factors would then have evolved to stabilize the interaction of condensin with specific loci, as shown previously (reviewed in [4]).
In this study we specifically considered two transcription by-products as potential condensin-attracting features: R-Loop formation and transcription-associated topological stress. Both features have been described both in pro - and eukaryotes and they generate structures (single-stranded DNA and positive supercoiling) for which condensin has been shown to display high affinity in vitro. Our data are not consistent with the idea that stable R-Loops could be involved in recruiting condensin. Similarly, topological stress on its own was not sufficient to disrupt the localization pattern of condensin. However, our data show that topological stress facilitated the association of condensin at Pol III-transcribed genes when Swd2.2 and Sen1 were missing. These observations are consistent with the recent demonstration that supercoiling at highly expressed genes contributes to the establishment of topological domains and small-range chromosome compaction in Caulaboacter crescentus [34].
How could topological stress create a better binding site for condensin at Pol III-transcribed genes in the absence of Swd2.2 and Sen1? First, condensin might simply have a higher affinity for supercoiled chromatin, as suggested by the observation that condensin associates preferentially in vitro with positively supercoiled plasmids [21]. Alternatively, or in addition, topological stress might work by facilitating nucleosome eviction [19]. Consistent with the latter, budding yeast condensin associates preferentially with nucleosome-free regions, especially at Pol III-transcribed genes [12]. To explain that lack of Top1 only facilitates the association of condensin at Pol III-transcribed genes when Swd2.2 and Sen1 are missing, we speculate that the level of topological stress has to go over a certain threshold in order to attract/stabilize condensin. This threshold would be reached in the chromatin around Pol III-transcribed genes when Swd2.2 and Sen1 are missing but not when Top1 only is missing.
Two reliable tools to map R-Loop forming regions in fission yeast
The biology of R-Loops is a rapidly expanding field of investigation, and many observations now demonstrate that R-Loops control genome stability and gene expression in multiple ways (reviewed in [35]). It is therefore essential to establish reliable methods to map R-Loop forming regions in genetically tractable organisms such as yeast to address the many functions of R-Loops in vivo. We presented evidence that the commonly used S9.6 ChIP method to map R-Loop forming regions in yeast is challenged by the fact that R-Loops, or at least their recognition by the S9.6 antibody, are partly sensitive to formaldehyde cross-linking and sonication. To circumvent this problem, we have developed two reliable alternatives to map R-Loop forming regions in fission yeast. Both of our methods concur to demonstrate that RNA-Pol III transcribed genes are major R-Loop forming regions in fission yeast. R-Loops have also been detected at Pol III-transcribed genes in budding yeast ([28]), suggesting that R-Loop formation is a conserved feature of Pol III transcription, at least in yeast.
We would like to argue that the two methods we have set up are complementary: not only do they map R-Loop forming regions but their use in parallel can also give information regarding the stability of R-Loops formed at different loci. Our data show that RNase H1 is most abundant at Pol III-transcribed genes throughout the cell-cycle, suggesting that R-Loops are constantly formed and detected by RNase H1 there. Our data also show that over-expression of RnhA in vivo counter-acts R-Loop formation more efficiently at Pol III-transcribed genes than within the rDNA for example (18S, Figure 3B). On the contrary, DRIP only yields significant signals at Pol III-transcribed genes when RNase H1 and RNase H2 are missing (rnh1Δrnh201Δ cells), whilst the DRIP signals at the rDNA (18S) are significant in wild-type cells, when RNase H1 and RNase H2 are fully active. At Pol III-transcribed genes, DRIP signals increase 10-20 fold in rnh1Δrnh201Δ cells, whilst they only increase ∼3-fold at the rDNA (18S). Our interpretation of these data is that R-Loops formed at 18S are stable and a relatively poor substrate for RNase H1, whilst R-Loops formed at Pol III-transcribed genes are unstable and a good substrate for RNase H1. A corollary to these observations is that DRIP is probably better suited to detect long-lived, stable R-Loops. This might explain why DRIP did not detect significant R-Loop formation at Pol III-transcribed genes in human cells ([16], [36]). We conclude that using both R-Loop mapping methods in parallel could provide indications of the relative stability of R-Loops at different loci.
The reasons why R-Loops formed at Pol III-transcribed genes are labile are still unclear but we speculate that R-Loops formed at Pol III-transcribed genes might be smaller than those formed at the 18S because the Pol III transcription units are much smaller. Further studies will be required to understand the consequences of R-Loop formation at Pol III-transcribed genes and how the half-life of an R-Loop might influence its function.
R-Loop-mediated chromosome compaction versus condensin-mediated chromosome condensation
R-Loop formation has been shown to be associated with increased phosphorylation of histone H3 on Serine 10 and reduced chromatin accessibility [14]. In turn, the phosphorylation of histone H3 on Serine 10 facilitates the interaction between adjacent nucleosomes, thereby promoting chromatin compaction [37]. We showed previously that to constitutively increase the levels of histone H3 phosphorylated on Serine 10 by deleting PP1 phosphatase (dis2Δ) was not sufficient to significantly improve chromosome segregation when condensin was deficient [22], suggesting that H3-S10-mediated chromatin compaction cannot compensate for the deficiency of condensin. Here we presented evidence that stable R-Loops do not significantly contribute to the recruitment of condensin. Taken together, these observations concur to establish that R-Loop-mediated chromatin compaction is distinct from condensin-mediated chromosome condensation. Our data also suggest that the action of condensin is more fundamental to building a mitotic chromosome than R-Loop-mediated chromatin compaction.
RNA processing factors and genome stability
Our data have highlighted unexpected ways by which proteins involved in the metabolism of RNA can affect chromosome segregation and genome integrity. Published data demonstrated conclusively that mutations in such factors in general and in Sen1 in particular resulted in chromosome instability (CIN) in yeast, in a mechanism involving R-Loop formation antagonizing replication fork progression ([38], [39] and reviewed in [35]). Here on the contrary, our data show that deletions of two such factors, Swd2.2 and Sen1, facilitate the segregation and stability of chromosomes when condensin is deficient, in a mechanism that does not require stable R-Loop formation.
In addition, our data show that Swd2.2 and Sen1 keep topological stress under control at Pol III-transcribed genes. We speculate that the enhanced transcription at Pol III-transcription associated with lack of Swd2.2 and Sen1 could contribute to such stress. However, we cannot exclude the possibility that RNA Pol III-dependent transcription is also defective in other ways that could explain the accumulation of topological stress when Swd2.2 and Sen1 are missing. The answer to this question will require further studies.
Sen1 antagonizes RNA Pol III-dependent transcription in fission yeast
Beautiful in vitro approaches demonstrated unequivocally that budding yeast Sen1 contributes to transcription termination of some RNA Pol II transcripts ([26]). It is not yet known whether fission yeast Sen1 has the same function. As fission yeast Sen1 is not essential for viability whilst its budding yeast counterpart is, it is possible that the function of Sen1 has diverged in fission yeast. This idea is supported by our data showing that RNA Pol III is likely to be the most stable binding partner of Sen1 in fission yeast and that Sen1 antagonizes Pol III-dependent transcription. On the contrary, a recent study aimed at identifying the binding partners of RNA Pol III in budding yeast did not identify Sen1, suggesting that the interaction between Sen1 and RNA Pol III is not as stable and/or abundant in budding yeast [40]. Further work is required to understand the function of fission yeast Sen1 at Pol III-transcribed genes.
Conclusion
Previous studies had concluded that the inhibition of RNA Pol I or RNA Pol II in mitosis was a pre-requisite for the binding of condensin at repetitive sequences [9],[10], suggesting that a processive RNA polymerase is a hindrance to the binding of condensin on chromatin. Here we challenge this idea by showing that an enhanced recruitment of condensin at Pol III-transcribed genes is associated with an increase in the expression of the same genes. These data show that, at least at Pol III-transcribed genes, an active polymerase is not an obstacle for the binding of condensin.
Materials and Methods
Fission yeast strains
A complete list of all of the strains used in this study is given in Table S3. Standard genetic crosses were employed to construct all strains. Rnh1-GFP, Sen1-GFP, and Top1-3flag were generated using a standard PCR procedure. To obtain Rnh1D129N, Rnh1 was PCR amplified and cloned into pCRII (Life technologies). Site-directed mutagenesis was then used to mutate the residue D129 into N (GAC to AAC) using Quickchange protocols (Stratagene). Overlapping PCR was used to add a C-terminus GFP tag and a cassette of resistance to kanamycin (KanR) to the mutagenized Rnh1 in order to integrate the mutagenized Rnh1 at the endogenous Rnh1 locus. After yeast transformation, proper integrants were selected by PCR and western blot and were sequenced to verify the presence of the mutation. The plasmid over-expressing RnhA tagged with 1xFLAG at its N-terminus was obtained from Eun Shik Choi and Robin Allshire (WTCCB, Edinburgh, UK). In order to stably integrate the plasmid in the genome, it was linearized by digestion with MluI and then transformed in to yeast according to standard procedures.
Chromatin immunoprecipitation
1,5.108 cells were treated with 1% formaldehyde (Sigma) at 17°C for 30′. After extensive washes with cold PBS, cells were frozen in liquid Nitrogen. Frozen cells were then broken open using a RETSCH MM400 Mill and then resuspended in cold lysis buffer (Hepes-KOH 50 mM pH 7,5, NaCl 140 mM, EDTA 1 mM, Triton 1%, Na-deoxycholate 0,1%, PMSF 1 mM). The lysats were then sonicated at 4°C using a Diagenode sonicator. Immuno-precipitation was done overnight at 4°C using Protein A-coupled Dynabeads previously incubated with the anti-GFP A11122 antibody (Invitrogen) or using Protein G-coupled Dynabeads previously incubated with the anti-myc 9E10 antibody (Sigma) according to the manufacturer's instructions. Beads were washed successively with (5′ incubation on rotating wheel): Wash I buffer (20 mM Tris pH 8, 150 mM NaCl, 2 mM EDTA, 1% Triton-X100, 0,1% SDS), Wash II buffer (20 mM Tris pH 8, 500 mM NaCl, 2 mM EDTA, 1% Triton-X100, 0,1% SDS) and Wash III buffer (20 mM Tris pH 8, 1 mM EDTA, 0,5% Na-deoxycholate, 1% Igepal, 250 mM LiCl). After two additional washes in TE pH 8, the beads were resuspended in 10% Chelex resin (Biorad) and incubated at 98°C for 10′. After addition of 2 µL of 10 mg/mL of proteinase K, the mixture was incubated at 43°C for 1 hour, then for another 10 mn at 98°C. After centrifugation, the supernatant was collected and analyzed by qPCR.
DRIP
8.108 cells were frozen in liquid nitrogen, broken open using a RETSCH MM400 Mill and then resuspended in cold lysis buffer (Hepes-KOH 50 mM pH 7,5, NaCl 140 mM, EDTA 1 mM, Triton 1%, Na-deoxycholate 0,1%). After phenol/chloroform purification and ethanol precipitation, the DNA was resuspended in TE pH 8 and split into two samples. Both samples were digested with BsrGI, EcoRI, HindIII, SspI and XbaI according to the manufacturer's instructions and RNase H was added to one of the two samples. After digestion, each sample was divided into two and incubated overnight at 4°C in IP buffer (100 mM MES pH 6,6, NaCl 500 mM, 0,05% Triton, 2 mg/mL BSA) in the presence of either Protein A-coupled Dynabeads or Protein A-coupled Dynabeads previously incubated with the S9.6 antibody according to the manufacturer's instructions. The beads were then washed three times in IP buffer. After two additional washes in TE pH 8, the beads were resuspended in 10% Chelex resin (Biorad) and incubated at 98°C for 5′. After addition of 2 µL of 10 mg/mL of proteinase K, the mixture was incubated at 43°C for 30′, then for another 5′ at 98°C. After centrifugation, the supernatant was collected and analyzed by qPCR.
Immunoprecipitation
Immunoprecipitation was carried out as described previously [22], except that cells were broken open using a RETSCH MM400 Mill. To purify Sen1-associated proteins (Table S2), a protein extract was prepared from 109 cells expressing GFP-tagged Sen1 from the endogenous locus. After immuno-precipitation with 15 µL of magnetic beads, the beads were washed three times with 1 mL of lysis buffer and twice with 1 mL of PBS containing 0,02% Tween. The beads samples were then subjected to in-solution reduction, carbamidomethylation and tryptic digestion. After acidification with 10%Trifluoroacetic Acid the samples were centrifuged 3 times to eliminate the beads.
Mass-spectrometry analysis
Peptide sequences were determined by mass spectrometry performed using a LTQ Velos instrument (Dual Pressure Linear Ion Trap) equipped with a nanospray source (Thermo Fisher Scientific) and coupled to a U3000 nanoLC system (Thermo Fisher Scientific). A MS survey scan was acquired over the m/z range 400–1600 in Enhanced resolution mode. The MS/MS scans were acquired in Normal resolution mode over the m/z range 65–2000 for the 20 most intense MS ions with a charge of 2 or more and with a collision energy set to 35eV. The spectra were recorded using dynamic exclusion of previously analyzed ions for 0.5 min with 50 millimass units (mmu) of mass tolerance. The peptide separation was obtained on a C18 PepMap micro-precolumn (5 µm; 100 Å; 300 µm×5 mm; Dionex) and a C18 PepMap nanocolumn (3 µm; 100 Å; 75 µm×200 mm; Dionex) using a linear 90 min gradient from 0 to 40%, where solvent A was 0.1% HCOOH in H2O/CH3CN (95/5) and solvent B was 0.1% HCOOH in H2O/CH3CN (20/80) at 300 nL/min flow rate.
Protein identification was performed using the MASCOT Algorithm from the Proteome Discoverer software v1.1 (Thermo Fisher Scientific) against the UniProtKB database reduced to Schizosaccharomyces pombe species [UniProt release 2013_12].
RNA extraction and RT-qPCR
These were performed as previously described [22].
Supporting Information
Zdroje
1. HiranoT, MitchisonTJ (1994) A heterodimeric coiled-coil protein required for mitotic chromosome condensation in vitro. Cell 79 : 449–458.
2. KimuraK, HiranoT (1997) ATP-dependent positive supercoiling of DNA by 13S condensin: a biochemical implication for chromosome condensation. Cell 90 : 625–634.
3. St-PierreJ, DouziechM, BazileF, PascariuM, BonneilE, et al. (2009) Polo kinase regulates mitotic chromosome condensation by hyperactivation of condensin DNA supercoiling activity. Mol Cell 34 : 416–426.
4. PiazzaI, HaeringCH, RutkowskaA (2013) Condensin: crafting the chromosome landscape. Chromosoma 122 : 175–190.
5. D'AmbrosioC, SchmidtCK, KatouY, KellyG, ItohT, et al. (2008) Identification of cis-acting sites for condensin loading onto budding yeast chromosomes. Genes Dev 22 : 2215–2227.
6. KimJH, ZhangT, WongNC, DavidsonN, MaksimovicJ, et al. (2013) Condensin I associates with structural and gene regulatory regions in vertebrate chromosomes. Nat Commun 4 : 2537.
7. GruberS, ErringtonJ (2009) Recruitment of condensin to replication origin regions by ParB/SpoOJ promotes chromosome segregation in B. subtilis. Cell 137 : 685–696.
8. KranzAL, JiaoCY, WinterkornLH, AlbrittonSE, KramerM, et al. (2013) Genome-wide analysis of condensin binding in Caenorhabditis elegans. Genome Biol 14: R112.
9. Clemente-BlancoA, SenN, Mayan-SantosM, SacristanMP, GrahamB, et al. (2011) Cdc14 phosphatase promotes segregation of telomeres through repression of RNA polymerase II transcription. Nat Cell Biol 13 : 1450–1456.
10. Clemente-BlancoA, Mayan-SantosM, SchneiderDA, MachinF, JarmuzA, et al. (2009) Cdc14 inhibits transcription by RNA polymerase I during anaphase. Nature 458 : 219–222.
11. AguileraA, Garcia-MuseT (2012) R loops: from transcription byproducts to threats to genome stability. Mol Cell 46 : 115–124.
12. PiazzaI, RutkowskaA, OriA, WalczakM, MetzJ, et al. (2014) Association of condensin with chromosomes depends on DNA binding by its HEAT-repeat subunits. Nat Struct Mol Biol 21 : 560–568.
13. GrieseJJ, WitteG, HopfnerKP (2010) Structure and DNA binding activity of the mouse condensin hinge domain highlight common and diverse features of SMC proteins. Nucleic Acids Res 38 : 3454–3465.
14. Castellano-PozoM, Santos-PereiraJM, RondonAG, BarrosoS, AndujarE, et al. (2013) R loops are linked to histone h3 s10 phosphorylation and chromatin condensation. Mol Cell 52 : 583–590.
15. AkaiY, KurokawaY, NakazawaN, Tonami-MurakamiY, SuzukiY, et al. (2011) Opposing role of condensin hinge against replication protein A in mitosis and interphase through promoting DNA annealing. Open Biol 1 : 110023.
16. GinnoPA, LottPL, ChristensenHC, KorfI, ChedinF (2012) R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol Cell 45 : 814–825.
17. TsaoYP, WuHY, LiuLF (1989) Transcription-driven supercoiling of DNA: direct biochemical evidence from in vitro studies. Cell 56 : 111–118.
18. NaughtonC, AvlonitisN, CorlessS, PrendergastJG, MatiIK, et al. (2013) Transcription forms and remodels supercoiling domains unfolding large-scale chromatin structures. Nat Struct Mol Biol 20 : 387–395.
19. TevesSS, HenikoffS (2014) Transcription-generated torsional stress destabilizes nucleosomes. Nat Struct Mol Biol 21 : 88–94.
20. KouzineF, GuptaA, BaranelloL, WojtowiczD, Ben-AissaK, et al. (2013) Transcription-dependent dynamic supercoiling is a short-range genomic force. Nat Struct Mol Biol 20 : 396–403.
21. KimuraK, RybenkovVV, CrisonaNJ, HiranoT, CozzarelliNR (1999) 13S condensin actively reconfigures DNA by introducing global positive writhe: implications for chromosome condensation. Cell 98 : 239–248.
22. VanoosthuyseV, LegrosP, van der SarSJA, YvertG, TodaK, et al. (2014) CPF-associated phosphatase activity opposes condensin-mediated chromosome condensation. PLoS Genet 10: e1004415.
23. SakaY, SutaniT, YamashitaY, SaitohS, TakeuchiM, et al. (1994) Fission yeast cut3 and cut14, members of a ubiquitous protein family, are required for chromosome condensation and segregation in mitosis. EMBO J 13 : 4938–4952.
24. RichardP, ManleyJL (2009) Transcription termination by nuclear RNA polymerases. Genes Dev 23 : 1247–1269.
25. KimHD, ChoeJ, SeoYS (1999) The sen1(+) gene of Schizosaccharomyces pombe, a homologue of budding yeast SEN1, encodes an RNA and DNA helicase. Biochemistry 38 : 14697–14710.
26. PorruaO, LibriD (2013) A bacterial-like mechanism for transcription termination by the Sen1p helicase in budding yeast. Nat Struct Mol Biol 20 : 884–891.
27. SchrammL, HernandezN (2002) Recruitment of RNA polymerase III to its target promoters. Genes Dev 16 : 2593–2620.
28. ChanYA, AristizabalMJ, LuPY, LuoZ, HamzaA, et al. (2014) Genome-Wide Profiling of Yeast DNA:RNA Hybrid Prone Sites with DRIP-Chip. PLoS Genet 10: e1004288.
29. WuH, LimaWF, CrookeST (2001) Investigating the structure of human RNase H1 by site-directed mutagenesis. J Biol Chem 276 : 23547–23553.
30. El HageA, FrenchSL, BeyerAL, TollerveyD (2010) Loss of Topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes Dev 24 : 1546–1558.
31. PhillipsDD, GarbocziDN, SinghK, HuZ, LepplaSH, et al. (2013) The sub-nanomolar binding of DNA-RNA hybrids by the single-chain Fv fragment of antibody S9.6. J Mol Recognit 26 : 376–381.
32. LemieuxC, BachandF (2009) Cotranscriptional recruitment of the nuclear poly(A)-binding protein Pab2 to nascent transcripts and association with translating mRNPs. Nucleic Acids Res 37 : 3418–3430.
33. UemuraT, OhkuraH, AdachiY, MorinoK, ShiozakiK, et al. (1987) DNA topoisomerase II is required for condensation and separation of mitotic chromosomes in S. pombe. Cell 50 : 917–925.
34. LeTB, ImakaevMV, MirnyLA, LaubMT (2013) High-resolution mapping of the spatial organization of a bacterial chromosome. Science 342 : 731–734.
35. Hamperl S, Cimprich KA (2014) The contribution of co-transcriptional RNA:DNA hybrid structures to DNA damage and genome instability. DNA Repair (Amst).
36. GinnoPA, LimYW, LottPL, KorfI, ChedinF (2013) GC skew at the 5′ and 3′ ends of human genes links R-loop formation to epigenetic regulation and transcription termination. Genome Res 23 : 1590–1600.
37. WilkinsBJ, RallNA, OstwalY, KruitwagenT, Hiragami-HamadaK, et al. (2014) A cascade of histone modifications induces chromatin condensation in mitosis. Science 343 : 77–80.
38. StirlingPC, ChanYA, MinakerSW, AristizabalMJ, BarrettI, et al. (2012) R-loop-mediated genome instability in mRNA cleavage and polyadenylation mutants. Genes Dev 26 : 163–175.
39. MischoHE, Gomez-GonzalezB, GrzechnikP, RondonAG, WeiW, et al. (2011) Yeast Sen1 helicase protects the genome from transcription-associated instability. Mol Cell 41 : 21–32.
40. Nguyen NT, Saguez C, Conesa C, Lefebvre O, Acker J (2014) Identification of proteins associated with RNA polymerase III using a modified tandem chromatin affinity purification. Gene. E-pub ahead of print. doi:10.1016/j.gene.2014.07.070
41. DroletM (2006) Growth inhibition mediated by excess negative supercoiling: the interplay between transcription elongation, R-loop formation and DNA topology. Mol Microbiol 59 : 723–730.
Štítky
Genetika Reprodukční medicína
Článek The COP9 Signalosome Converts Temporal Hormone Signaling to Spatial Restriction on Neural CompetenceČlánek Coordinate Regulation of Stem Cell Competition by Slit-Robo and JAK-STAT Signaling in the TestisČlánek The CSN/COP9 Signalosome Regulates Synaptonemal Complex Assembly during Meiotic Prophase I ofČlánek GPA: A Statistical Approach to Prioritizing GWAS Results by Integrating Pleiotropy and AnnotationČlánek Functional Diversity of Carbohydrate-Active Enzymes Enabling a Bacterium to Ferment Plant BiomassČlánek Heat-Induced Release of Epigenetic Silencing Reveals the Concealed Role of an Imprinted Plant GeneČlánek p53- and ERK7-Dependent Ribosome Surveillance Response Regulates Insulin-Like Peptide SecretionČlánek The Complex I Subunit Selectively Rescues Mutants through a Mechanism Independent of MitophagyČlánek Rad59-Facilitated Acquisition of Y′ Elements by Short Telomeres Delays the Onset of SenescenceČlánek ARTIST: High-Resolution Genome-Wide Assessment of Fitness Using Transposon-Insertion Sequencing
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 11
-
Všechny články tohoto čísla
- Establishing a Multidisciplinary Context for Modeling 3D Facial Shape from DNA
- RNA Processing Factors Swd2.2 and Sen1 Antagonize RNA Pol III-Dependent Transcription and the Localization of Condensin at Pol III Genes
- Inversion of the Chromosomal Region between Two Mating Type Loci Switches the Mating Type in
- A Thermolabile Aldolase A Mutant Causes Fever-Induced Recurrent Rhabdomyolysis without Hemolytic Anemia
- The Role of Regulatory Evolution in Maize Domestication
- Stress Granule-Defective Mutants Deregulate Stress Responsive Transcripts
- 24-Hour Rhythms of DNA Methylation and Their Relation with Rhythms of RNA Expression in the Human Dorsolateral Prefrontal Cortex
- Pseudoautosomal Region 1 Length Polymorphism in the Human Population
- Fungal Communication Requires the MAK-2 Pathway Elements STE-20 and RAS-2, the NRC-1 Adapter STE-50 and the MAP Kinase Scaffold HAM-5
- The COP9 Signalosome Converts Temporal Hormone Signaling to Spatial Restriction on Neural Competence
- The Protein -glucosyltransferase Rumi Modifies Eyes Shut to Promote Rhabdomere Separation in
- The Talin Head Domain Reinforces Integrin-Mediated Adhesion by Promoting Adhesion Complex Stability and Clustering
- Quantitative Genetics of CTCF Binding Reveal Local Sequence Effects and Different Modes of X-Chromosome Association
- Coordinate Regulation of Stem Cell Competition by Slit-Robo and JAK-STAT Signaling in the Testis
- Genetic Analysis of a Novel Tubulin Mutation That Redirects Synaptic Vesicle Targeting and Causes Neurite Degeneration in
- A Systems Genetics Approach Identifies , , and as Novel Aggressive Prostate Cancer Susceptibility Genes
- Three RNA Binding Proteins Form a Complex to Promote Differentiation of Germline Stem Cell Lineage in
- Approximation to the Distribution of Fitness Effects across Functional Categories in Human Segregating Polymorphisms
- The CSN/COP9 Signalosome Regulates Synaptonemal Complex Assembly during Meiotic Prophase I of
- SAS-1 Is a C2 Domain Protein Critical for Centriole Integrity in
- An RNA-Seq Screen of the Antenna Identifies a Transporter Necessary for Ammonia Detection
- GPA: A Statistical Approach to Prioritizing GWAS Results by Integrating Pleiotropy and Annotation
- Let's Face It—Complex Traits Are Just Not That Simple
- Glutamate Receptor Gene , Coffee, and Parkinson Disease
- The Red Queen Model of Recombination Hotspots Evolution in the Light of Archaic and Modern Human Genomes
- The Ethics of Our Inquiry: An Interview with Hank Greely
- Functional Diversity of Carbohydrate-Active Enzymes Enabling a Bacterium to Ferment Plant Biomass
- Regularized Machine Learning in the Genetic Prediction of Complex Traits
- Phylogenetically Driven Sequencing of Extremely Halophilic Archaea Reveals Strategies for Static and Dynamic Osmo-response
- Lack of Replication of the -by-Coffee Interaction in Parkinson Disease
- Natural Polymorphisms in Human APOBEC3H and HIV-1 Vif Combine in Primary T Lymphocytes to Affect Viral G-to-A Mutation Levels and Infectivity
- A Germline Polymorphism of Thymine DNA Glycosylase Induces Genomic Instability and Cellular Transformation
- Heat-Induced Release of Epigenetic Silencing Reveals the Concealed Role of an Imprinted Plant Gene
- ATPase-Independent Type-III Protein Secretion in
- p53- and ERK7-Dependent Ribosome Surveillance Response Regulates Insulin-Like Peptide Secretion
- The Complex I Subunit Selectively Rescues Mutants through a Mechanism Independent of Mitophagy
- Evolution of DNA Methylation Patterns in the Brassicaceae is Driven by Differences in Genome Organization
- Regulation of mRNA Abundance by Polypyrimidine Tract-Binding Protein-Controlled Alternate 5′ Splice Site Choice
- Systematic Comparison of the Effects of Alpha-synuclein Mutations on Its Oligomerization and Aggregation
- Rad59-Facilitated Acquisition of Y′ Elements by Short Telomeres Delays the Onset of Senescence
- A Functional Portrait of Med7 and the Mediator Complex in
- Systematic Analysis of the Role of RNA-Binding Proteins in the Regulation of RNA Stability
- ARTIST: High-Resolution Genome-Wide Assessment of Fitness Using Transposon-Insertion Sequencing
- Genomic Evidence of Rapid and Stable Adaptive Oscillations over Seasonal Time Scales in Drosophila
- Genome-Wide Associations between Genetic and Epigenetic Variation Influence mRNA Expression and Insulin Secretion in Human Pancreatic Islets
- HAM-5 Functions As a MAP Kinase Scaffold during Cell Fusion in
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- An RNA-Seq Screen of the Antenna Identifies a Transporter Necessary for Ammonia Detection
- Systematic Comparison of the Effects of Alpha-synuclein Mutations on Its Oligomerization and Aggregation
- Functional Diversity of Carbohydrate-Active Enzymes Enabling a Bacterium to Ferment Plant Biomass
- Regularized Machine Learning in the Genetic Prediction of Complex Traits
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



