-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaLet's Face It—Complex Traits Are Just Not That Simple
article has not abstract
Published in the journal: . PLoS Genet 10(11): e32767. doi:10.1371/journal.pgen.1004724
Category: Formal Comment
doi: https://doi.org/10.1371/journal.pgen.1004724Summary
article has not abstract
The idea that we can reconstruct a human face from a DNA sample has great appeal: DNA from a crime scene could be used to create a facial image of a suspect; the faces of prehistoric peoples could be reconstructed from their remains; the face of a child could be predicted in utero from amniocentesis. This is the promise implicit in the study of Claes et al. [1]. In their own words:
“…our methods provide the means of identifying the genes that affect facial shape and for modeling the effects of these genes to generate a predicted face.” (pg. 10)
Unfortunately, this promise greatly overreaches the data and analyses represented by the study, and it misrepresents our current understanding of the genetics of complex morphological traits. Worse, this claim, and the fairly sensational media reports that have stemmed from it, detract significantly from what is otherwise an important paper that highlights the potential of an interesting new technique to investigate the genetic basis for variation in the shape of the human face.
Claes et al. [1] base their analysis on a mixed ancestry sample of 592 people genotyped with a 540,000 single nucleotide polymorphism (SNP) array. The primary analyses conducted deal with the effects of sex and ancestry on facial shape. Sexual dimorphism (when taking ancestry into account) explains 13% of the total shape variation in the face, while ancestry (when taking sex into account) explains 10%. These results are interesting, particularly as the contribution of ancestry is unexpectedly high. This surprising result has tremendous implications for the microevolution of facial shape and, potentially, the role of sexual selection. Could the amount of facial shape variation attributed to ancestry have accumulated through genetic drift? If not, what might explain the surprising contribution of ancestry to facial shape? These questions are intriguing and important regardless of the extent facial shape, or even facial shape characteristics, can be reliably predicted from DNA.
But why is the claim that facial shape can be predicted from DNA so troubling? The first reason is that it isn't actually supported by the work done in this study. Claes et al. [1] examine the effects of candidate genes for normal craniofacial variation. They select 46 genes based on evidence of accelerated evolution and evidence from animal models that these genes are expressed in the head. Here, they argue that, because the genes in question are already known to contribute to head development, no adjustment for multiple comparisons is necessary. This assumption is highly problematic. Lots of genes are involved in the development of the head, but that does not mean that those genes contribute to normal variation in the face. Large-scale genome wide association studies (GWAS) in humans often fail to demonstrate significant roles for genes that are known to produce relevant phenotypes in experimental models. In fact, it is also quite possible that many genes not known to play important roles in craniofacial development contribute to normal variation in the face. Many genes that have known developmental roles may be so important that variation in their function or expression is highly selected against. For example, Shh plays crucial roles in forming the face [2], and yet this gene does not appear in GWAS studies of human facial shape variation [3]. Alternatively, variation in a characteristic like facial shape may come from unexpected sources such as genetic variation in gap junction proteins that influence the ability of bone to respond to mechanical load during chewing [4]. Examination of Table S2 reveals that only one of the 46 candidate genes tested would have survived Bonferroni adjustment for multiple testing. Even if one accepts the rationale for avoiding adjustment for multiple testing, the authors could have compared the p values obtained for the 46 genes from random samples of SNPs drawn from the 55,000 tested. In the absence of such a test, the study contributes nothing new to our understanding of how genes influence the shape of the face since the genes tested may or may not actually contribute anything to normal variation in the shape of the face.
The second, and deeper, issue, though, is whether genomic prediction of complex morphologies is even feasible. Obviously, variation in genes causes variation at the phenotypic level. This does not mean, though, that a complex phenotype can be accurately predicted from genetic data. For a trait such as coronary heart disease (CHD), prediction of risk is highly problematic, even though quite a lot is known about the underlying genetics [5]. Much less is known about the genetics of complex morphological traits like the shape of the face. The problem is that the genotype–phenotype map for morphological traits is incredibly complicated (Figure 1). It is not just that variation in genes exhibit many relationships with phenotypic outcomes (Figure 1B). It is, rather, that phenotypic variation in morphological traits is structured by developmental processes at multiple levels and times in development. These processes and their interactions are complex but modular in their organization [6], [7]. This architecture of development is responsible for the modulation of phenotypic variance [8] and covariation structure [6], [7] (Figure 1C). Changes to developmental processes that influence the shape of the head tend to produce highly integrated and often unexpected effects on global shape [7], [9]–[11]. Even subtle effects, such as those produced by enhancers to craniofacial genes acting in spatiotemporally specific ways during development, produce global rather than localized shape transformations of the head [12].
Fig. 1. Two complementary depictions of the developmental architecture underlying the genotype–phenotype map for complex traits. 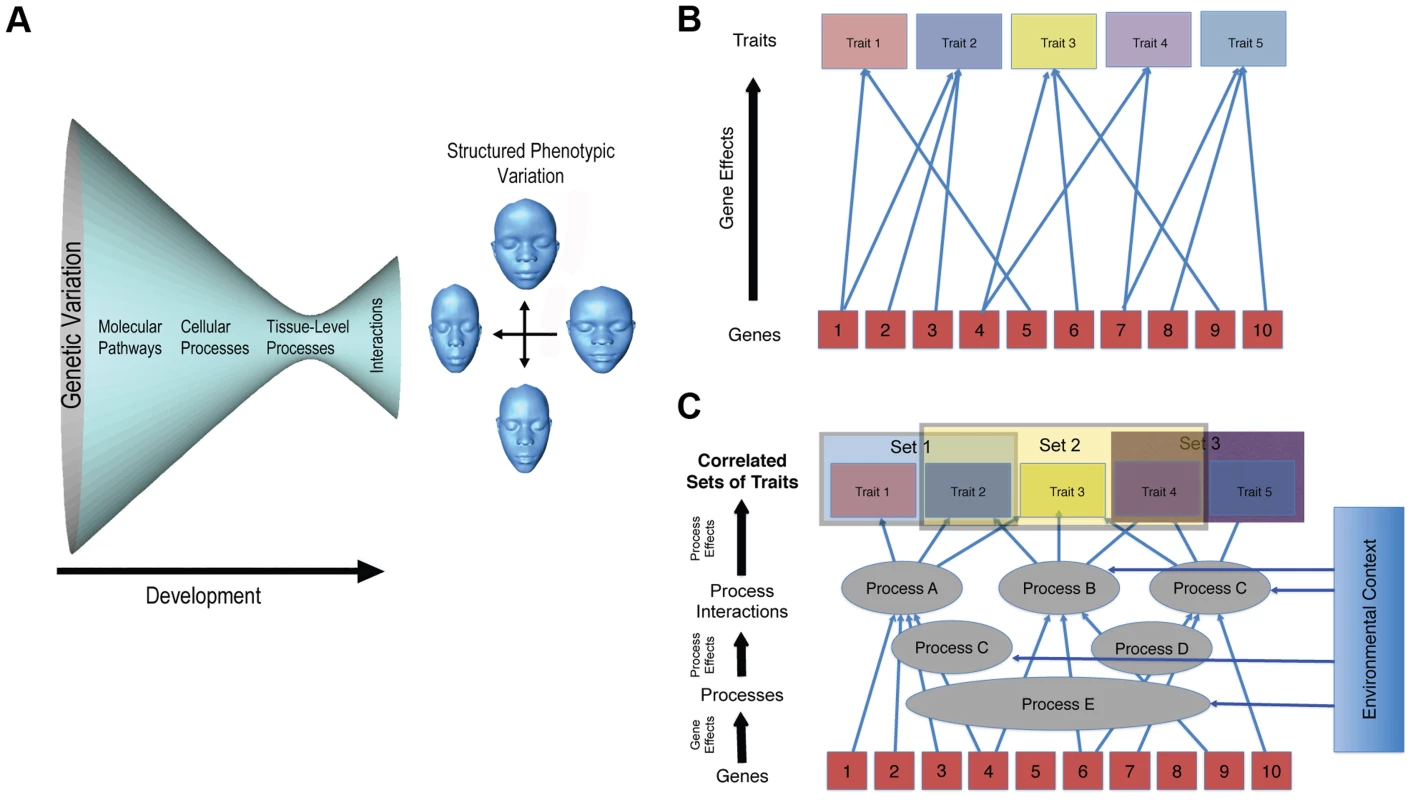
A captures the idea that large amounts of genetic variation funnel to smaller numbers of pathways and processes. These processes then interact to produce structured and modulated phenotypic variation in a complex trait. B, which derives from Wagner [20], shows the many-to-many mapping of genes to traits; while both Cs show the modular pattern of gene effects on processes and the effects of processes on sets of phenotypic traits. These depictions illustrate some of the complexity involved in constructing models of genotype–phenotype relations in complex traits. Complex patterns of interactions among developmental processes can also generate unexpected patterns of heritability. Although genetic variance may be predominantly additive for complex traits such as oil content in maize or body mass in mice [13], [14], this may not be the case for multidimensional and modularly organized morphological traits like facial shape [15], [16]. If a significant proportion of the genetic variance for facial shape is non-additive, which remains an empirical question, prediction of facial shape from genotype is greatly complicated. The genetic basis for facial shape variation may be as much, if not more, about epistatic interactions and context-specific developmental interactions than about the additive effects of individual genes. The fact that a large, recent GWAS study of facial shape revealed few causative loci is suggestive of a very complex genetic architecture for this trait [3].
The overselling of these results is unfortunate and unnecessary because it detracts from what is otherwise a very interesting study. The bootstrapped response-based imputation modeling (BRIM) technique, for example, is an intriguing extension of the shape analysis toolkit. Like other geometric morphometric methods, it is based on Procrustes superimposition and multivariate data reduction of variation in landmark position. The method allows for estimation of a single quantitative axis of variation that would correspond to a multidimensional factor such as ancestry, or a discrete variable such as sex. The method relies on machine learning algorithms to define shape axes that best correspond to such variables. As such, the method has great potential for furthering quantitative analyses of the genetics of complex morphologies. The use of dense landmark representation and machine learning algorithms similarly has potential in the analysis of complex morphologies, and this study points the way towards future applications of such techniques. This aspect of the paper would be stronger had they validated BRIM by comparison to existing methods. Estimation of the effects of single genes, ancestry, sex or any other factor of interest on total shape variation and local shape variation can already be done using the current GM toolkit [17] or dense correspondence analysis [18], [19]. Although, how much better BRIM performs than existing methods is hard to tell without validation, despite the seemingly encouraging results presented here.
Developing a mechanistic understanding of the genotype–phenotype map is undoubtedly one of the greatest challenges of modern biology. Claes et al. offer us a new and valuable tool to apply to this grand challenge. We should not be fooled, however, into thinking that we are anywhere close to understanding developmental genetics at the level where prediction of complex morphological traits is feasible. Overselling and overpromising in science is dangerous because it creates unreasonable expectations both at the public and policymaker levels. Ultimately, this runs the risk of diverting valuable scientific resources away from the important task of understanding how variation in genes plays through developmental processes to produce the amazing diversity of organismal form.
Zdroje
1. ClaesP, LibertonDK, DanielsK, RosanaKM, QuillenEE, et al. (2014) Modeling 3D Facial Shape from DNA. PLoS Genet 10: e1004224.
2. HuD, MarcucioRS (2009) A SHH-responsive signaling center in the forebrain regulates craniofacial morphogenesis via the facial ectoderm. Development 136 : 107–116.
3. LiuF, van der LijnF, SchurmannC, ZhuG, ChakravartyMM, et al. (2012) A genome-wide association study identifies five loci influencing facial morphology in Europeans. PLoS Genet 8: e1002932.
4. GrimstonSK, GoldbergDB, WatkinsM, BrodtMD, SilvaMJ, et al. (2011) Connexin43 deficiency reduces the sensitivity of cortical bone to the effects of muscle paralysis. J Bone Miner Res 26 : 2151–2160.
5. HumphriesSE, DrenosF, Ken-DrorG, TalmudPJ (2010) Coronary heart disease risk prediction in the era of genome-wide association studies: current status and what the future holds. Circulation 121 : 2235–2248.
6. WagnerGP, PavlicevM, CheverudJM (2007) The road to modularity. Nat Rev Genet 8 : 921–931.
7. HallgrimssonB, JamniczkyH, YoungNM, RolianC, ParsonsTE, et al. (2009) Deciphering the Palimpsest: Studying the Relationship Between Morphological Integration and Phenotypic Covariation. Evol Biol 36 : 355–376.
8. HermissonJ, WagnerGP (2004) The population genetic theory of hidden variation and genetic robustness. Genetics 168 : 2271–2284.
9. Martinez-AbadiasN, MitteroeckerP, ParsonsTE, EsparzaM, SjovoldT, et al. (2012) The Developmental Basis of Quantitative Craniofacial Variation in Humans and Mice. Evol Biol 39 : 554–567.
10. HeuzeY, SinghN, BasilicoC, JabsEW, HolmesG, et al. (2014) Morphological comparison of the craniofacial phenotypes of mouse models expressing the Apert FGFR2 S252W mutation in neural crest - or mesoderm-derived tissues. Bone 63 : 101–109 doi:10.1016/j.bone.2014.03.003
11. YoungNM, ChongHJ, HuD, HallgrimssonB, MarcucioRS (2010) Quantitative analyses link modulation of sonic hedgehog signaling to continuous variation in facial growth and shape. Development 137 : 3405–3409.
12. AttanasioC, NordAS, ZhuY, BlowMJ, LiZ, et al. (2013) Fine tuning of craniofacial morphology by distant-acting enhancers. Science 342 : 1241006.
13. HillWG, GoddardME, VisscherPM (2008) Data and theory point to mainly additive genetic variance for complex traits. PLoS Genet 4: e1000008.
14. YangJ, BenyaminB, McEvoyBP, GordonS, HendersAK, et al. (2010) Common SNPs explain a large proportion of the heritability for human height. Nat Genet 42 : 565–569.
15. WolfJB, PompD, EisenEJ, CheverudJM, LeamyLJ (2006) The contribution of epistatic pleiotropy to the genetic architecture of covariation among polygenic traits in mice. Evol Dev 8 : 468–476.
16. WolfJB, FrankinoWA, AgrawalAF, BrodieED3rd, MooreAJ (2001) Developmental interactions and the constituents of quantitative variation. Evolution Int J Org Evolution 55 : 232–245.
17. MitteroeckerP, BooksteinF (2011) Linear Discrimination, Ordination, and the Visualization of Selection Gradients in Modern Morphometrics. Evol Biol 38 : 100–114.
18. HammondP, HuttonTJ, AllansonJE, BuxtonB, CampbellLE, et al. (2005) Discriminating power of localized three-dimensional facial morphology. Am J Hum Genet 77 : 999–1010.
19. HammondP, HuttonTJ, AllansonJE, CampbellLE, HennekamRC, et al. (2004) 3D analysis of facial morphology. Am J Med Genet A 126A: 339–348.
20. WagnerGP (1996) Homologues, natural kinds and the evolution of modularity. Am Zool 36 : 36–43.
Štítky
Genetika Reprodukční medicína
Článek The COP9 Signalosome Converts Temporal Hormone Signaling to Spatial Restriction on Neural CompetenceČlánek Coordinate Regulation of Stem Cell Competition by Slit-Robo and JAK-STAT Signaling in the TestisČlánek The CSN/COP9 Signalosome Regulates Synaptonemal Complex Assembly during Meiotic Prophase I ofČlánek GPA: A Statistical Approach to Prioritizing GWAS Results by Integrating Pleiotropy and AnnotationČlánek Functional Diversity of Carbohydrate-Active Enzymes Enabling a Bacterium to Ferment Plant BiomassČlánek Heat-Induced Release of Epigenetic Silencing Reveals the Concealed Role of an Imprinted Plant GeneČlánek p53- and ERK7-Dependent Ribosome Surveillance Response Regulates Insulin-Like Peptide SecretionČlánek The Complex I Subunit Selectively Rescues Mutants through a Mechanism Independent of MitophagyČlánek Rad59-Facilitated Acquisition of Y′ Elements by Short Telomeres Delays the Onset of SenescenceČlánek ARTIST: High-Resolution Genome-Wide Assessment of Fitness Using Transposon-Insertion Sequencing
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 11
-
Všechny články tohoto čísla
- Establishing a Multidisciplinary Context for Modeling 3D Facial Shape from DNA
- RNA Processing Factors Swd2.2 and Sen1 Antagonize RNA Pol III-Dependent Transcription and the Localization of Condensin at Pol III Genes
- Inversion of the Chromosomal Region between Two Mating Type Loci Switches the Mating Type in
- A Thermolabile Aldolase A Mutant Causes Fever-Induced Recurrent Rhabdomyolysis without Hemolytic Anemia
- The Role of Regulatory Evolution in Maize Domestication
- Stress Granule-Defective Mutants Deregulate Stress Responsive Transcripts
- 24-Hour Rhythms of DNA Methylation and Their Relation with Rhythms of RNA Expression in the Human Dorsolateral Prefrontal Cortex
- Pseudoautosomal Region 1 Length Polymorphism in the Human Population
- Fungal Communication Requires the MAK-2 Pathway Elements STE-20 and RAS-2, the NRC-1 Adapter STE-50 and the MAP Kinase Scaffold HAM-5
- The COP9 Signalosome Converts Temporal Hormone Signaling to Spatial Restriction on Neural Competence
- The Protein -glucosyltransferase Rumi Modifies Eyes Shut to Promote Rhabdomere Separation in
- The Talin Head Domain Reinforces Integrin-Mediated Adhesion by Promoting Adhesion Complex Stability and Clustering
- Quantitative Genetics of CTCF Binding Reveal Local Sequence Effects and Different Modes of X-Chromosome Association
- Coordinate Regulation of Stem Cell Competition by Slit-Robo and JAK-STAT Signaling in the Testis
- Genetic Analysis of a Novel Tubulin Mutation That Redirects Synaptic Vesicle Targeting and Causes Neurite Degeneration in
- A Systems Genetics Approach Identifies , , and as Novel Aggressive Prostate Cancer Susceptibility Genes
- Three RNA Binding Proteins Form a Complex to Promote Differentiation of Germline Stem Cell Lineage in
- Approximation to the Distribution of Fitness Effects across Functional Categories in Human Segregating Polymorphisms
- The CSN/COP9 Signalosome Regulates Synaptonemal Complex Assembly during Meiotic Prophase I of
- SAS-1 Is a C2 Domain Protein Critical for Centriole Integrity in
- An RNA-Seq Screen of the Antenna Identifies a Transporter Necessary for Ammonia Detection
- GPA: A Statistical Approach to Prioritizing GWAS Results by Integrating Pleiotropy and Annotation
- Let's Face It—Complex Traits Are Just Not That Simple
- Glutamate Receptor Gene , Coffee, and Parkinson Disease
- The Red Queen Model of Recombination Hotspots Evolution in the Light of Archaic and Modern Human Genomes
- The Ethics of Our Inquiry: An Interview with Hank Greely
- Functional Diversity of Carbohydrate-Active Enzymes Enabling a Bacterium to Ferment Plant Biomass
- Regularized Machine Learning in the Genetic Prediction of Complex Traits
- Phylogenetically Driven Sequencing of Extremely Halophilic Archaea Reveals Strategies for Static and Dynamic Osmo-response
- Lack of Replication of the -by-Coffee Interaction in Parkinson Disease
- Natural Polymorphisms in Human APOBEC3H and HIV-1 Vif Combine in Primary T Lymphocytes to Affect Viral G-to-A Mutation Levels and Infectivity
- A Germline Polymorphism of Thymine DNA Glycosylase Induces Genomic Instability and Cellular Transformation
- Heat-Induced Release of Epigenetic Silencing Reveals the Concealed Role of an Imprinted Plant Gene
- ATPase-Independent Type-III Protein Secretion in
- p53- and ERK7-Dependent Ribosome Surveillance Response Regulates Insulin-Like Peptide Secretion
- The Complex I Subunit Selectively Rescues Mutants through a Mechanism Independent of Mitophagy
- Evolution of DNA Methylation Patterns in the Brassicaceae is Driven by Differences in Genome Organization
- Regulation of mRNA Abundance by Polypyrimidine Tract-Binding Protein-Controlled Alternate 5′ Splice Site Choice
- Systematic Comparison of the Effects of Alpha-synuclein Mutations on Its Oligomerization and Aggregation
- Rad59-Facilitated Acquisition of Y′ Elements by Short Telomeres Delays the Onset of Senescence
- A Functional Portrait of Med7 and the Mediator Complex in
- Systematic Analysis of the Role of RNA-Binding Proteins in the Regulation of RNA Stability
- ARTIST: High-Resolution Genome-Wide Assessment of Fitness Using Transposon-Insertion Sequencing
- Genomic Evidence of Rapid and Stable Adaptive Oscillations over Seasonal Time Scales in Drosophila
- Genome-Wide Associations between Genetic and Epigenetic Variation Influence mRNA Expression and Insulin Secretion in Human Pancreatic Islets
- HAM-5 Functions As a MAP Kinase Scaffold during Cell Fusion in
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- An RNA-Seq Screen of the Antenna Identifies a Transporter Necessary for Ammonia Detection
- Systematic Comparison of the Effects of Alpha-synuclein Mutations on Its Oligomerization and Aggregation
- Functional Diversity of Carbohydrate-Active Enzymes Enabling a Bacterium to Ferment Plant Biomass
- Regularized Machine Learning in the Genetic Prediction of Complex Traits
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



