-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaEstablishing a Multidisciplinary Context for Modeling 3D Facial Shape from DNA
article has not abstract
Published in the journal: . PLoS Genet 10(11): e32767. doi:10.1371/journal.pgen.1004725
Category: Formal Comment
doi: https://doi.org/10.1371/journal.pgen.1004725Summary
article has not abstract
In their perspective piece on Claes et al. [1], Hallgrimsson and colleagues [2] make some points worthy of discussion, but do so largely in the context of a series of strong opinions that they incorrectly attribute to us. The pervasive straw man that is set up in the title and throughout their piece is that we think faces are simple traits, and that predicting facial shape from genotype is already practicable, consequentially overreaching the science. The point of our statement, quoted by these authors, “…our methods provide the means of identifying the genes that affect facial shape and for modeling the effects of these genes to generate a predicted face.” was to highlight the conceptual and methodological advances reported in that work (more on this below). The very next and final sentence of Claes et al., 2014 [1] frames the context of this sentence and is what we meant and by which we continue to stand, and reads, “Although much more work is needed before we can know how many genes will be required to estimate the shape of a face in some useful way, and many more populations need to be studied before we can know how generalizable the results are, these results provide both the impetus and analytical framework for these studies.” This concluding sentence clearly emphasizes that additional work is required and that we only claim to have provided a methodological framework and motivation. In a recent paper [3], we investigated a means of combining the effects of independent factors (namely, sex, genomics ancestry, and genotypes for the 24 single nucleotide polymorphisms (SNPs) from Claes et al.) into a single predicted face. We also explored considerations for how to judge the accuracy of these predicted faces. In short, although we find that sex and ancestry provide much more precision in estimating facial shape from these data, the 24 SNPs do add a small, but statistically significant, level of improvement in facial distinctiveness.
Although it remains to be seen how many alleles and loci affecting normal-range variation in facial features will be discovered, we are encouraged not only by the results presented in Claes et al. but by five rather common observations that are slowly, but surely being formally supported using modern morphometric methods: 1) identical twins are strikingly similar [4], 2) genetic relatives often show particular distinctive features [5], 3) conditions of atypical facial development are often distinctive and easily recognizable [6], 4) human population groups show observable differences [7], [8], and 5) men and women are facially distinctive [9], [10]. Despite the complexity of craniofacial development and the largely unknown mechanisms by which genetic variation affects facial features, these observations compellingly support the assertion that at least some genetic variants have consistent and thus predicable effects on the human face. Such a connection can provide sufficient impetus to apply human genetics methods to both discover which alleles and loci affect variation in the face and to attempt to model facial phenotype from genotype [11]–[13].
Hallgrimsson twice cites one genome-wide association study (GWAS) on facial features [14] as evidence that the SHH gene plays no role in normal-range facial features and, because these authors found so few genes, as evidence that the genetic architecture of facial variation has a “very complex architecture.” Although these two points may well be proven true in time, negative evidence from one study is not very compelling support for either conclusion. Although we are cautious of strong conclusions based on analogies with other traits, such as the coronary heart disease example presented by Hallgrimsson, we do expect that different genetic and genomic [15] methods will be useful in identifying different types of variants. For example, rare variants with large effects, like those causing Mendelian conditions presenting with atypical craniofacial development, will most likely be discoverable using linkage analysis in families [16]. Alternatively, common alleles with smaller effect sizes will likely be easier to map using genetic association [17]. Alleles leading to facial differences between populations can be specifically targeted and thus most efficiently identified using admixture mapping [18]. There are a number of other sources of information beyond human–genetic methods that can and should contribute to facial feature gene identification efforts (Figure 1). Recent work by Hallgrimsson's group, for example, provided an interesting combination of functional genomic and animal model approaches using the mouse. Ideally, researchers will emerge who can make the most of several types of information to help understand the developmental genetic architecture of the human face. Indeed, the face is complex and we fully expect that a combination of all of these efforts will most constructively contribute to a more complete understanding of both its evolution and development.
Fig. 1. Diagram of a framework for research on modeling facial features from DNA. 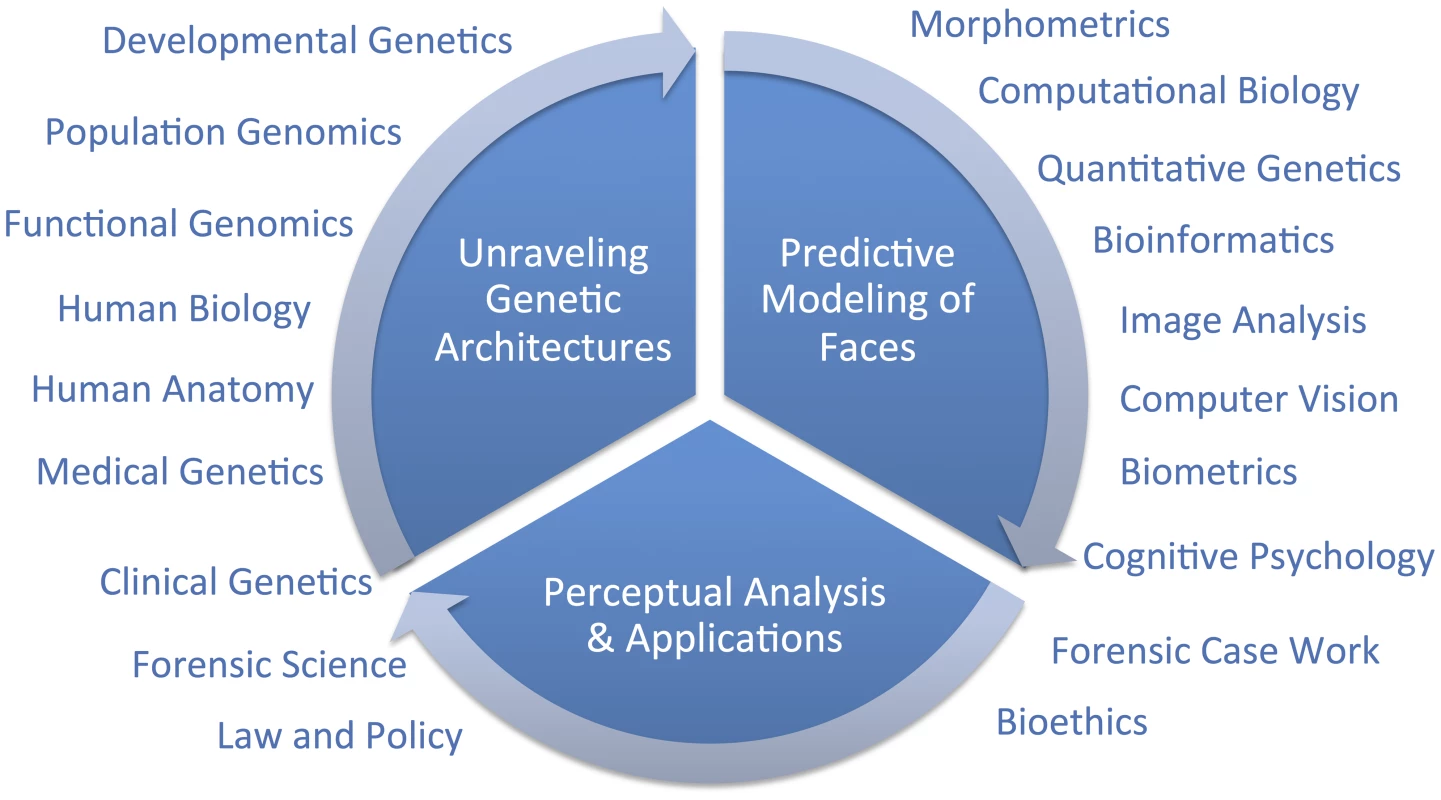
One key aspect of facial research is how to systematically measure and model facial variation. In most prior facial feature mapping analyses, researchers focused on using individual interlandmark distances and principal component scores as traits [14], [19]–[21]. The primary drawback of these univariate approaches is that the response variables used represent only either some arbitrary or a priori subset of the total facial variation, which will not necessarily correspond to the facial effects of genes or any other factors. Additionally, univariate methods are statistically underpowered when used to map multivariate traits in GWAS [22], adding, for example, a layer of multiple testing reducing statistical power by a factor equal to the number of traits analyzed. Although most of the normal-range gene mapping studies have focused on univariate analyses, one recent paper used dense-correspondence based methods, which allow all parts of the face to be modeled together [23]. Dense correspondence methods have proven useful in investigating conditions of atypical facial development [24] and can be used to create average or consensus faces for cases and controls or, as demonstrated by Peng et al., by genotype. Although these genotype-average faces do allow any part of the face to be affected, it is currently unclear how to condition for confounding variables in these analyses, or how to accommodate continuously distributed independent factors, like genomic ancestry. Peng and colleagues overcame these limitations by focusing on a study population showing limited variation in genomic ancestry and by stratifying their analyses by sex, in effect matching males with males and females with females.
The approach we explored in Claes et al. is fundamentally different from the other methods being used to study human facial variation and facilitates both conditioning for confounding variables and the inclusion of all facial regions. Briefly, we applied partial least squares regression (PLSR) and multidimensional scoring in a novel forced imputation framework. This approach allows any set of facial regions to be combined into a single numerical score of that factor's effect on each face. These scores are essentially the predicted value of the independent (predictor) variable, e.g., sex, genomic ancestry, or genotype, given the relationship between facial variation and that variable observed in the sample. We called this new type of variable the response-based imputed predictor (RIP) variable and, given empirically observed improvements through multiple iterations, have called the method, generally, bootstrapped response-based imputation modeling (BRIM). The ability of BRIM to model facial sex and facial ancestry was assessed using a series of analytical experiments and human perception experiments [1].
Additionally, univariate methods provide no obvious means for visualizing facial modeling analyses as images. As shown in Claes et al., such images can be used in post hoc comparisons between normal-range effects and clinically significant effects [1]. Visualizing the effects also opens the door for systematic transformations of particular faces, which could be useful in experiments on the psychology of facial perception. Finally, without a means of visualizing the effects of genes and other factors, methods for assembling composite faces, like the one explored in our recent paper [3], would not be possible.
The important question that remains is, what is a suitable scientific context for modeling 3D facial shape from DNA? We do share Hallgrimsson and colleagues' perspective that when publishing novel scientific methods, it is important to establish reasonable expectations to policymakers and the public. The full context will only be known in time; overpromising results is certainly not the right framework for progress, but neither is diminishing novel synthetic efforts. Unraveling the genetic architecture of facial morphology is only one aspect of a comprehensive predictive modeling effort. The creation of usefully accurate DNA-based facial composites, as discussed in [3], involves at least two other aspects which are also quite multidisciplinary; namely, 1) predictive modeling of faces, and 2) perceptual analysis and applications. In the figure, we diagram these three primary components and indicate broadly which are some of the fields that can, and should, be drawn on to address these three components. We believe that the most constructive, and thus useful, context for facial feature genetics will be possible after adopting a multidisciplinary point of view.
Zdroje
1. ClaesP, LibertonDK, DanielsK, RosanaKM, QuillenEE, et al. (2014) Modeling 3D Facial Shape from DNA. PLoS Genet 10: e1004224 doi:10.1371/journal.pgen.1004224
2. HallgrimssonB, MioW, MarcucioR, SpritzRA (2014) Let's Face it - Complex traits are just not that simple. PLoS Genet 10: e1004724.
3. ClaesP, HillH, ShriverMD (2014) Towards DNA-based facial composites: Preliminary results and validation. Forensic Science International: Genetics 13 : 208–216 doi:10.1016/j.fsigen.2014.08.008
4. WeinbergSM, ParsonsTE, MarazitaML, MaherBS (2013) Heritability of face shape in twins: a preliminary study using 3D stereophotogrammetry and geometric morphometrics. Dent 3000 1 : 14 doi:10.5195/d3000.2013.14
5. KimHJ, ImSW, JargalG, LeeS, YiJH (2013) Heritabilities of Facial Measurements and Their Latent Factors in Korean Families. Genomics Inform 11 : 83–92.
6. HammondP (2007) The use of 3D face shape modelling in dysmorphology. Arch Dis Child 92 : 1120–1126 doi:10.1136/adc.2006.103507
7. FarkasLG, KaticMJ, ForrestCR (2005) International Anthropometric Study of Facial Morphology in Various Ethnic Groups/Races. Journal of Craniofacial Surgery 16 : 615–646 doi:10.1097/01.scs.0000171847.58031.9e
8. HopmanS, MerksJ, SuttieM (2014) Face shape differs in phylogenetically related populations. Eur J Hum Genet E-pub ahead of print. doi: 10.1038/ejhg.2013.289
9. ClaesP, WaltersM, ShriverMD, PutsD, GibsonG, et al. (2012) Sexual dimorphism in multiple aspects of 3D facial symmetry and asymmetry defined by spatially dense geometric morphometrics. Journal of Anatomy 221 : 97–114 doi:10.1111/j.1469-7580.2012.01528.x
10. GilaniSZ, RooneyK, ShafaitF, WaltersM, MianA (2014) Geometric Facial Gender Scoring: Objectivity of Perception. PLoS ONE 9: e99483 doi:10.1371/journal.pone.0099483
11. OberU, ErbeM, LongN, PorcuE, SchlatherM, et al. (2011) Predicting Genetic Values: A Kernel-Based Best Linear Unbiased Prediction With Genomic Data. Genetics 188 : 695–708 doi:10.1534/genetics.111.128694
12. OberU, AyrolesJF, StoneEA, RichardsS, ZhuD, et al. (2012) Using Whole-Genome Sequence Data to Predict Quantitative Trait Phenotypes in Drosophila melanogaster. PLoS Genet 8: e1002685 doi:10.1371/journal.pgen.1002685
13. Ben JHayes, PryceJ, ChamberlainAJ, BowmanPJ, GoddardME (2010) Genetic Architecture of Complex Traits and Accuracy of Genomic Prediction: Coat Colour, Milk-Fat Percentage, and Type in Holstein Cattle as Contrasting Model Traits. PLoS Genet 6: e1001139 doi:10.1371/journal.pgen.1001139
14. LiuF, van der LijnF, SchurmannC, ZhuG, ChakravartyMM, et al. (2012) A Genome-Wide Association Study Identifies Five Loci Influencing Facial Morphology in Europeans. PLoS Genet 8: e1002932 doi:10.1371/journal.pgen.1002932
15. KhandelwalKD, van BokhovenH, RoscioliT, CarelsCEL, ZhouH (2013) Genomic approaches for studying craniofacial disorders. Am J Med Genet 163 : 218–231 doi:10.1002/ajmg.c.31379
16. WilkieAOM, Morriss-KayGM (2001) Genetics of craniofacial development and malformation. Nat Rev Genet 2 : 458–468 doi:10.1038/35076601
17. RischN, MerikangasK (1996) The future of genetic studies of complex human diseases. Science 273 : 1516–1517.
18. McKeiguePM (2005) Prospects for Admixture Mapping of Complex Traits. The American Journal of Human Genetics 76 : 1–7 doi:10.1086/426949
19. BoehringerS, van der LijnF, LiuF, GuntherMGU, SinigerovaS, et al. (2011) Genetic determination of human facial morphology: links between cleft-lips and normal variation. Eur J Hum Genet 19 : 1192–1197 doi:10.1038/ejhg.2011.110
20. PaternosterL, ZhurovAI, TomaAM, KempJP, PourcainBS, et al. (2012) REPOR TGenome-wide Association Study of Three-Dimensional Facial Morphology Identifies a Variantin PAX3 Associated with Nasion Position. Am J Hum Genet 90 : 478–485 doi:10.1016/j.ajhg.2011.12.021
21. FatemifarG, HoggartCJ, PaternosterL, KempJP, ProkopenkoI, et al. (2013) Genome-wide association study of primary tooth eruption identifies pleiotropic loci associated with height and craniofacial distances. Human Molecular Genetics 22 : 3807–3817 doi:10.1093/hmg/ddt231
22. GaleslootTE, van SteenK, KiemeneyLALM, JanssLL, VermeulenSH (2014) A Comparison of Multivariate Genome-Wide Association Methods. PLoS ONE 9: e95923 doi:10.1371/journal.pone.0095923
23. PengS, TanJ, HuS, ZhouH, GuoJ, et al. (2013) Detecting Genetic Association of Common Human Facial Morphological Variation Using High Density 3D Image Registration. PLoS Comput Biol 9: e1003375 doi:10.1371/journal.pcbi.1003375
24. HammondP, SuttieM (2012) Large-scale objective phenotyping of 3D facial morphology. Hum Mutat 33 : 817–825 doi:10.1002/humu.22054
Štítky
Genetika Reprodukční medicína
Článek The COP9 Signalosome Converts Temporal Hormone Signaling to Spatial Restriction on Neural CompetenceČlánek Coordinate Regulation of Stem Cell Competition by Slit-Robo and JAK-STAT Signaling in the TestisČlánek The CSN/COP9 Signalosome Regulates Synaptonemal Complex Assembly during Meiotic Prophase I ofČlánek GPA: A Statistical Approach to Prioritizing GWAS Results by Integrating Pleiotropy and AnnotationČlánek Functional Diversity of Carbohydrate-Active Enzymes Enabling a Bacterium to Ferment Plant BiomassČlánek Heat-Induced Release of Epigenetic Silencing Reveals the Concealed Role of an Imprinted Plant GeneČlánek p53- and ERK7-Dependent Ribosome Surveillance Response Regulates Insulin-Like Peptide SecretionČlánek The Complex I Subunit Selectively Rescues Mutants through a Mechanism Independent of MitophagyČlánek Rad59-Facilitated Acquisition of Y′ Elements by Short Telomeres Delays the Onset of SenescenceČlánek ARTIST: High-Resolution Genome-Wide Assessment of Fitness Using Transposon-Insertion Sequencing
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 11
-
Všechny články tohoto čísla
- Establishing a Multidisciplinary Context for Modeling 3D Facial Shape from DNA
- RNA Processing Factors Swd2.2 and Sen1 Antagonize RNA Pol III-Dependent Transcription and the Localization of Condensin at Pol III Genes
- Inversion of the Chromosomal Region between Two Mating Type Loci Switches the Mating Type in
- A Thermolabile Aldolase A Mutant Causes Fever-Induced Recurrent Rhabdomyolysis without Hemolytic Anemia
- The Role of Regulatory Evolution in Maize Domestication
- Stress Granule-Defective Mutants Deregulate Stress Responsive Transcripts
- 24-Hour Rhythms of DNA Methylation and Their Relation with Rhythms of RNA Expression in the Human Dorsolateral Prefrontal Cortex
- Pseudoautosomal Region 1 Length Polymorphism in the Human Population
- Fungal Communication Requires the MAK-2 Pathway Elements STE-20 and RAS-2, the NRC-1 Adapter STE-50 and the MAP Kinase Scaffold HAM-5
- The COP9 Signalosome Converts Temporal Hormone Signaling to Spatial Restriction on Neural Competence
- The Protein -glucosyltransferase Rumi Modifies Eyes Shut to Promote Rhabdomere Separation in
- The Talin Head Domain Reinforces Integrin-Mediated Adhesion by Promoting Adhesion Complex Stability and Clustering
- Quantitative Genetics of CTCF Binding Reveal Local Sequence Effects and Different Modes of X-Chromosome Association
- Coordinate Regulation of Stem Cell Competition by Slit-Robo and JAK-STAT Signaling in the Testis
- Genetic Analysis of a Novel Tubulin Mutation That Redirects Synaptic Vesicle Targeting and Causes Neurite Degeneration in
- A Systems Genetics Approach Identifies , , and as Novel Aggressive Prostate Cancer Susceptibility Genes
- Three RNA Binding Proteins Form a Complex to Promote Differentiation of Germline Stem Cell Lineage in
- Approximation to the Distribution of Fitness Effects across Functional Categories in Human Segregating Polymorphisms
- The CSN/COP9 Signalosome Regulates Synaptonemal Complex Assembly during Meiotic Prophase I of
- SAS-1 Is a C2 Domain Protein Critical for Centriole Integrity in
- An RNA-Seq Screen of the Antenna Identifies a Transporter Necessary for Ammonia Detection
- GPA: A Statistical Approach to Prioritizing GWAS Results by Integrating Pleiotropy and Annotation
- Let's Face It—Complex Traits Are Just Not That Simple
- Glutamate Receptor Gene , Coffee, and Parkinson Disease
- The Red Queen Model of Recombination Hotspots Evolution in the Light of Archaic and Modern Human Genomes
- The Ethics of Our Inquiry: An Interview with Hank Greely
- Functional Diversity of Carbohydrate-Active Enzymes Enabling a Bacterium to Ferment Plant Biomass
- Regularized Machine Learning in the Genetic Prediction of Complex Traits
- Phylogenetically Driven Sequencing of Extremely Halophilic Archaea Reveals Strategies for Static and Dynamic Osmo-response
- Lack of Replication of the -by-Coffee Interaction in Parkinson Disease
- Natural Polymorphisms in Human APOBEC3H and HIV-1 Vif Combine in Primary T Lymphocytes to Affect Viral G-to-A Mutation Levels and Infectivity
- A Germline Polymorphism of Thymine DNA Glycosylase Induces Genomic Instability and Cellular Transformation
- Heat-Induced Release of Epigenetic Silencing Reveals the Concealed Role of an Imprinted Plant Gene
- ATPase-Independent Type-III Protein Secretion in
- p53- and ERK7-Dependent Ribosome Surveillance Response Regulates Insulin-Like Peptide Secretion
- The Complex I Subunit Selectively Rescues Mutants through a Mechanism Independent of Mitophagy
- Evolution of DNA Methylation Patterns in the Brassicaceae is Driven by Differences in Genome Organization
- Regulation of mRNA Abundance by Polypyrimidine Tract-Binding Protein-Controlled Alternate 5′ Splice Site Choice
- Systematic Comparison of the Effects of Alpha-synuclein Mutations on Its Oligomerization and Aggregation
- Rad59-Facilitated Acquisition of Y′ Elements by Short Telomeres Delays the Onset of Senescence
- A Functional Portrait of Med7 and the Mediator Complex in
- Systematic Analysis of the Role of RNA-Binding Proteins in the Regulation of RNA Stability
- ARTIST: High-Resolution Genome-Wide Assessment of Fitness Using Transposon-Insertion Sequencing
- Genomic Evidence of Rapid and Stable Adaptive Oscillations over Seasonal Time Scales in Drosophila
- Genome-Wide Associations between Genetic and Epigenetic Variation Influence mRNA Expression and Insulin Secretion in Human Pancreatic Islets
- HAM-5 Functions As a MAP Kinase Scaffold during Cell Fusion in
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- An RNA-Seq Screen of the Antenna Identifies a Transporter Necessary for Ammonia Detection
- Systematic Comparison of the Effects of Alpha-synuclein Mutations on Its Oligomerization and Aggregation
- Functional Diversity of Carbohydrate-Active Enzymes Enabling a Bacterium to Ferment Plant Biomass
- Regularized Machine Learning in the Genetic Prediction of Complex Traits
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



