-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaGenes That Act Downstream of Sensory Neurons to Influence Longevity, Dauer Formation, and Pathogen Responses in
The sensory systems of multicellular organisms are designed to provide information about the environment and thus elicit appropriate changes in physiology and behavior. In the nematode Caenorhabditis elegans, sensory neurons affect the decision to arrest during development in a diapause state, the dauer larva, and modulate the lifespan of the animals in adulthood. However, the mechanisms underlying these effects are incompletely understood. Using whole-genome microarray analysis, we identified transcripts whose levels are altered by mutations in the intraflagellar transport protein daf-10, which result in impaired development and function of many sensory neurons in C. elegans. In agreement with existing genetic data, the expression of genes regulated by the transcription factor DAF-16/FOXO was affected by daf-10 mutations. In addition, we found altered expression of transcriptional targets of the DAF-12/nuclear hormone receptor in the daf-10 mutants and showed that this pathway influences specifically the dauer formation phenotype of these animals. Unexpectedly, pathogen-responsive genes were repressed in daf-10 mutant animals, and these sensory mutants exhibited altered susceptibility to and behavioral avoidance of bacterial pathogens. Moreover, we found that a solute transporter gene mct-1/2, which was induced by daf-10 mutations, was necessary and sufficient for longevity. Thus, sensory input seems to influence an extensive transcriptional network that modulates basic biological processes in C. elegans. This situation is reminiscent of the complex regulation of physiology by the mammalian hypothalamus, which also receives innervations from sensory systems, most notably the visual and olfactory systems.
Published in the journal: . PLoS Genet 8(12): e32767. doi:10.1371/journal.pgen.1003133
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003133Summary
The sensory systems of multicellular organisms are designed to provide information about the environment and thus elicit appropriate changes in physiology and behavior. In the nematode Caenorhabditis elegans, sensory neurons affect the decision to arrest during development in a diapause state, the dauer larva, and modulate the lifespan of the animals in adulthood. However, the mechanisms underlying these effects are incompletely understood. Using whole-genome microarray analysis, we identified transcripts whose levels are altered by mutations in the intraflagellar transport protein daf-10, which result in impaired development and function of many sensory neurons in C. elegans. In agreement with existing genetic data, the expression of genes regulated by the transcription factor DAF-16/FOXO was affected by daf-10 mutations. In addition, we found altered expression of transcriptional targets of the DAF-12/nuclear hormone receptor in the daf-10 mutants and showed that this pathway influences specifically the dauer formation phenotype of these animals. Unexpectedly, pathogen-responsive genes were repressed in daf-10 mutant animals, and these sensory mutants exhibited altered susceptibility to and behavioral avoidance of bacterial pathogens. Moreover, we found that a solute transporter gene mct-1/2, which was induced by daf-10 mutations, was necessary and sufficient for longevity. Thus, sensory input seems to influence an extensive transcriptional network that modulates basic biological processes in C. elegans. This situation is reminiscent of the complex regulation of physiology by the mammalian hypothalamus, which also receives innervations from sensory systems, most notably the visual and olfactory systems.
Introduction
Organisms are constantly interacting with their environment. Behavioral responses as well as physiological processes such as energy homeostasis, development and immune homeostasis need to be modulated depending on the environmental situation. For example, an animal's feeding and development are crucial to survival in general, but may need to be reduced or delayed under certain environmental conditions to allow efficient allocation of resources for survival. To achieve such modulation, animals have developed complex sensory systems that acquire and integrate various sorts of information about their environment and their internal state. However, the mechanisms by which sensory neurons influence complex physiological processes are still incompletely understood.
Because of its relatively simple nervous system and genetic tractability, the nematode Caenorhabditis elegans has been studied extensively as an experimental organism to dissect the molecular mechanisms regulating sensory control of behavior. A small number of ciliated sensory neurons located mainly near the head and tail of the animal detect environmental signals, including soluble and volatile compounds, gases, osmolarity, and mechanosensory and noxious stimuli (reviewed in [1]). As in other organisms, it is now clear that the sensory system of C. elegans regulates physiological functions of the animals as well as its behavior. When certain sensory neurons are compromised, for example, worms are more likely to arrest in an alternative developmental state of diapause called dauer in response to higher temperatures [2]–[4]. In turn, dauer arrest results in modulation of behavioral output and reduced response to stimuli [5]. In addition, various signaling pathways in the nervous system contribute to lifespan regulation [3], [6]–[8] and the response to pathogenic insults [9]–[13]. One possible model is that sensory input is translated into whole-organism physiological alterations by the regulation of transcriptional programs. However, to date only a few genes have been reported to regulate physiological changes downstream of sensory perception, most notably the Forkhead transcription factor daf-16/FOXO [3], [14]–[16], an important regulator of lifespan, dauer formation and immune response downstream of the daf-2/insulin/IGF1-like receptor (InsR). Thus, much remains to be learned about how the disruption of sensory neurons results in changes in the physiology of the whole organism.
In this study, we examine the transcriptional profile of daf-10 sensory mutants to identify genes and signaling pathways that may be targeted by the sensory system to regulate the physiology of the animal. The daf-10 gene encodes the C. elegans homolog of the intraflagellar transport protein IFT222 and its mutation results in altered development of a number of sensory neurons [17], [18], leading to defects in sensory perception. daf-10 mutants also exhibit an increase in longevity and an increase in spontaneous dauer formation at high temperature (27°C) [2], [3]. In daf-10 mutant animals we find evidence of activation of DAF-16/FOXO, as expected, as well as activation of the nuclear hormone receptor (NHR) DAF-12, which we demonstrate is required for the increased dauer formation, but not for the extended longevity of sensory mutant animals. In addition, we show that the response to pathogenic bacteria is altered in the sensory daf-10 mutant animals and that both behavioral and physiological responses to pathogens are affected. Furthermore, we examine the functional significance of genes that are up-regulated by daf-10 mutations and find that mct-1/2, a putative monocarboxylate transporter, is required for the extended lifespan of daf-10 mutants. Taken together, our data suggest that the sensory system modulates several transcriptional programs to exert its effects on lifespan, dauer formation and innate immunity in response to the environmental changes.
Results
Microarray analysis reveals differentially expressed genes in the long-lived daf-10(m79) mutant
To identify the genes and pathways that are downstream of the sensory system, we used whole-genome oligonucleotide-based microarrays to compare gene expression in young adult animals of a long-lived sensory mutant strain, daf-10(m79), with that of wild-type young adult worms. Many of the mutations that compromise the development and function of the sensory neurons of C. elegans also lead to altered longevity and dauer formation at high temperature [3], [8], [16]. We chose this particular mutant strain because daf-10 has been shown to be expressed in all ciliated neurons [19] and multiple alleles of daf-10 cause a significant longevity phenotype [3], showing that the effect of daf-10 sensory mutations is robust.
Our microarray analysis identified 14 genes that were reliably up-regulated in the daf-10(m79) animals and 56 genes that were down-regulated (Table 1 and Table 2). We further re-tested 5 of the up-regulated and 17 of the down-regulated genes using quantitative RT-PCR (qRT-PCR) analysis and confirmed that 18 out of the 22 genes that we tested showed similar changes using qRT-PCR and microarray analysis (Figure 1A and 1B). In addition, our qRT-PCR analysis showed that 9 out of the 22 genes exhibited a similar trend in expression in osm-5(p813), another mutant with defective ciliated neurons and extended longevity [3] (Figure S1A and S1B). The osm-5 gene encodes another component of the intraflagellar transport complex [20], [21], which becomes truncated by a premature stop codon in p813. Thus, our microarray analysis has identified a small but reliable set of genes that are differentially expressed in response to defects in sensory perception.
Fig. 1. qRT–PCR analysis confirms differential regulation by daf-10(m79) mutations of genes identified through microarray analysis. 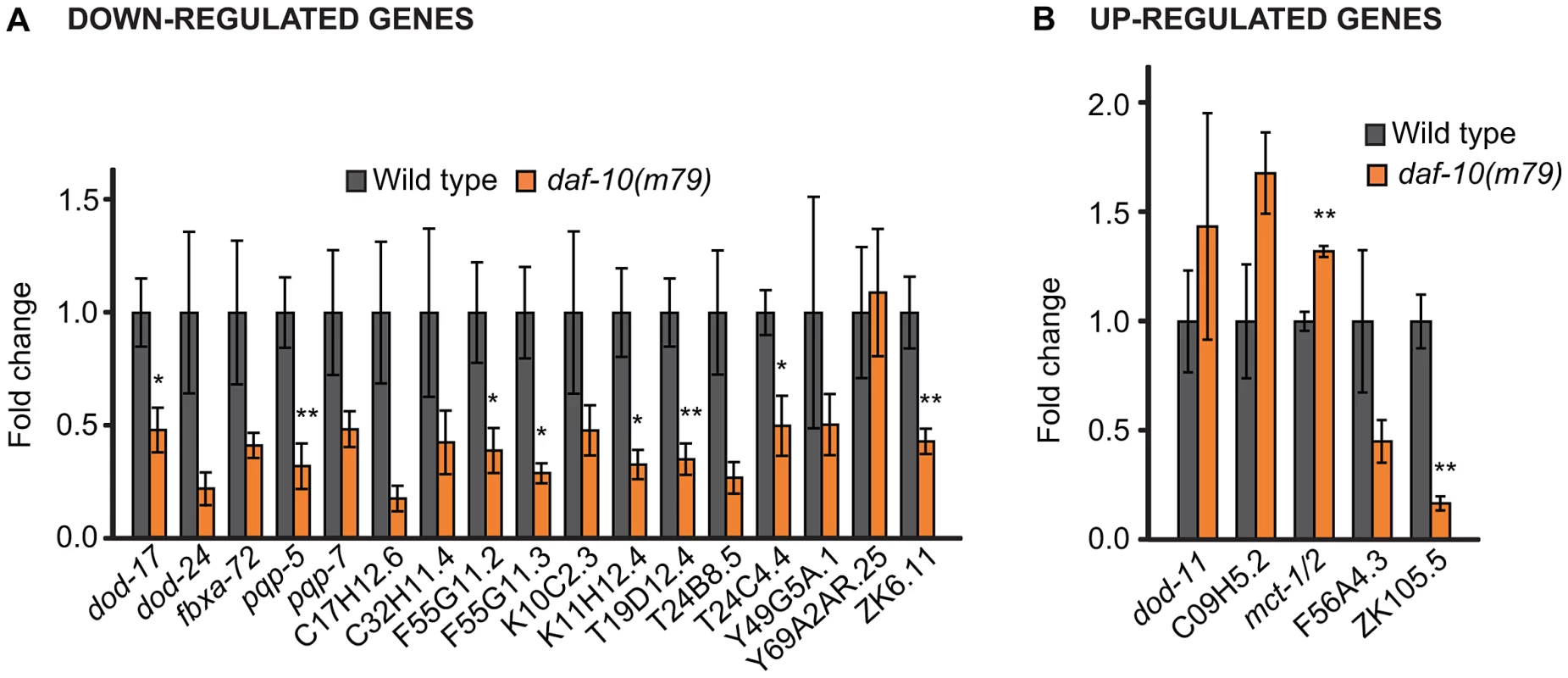
qRT-PCR was used to examine changes in the expression of genes that were down-regulated (A) or up-regulated (B) in the microarray analysis. Error bars represent s.e.m. (* p<0.05, ** p<0.01, Student's t-test). Tab. 1. Genes up-regulated in daf-10(m79) mutants versus wild-type animals. 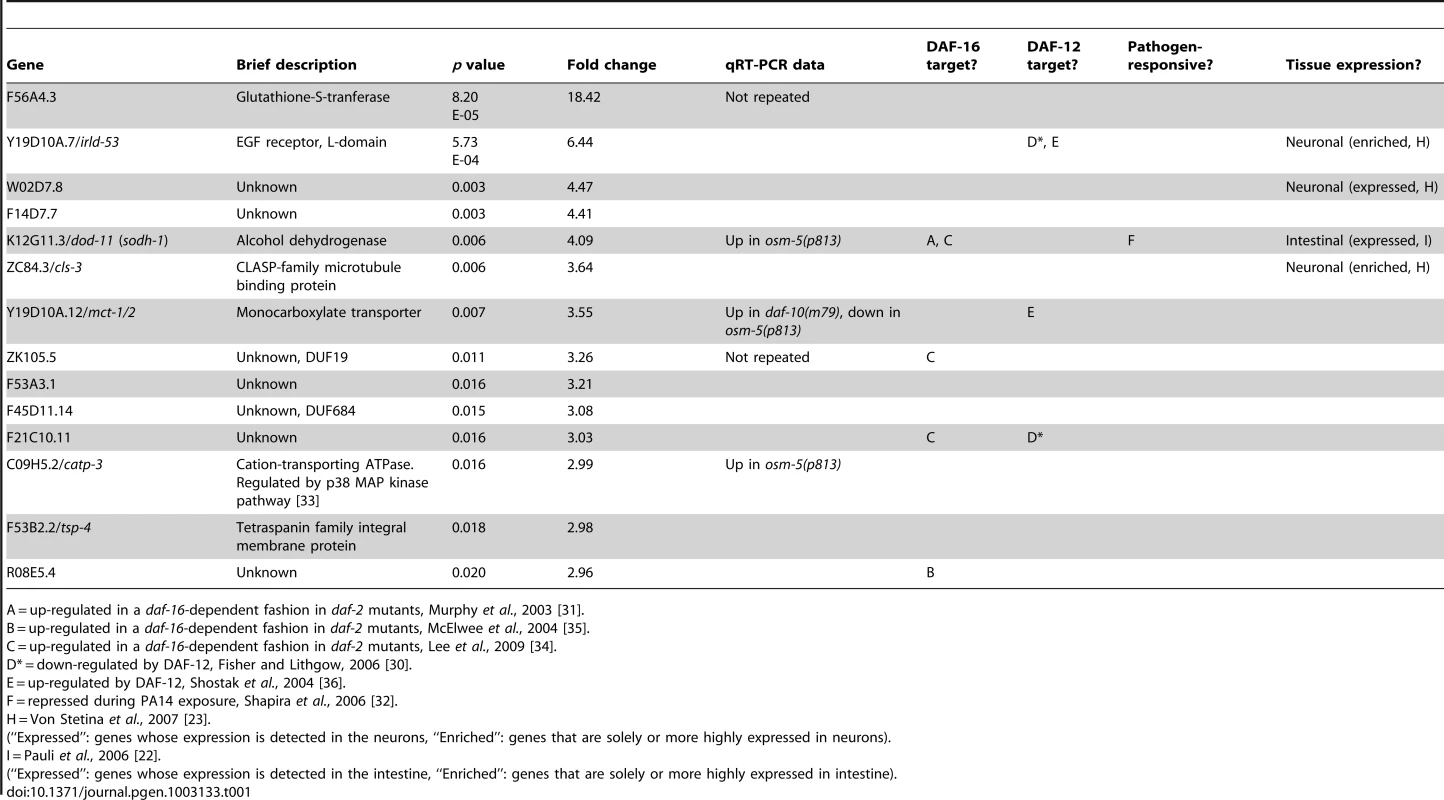
A = up-regulated in a daf-16-dependent fashion in daf-2 mutants, Murphy et al., 2003 [31]. Tab. 2. Genes down-regulated in daf-10(m79) mutants versus wild-type animals. 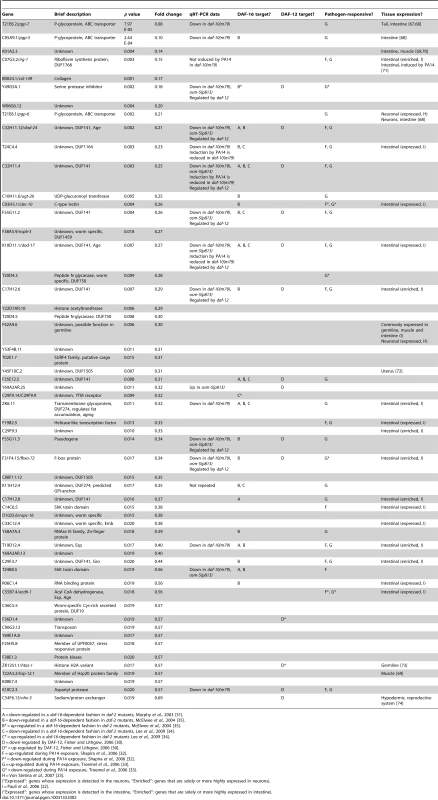
A = down-regulated in a daf-16-dependent fashion in daf-2 mutants, Murphy et al., 2003 [31]. Comparative analysis of the genes that are differentially expressed in daf-10(m79) worms
We used several previously published microarray datasets to determine the tissue-specific expression and the potential function of the genes we had identified as differentially expressed in the daf-10(m79) mutant animals. First we compared our gene list to those of genes enriched or solely expressed in intestine [22], neurons [23] or muscle [22]. Of the 70 genes that we identified, 22 are likely expressed in the intestine and 5 in neurons (Table 1 and Table 2). Six genes are expressed in other tissues. We also compared our genes to a C. elegans global expression map created using 553 microarray data sets (http://nemates.org/gl/cgi-bin/gene_list.cgi?set=20002) [24]. This map represents the correlation in gene expression mapped against gene density in three dimensions and can be used as a tool to assign gene function based on co-regulation with known sets of genes. The genes differentially regulated in daf-10(m79) animals mapped predominantly to mountains 19 and 21. Mountain 19 includes genes that are also changed in response to mutations in the daf-2/InsR pathway in a daf-16/FOXO-dependent manner or in response to mutations in daf-12/NHR. Four out of five of the genes that overlap with mountain 21 may be involved in detoxification: the UDP-glucuronosyltransferase (UGT) ugt-26 and the P-glycoproteins pgp-5, -6, -7. UGTs metabolize foreign substances and endogenous toxins by glycosylating these molecules and facilitating their elimination. P-glycoproteins are members of the ABC transporter family and function to extrude large hydrophobic molecules from cells [25], [26]. pgp-5 may also have a role in immune responses in C. elegans [27]. Interestingly, classification of the differentially expressed genes by GO-term annotation using the DAVID program [28], [29] also highlighted genes with ATPase or transport activity (which include pgp genes), as well as genes involved in aging or determination of lifespan (which include mountain 19 genes). When we used the DAVID program to classify genes based on protein domains as defined by the Interpro database [29], proteins with a CUB-like domain (formerly known as DUF141) emerged as overrepresented. CUB-like domains are C. elegans-specific domains that resemble CUB domains, which are found on extracellular and membrane proteins such as complement proteins. CUB-like domains may be found predominantly within secreted proteins [30]. In addition, proteins with a CUB-like domain may have important, though unknown, roles in longevity. Two such proteins, dod-24 and dod-17, are down-regulated in long-lived daf-2/InsR mutant animals and knockdown of these genes by RNA interference increases lifespan [31]. CUB-like domain proteins were also identified among the transcriptional targets of daf-12/NHR [30] and among genes that are activated by exposure to the pathogen Pseudomonas aeruginosa [32], [33] (see below).
Because the extended lifespan of daf-10(m79) animals is dependent on the daf-16/FOXO transcription factor and because of the mapping to mountain 19, we compared our list with genes whose expression changes in daf-2 mutants in a daf-16/FOXO-dependent fashion. We found a significant overlap between our gene set and three independent DAF-16/FOXO target lists (Table 3, p<0.0001 in all three cases, hypergeometric probability) [31], [34], [35]. Specifically, of 70 differentially regulated genes in our array list, 9 were also identified by Murphy et al. [31], 21 by McElwee et al. [35] and 10 by Lee et al. [34]. This is consistent with genetic data that has implicated daf-16/FOXO and presumably insulin signaling in the regulation of lifespan downstream of the sensory system [3], [15]. It is notable that the overlaps among these three studies are of comparable size (for example, 36% of the 506 genes identified by Murphy et al. and 23% of the 457 genes identified by Lee at al. were also detected in the study by McElwee et al., compared to 30% of the 70 genes identified in our study). Only 31 genes were identified in all three studies, and of these 4 genes were also identified as differentially regulated in daf-10(m79) animals.
Tab. 3. Overlap between the genes differentially regulated in daf-10(m79) animals and previous gene expression analysis results. 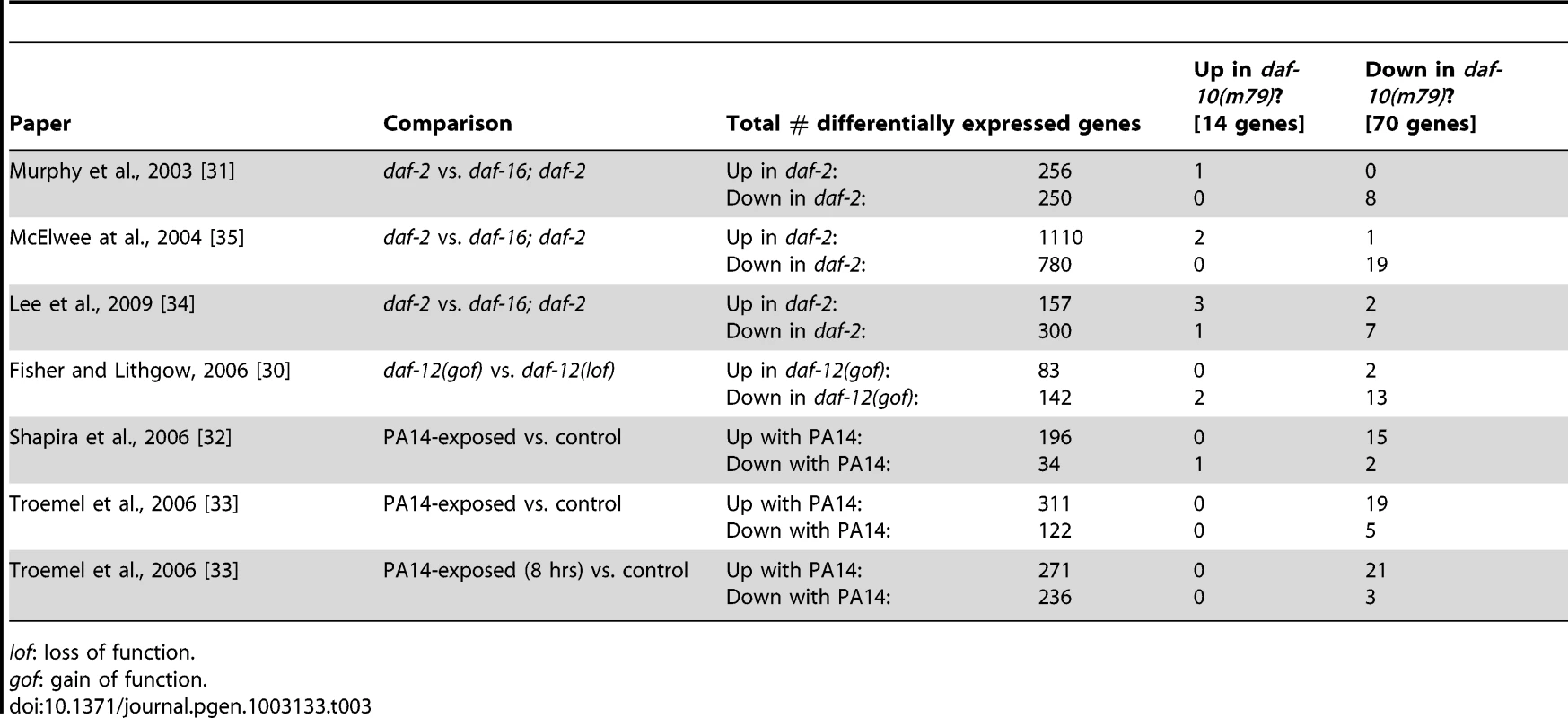
lof: loss of function. In summary, unbiased analysis using the C. elegans expression map and classification by DAVID as well as directed comparison with daf-2/InsR - and daf-16/FOXO-regulated genes reveal that the genes differentially regulated in daf-10(m79) sensory mutants are likely involved in lifespan regulation and insulin-like signaling in the worm. Additionally, these analyses uncovered the overrepresentation of a second class of genes, those involved in detoxification, in the dataset.
The sensory system works through daf-12/NHR to control dauer formation, but not lifespan
Because both putative daf-12/NHR targets and genes differentially expressed in daf-10(m79) sensory mutant worms mapped to mountain 19 and included a number of CUB-like genes [29], we hypothesized that DAF-12/NHR targets may also be regulated by the sensory system. Therefore, we compared our dataset with a list of putative DAF-12/NHR targets identified in a comparative study of daf-12/NHR gain-of-function and loss-of-function mutant animals [30] and found a very significant overlap (Table 3, p<0.0001, hypergeometric probability; genes that were reported as DAF-12/NHR-binding target genes in another study [36] are also indicated in Table 1 and Table 2). We then performed qRT-PCR to compare the expression of these putative DAF-12/NHR target genes in the presence and absence of daf-12/NHR, both in daf-10 sensory mutants and in otherwise wild-type animals (Figure 2A). We found that of the nine genes tested, eight were no longer down-regulated by daf-10 mutations in the daf-12(rh61rh411) null mutant background (Figure 2A), confirming that these genes are indeed regulated by DAF-12/NHR in the context of daf-10 sensory mutations. Interestingly, loss of DAF-12/NHR alone did not alter the expression of most of the genes tested, with the exception of Y49AG5A.1 and dod-17 (in the latter case, the effect was opposite to what was expected). Together with the microarray results, these data indicate that the activity of DAF-12/NHR is altered as a result of sensory system mutations. Because the basal level of the target genes was largely unchanged in daf-12(rh61rh411) animals, DAF-12/NHR is likely to be specifically activated in sensory mutant worms.
Fig. 2. daf-10 mutations influence the expression of DAF-12/NHR-regulated genes. 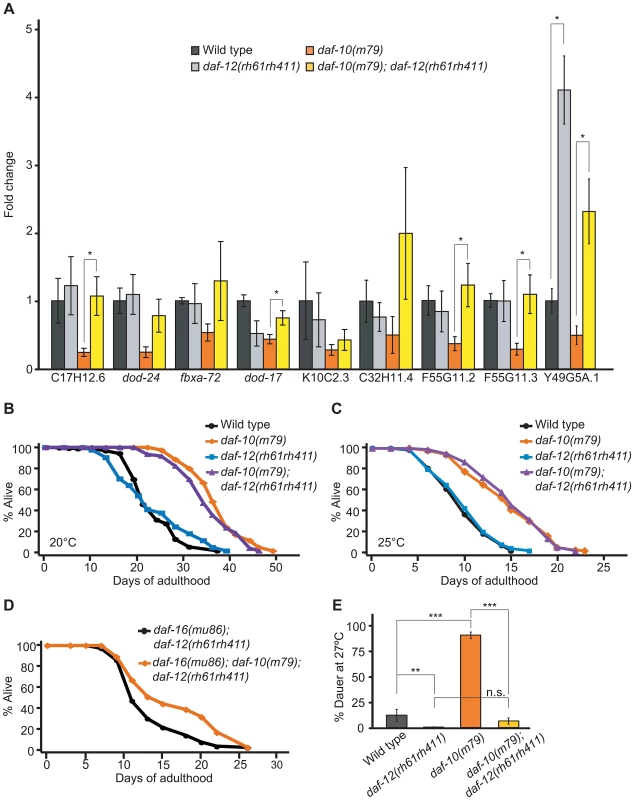
A. qRT-PCR was used to determine whether the putative DAF-12 targets were regulated in a daf-12/NHR-dependent fashion in the daf-10(m79) background. B–C. daf-12/NHR was not required for daf-10(m79) mutant animals to live long at 20°C (B) or at 25°C (C). D. Mutations in daf-10 can still extend the lifespan of daf-16(mu86); daf-12(rh61rh411) mutant animals. A summary of the data presented in these panels and additional repeats is included in Table S1A. E. daf-12/NHR was required for daf-10(m79) mutants to arrest at the dauer stage when grown at 27°C. Note that the difference between daf-10(m79); daf-12(rh61rh411) and daf-12(rh61rh411) animals was not statistically significant (p = 0.07, n.s.). Error bars represent s.e.m. (* p<0.05, ** p<0.01, *** p<0.001, Student's t-test). Since daf-12/NHR is important for both lifespan regulation and development into the arrested dauer state [37], [38] and both of these processes become misregulated in sensory mutants, we tested whether daf-12/NHR was required for increased lifespan or dauer formation in daf-10(m79) animals. daf-12/NHR appeared to be dispensable for the extended longevity of daf-10(m79) animals at 20°C (Figure 2B). We previously showed that daf-12/NHR was required for the influence of thermosensory neurons on lifespan at 25°C [8]. We therefore tested whether daf-12/NHR was required for the longevity caused by daf-10(m79) sensory mutation at 25°C, but found it was not (Figure 2C). In addition, since daf-16(mu86); daf-10(m79) animals have a small residual increase in lifespan compared to daf-16(mu86) animals [3], we also tested specifically whether daf-12/NHR was required for the daf-16/FOXO-independent portion of the lifespan increase. However, daf-10 mutations still extended lifespan slightly in the daf-16(mu86); daf-12(rh61rh411) background (Figure 2D). In contrast, daf-12/NHR was required for the increased high-temperature dauer formation due to daf-10 mutations (Figure 2E, percentage dauers, mean ± s.e.m.: wild type = 11.8%±5.7%, daf-12(rh61rh411) = 0.4%±0.3%, daf-10(m79) = 88.8%±3.2%, daf-10(m79); daf-12(rh61rh411) = 6.3%±3.0%). This is consistent with what was observed with tax-4 mutations, which eliminate the function of a cyclic nucleotide-gated channel required for sensory transduction [39] and also cause increased dauer formation at 27°C in a daf-12/NHR-dependent manner [2]. Together these findings suggest that daf-10 mutants tend to become dauers at high temperature because they have basally altered activity of DAF-12/NHR, as well as DAF-16/FOXO.
Sensory mutations lead to down-regulation of pathogen-response genes
Because some of the genes in our dataset were annotated as pathogen-responsive genes, we compared the genes differentially expressed in sensory mutants with the transcriptional profiles of C. elegans exposed to the bacterial pathogen P. aeruginosa PA14 [32], [33]. We found that a significant number of the genes down-regulated in daf-10 mutants on normal E. coli OP50 diet were up-regulated in response to exposure to PA14 in adult wild-type animals (Out of 56 down-regulated genes, 15 were classified as up-regulated by PA14 exposure in Shapira et al. [32] and 21 in Troemel et al. [33], p<0.0001 in both cases, hypergeometric probability, Table 3). Using quantitative RT-PCR analysis, we confirmed that four out of five PA14-responsive genes selected from our microarray analysis were down-regulated in daf-10 mutants on an OP50 diet (Figure 3A). In addition, we also found that daf-10 mutations resulted in a severe reduction in the induction of four out of the five genes following PA14 exposure (Figure 3A). These data suggest that the sensory system is required not only for the basal expression of these pathogen-responsive genes when animals are fed the normal laboratory food, OP50, but also for their full induction upon PA14 pathogen exposure. We then tested whether the daf-10 sensory mutants had an altered response to pathogens by measuring their survival on P. aeruginosa. Under standard testing conditions, we found results to be highly variable, but could detect a significant decrease in the survival of daf-10(m79) animals on PA14 in four out of ten trials (Figure 3B, Table S2).
Fig. 3. daf-10 mutations alter the worm's response to pathogens. 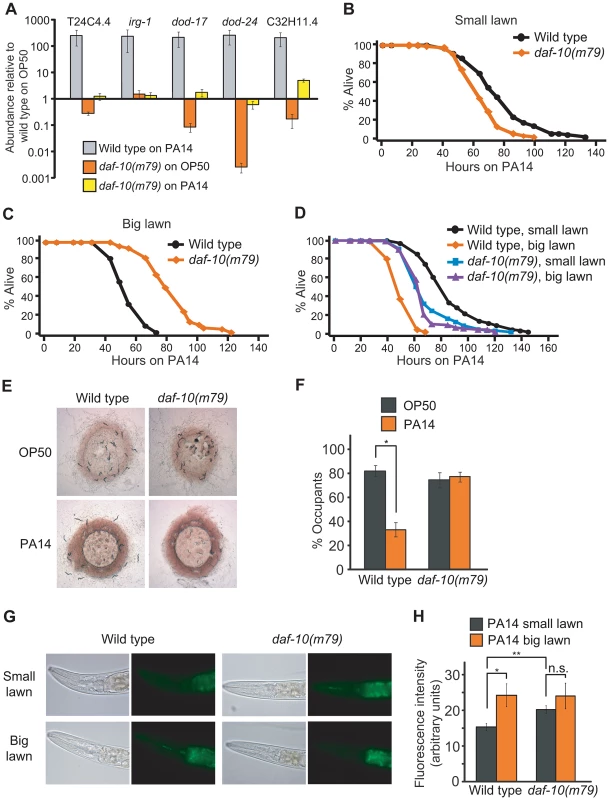
A. qRT-PCR analysis was performed to determine how five pathogen (PA14)-responsive genes were regulated in daf-10(m79) mutants in the presence of E. coli OP50, the normal laboratory food, or of pathogenic P. aeruginosa PA14 (big lawn). B–D. daf-10(m79) animals showed opposite phenotypes when fed P. aeruginosa in a small lawn assay (B) or in a big lawn assay (C). When the two assays were done in parallel (D), the survival of daf-10(m79) animals was similar in both conditions. In contrast, the survival of wild-type animals was dramatically reduced when a big lawn was used. A summary of the data presented in these panels and additional repeats is included in Table S2. E–F. Whereas wild-type animals avoided PA14, daf-10(m79) animals occupied lawns of E. coli OP50 and P. aeruginosa PA14 bacteria to a similar degree. Approximately 100 wild-type and daf-10(m79) worms were placed on lawns of PA14 and OP50 and imaged after 16 hrs (E). The % of worms that occupied (% occupants) the bacterial lawn was also determined (F). G–H. Representative pictures of the fluorescence signal from GFP-labeled PA14 bacteria in the intestine of worms (G) and quantitation of fluorescence signal (n≥26 worms per strain/condition) (H), showing the amount of PA14 ingested in small vs. big lawn assays by wild-type and daf-10(m79) worms. Error bars represent s.e.m. (n.s. p>0.05, * p<0.05, ** p<0.01, Student's t-test). Interestingly, a seemingly unrelated sensory system mutant, the npr-1 mutant, which has a defect in sensing oxygen concentrations, also presents an increased sensitivity to pathogens [9], [11]. Indeed, the shorter survival of daf-10(m79) mutant animals on P. aeruginosa, like that of npr-1 mutant animals [11], was dependent on the oxygen-sensing guanylate cyclase gcy-35, because gcy-35(ok769); daf-10(m79) animals were as resistant to P. aeruginosa as gcy-35(ok769) animals (Figure S2A). Although npr-1 mutant animals are more sensitive than wild-type animals under the usual “small lawn” assay conditions, their survival is similar to that of wild-type animals if assayed on a plate completely covered with bacteria (big lawn) [9]. This is likely because wild-type animals actively avoid the pathogenic bacteria in small lawn assays, but cannot do so in big lawn assays, whereas npr-1 mutants actively seek the lower oxygen concentrations provided by the thick P. aeruginosa bacterial lawn even in the small lawn assay [9]. When we used a big lawn to test the survival of daf-10(m79) animals on PA14, we found that not only daf-10(m79) animals were not short lived under these conditions, but they were in fact longer lived than wild-type animals (Figure 3C). This may indicate that daf-10(m79) animals are physiologically more resistant to pathogens, but are behaviorally unable to avoid them. When the assays were done in parallel, the survival of wild-type animals was 40% shorter in the big lawn assay than in the small lawn assay, whereas the survival of daf-10 mutants was not significantly different between the two conditions (Figure 3D). Consistent with these data, wild-type animals avoided PA14 and had lower occupancy of the PA14 bacterial lawn compared to the OP50 lawn, whereas daf-10 mutants displayed defects in avoiding PA14 (Figure 3E and 3F and Figure S2B). In addition, daf-10(m79) mutants ingested more GFP-PA14 than wild-type animals in the small lawn assay (Figure 3G and 3H).
Increased DAF-16/FOXO confers resistance to various pathogenic bacteria including PA14 [14], and we found that DAF-16/FOXO was required for the pathogen resistance of daf-10(m79) animals in the big lawn assay (Figure 4A). In contrast, daf-12/NHR mutation did not significantly affect the average survival time of daf-10 mutants on PA14 (Figure 4B). However, daf-12 mutations shortened the maximal survival time of daf-10(m79) animals on PA14, while increasing their survival at early time points. The difference in the survival curves was significant in three out of five trials when using the Wilcoxon test, which does not assume constant hazard ratios [40] (Figure S3 and Table S2). This may indicate a more subtle and complex effect of DAF-12/NHR on the response to pathogens, perhaps aiding in long-term survival but negatively impacting responses after short exposures. Alternatively, loss of daf-12 could simultaneously affect the activity of two different neuronal populations with opposing roles in regulating sensitivity to PA14, and thus result in a complex phenotype.
Fig. 4. The increased PA14 resistance of daf-10 mutants requires DAF-16/FOXO. 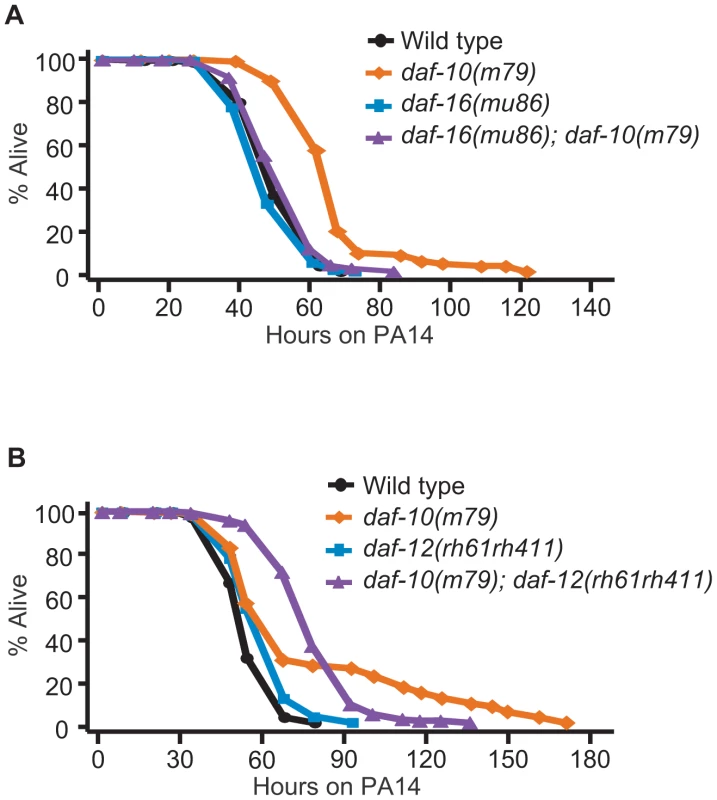
daf-16(mu86) mutations completely suppressed the PA14 resistance caused by daf-10(m79) mutations in the big lawn assay (A), whereas daf-12(rh61rh411) mutations did not affect mean survival (B). However, daf-12 mutations altered the survival curve of daf-10(m79) mutants on PA14 in a statistically significant manner (see Figure S3 for additional repeats). A summary of the data presented in these panels and additional repeats is included in Table S2. We have thus shown that daf-10(m79) animals have decreased expression of pathogen-responsive genes even when exposed to E. coli, but are in fact physiologically more resistant to pathogenic bacteria such as P. aeruginosa due to increased DAF-16/FOXO activity. In contrast, behavioral avoidance of P. aeruginosa is impaired in daf-10(m79) mutant worms, underscoring the importance of sensory input in the behavioral response to pathogens.
The up-regulated gene mct-1/2 is required for the lifespan increase of daf-10(m79) sensory mutant animals
Genes that are up-regulated in response to sensory system mutations may also be required for their effects on physiology. We focused on the role of these genes in lifespan regulation and tested whether knocking them down by RNA interference (RNAi) affected the lifespan of rrf-3(pk1426); daf-10(m79) animals. As neurons are usually refractory to RNAi, the rrf-3 mutation was used to increase neuronal sensitivity to the treatment [41], [42]. We were able to test 9 of the 14 up-regulated genes (for the remaining 5, RNAi clones were not available or did not grow). We identified one that was required for the extended longevity of rrf-3(pk1426); daf-10(m79) animals, the uncharacterized gene Y19D10A.12, which we named mct-1 since it encodes a putative monocarboxylate transporter (Figure 5A and 5B). mct-1 is located within a region of chromosome V that has been duplicated [43]. Therefore, chromosome V contains a second gene, C01B4.9 or mct-2, which is almost identical to mct-1 (Figure S4). Based on the reference sequences and gene models available in Wormbase WS230, mct-2 has a 319 base pair (bp) insertion in the intronic region between exon 2 and exon 3. In addition, the splicing of the 3rd exon of mct-1 and mct-2 differs, so that the mct-2 3rd exon likely contains 81 additional nucleotides at its 3′ end (Figure S4). The promoter sequence of the two genes also appears to be almost identical, the only difference being a single guanosine insertion in a G stretch 1629 bp 5′ of the start codon in mct-1 (Figure S4). This suggests that the expression and regulation of mct-1 and mct-2 is likely to be very similar. Because the RNAi treatment is likely to affect both genes and our qPCR primers do not distinguish between them (Figure S4), we are unable to separate their actions and we will henceforth refer to these genes collectively as mct-1/2.
Fig. 5. mct-1/2 is required for the long lifespan and dauer formation of daf-10 mutants. 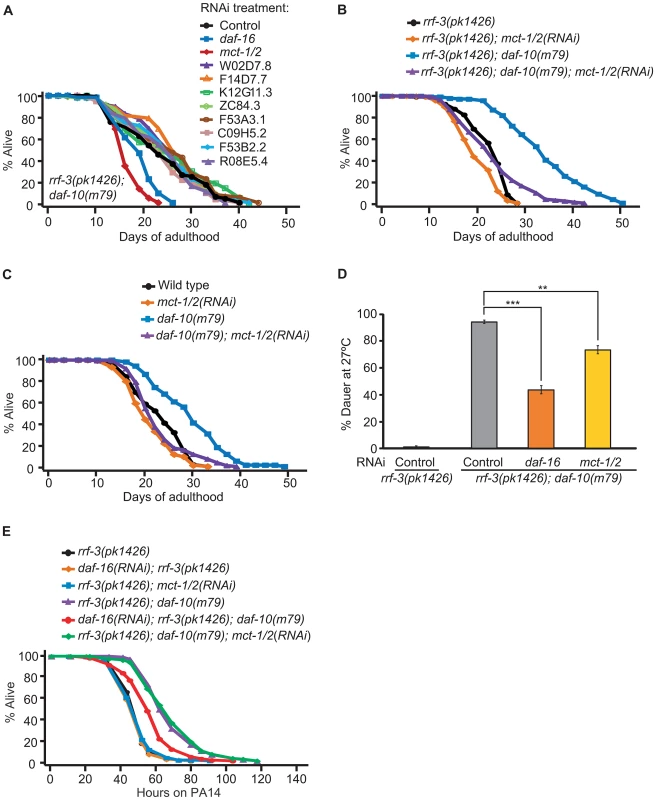
A. RNAi bacteria for 9 of the 14 up-regulated genes were obtained from the C. elegans RNAi libraries [65], [66]. rrf-3(pk1426); daf-10(m79) animals were treated with RNAi targeting the nine genes, control RNAi (containing empty vector) or daf-16 RNAi bacteria. The latter, as shown previously [3], shortened lifespan of the sensory mutant animals and served as a positive control. RNAi against mct-1/2 significantly decreased the long lifespan of rrf-3(pk1426); daf-10(m79) animals. B. RNAi targeting mct-1/2 almost completely suppressed the longevity of rrf-3(pk1426); daf-10(m79) animals, while having little effect on that of rrf-3(pk1426) animals. C. mct-1/2 was knocked down by RNAi in wild-type and daf-10(m79) animals. When grown on mct-1/2 RNAi, the life-extending effect of daf-10(m79) mutations was significantly reduced. D. mct-1/2 RNAi partially but significantly suppressed the constitutive dauer formation phenotype of daf-10(m79) mutants at 27°C. daf-16 RNAi was used as a positive control. Error bars represent s.e.m. (** p<0.01, *** p<0.001, Student's t-test). E. mct-1/2 RNAi did not affect the PA14 resistance due to daf-10(m79) mutations in the big lawn assay, whereas daf-16 RNAi did. The RNAi-hypersensitive rrf-3(pk1426) mutant background was used to potentiate the RNAi effect. A summary of the data presented in this figure is included in Tables S1B and S2. Given our initial finding, we decided to further characterize the effects of mct-1/2 and found that mct-1/2 knockdown also shortened the long lifespan of rrf-3(+); daf-10(m79) strains, while wild-type animals were largely unaffected (Figure 5C). mct-1/2 was also required for the increased dauer formation of daf-10(m79) animals at high temperature (Figure 5D). In contrast, knockdown of mct-1/2 did not affect the increased PA14 resistance of daf-10(m79) animals in the big lawn assay (Figure 5E).
To determine the role of mct-1/2 in other lifespan-extending pathways, we tested whether mct-1/2 RNAi could influence the lifespan of other long-lived C. elegans mutants. We found that the longevity of another long-lived sensory mutant strain, osm-5(p813), was not influenced by reduction in mct-1/2 levels (Figure 6A). This is consistent with our quantitative RT-PCR results showing that the mRNA levels of mct-1/2 were increased in daf-10 mutants but not in osm-5 mutants (Figure 1B and Figure S1B). Moreover, it suggests previously unappreciated differences in longevity regulation between these two sensory mutants. We also observed that RNAi knock-down of mct-1/2 had little or no effect on the long lifespan of daf-2/InsR mutants in either rrf-3(+) or rrf-3(−) backgrounds (Figure 6B and 6C), despite the fact that both daf-2/InsR and daf-10 mutant animals require DAF-16/FOXO for their longevity. Lastly, the extended longevity of dietary-restricted eat-2 mutants or mitochondrial respiration-defective isp-1 mutants was largely unaffected by mct-1/2 RNAi treatment (Figure 6D and 6E). These data suggest that the requirement of mct-1/2 for longevity regulation exhibits specificity for daf-10 sensory mutants.
Fig. 6. mct-1/2 is dispensable for the longevity of various long-lived mutants. 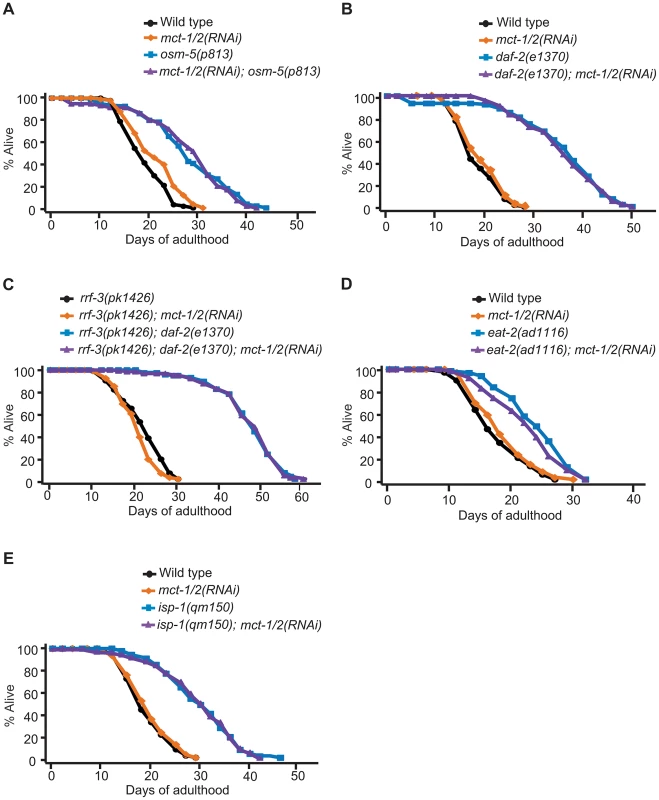
Reduction of mct-1/2 levels by RNAi did not shorten the lifespan of osm-5(p813) sensory mutants (3 out of 4 times) (A), daf-2(e1370) mutants in either rrf-3(+) (B) or rrf-3(−) (C) backgrounds, dietary-restricted eat-2(ad1116) animals (D) or mitochondrial respiration-impaired isp-1(qm150) mutants (E). A summary of the data presented in this figure and additional repeats is included in Table S1B. We decided to examine the expression pattern of mct-1/2 and to test whether mct-1/2 overexpression was sufficient to extend the lifespan of otherwise wild-type worms. We designed primers that would amplify the promoter and the cDNA of both genes, and we were able to clone promoter and cDNA sequences matching the gene model for mct-1. We then generated transgenic animals that express GFP-tagged MCT-1 protein under the mct-1 promoter (mct-1p::mct-1::GFP) and found that mct-1 was expressed in the pharynx (Figure 7A–7D). The expression pattern persisted from hatching to adulthood (data not shown), and GFP-tagged MCT-1 was not detected in six amphid sensory neurons that are labeled by DiI dye (Figure 7E–7H). As expected from the microarray and quantitative RT-PCR data, we found that daf-10(m79) mutations increased expression of the mct-1p::mct-1::GFP transgene in the pharynx, whereas the expression pattern of MCT-1::GFP was not altered by daf-10(m79) mutations (Figure 7I and 7J). These data imply that the sensory neuronal defects of the daf-10 mutants may increase the expression of mct-1 in the pharynx to promote longevity and dauer formation. In agreement with this hypothesis, overexpression of the MCT-1 transporter slightly but significantly extended the lifespan of otherwise normal animals in two different trials using three independent transgenic lines (Figure 7K and Table S1B). Collectively, these data indicate that a previously uncharacterized gene, mct-1/2 is specifically induced by certain sensory system alterations and contributes to the longevity associated with sensory system defects.
Fig. 7. Overexpression of mct-1 extends lifespan. 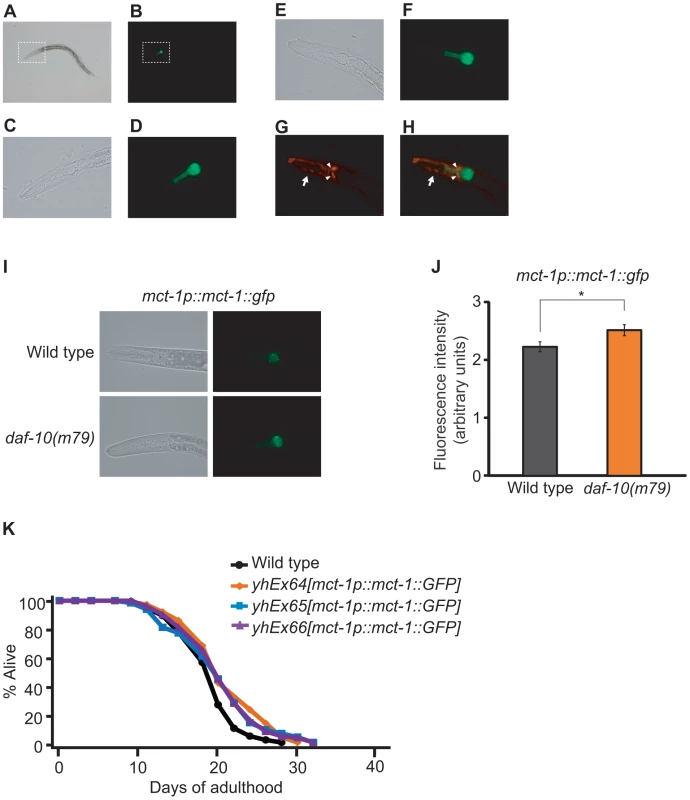
A–D. Expression pattern of GFP-fused MCT-1 protein in mct-1p::mct-1::GFP transgenic animals. Bright field image (A) and green fluorescence image (B) of an L4 mct-1p::mct-1::GFP transgenic larva. Magnified bright field image (C) and green fluorescence image (D) of dotted box regions in A and B respectively. E–H. The expression of GFP-fused MCT-1 protein in the mct-1p::mct-1::GFP transgenic animal did not overlap with the signal from amphid sensory neurons labeled with the DiI dye. Bright field image (E), green fluorescence image (F), red fluorescence image (G) and overlay of green and red fluorescence images (H) of the head of an L3 mct-1p::mct-1::GFP transgenic animal. Arrowheads indicate cell bodies of amphid sensory neurons that are stained with DiI dye. The arrow indicates body wall muscle that expresses a muscle-specific myo-3p::RFP transgene, used as a marker for the generation of the transgenic animals. I–J. Expression pattern of GFP-fused MCT-1 protein in daf-10(m79) mutants, including representative green fluorescence and bright field images (I) and quantification of fluorescence intensity (J). Error bars represent s.e.m. (* p<0.05, Student's t-test). K. Overexpression of mct-1::GFP lengthens lifespan. Three independent lines of mct-1 transgenic animals lived slightly but significantly longer than control animals. Statistical analysis of the lifespan data presented in this figure and additional repeats is included in Table S1B. Discussion
In this study, we sought to understand how changes in transcriptional programs downstream of alterations in sensory perception may lead to changes in organismal physiology such as extended longevity and altered developmental programs. We examined the whole-organism transcriptional profile of worms carrying a mutation in a gene encoding the worm homolog of the intraflagellar transport protein IFT222, daf-10. daf-10 mutations cause broad defects in the sensory system of the worms. As expected based on previous genetic data, the expression of targets of the transcription factor DAF-16/FOXO was altered in daf-10(m79) animals, confirming the validity of our approach. Moreover, we showed that the activity of another transcription factor involved in the regulation of lifespan and dauer formation, DAF-12/NHR, was also affected in daf-10 mutant worms and that DAF-12/NHR was specifically required for dauer formation. We also found that one of the genes that were up-regulated in daf-10(m79) animals, the monocarboxylate transporter homolog mct-1/2, was required for the longevity of sensory mutant animals. Lastly, we uncovered a high representation of immune response genes among the genes whose expression was reduced by the sensory mutation, which was likely related to the reduced pathogen avoidance of sensory mutant animals.
The up-regulation of DAF-16/FOXO targets in daf-10(m79) mutants is in accord with the fact that daf-16/FOXO is largely required for their longevity and their increased dauer formation. Our data further support the idea that one or multiple insulin-like peptides are misregulated in response to sensory system mutations contributing to the lifespan increase of worms with defective sensory neurons. It is interesting, however, that some genes that are typically used as reporters of DAF-16/FOXO activity, such as sod-3 and mtl-1, were not up-regulated in daf-10(m79) mutants based on our microarray data. qPCR analysis confirmed this observation (Figure S5). This hints at an unappreciated complexity in the relationship between the sensory system and insulin/IGF-1 signaling in C. elegans.
Our microarray data also support regulation of DAF-12/NHR activity by the sensory system because of the reduced expression of DAF-12/NHR target genes in daf-10(m79) mutant animals. This interpretation is consistent with the observation that daf-12/NHR is required for the increased dauer formation at high temperatures of the daf-10 (Figure 2E) and osm-5 sensory mutants (data not shown), as well as the tax-4 sensory mutant as previously reported [2]. In fact, daf-12/NHR is necessary for dauer formation under a range of conditions, which suggests either that many different dauer-inducing conditions affect the activity of DAF-12/NHR, or that basal expression of DAF-12/NHR target genes is required for entry in the dauer state. Because we observe that the DAF-12/NHR target genes are mostly unchanged in daf-12(rh61rh411) mutant animals relative to wild-type animals, we conclude that the first possibility is more likely, at least as far as sensory mutant worms are concerned, and that the sensory system actively regulates the daf-12/NHR signaling pathway.
The fact that lifespan, dauer formation and immune response (see below) are whole-organism events implies that hormonal signaling downstream of the sensory system may be necessary to coordinate whole-body changes. Whereas steroid signals through DAF-12/NHR may fulfill that role for dauer formation, it is unclear at present what such a signal may be for the lifespan phenotype. Among the genes that were up-regulated in daf-10(m79) animals, one gene, mct-1/2, was also required for the extended longevity of these mutants. mct-1 and its nearly identical paralog, mct-2, are homologous to monocarboxylate transporters, which can transport small molecule hormones such as thyroid hormones in mammalian systems [44]. Moreover, expression of a Drosophila melanogaster homolog of monocarboxylate transporters is altered in flies after glucose feeding [45], suggesting a potential link to insulin/IGF-1-like signaling. Thus, it is possible that the specific role of mct-1/2 in the lifespan of sensory mutant animals is linked to the signals that help coordinate the development and aging of different tissues. In addition, mct-1/2 is one of the first genes to be identified as a specific regulator of longevity downstream of the sensory system. Further analysis of the biochemical function of this gene will thus provide crucial information on how sensory perception regulates lifespan. Our finding that mct-1/2 is not required for the longevity of osm-5 mutant animals suggests that daf-10 and osm-5 sensory mutations control divergent downstream pathways, rather than converging on the same regulatory mechanism. This is consistent with the differential effect these mutations have on subsets of sensory neurons [18], despite the fact that they both cause pervasive defects in the development of amphid sensory cilia. For example, the cilia of some amphid neurons (ADF, ASH, ALD) are still partially intact in daf-10(m79) mutants, and these animals are not defective in chemotaxis toward NaCl, unlike osm-5 animals [18]. Conversely, daf-10(m79) animals are defective in male mating, whereas osm-5(p813) animals are not, suggesting the daf-10 mutation leads to stronger impairment of neurons involved in mating [18]. This is also consistent with previous studies showing that multiple sensory neurons control different pathways that regulate lifespan, and can influence longevity in both a positive and a negative direction [3], [6], [8]. Moreover, we cannot exclude that the mutations have different effects on signaling even in those neurons where the morphological effects are similar.
The microarray data also point at a role for the neurons affected by daf-10(m79) mutations in regulating the response to bacterial pathogens such as Pseudomonas aeruginosa. daf-10 mutations not only alter the expression of genes that are normally responsive to P. aeruginosa exposure, but also prevent worms from leaving the lawn of pathogenic bacteria and simultaneously increase the resistance of the animals to P. aeruginosa. In recent studies, several different components of the sensory system, including oxygen-sensing neurons [9], [11], serotonin pathways [10], [13] and neuropeptide pathways [46], have been implicated in the two main protective responses to P. aeruginosa exposure: up-regulation of protective anti-microbial genes and decreased pathogen consumption due to avoidance of the bacterial lawn. Because the daf-10 mutations affect the function of a number of ciliated neurons, it is unclear at present how different sensory modalities may contribute to the phenotype of daf-10(m79) animals. For example, the oxygen-sensing gcy-35-expressing neurons AQR and PQR [47], [48] are ciliated neurons, and the daf-10 survival phenotype on small PA14 lawns resembles that of the npr-1 mutant, in which these neurons are hyperactive [49]. It is possible that in daf-10(m79) animals, lack of proper sensory cilia in AQR and PQR results in hyperactive signaling from the neurons, preventing the animals from leaving the lawn. The subsequent increased pathogen consumption would then mask the physiological resistance to pathogens that becomes apparent in the big lawn assay. This interpretation would be consistent with the fact that mutations in the oxygen-sensing guanylate cyclase gcy-35 suppress the increased pathogen sensitivity of daf-10(m79) mutants in the small lawn assay (Figure S2A). Hyperactivation of defective sensory neurons is also consistent with the paradoxical observation that daf-10 mutants are considered Daf-d (dauer defective) because they are insensitive to dauer pheromone [50], and yet these animal arrest more readily as dauers at high temperature [2], [3]. In contrast, the decreased sensitivity of daf-10(m79) animals to PA14 in the big lawn assay likely results from the activation of the DAF-16/FOXO transcription factor. Although this “competing signals” model is consistent with the data, we also noticed that the survival of daf-10(m79) animals was similar between the two types of PA14 resistance assays, while the survival of wild-type animals was shorter in the big lawn assay. This suggests an alternative model whereby reduced pathogen perception due to sensory mutations results in the inability of daf-10(m79) mutant animals to modulate their physiology and their behavior in the presence of different bacterial strains. The altered expression of pathogen response genes may also stem from an inability of daf-10(m79) animals to perceive the presence of Escherichia coli, which may be slightly pathogenic for worms [51], [52].
The importance of the neuronal regulation of many basic physiological processes has been appreciated only recently. In mammals, hormonal release regulated by the hypothalamus has an important role in energy homeostasis, fat storage, immune responses and possibly aging (reviewed in [53]). For example, recent studies have shown that loss of the IGF-1 receptor specifically in the brain has profound effects on energy homeostasis and results in increased lifespan by changing the growth hormone axis [54]. Interestingly, the hypothalamus receives direct innervation from many cortical areas devoted to sensory processing [55], including the olfactory cortex and the insular cortex, which processes gustatory information, pointing at a clear role for environmental input in the regulation of physiology. Although the C. elegans circuitry is much more compact than that of mammals, studying the signal relay from the nervous system to peripheral tissues in this simple, genetically tractable organism, may give insights into what pathways may be used in higher organisms to mediate similar signals. In this context, our study highlights several transcriptional changes that correspond to precise phenotypes, providing examples of how neuronal signaling changes may be translated into an organismal physiological output.
Materials and Methods
Strains
Nematodes were raised under standard laboratory conditions on agar plates containing a lawn of Escherichia coli strain OP50, as described previously [56]. “Wild type” was the C. elegans strain N2. The mutant and transgenic strains used were as follows: CF2100 daf-10(m79), CF2479 daf-12(rh61rh411), CF2553 osm-5(p813), CF3152 rrf-3(pk1426), CF3295 daf-10(m79); daf-12(rh61rh411), CF3302 daf-16(mu86); daf-10(m79), CF3369 daf-16(mu86); daf-10(m79); daf-12(rh61rh411), CF3389 gcy-35(ok769), CF3390 gcy-35(ok769); daf-10(m79), CF3391 rrf-3(pk1426); daf-10(m79), CF1814 rrf-3(pk1426); daf-2(e1370), CF1041 daf-2(e1370), CF2172 isp-1(qm150), CF1908 eat-2(ad1116), IJ294 yhEx64[mct-1p::mct-1::gfp, myo-3p::rfp], IJ295 yhEx65[mct-1p::mct-1::gfp, myo-3p::rfp], IJ296 yhEx66 [mct-1p::mct-1::gfp, myo-3p::rfp], IJ374 daf-10(m79); yhEx64 [mct-1p::mct-1cDNA::gfp, myo-3p::rfp]. All strains were outcrossed to an isogenic N2 strain at least three times.
Microarray analysis
To make sure that background differences did not affect our results, prior to doing microarray analysis we outcrossed daf-10(m79) animals four times to our N2 strain. We confirmed that the outcrossed daf-10(m79) animals lived approximately 30% longer than wild-type animals and that this increase was largely dependent on daf-16/FOXO (data not shown). Wild-type (N2) and daf-10(m79) worms were synchronized by arresting at L1 overnight in M9 buffer. They were then grown at 20°C and collected as young adults. Total RNA was purified using TriZol reagent (Invitrogen), and mRNA was purified using Oligotex kit (Qiagen). cDNA was generated, coupled to Cy3/Cy5 dyes and hybridized to single-stranded DNA nucleotide arrays printed in-house using standard techniques. The oligonucleotides were purchased from Illumina and represented 20,374 unique C. elegans genes. Three repeats of a direct comparison between wild-type and daf-10(m79) animals were carried out. Chips were scanned using a GenePix 4000B scanner, and initial quality check and identification of spots was done using Genepix 6.0 software. Linear normalization was carried out with the Acuity 4.0 software and significance analysis using the Cyber T-test program [57]. Genes with a p value lower than 0.02 were considered significant. The significant gene list was compared to known gene lists using the hypergeometric probability. Microarray data have been deposited in the Gene Expression Omnibus database under accession number GSE41943.
Quantitative RT–PCR
RNA extraction using TriZol reagent (Invitrogen), purification using RNeasy kit (Qiagen) and reverse transcription using Proto-Script 1st strand cDNA synthesis kit (NEB) were performed as described [58]. Quantitative RT-PCR was performed using a 7300 Real Time PCR System (Applied Biosystems) and analyzed by the Ct method (Applied Biosystems Prism 7700 Users Bulletin No. 2 http://docs.appliedbiosystems.com/pebiodocs/04303859.pdf). mRNA levels of act-1, nhr-23 and ama-1 were used for normalization. Raw values were normalized to each of the three control genes and the average of the normalized values was used as the data point. The average of at least two technical repeats was used for each biological repeat. The data shown in the figures are the average of at least three biological repeats. Primer sequences are listed in Table S3.
Lifespan analysis
Lifespan analysis was conducted as described previously [59]. All assays were carried out at 20°C. For RNAi experiments, eggs were placed on lawn of double-stranded RNA-expressing E. coli HT115. The lifespan of the progeny of these animals was assayed on RNAi bacteria. A bacterial strain containing an empty vector was used as control. Some assays were carried out in the presence of 75 µM 5′-fluorodeoxyuridine (Sigma). OASIS (online application for the survival analysis, http://sbi.postech.ac.kr/oasis) was used to analyze the data [60], and p values were calculated using the log-rank test (Mantel-Cox method).
Pathogen resistance analysis
Pathogen resistance assays were carried out as described with minor modifications [61]. Pseudomonas aeruginosa strain PA14-coated plates were made by seeding 5 µl of an overnight culture of PA14 in Terrific broth and incubating at 37°C overnight. For big lawn assays, 15 µl of cultured PA14 overnight was seeded and spread in order to cover the whole surface of the plate, and incubated 37°C overnight [9]. Plates were subsequently kept in a 25°C incubator. Worms were grown on E. coli OP50 and placed on PA14 plates at day 1 of adulthood. Plates were examined one to three times per day, and animals were scored as alive if they moved when prodded. Animals that crawled onto the sides of the plates (where they died of desiccation) were excluded from the analysis. OASIS (Online Application for the Survival Analysis, http://sbi.postech.ac.kr/oasis) [60] was used to analyze the data, and p values were calculated using the log-rank test (Mantel-Cox method) and the Wilcoxon test, which does not assume constant hazard ratios [40], [62], because differences in the shape of the survival curves for one set of data [daf-10(m79) vs. daf-10(m79); daf-12(rh61rh411)] suggested the latter test may be more appropriate for some of the comparisons.
GFP-PA14 intake assays
The assays were performed as described [9] using GFP-expressing Pseudomonas aeruginosa (GFP-PA14) to measure PA14 intake. Worms were grown on OP50 until they reached adulthood, and then placed on plates seeded with small and big lawns of GFP-PA14 for 24 hours. Images of GFP-PA14-fed worms were captured using AxioCam (Zeiss Corporation, Germany) with HRc Zeiss Axioscope A.1 (Zeiss Corporation, Germany). GFP fluorescence was quantified using ImageJ (http://rsbweb.nih.gov/ij/).
Pathogen avoidance assays
The assays were performed as described with minor modifications [63]. Five µl of cultured PA14 or OP50 was seeded on high peptone (0.35%) agar plates to prepare plates with small lawns of bacteria. After incubation for 24 hours at 37°C, plates were moved to 25°C and used for the avoidance assay within 24 hours. Approximately 100 young (day 1) adult worms were transferred onto plates seeded with PA14 or OP50. The number of occupants was counted every 4 hours for 16 hours after the transfer. Images of the plates were captured using DIMIS-M (Siwon Optical Technology, South Korea) camera.
Dauer assays
Embryos were incubated at 27°C for 2 days, and plates were then examined for the presence of dauer larvae. For the RNAi experiment in Figure 5D, rrf-3(pk1426); daf-10(m79) mutant embryos were placed on plates seeded with control bacteria (containing empty vector), or bacteria expressing double-stranded RNA targeting daf-16/FOXO or mct-1/2 at 20°C. The progeny of these animals were transferred to 27°C as embryos, and scored for the presence of dauer larvae two days later. The experiments were repeated eight times.
Generation and analysis of transgenic animals
To generate transgenic animals overexpressing GFP-fused MCT-1, approximately 2 kb upstream of the mct-1 coding region and the cDNA of mct-1 were PCR amplified. The PCR products were then fused to Gateway donor vectors using BP clonase (Invitrogen) followed by recombination with a Gateway destination vector containing GFP and unc-54 3′UTR using LR clonase (Invitrogen). The DNA construct was injected into the gonads of young adults at a concentration of 25 ng/µl with 75 ng/µl of a myo-3p::RFP marker. Images of transgenic worms were captured using AxioCam (Zeiss Corporation, Germany) with HRc Zeiss Axioscope A.1 (Zeiss Corporation, Germany). GFP fluorescence was quantified using ImageJ (http://rsbweb.nih.gov/ij/).
Di<$>\scale 130%\raster="rg1"\<$> staining
DiI staining was performed as described previously with some modifications [64]. DiI staining solution was prepared by diluting DiI (1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate, Invitrogen) in M9 buffer to a final concentration of 40 µg/ml. Approximately 30 L4 larvae were washed with M9 buffer three times and incubated in the DiI staining solution for 2 hours at 20°C while shaking (200 rpm). The worms were then washed with M9 buffer three times and placed on OP50-seeded NGM plates for 10 minutes. They were subsequently mounted on 2% agarose pads and anesthetized with 100 mM sodium azide. Images of the stained worms were captured using an AxioCam HRc (Zeiss Corporation, Germany) camera attached to a Zeiss Axioscope A.1 microscope (Zeiss Corporation, Germany).
Supporting Information
Zdroje
1. InglisPN, OuG, LerouxMR, ScholeyJM (2007) The sensory cilia of Caenorhabditis elegans. WormBook 1–22.
2. AilionM, ThomasJH (2000) Dauer formation induced by high temperatures in Caenorhabditis elegans. Genetics 156 : 1047–1067.
3. ApfeldJ, KenyonC (1999) Regulation of lifespan by sensory perception in Caenorhabditis elegans. Nature 402 : 804–809.
4. BargmannCI, HorvitzHR (1991) Control of larval development by chemosensory neurons in Caenorhabditis elegans. Science 251 : 1243–1246.
5. GagliaMM, KenyonC (2009) Stimulation of movement in a quiescent, hibernation-like form of Caenorhabditis elegans by dopamine signaling. J Neurosci 29 : 7302–7314.
6. AlcedoJ, KenyonC (2004) Regulation of C. elegans longevity by specific gustatory and olfactory neurons. Neuron 41 : 45–55.
7. JeongDE, ArtanM, SeoK, LeeSJ (2012) Regulation of lifespan by chemosensory and thermosensory systems: findings in invertebrates and their implications in mammalian aging. Front Genet 3 : 218.
8. LeeSJ, KenyonC (2009) Regulation of the longevity response to temperature by thermosensory neurons in Caenorhabditis elegans. Curr Biol 19 : 715–722.
9. ReddyKC, AndersenEC, KruglyakL, KimDH (2009) A polymorphism in npr-1 is a behavioral determinant of pathogen susceptibility in C. elegans. Science 323 : 382–384.
10. ShiversRP, KooistraT, ChuSW, PaganoDJ, KimDH (2009) Tissue-specific activities of an immune signaling module regulate physiological responses to pathogenic and nutritional bacteria in C. elegans. Cell Host Microbe 6 : 321–330.
11. StyerKL, SinghV, MacoskoE, SteeleSE, BargmannCI, et al. (2008) Innate immunity in Caenorhabditis elegans is regulated by neurons expressing NPR-1/GPCR. Science 322 : 460–464.
12. SunJ, SinghV, Kajino-SakamotoR, AballayA (2011) Neuronal GPCR controls innate immunity by regulating noncanonical unfolded protein response genes. Science 332 : 729–732.
13. ZhangY, LuH, BargmannCI (2005) Pathogenic bacteria induce aversive olfactory learning in Caenorhabditis elegans. Nature 438 : 179–184.
14. GarsinDA, VillanuevaJM, BegunJ, KimDH, SifriCD, et al. (2003) Long-lived C. elegans daf-2 mutants are resistant to bacterial pathogens. Science 300 : 1921.
15. LinK, HsinH, LibinaN, KenyonC (2001) Regulation of the Caenorhabditis elegans longevity protein DAF-16 by insulin/IGF-1 and germline signaling. Nat Genet 28 : 139–145.
16. VowelsJJ, ThomasJH (1992) Genetic analysis of chemosensory control of dauer formation in Caenorhabditis elegans. Genetics 130 : 105–123.
17. BellLR, StoneS, YochemJ, ShawJE, HermanRK (2006) The molecular identities of the Caenorhabditis elegans intraflagellar transport genes dyf-6, daf-10 and osm-1. Genetics 173 : 1275–1286.
18. PerkinsLA, HedgecockEM, ThomsonJN, CulottiJG (1986) Mutant sensory cilia in the nematode Caenorhabditis elegans. Dev Biol 117 : 456–487.
19. WangJ, SchwartzHT, BarrMM (2010) Functional specialization of sensory cilia by an RFX transcription factor isoform. Genetics 186 : 1295–1307.
20. HaycraftCJ, SwobodaP, TaulmanPD, ThomasJH, YoderBK (2001) The C. elegans homolog of the murine cystic kidney disease gene Tg737 functions in a ciliogenic pathway and is disrupted in osm-5 mutant worms. Development 128 : 1493–1505.
21. QinH, RosenbaumJL, BarrMM (2001) An autosomal recessive polycystic kidney disease gene homolog is involved in intraflagellar transport in C. elegans ciliated sensory neurons. Curr Biol 11 : 457–461.
22. PauliF, LiuY, KimYA, ChenPJ, KimSK (2006) Chromosomal clustering and GATA transcriptional regulation of intestine-expressed genes in C. elegans. Development 133 : 287–295.
23. Von StetinaSE, WatsonJD, FoxRM, OlszewskiKL, SpencerWC, et al. (2007) Cell-specific microarray profiling experiments reveal a comprehensive picture of gene expression in the C. elegans nervous system. Genome Biol 8: R135.
24. KimSK, LundJ, KiralyM, DukeK, JiangM, et al. (2001) A gene expression map for Caenorhabditis elegans. Science 293 : 2087–2092.
25. SchinkelAH (1997) The physiological function of drug-transporting P-glycoproteins. Semin Cancer Biol 8 : 161–170.
26. ShepsJA, RalphS, ZhaoZ, BaillieDL, LingV (2004) The ABC transporter gene family of Caenorhabditis elegans has implications for the evolutionary dynamics of multidrug resistance in eukaryotes. Genome Biol 5: R15.
27. KurzCL, ShapiraM, ChenK, BaillieDL, TanMW (2007) Caenorhabditis elegans pgp-5 is involved in resistance to bacterial infection and heavy metal and its regulation requires TIR-1 and a p38 map kinase cascade. Biochem Biophys Res Commun 363 : 438–443.
28. Huang daW, ShermanBT, LempickiRA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4 : 44–57.
29. Huang daW, ShermanBT, LempickiRA (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37 : 1–13.
30. FisherAL, LithgowGJ (2006) The nuclear hormone receptor DAF-12 has opposing effects on Caenorhabditis elegans lifespan and regulates genes repressed in multiple long-lived worms. Aging Cell 5 : 127–138.
31. MurphyCT, McCarrollSA, BargmannCI, FraserA, KamathRS, et al. (2003) Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature 424 : 277–283.
32. ShapiraM, HamlinBJ, RongJ, ChenK, RonenM, et al. (2006) A conserved role for a GATA transcription factor in regulating epithelial innate immune responses. Proc Natl Acad Sci U S A 103 : 14086–14091.
33. TroemelER, ChuSW, ReinkeV, LeeSS, AusubelFM, et al. (2006) p38 MAPK regulates expression of immune response genes and contributes to longevity in C. elegans. PLoS Genet 2: e183 doi:10.1371/journal.pgen.0020183.
34. LeeSJ, MurphyCT, KenyonC (2009) Glucose shortens the life span of C. elegans by downregulating DAF-16/FOXO activity and aquaporin gene expression. Cell Metab 10 : 379–391.
35. McElweeJJ, SchusterE, BlancE, ThomasJH, GemsD (2004) Shared transcriptional signature in Caenorhabditis elegans Dauer larvae and long-lived daf-2 mutants implicates detoxification system in longevity assurance. J Biol Chem 279 : 44533–44543.
36. ShostakY, Van GilstMR, AntebiA, YamamotoKR (2004) Identification of C. elegans DAF-12-binding sites, response elements, and target genes. Genes Dev 18 : 2529–2544.
37. GerischB, WeitzelC, Kober-EisermannC, RottiersV, AntebiA (2001) A hormonal signaling pathway influencing C. elegans metabolism, reproductive development, and life span. Dev Cell 1 : 841–851.
38. HsinH, KenyonC (1999) Signals from the reproductive system regulate the lifespan of C. elegans. Nature 399 : 362–366.
39. KomatsuH, MoriI, RheeJS, AkaikeN, OhshimaY (1996) Mutations in a cyclic nucleotide-gated channel lead to abnormal thermosensation and chemosensation in C. elegans. Neuron 17 : 707–718.
40. HarringtonDP, FlemingTR (1982) A class of rank test procedures for censored survival data. Biometrika 69 : 553–566.
41. TimmonsL, CourtDL, FireA (2001) Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 263 : 103–112.
42. SimmerF, TijstermanM, ParrishS, KoushikaSP, NonetML, et al. (2002) Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr Biol 12 : 1317–1319.
43. VergaraIA, MahAK, HuangJC, Tarailo-GraovacM, JohnsenRC, et al. (2009) Polymorphic segmental duplication in the nematode Caenorhabditis elegans. BMC Genomics 10 : 329.
44. VisserWE, FriesemaEC, VisserTJ (2011) Minireview: thyroid hormone transporters: the knowns and the unknowns. Mol Endocrinol 25 : 1–14.
45. ZinkeI, SchutzCS, KatzenbergerJD, BauerM, PankratzMJ (2002) Nutrient control of gene expression in Drosophila: microarray analysis of starvation and sugar-dependent response. EMBO J 21 : 6162–6173.
46. KawliT, TanMW (2008) Neuroendocrine signals modulate the innate immunity of Caenorhabditis elegans through insulin signaling. Nat Immunol 9 : 1415–1424.
47. CheungBH, Arellano-CarbajalF, RybickiI, de BonoM (2004) Soluble guanylate cyclases act in neurons exposed to the body fluid to promote C. elegans aggregation behavior. Curr Biol 14 : 1105–1111.
48. GrayJM, KarowDS, LuH, ChangAJ, ChangJS, et al. (2004) Oxygen sensation and social feeding mediated by a C. elegans guanylate cyclase homologue. Nature 430 : 317–322.
49. CoatesJC, de BonoM (2002) Antagonistic pathways in neurons exposed to body fluid regulate social feeding in Caenorhabditis elegans. Nature 419 : 925–929.
50. AlbertPS, BrownSJ, RiddleDL (1981) Sensory control of dauer larva formation in Caenorhabditis elegans. J Comp Neurol 198 : 435–451.
51. GariganD, HsuAL, FraserAG, KamathRS, AhringerJ, et al. (2002) Genetic analysis of tissue aging in Caenorhabditis elegans: a role for heat-shock factor and bacterial proliferation. Genetics 161 : 1101–1112.
52. GemsD, RiddleDL (2000) Genetic, behavioral and environmental determinants of male longevity in Caenorhabditis elegans. Genetics 154 : 1597–1610.
53. Purves DA, George J., Fitzpatrick, David, Katz, Lawrence C., LaMantia, Anthony-Samuel, McNamara, James O., Williams S. Mark. (2001) Neuroscience. Sunderland (MA): Sinauer Associates, Inc.
54. KappelerL, Filho CdeM, DupontJ, LeneuveP, CerveraP, et al. (2008) Brain IGF-1 receptors control mammalian growth and lifespan through a neuroendocrine mechanism. PLoS Biol 6: e254 doi:10.1371/journal.pbio.0060254.
55. RisoldPY, ThompsonRH, SwansonLW (1997) The structural organization of connections between hypothalamus and cerebral cortex. Brain Res Brain Res Rev 24 : 197–254.
56. BrennerS (1974) The genetics of Caenorhabditis elegans. Genetics 77 : 71–94.
57. BaldiP, LongAD (2001) A Bayesian framework for the analysis of microarray expression data: regularized t -test and statistical inferences of gene changes. Bioinformatics 17 : 509–519.
58. TaubertS, Van GilstMR, HansenM, YamamotoKR (2006) A Mediator subunit, MDT-15, integrates regulation of fatty acid metabolism by NHR-49-dependent and -independent pathways in C. elegans. Genes Dev 20 : 1137–1149.
59. LeeSJ, HwangAB, KenyonC (2010) Inhibition of respiration extends C. elegans life span via reactive oxygen species that increase HIF-1 activity. Curr Biol 20 : 2131–2136.
60. YangJS, NamHJ, SeoM, HanSK, ChoiY, et al. (2011) OASIS: online application for the survival analysis of lifespan assays performed in aging research. PLoS ONE 6: e23525 doi:10.1371/journal.pone.0023525.
61. TanMW, RahmeLG, SternbergJA, TompkinsRG, AusubelFM (1999) Pseudomonas aeruginosa killing of Caenorhabditis elegans used to identify P. aeruginosa virulence factors. Proc Natl Acad Sci U S A 96 : 2408–2413.
62. LeeET, GoOT (1997) Survival analysis in public health research. Annu Rev Public Health 18 : 105–134.
63. PradelE, ZhangY, PujolN, MatsuyamaT, BargmannCI, et al. (2007) Detection and avoidance of a natural product from the pathogenic bacterium Serratia marcescens by Caenorhabditis elegans. Proc Natl Acad Sci U S A 104 : 2295–2300.
64. Shaham S, ed. (2006) WormBook: Methods in Cell Biology. In: Community TCeR, editor. Wormbook.
65. KamathRS, FraserAG, DongY, PoulinG, DurbinR, et al. (2003) Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421 : 231–237.
66. RualJF, CeronJ, KorethJ, HaoT, NicotAS, et al. (2004) Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res 14 : 2162–2168.
67. Hunt-NewburyR, ViveirosR, JohnsenR, MahA, AnastasD, et al. (2007) High-throughput in vivo analysis of gene expression in Caenorhabditis elegans. PLoS Biol 5: e237 doi:10.1371/journal.pbio.0050237.
68. ZhaoZ, ShepsJA, LingV, FangLL, BaillieDL (2004) Expression analysis of ABC transporters reveals differential functions of tandemly duplicated genes in Caenorhabditis elegans. J Mol Biol 344 : 409–417.
69. MeissnerB, RogalskiT, ViveirosR, WarnerA, PlastinoL, et al. (2011) Determining the sub-cellular localization of proteins within Caenorhabditis elegans body wall muscle. PLoS ONE 6: e19937 doi:10.1371/journal.pone.0019937.
70. MochiiM, YoshidaS, MoritaK, KoharaY, UenoN (1999) Identification of transforming growth factor-beta - regulated genes in Caenorhabditis elegans by differential hybridization of arrayed cDNAs. Proc Natl Acad Sci U S A 96 : 15020–15025.
71. EstesKA, DunbarTL, PowellJR, AusubelFM, TroemelER (2010) bZIP transcription factor zip-2 mediates an early response to Pseudomonas aeruginosa infection in Caenorhabditis elegans. Proc Natl Acad Sci U S A 107 : 2153–2158.
72. HaoY, XuN, BoxAC, SchaeferL, KannanK, et al. (2011) Nuclear cGMP-dependent kinase regulates gene expression via activity-dependent recruitment of a conserved histone deacetylase complex. PLoS Genet 7: e1002065 doi:10.1371/journal.pgen.1002065.
73. ChuDS, LiuH, NixP, WuTF, RalstonEJ, et al. (2006) Sperm chromatin proteomics identifies evolutionarily conserved fertility factors. Nature 443 : 101–105.
74. NehrkeK, MelvinJE (2002) The NHX family of Na+-H+ exchangers in Caenorhabditis elegans. J Biol Chem 277 : 29036–29044.
Štítky
Genetika Reprodukční medicína
Článek Defining the Pathways Underlying the Prolonged PR Interval in Atrioventricular Conduction DiseaseČlánek Translation in Giant Viruses: A Unique Mixture of Bacterial and Eukaryotic Termination SchemesČlánek Controls Gliogenesis by Regulating the Transient Expression of the Gcm/Glide Fate Determinant
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 12
-
Všechny články tohoto čísla
- A Mystery Unraveled: Essentiality of RNase III in Is Caused by Resident Prophages
- Defining the Pathways Underlying the Prolonged PR Interval in Atrioventricular Conduction Disease
- Insertion/Deletion Polymorphisms in the Promoter Are a Risk Factor for Bladder Exstrophy Epispadias Complex
- Mi2β Is Required for γ-Globin Gene Silencing: Temporal Assembly of a GATA-1-FOG-1-Mi2 Repressor Complex in β-YAC Transgenic Mice
- Dissection of a Quantitative Trait Locus for PR Interval Duration Identifies as a Novel Modulator of Cardiac Conduction
- Dnmt3a Protects Active Chromosome Domains against Cancer-Associated Hypomethylation
- Construction of a Global Pain Systems Network Highlights Phospholipid Signaling as a Regulator of Heat Nociception
- Genes Contributing to Pain Sensitivity in the Normal Population: An Exome Sequencing Study
- Identification of , a Locus Controlling Dominant Resistance to Autoimmune Orchitis, as Kinesin Family Member 1C
- ATX1-Generated H3K4me3 Is Required for Efficient Elongation of Transcription, Not Initiation, at ATX1-Regulated Genes
- Dynamic and Differential Regulation of Stem Cell Factor FoxD3 in the Neural Crest Is Encrypted in the Genome
- Identification of Novel Type 2 Diabetes Candidate Genes Involved in the Crosstalk between the Mitochondrial and the Insulin Signaling Systems
- The Genetic Architecture of Adaptations to High Altitude in Ethiopia
- Population Genomics of the Endosymbiont in
- Translation in Giant Viruses: A Unique Mixture of Bacterial and Eukaryotic Termination Schemes
- Testicular Differentiation Occurs in Absence of R-spondin1 and Sox9 in Mouse Sex Reversals
- A Yeast GSK-3 Kinase Mck1 Promotes Cdc6 Degradation to Inhibit DNA Re-Replication
- Genetic Adaptation Associated with Genome-Doubling in Autotetraploid
- The Essential Function of RNase III Is to Silence Foreign Toxin Genes
- Long-Range Regulatory Polymorphisms Affecting a GABA Receptor Constitute a Quantitative Trait Locus (QTL) for Social Behavior in
- A New Isolation with Migration Model along Complete Genomes Infers Very Different Divergence Processes among Closely Related Great Ape Species
- Chromosome Fragile Sites in Harbor Matrix Attachment Regions That May Be Associated with Ancestral Chromosome Rearrangement Events
- Genome-Wide Association Study Implicates Testis-Sperm Specific as a Susceptibility Locus for Impaired Acrosome Reaction in Stallions
- A Mechanism of Gene Amplification Driven by Small DNA Fragments
- Base Damage within Single-Strand DNA Underlies Hypermutability Induced by a Ubiquitous Environmental Agent
- Integrative Analysis of a Cross-Loci Regulation Network Identifies as a Gene Regulating Insulin Secretion from Pancreatic Islets
- Reciprocal Signaling between the Ectoderm and a Mesendodermal Left-Right Organizer Directs Left-Right Determination in the Sea Urchin Embryo
- Population Genomics of Sub-Saharan : African Diversity and Non-African Admixture
- Genome-Wide Fine-Scale Recombination Rate Variation in
- Deciphering the Transcriptional-Regulatory Network of Flocculation in
- On Lung Function and Interactions Using Genome-Wide Data
- Genome-Wide Joint Meta-Analysis of SNP and SNP-by-Smoking Interaction Identifies Novel Loci for Pulmonary Function
- The Effective Population Size of Malaria Mosquitoes: Large Impact of Vector Control
- Recessive Mutations in Implicate β-III Spectrin in Both Cognitive and Motor Development
- An Excess of Gene Expression Divergence on the X Chromosome in Embryos: Implications for the Faster-X Hypothesis
- Reduced Life- and Healthspan in Mice Carrying a Mono-Allelic MVA Mutation
- Natural Variation at the MATE Transporter Locus Reveals Cross-Talk between Fe Homeostasis and Zn Tolerance in
- Histone Deacetylase HDA6 Is Functionally Associated with AS1 in Repression of Genes in
- A Framework for the Establishment of a Cnidarian Gene Regulatory Network for “Endomesoderm” Specification: The Inputs of ß-Catenin/TCF Signaling
- A Polycomb Group Protein Is Retained at Specific Sites on Chromatin in Mitosis
- Diapause Formation and Downregulation of Insulin-Like Signaling via DAF-16/FOXO Delays Axonal Degeneration and Neuronal Loss
- Genes That Act Downstream of Sensory Neurons to Influence Longevity, Dauer Formation, and Pathogen Responses in
- A Genome-Wide RNAi Screen Reveals MAP Kinase Phosphatases as Key ERK Pathway Regulators during Embryonic Stem Cell Differentiation
- Recurrent Targeted Genes of Hepatitis B Virus in the Liver Cancer Genomes Identified by a Next-Generation Sequencing–Based Approach
- Excessive Astrocyte-Derived Neurotrophin-3 Contributes to the Abnormal Neuronal Dendritic Development in a Mouse Model of Fragile X Syndrome
- Pre-Disposition and Epigenetics Govern Variation in Bacterial Survival upon Stress
- Controls Gliogenesis by Regulating the Transient Expression of the Gcm/Glide Fate Determinant
- Systems Genetic Analysis of Osteoblast-Lineage Cells
- Population Genomics of Inversion Polymorphisms in
- Spreading of Heterochromatin Is Limited to Specific Families of Maize Retrotransposons
- DNA Topoisomerases Maintain Promoters in a State Competent for Transcriptional Activation in
- A Histone Deacetylase Adjusts Transcription Kinetics at Coding Sequences during Morphogenesis
- Approaching the Functional Annotation of Fungal Virulence Factors Using Cross-Species Genetic Interaction Profiling
- Evidence for the Robustness of Protein Complexes to Inter-Species Hybridization
- Systematic Identification of Rhythmic Genes Reveals as a New Element in the Circadian Clockwork
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Dnmt3a Protects Active Chromosome Domains against Cancer-Associated Hypomethylation
- Population Genomics of Sub-Saharan : African Diversity and Non-African Admixture
- Excessive Astrocyte-Derived Neurotrophin-3 Contributes to the Abnormal Neuronal Dendritic Development in a Mouse Model of Fragile X Syndrome
- Pre-Disposition and Epigenetics Govern Variation in Bacterial Survival upon Stress
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



