-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaThe Gene Contains Hotspots for L1 Endonuclease-Dependent Insertion
Long interspersed (L1) and Alu elements are actively amplified in the human genome through retrotransposition of their RNA intermediates by the ∼100 still retrotranspositionally fully competent L1 elements. Retrotransposition can cause inherited disease if such an element is inserted near or within a functional gene. Using direct cDNA sequencing as the primary assay for comprehensive NF1 mutation analysis, we uncovered in 18 unrelated index patients splicing alterations not readily explained at the genomic level by an underlying point-mutation or deletion. Improved PCR protocols avoiding allelic drop-out of the mutant alleles uncovered insertions of fourteen Alu elements, three L1 elements, and one poly(T) stretch to cause these splicing defects. Taken together, the 18 pathogenic L1 endonuclease-mediated de novo insertions represent the largest number of this type of mutations characterized in a single human gene. Our findings show that retrotransposon insertions account for as many as ∼0.4% of all NF1 mutations. Since altered splicing was the main effect of the inserted elements, the current finding was facilitated by the use of RNA–based mutation analysis protocols, resulting in improved detection compared to gDNA–based approaches. Six different insertions clustered in a relatively small 1.5-kb region (NF1 exons 21(16)–23(18)) within the 280-kb NF1 gene. Furthermore, three different specific integration sites, one of them located in this cluster region, were each used twice, i.e. NM_000267.3(NF1):c.1642-1_1642 in intron 14(10c), NM_000267.3(NF1):c.2835_2836 in exon 21(16), and NM_000267.3(NF1):c.4319_4320 in exon 33(25). Identification of three loci that each served twice as integration site for independent retrotransposition events as well as 1.5-kb cluster region harboring six independent insertions supports the notion of non-random insertion of retrotransposons in the human genome. Currently, little is known about which features make sites particularly vulnerable to L1 EN-mediated insertions. The here identified integration sites may serve to elucidate these features in future studies.
Published in the journal: . PLoS Genet 7(11): e32767. doi:10.1371/journal.pgen.1002371
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002371Summary
Long interspersed (L1) and Alu elements are actively amplified in the human genome through retrotransposition of their RNA intermediates by the ∼100 still retrotranspositionally fully competent L1 elements. Retrotransposition can cause inherited disease if such an element is inserted near or within a functional gene. Using direct cDNA sequencing as the primary assay for comprehensive NF1 mutation analysis, we uncovered in 18 unrelated index patients splicing alterations not readily explained at the genomic level by an underlying point-mutation or deletion. Improved PCR protocols avoiding allelic drop-out of the mutant alleles uncovered insertions of fourteen Alu elements, three L1 elements, and one poly(T) stretch to cause these splicing defects. Taken together, the 18 pathogenic L1 endonuclease-mediated de novo insertions represent the largest number of this type of mutations characterized in a single human gene. Our findings show that retrotransposon insertions account for as many as ∼0.4% of all NF1 mutations. Since altered splicing was the main effect of the inserted elements, the current finding was facilitated by the use of RNA–based mutation analysis protocols, resulting in improved detection compared to gDNA–based approaches. Six different insertions clustered in a relatively small 1.5-kb region (NF1 exons 21(16)–23(18)) within the 280-kb NF1 gene. Furthermore, three different specific integration sites, one of them located in this cluster region, were each used twice, i.e. NM_000267.3(NF1):c.1642-1_1642 in intron 14(10c), NM_000267.3(NF1):c.2835_2836 in exon 21(16), and NM_000267.3(NF1):c.4319_4320 in exon 33(25). Identification of three loci that each served twice as integration site for independent retrotransposition events as well as 1.5-kb cluster region harboring six independent insertions supports the notion of non-random insertion of retrotransposons in the human genome. Currently, little is known about which features make sites particularly vulnerable to L1 EN-mediated insertions. The here identified integration sites may serve to elucidate these features in future studies.
Introduction
Long interspersed nuclear elements (LINE-1 or L1 elements) and Alu sequences belonging to the family of short interspersed nuclear elements (SINEs) still actively amplify in the human genome, by a process called retrotransposition. L1 elements comprise ∼17% of the human genome sequence [1] but of the ∼500.000 L1 copies only ∼80–100 are still fully capable of active retrotransposition [2]. Equally, only a small minority of the >1 million Alu elements comprising more than 10% of the human genome can retrotranspose in a non-autonomous process, using proteins encoded by L1 elements to mediate their mobility [3]. The active Alu elements are named master or source Alu elements [4]–[5]. L1 elements are transcribed by RNA polymerase II whereas polymerase III transcribes Alu elements. Both elements are transcribed from an internal promoter [6]–[7]. While L1 transcripts are polyadenylated after transcription, the poly(A) tail of Alu transcripts may be encoded directly from the genomic site of transcription [8]. Alu transcripts are then terminated at the 3′ end with a short run of U's [8]. L1 elements are autonomous retrotransposons. Active L1 elements are typically 6 kb in length and contain two non-overlapping open reading frames ORF1 and ORF2 [9]–[10]. The latter encodes a protein with endonuclease (L1 EN) and reverse transcriptase (L1 RT) activities [11]–[12]. It is generally accepted that L1 EN forms a nick at the insertion site of L1 elements and the L1 transcripts are reverse transcribed using the 3′ overhang of the nick as a primer [11]. The consensus cleavage site of L1 EN 3′-AA/TTTT-5′ (and derivates thereof) [11], [13] which usually cleaves at the bottom strand allows the T's at the 3′ terminus of the nick to prime reverse transcription from the poly(A) end of a L1 transcript. There is evidence that Alu elements are reverse transcribed by the same process called “target primed reverse transcription” (TPRT), but they need to “borrow” the factors for TPRT from L1 elements [14] and are, hence, called non-autonomous retrotransposons. Integration of the generated cDNA is less well understood. Generally, cleavage of the second DNA strand occurs a few base pairs (typically 7–20 bp) downstream of the first nick, causing target site duplications (TSD) and the free 3′ end at the second cleavage site is used to prime the second strand cDNA synthesis. The whole process results in the formation of a new DNA copy of an L1 or Alu element including the poly(A) tail flanked by short direct repeats of the duplicated target site. For a detailed description of the mechanisms of autonomous and non-autonomous retrotransposition see [8], [15] and papers cited therein.
Although retrotransposable elements have no immediate function in the cell, their motility can be important for genome plasticity and the creation of genetic variation [16]. Most recent studies [17]–[20] demonstrate that L1 and Alu elements dimorphic with respect to presence/absence at a given site contribute significantly to structural variation of the human genome. Furthermore, these studies show that current activity of mobile elements is likely to be higher than previously appreciated.
L1 and Alu elements are believed to insert randomly into the human genome. Hence, de novo transposition of an element may occasionally cause an inherited disease when the element is inserted into (or at proximity of) the coding region of a functional gene [21]. To date, only ∼65 cases of de novo L1 EN-mediated insertions of retrotransposable elements causing genetic diseases have been reported [22]–[23]. A systematic analysis of retrotranspositional events causing genetic disorders lists 48 such mutations consisting of 26 Alu, 15 L1, four SINE/VNTR/Alu (SVA) composite element insertions as well as three simple poly(A) insertions [24]. These de novo insertions were found in 31 different genes including one insertion in the NF1 gene [25]. It has long been suspected that L1 EN-mediated retrotranspositional events are underreported as disease causing mutations, since they may be overlooked by the most commonly used mutation detection methods that rely on PCR amplification of small amplicons, i.e. exons from genomic DNA [26] (and reviewed later on in [27]–[28]). Here we report 18 novel L1 EN-mediated insertions in the NF1 gene: one poly(T), three L1 and 14 Alu insertions were all identified using an RNA-based core assay as the starting point for comprehensive NF1-gene analysis illustrating the strength of this approach in identifying also this complex type of mutations. Of note, six of the integration sites were located within a relatively small genomic region of 1500 bp between NF1 exons 21 (16) and 23 (18) (exons are numbered consecutively according to the NCBI reference sequence NM_000267.3; in addition, the more widely known historical legacy numbers, originally designated by the international NF1 consortium, are given in parentheses). Even more striking, three of the integration sites within the NF1 gene were used twice. Our results indicate and confirm that some genomic locations may be especially prone to L1 EN mediated retrotransposition.
Results
Identification of 18 L1 EN-dependent de novo insertions into the NF1 gene
Direct cDNA sequencing used as the core assay of comprehensive NF1 mutation analysis in the two centers (UAB and MUI) uncovered heterozygous (∼50% of the transcripts affected) splicing alterations in 18 NF1 index patients (Table 1) that could neither be explained by a heterozygous mutation of the splice regulatory elements of the affected exon and/or the flanking intronic sequences nor by a deletion of the genomic DNA. However, fragment analysis of the gDNA amplicons of the affected exons showed in some cases a faint extra band of larger size absent in the PCR products of a control DNA (Figure S2A). Sequence analysis of these PCR products revealed a faint additional sequence, besides the wild type sequence, starting at the position where the retroelement was inserted (Figure S2B). Manual reading of the background sequences revealed the presence of Alu elements inserted within the exonic sequences. In addition to the approximately 280-bp long Alu elements a 60–120-bp long poly(A) tail -or a poly(T) tail - was inserted depending on whether the Alu element was inserted in the sense or antisense orientation with respect to the NF1 reading frame (Figure S2B). As expected for de novo retrotransposed Alu elements the inserted sequences were flanked by short duplicated sequences derived from the insertion sites, i.e. target site duplication (TSD). We reasoned that the substantial increase in size of the mutant exons containing the Alu/L1 insertions, compared to the wild type exons caused allelic drop-out under standard PCR conditions. We increased the extension time of the PCR reactions and/or increased the size of the PCR amplicons to enhance the amplification of the larger mutant alleles and, hence, facilitate the detection of possible de novo Alu insertions. Amplification of all exons with so far unexplained splicing alterations using PCR conditions optimized to amplify larger PCR products (Figure 1A and 1B) led to the identification of 14 different de novo Alu insertions (Table 1) as well as an approximately 120-bp poly(T) stretch in exon 25 (19b) resulting in skipping of the last 40 nucleotides of this exon in the mRNA transcripts.
Fig. 1. Example of de novo Alu insertion detection by improved PCR conditions. 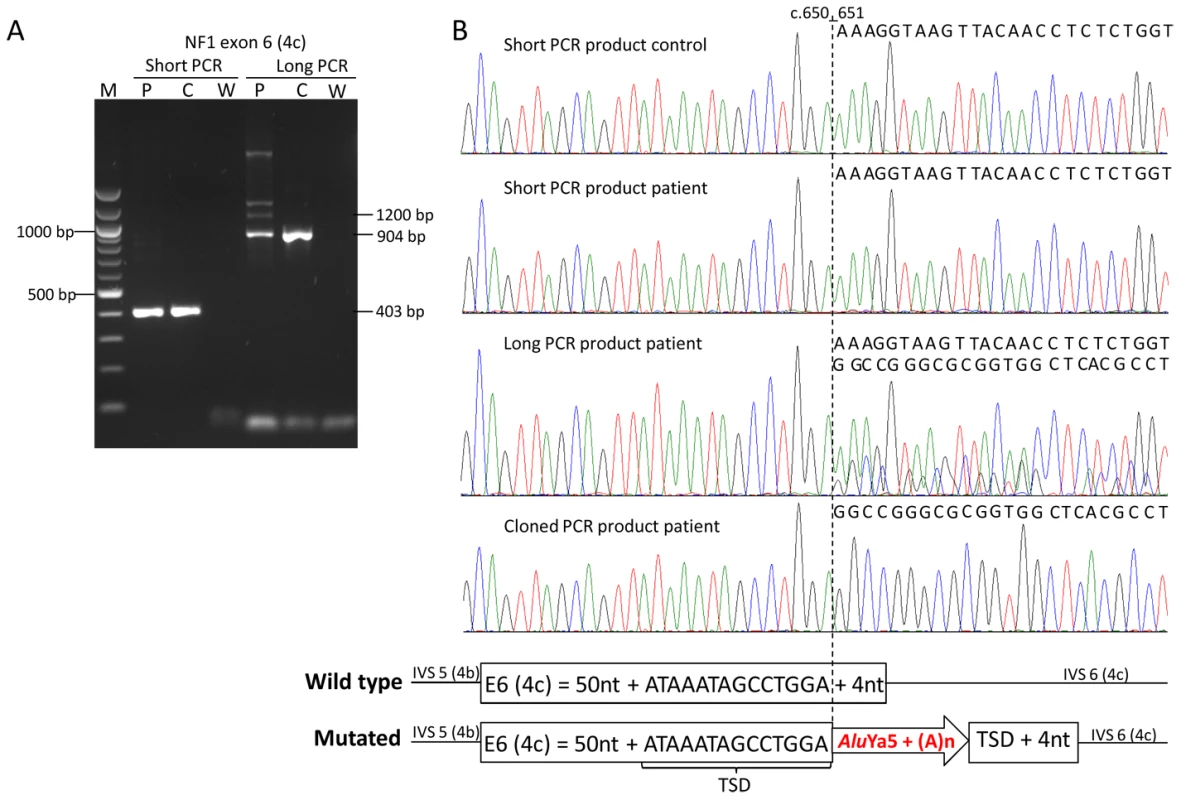
A) Agarose gel with PCR products of exon 6 (4c) generated with two different primer pairs and PCR conditions generating a 403-bp (short) and a 904-bp (long) wild type PCR product (see Table S1). PCR products were generated from a control individual (C) and the patient (P) harboring an AluYa5 insertion in exon 6 (4c) (W = water control). Amplification of the large PCR product from the patient revealed the presence of extra bands of increased size, i.e. a ∼1200-bp band representing the homoduplex of the PCR product from the mutant allele and two larger bands probably derived from heteroduplexes of this PCR product with the PCR product from the wild type allele, which are neither present in the control nor in the shorter amplified PCR products. (B) Sequences of the short and long PCR products from the control individual and the patient; the sequence of the long PCR product but not of the short PCR product of the patient shows starting at nucleotide position c.651 (vertical dotted line) a background sequence coming from the inserted AluYa5 element (see scheme below the sequences). The sequence of a cloned long PCR product clearly shows the presence of the AluYa5 element within the NF1 exon 6 (4c). Tab. 1. List of all Alu, L1 and poly(T) insertions uncovered in the NF1 gene. 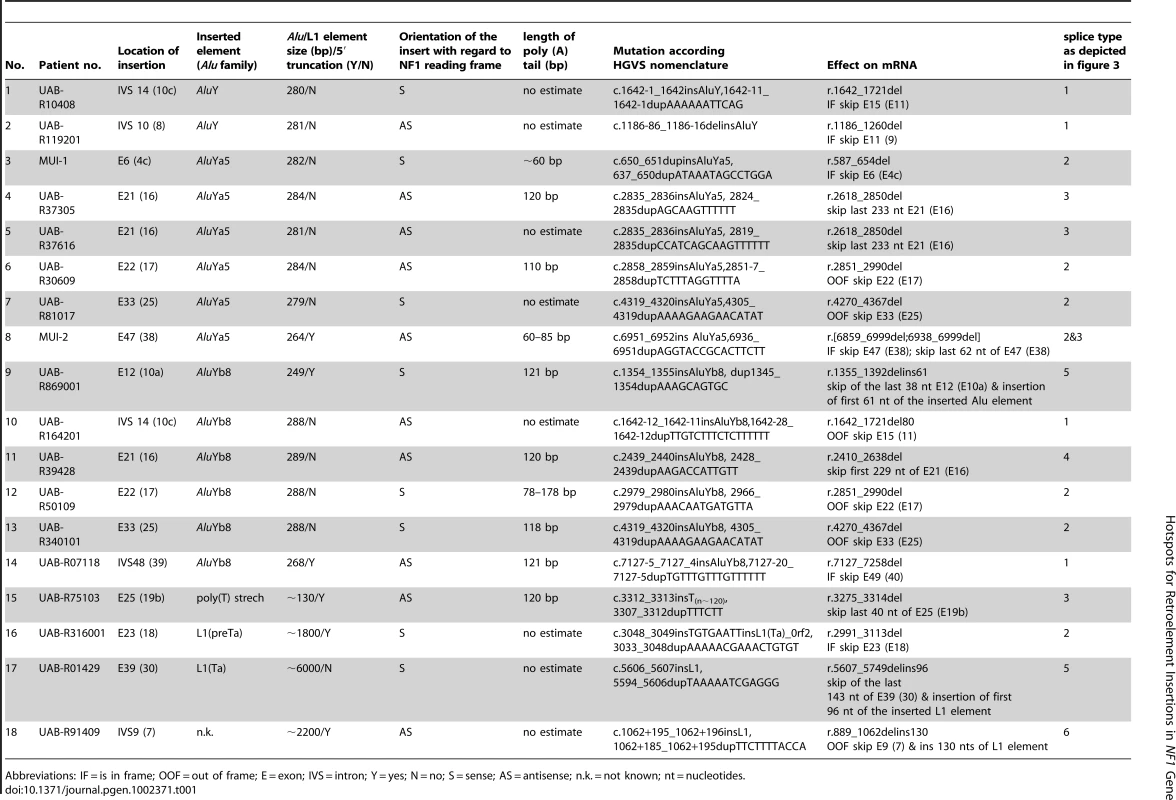
Abbreviations: IF = is in frame; OOF = out of frame; E = exon; IVS = intron; Y = yes; N = no; S = sense; AS = antisense; n.k. = not known; nt = nucleotides. To further confirm the presence of the Alu elements within the mutant NF1 alleles and to determine precisely the inserted sequence, the PCR products showing the extra bands of increased size were cloned and sequenced (Figure 1B). Additionally or alternatively, Alu-insertion-specific primers were used together with the regular exon primer at the opposite site of the exon to specifically amplify and subsequently sequence the mutant alleles (Figure S3). All 14 Alu insertions identified are listed in Table 1. Alignment of the inserted Alu sequences with the consensus sequence of the different Alu families (as deposited in Repbase Giri [29]) showed that two Alu elements belong to the AluY, six to the AluYa5 and six to the AluYb8 family (Figure S4). Three of the Alu insertions were truncated and lacked at their 5′ end 17, 20 and 39-bp, respectively, (see patients MUI-2, UAB-R07118 and UAB-R869001 in Figure S4). Six of the Alu elements were inserted in sense and eight in anti-sense orientation.
Increase of the amplicon size and the PCR extension time of exons 39 (30), 23 (18) and 9 (7) did not allow resolution of the cause of their mis-splicing in the patients UAB-R01429, UAB-R316001 and UAB-R91409, respectively. Use of a Taq polymerase enabling amplification of extremely long PCR products (up to >6 kb) was needed to amplify the mutant exons which had 10 - and 17-times increased in size compared to their wild type normal allele in patients UAB-R316001 and UAB-R01429, respectively. A 6-kb full-length L1 element inserted in sense orientation into exon 39 (30) was identified in patient UAB-R01429 (Figure 2A). Full sequencing of the inserted L1 element showed it to belong to the youngest L1 (Ta-1d) subset since it carried all sequence variants that distinguish this L1 (Ta) group from others [2], [30]. It contained 10 deviations from the consensus sequence of hot L1 sequences as given by [2] (Figure S5). Sequence alignment with all known intact L1s of the Ta-1d subset [31] showed that the closest related hot L1 (Ta-1d) element is contained in NCBI sequence AL356438. Only six nucleotides differentiate the full-length L1 identified in exon 39 (30) from this possible precursor sequence. A 5′-truncated L1 element was found to be inserted in sense into exon 23 (18) in patient UAB-R316001. The latter sequence contained 1753 bp of the 3′-half of ORF2 and the poly(A) tail but lacked the ORF1 and the promoter region of L1 elements. Furthermore, it was preceded by a short 8-bp sequence neither derived from the target sequence nor from the inserted L1 element, a finding that previously has been observed for L1 retrotransposition events in cultured cells [32]–[34]. This L1 element belongs to the L1(pre-Ta) subset since it carries a 3-bp ACG at the site discriminating the L1(Ta) and L1(pre-Ta) subsets [35]. In intron 9 (7) we found a sequence inserted that contained a poly(T) stretch at the 5′-end indicating that the poly(A) tail of the inserted retrotransposon transcript had annealed to the sense strand where reverse transcription from the template started. Sequencing from the reverse strand showed, however, that the 3′-end of the inserted sequence contained part of an L1 ORF2 sequence (see Figure S6). This sequence was inserted in sense orientation with regard to the NF1 coding sequence suggesting that during the process of retrotransposition the orientation of the reverse transcription from the L1-RNA template that started at the sense strand of the NF1 gene switched and continued from the anti-sense strand. Due to experimental difficulties and lack of sufficient patient's DNA it was not possible to assess the full sequence of the inserted L1 element and, therefore, the breakpoint of inversion could not be defined.
Fig. 2. Amplification of the mutant allele containing a full-length L1 element inserted into exon 39 (30). 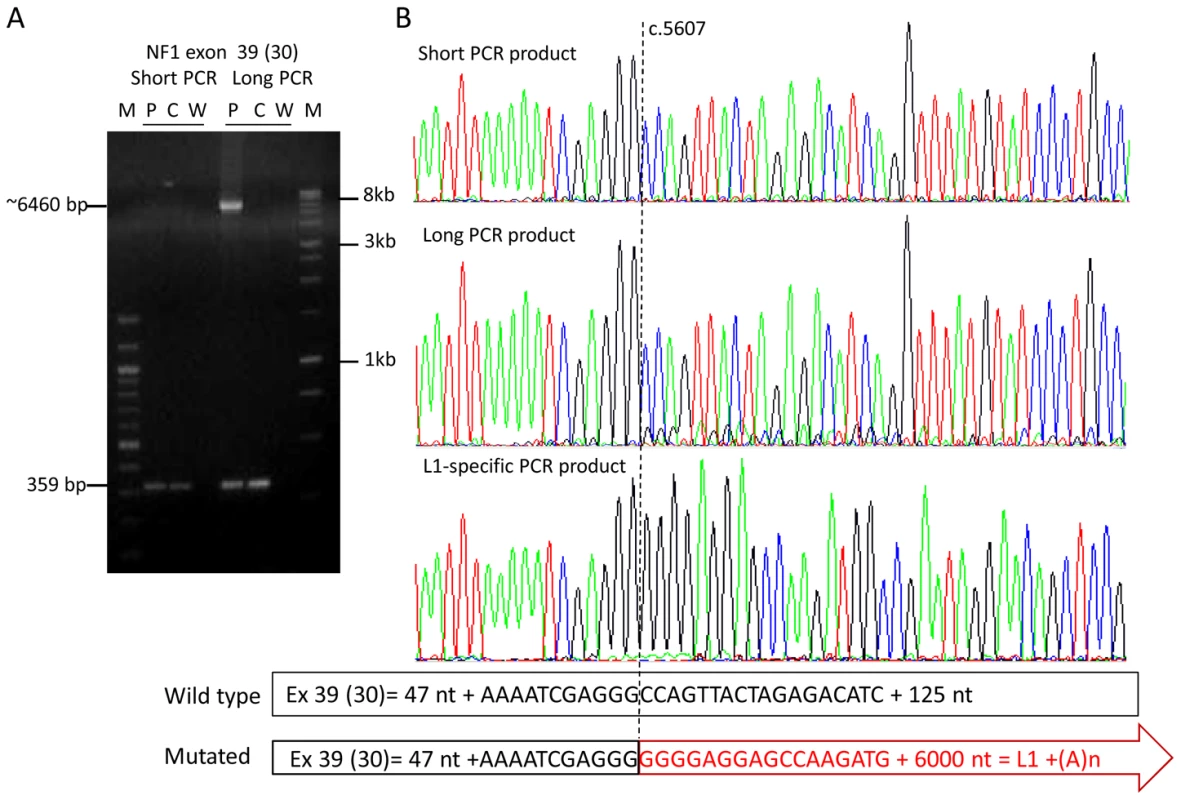
(A) Agarose gel with PCR products of exon 39 (30) generated with two different PCR conditions. PCR products were generated from a control individual (C) and the patient (P) harboring a full-length L1 element in exon 39 (30) (W = water control). The PCR conditions (short) used in the diagnostic laboratory generate only the 359-bp product from the wild type allele present in the control and the patient, whereas a >6000-bp PCR product derived from the mutant allele was amplified along with the wild type 359-bp PCR product using the Expand Long Template PCR system kit (Roche), buffer system 2 and 3 (long) (see Table S1). (B) Sequences of the PCR products from the patient generated with the PCR conditions “short” and “long”; the sequence of the long PCR product but not of the short PCR product of the patient shows starting at nucleotide position c.5607 (vertical dotted line), besides the main wild type sequence, minor traces of a sequence coming from the inserted L1 element (see scheme below the sequences). The presence of the L1 element within the NF1 exon 39 (30) was confirmed using a L1-specific PCR (primers: 39f and L1-5_39r). The entire 6021-bp sequence of this L1 (Ta-1d) element is deposited in Figure S5. Analysis of insertion sites
Alignment of the sequences at the integration sites (Table 2) shows that all integration sites match the reported L1 EN consensus cleavage site [13], indicating that all insertions arose via L1 EN-mediated retrotransposition. The exact insertion site of an AluY element into intron 10 (8) cannot be determined due to a 71-bp deletion associated with this insertion that causes also lack of a TSD. Nevertheless, the most likely insertion site matching a L1 EN cleavage site is tentatively given in Table 2. The alignment of the integration sites also shows that the 18 retroelements inserted in 15 different integration sites with three of them used twice independently. We found in two non-related patients (UAB-R10408 and UAB-R164201 in Table 1) an AluY and AluYb8 element inserted into the splice acceptor site of intron 14 (10c). The same L1 EN cleavage site was used once at the anti-sense strand to insert the AluY element in sense orientation with respect to the coding sequence of the NF1 gene (integration site according to HGVS nomenclature c.1642-1_1642) and in an unrelated patient once at the sense strand to insert the AluYb8 element in anti-sense orientation (integration site according HGVS nomenclature c.1642-12_1642-11). Furthermore, we found in two non-related patients (UAB-R81017 and UAB-R340101 in Table 1) an AluYa5 and an AluYb8 element, respectively, inserted at the identical integration site, c.4319_4320, in exon 33 (25). Both Alu elements were inserted in sense orientation and flanked by the same sized TSD (Table 2). Finally, in two non-related patients (UAB-R37616 and UAB-R37305 in Table 1) -one of them being a sporadic patient proven to have a de novo insertion - an AluYa5 element inserted at the identical position, c.2835_2836, in exon 21 (16). The length of TSDs flanking the two insertions differed between both patients (Table 2) confirming further that two independent events led to the formation of these two mutant alleles. This site was located within a 1.5-kb genomic region containing exons 21 (16), 22 (17) and 23 (18) that harbor a total of six independent retrotranspositions. The remaining eight integration sites of de novo insertions were distributed over the entire NF1 gene from exon 6 (4c) to intron 48 (39).
Tab. 2. Sequences at the integration sites aligned to the L1 endonuclease cleavage consensus site. 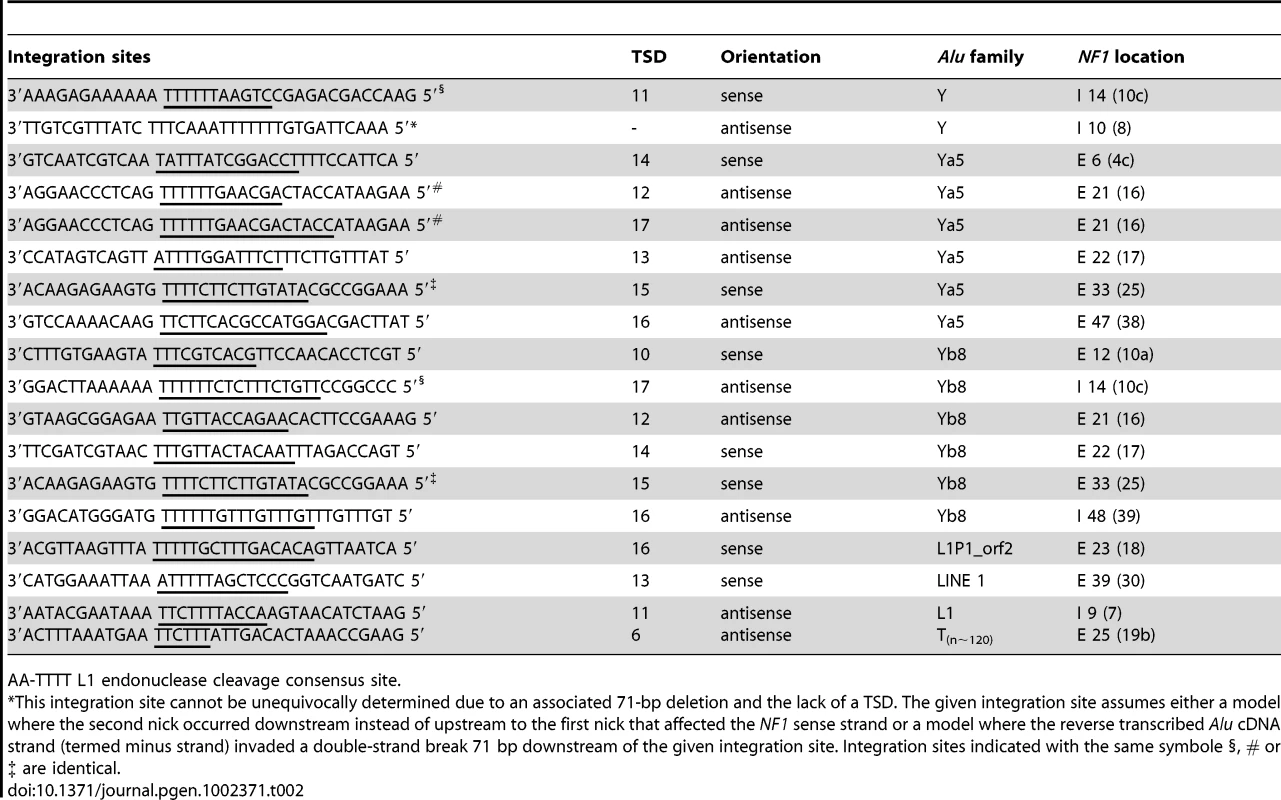
AA-TTTT L1 endonuclease cleavage consensus site. Different splicing effects are associated with de novo Alu and L1 insertions
Four out of 14 Alu elements (UAB-R10408, UAB-R164201, UAB-R07118 and UAB-R119201 in Table 1) were inserted into the canonical AG-dinucleotide (1/4) or the polypyrimidine tract (3/4) of the 3′ splice sites of introns 14 (10c), 48 (39), and 10 (8), respectively. Disruption of these splice sites readily explains the observed skipping of the downstream exons (type 1 splicing effect in Figure 3). Skipping of the affected exon was also observed in five other patients with Alu insertions, i.e. two insertions at the identical site in exon 33 (25), two insertions at different integration sites in exon 22 (17) and one insertion in exon 6 (4c) (type 2 effect in Figure 3). Two AluYa5 elements (UAB-R37305 and UAB-R37616 in Table 1) inserted at the identical integration site c.2835_2836 in exon 21 (16) in two unrelated patients causing skipping of the last 233 nucleotides of this 441-bp exon. This partial exon skipping results from the use of a cryptic 5′ splice site located upstream of the integration site (type 3 effect in Figure 3). Similarly, integration of an AluYb8 element at position c.2439_2440 in the 5′ half of exon 21 (16), 30 nucleotides downstream of the intron-exon 21 (16) border, resulted in the use of a cryptic 3′ splice site downstream of the integration site and loss of the first 229 bp of the exon 21 (16) (mutation UAB-R39428 in Table 1; type 4 effect in Figure 3). Insertion of the approximately 120-bp poly(T) stretch at c.3312_3313 in exon 25 (19b), 3 nucleotides upstream of the exon-intron border also led to use of a cryptic 5′ splice site and loss of the last 40 nucleotides of this exon (mutation UAB-R75103; type 3 effect in Figure 3). Insertion of an AluYa5 element into exon 47 (38) resulted in at least two different splicing effects, i.e. skipping of the exon and use of an exonic cryptic 5′ splice site upstream of the integration site (mutation MUI-2; type 2 and 3 splicing effect in Figure 3; see also Figure S1). Insertion of an AluYb8 element into exon 12 (10a) also resulted in a more complex splicing defect. This element lacks the first 39 bp at its 5′ end (UAB-R869001 in Table 1 and Figure S4). A cryptic 5′ splice site within this truncated Alu element is used instead of the natural one of the exon 12 (10a), leading to transcripts containing the first 61 nucleotides of this Alu element but lacking the 3′ half of exon 12 (10a), downstream of the Alu integration site (type 5 effect in Figure 3).
Fig. 3. Schematic presentation of the splicing effects caused by the identified Alu and L1 insertions. 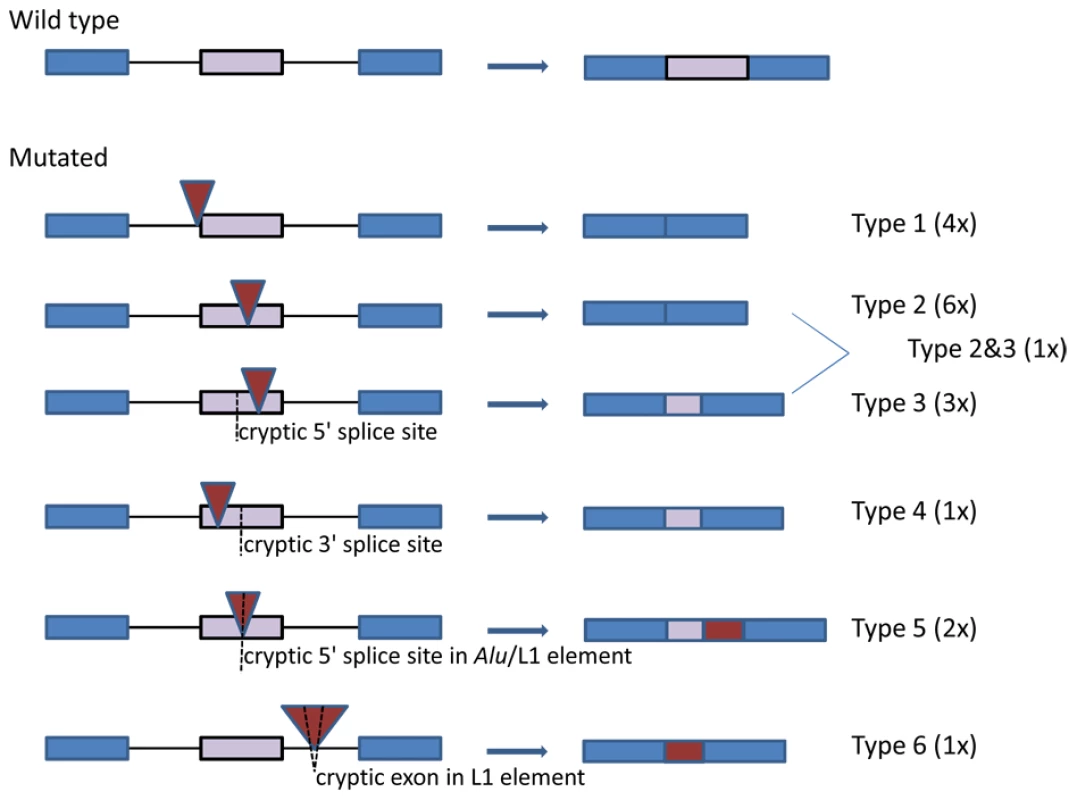
The affected exon is indicated with a purple bar and the two flanking exons with blue bars. Intronic sequences are indicated by black lines. Red triangles denote the inserted Alu and L1 sequences and cryptic splice sites within an exon or the retrotransposon are indicated by a dotted line. The genomic sequence and the ensuing mRNA transcripts are depicted at the left and right hand site, respectively. Insertion of the 5′-truncated L1 element into exon 23 (18) also caused simple exon skipping in the transcripts of the patient UAB-R316001 (type 2 splicing effect in Figure 3). The insertion of the 5′-inverted L1 element 195 nucleotides downstream of exon 9 (7) caused skipping of exon 9 (7) as well as inclusion of a 130-bp cryptic exon embedded within the inserted L1 element (patient UAB-R91409; type 6 effect in Figure 3; see also Figure S6). Finally, a 5′ splice site within the full-length L1 element inserted into exon 39 (30) is used instead of the natural 5′ splice site of exon 39 (30), leading to a transcript lacking the 143 nucleotides of the exon downstream of the integration site and containing instead the first 96 nucleotides of the L1 element (patient UAB-R01429; type 5 effect in Figure 3).
Taken together, all Alu and L1 elements as well as the insertion of a poly(T) element lead to altered splicing of the transcripts from the mutant allele. None of the inserted Alu or L1 elements was fully included into the transcripts.
Discussion
Here we report the identification of 18 pathogenic L1 EN-mediated insertions into the NF1 gene. This is the largest number of disease causing mutations of this type found in a single human gene. Sixteen of these insertions were found in a cohort of 4300 unrelated NF1 mutation-positive patients analyzed at UAB and two in a cohort of 200 unrelated NF1 mutation-positive patients analyzed at MUI. Taken together, this suggests that retrotransposon insertions account for ∼0.4% (18/4500) of NF1 mutations identified. This frequency is two to four times higher than previously anticipated [36] and expected from a database survey in human genetic diseases [28]. Two mutually non-exclusive explanations for this observation may be taken in consideration.
Improved detection of L1 EN-mediated de novo insertions by NF1 cDNA sequencing
Firstly, mutations due to retrotransposition have long been thought to be underestimated, since they may be missed by mutation detection methods relying on PCR amplification of small amplicons, i.e. exons, from genomic DNA [26]–[28] and references cited therein. Indeed, all insertions reported here were identified because they altered splicing of the transcripts as detected by direct cDNA sequencing [37]. In order to identify the cause of the splicing defects, i.e. the Alu, L1 and poly(T) stretch insertions, it was necessary in most of the cases to modify the PCR conditions to prevent allelic drop-out of the substantially larger mutant allele. Hence, most of these mutant alleles would be missed by an exon by exon-sequencing approach. Similar findings were reported for the BRCA1 and BRCA2 genes [38] and even for genes involved in X-linked diseases [39] although allelic drop-out is expected to be less severe in X-linked diseases due to hemizygosity of the mutant allele in the affected males. Thus, our results demonstrate that RNA-based mutation analysis protocols are more apt to uncover L1 EN-mediated retrotransposon insertions.
The NF1 gene may be a preferred target of retrotransposon integration in the human genome
Secondly, it is possible that the NF1 gene is particularly susceptible to retrotransposon integration. It is striking that three specific sites within the NF1 exons 21 (16) and 33 (25) and the splice acceptor site of intron 14 (10c) hosted each two independently retrotransposed elements. Moreover, one of these sites is located in a small region of approximately 1.5 kb within the ∼280-kb NF1 gene where six of the 18 insertions cluster. Together, these finding strongly support the notion of an non-random de novo insertion of retrotransposons in the human genome [24]. A systematic analysis of 48 previously reported simple L1 EN-dependent insertions showed that some genes, e.g. F8 and F9 mutated in hemophilia B, may have hotspot regions for retrotransposon integration [24]. Furthermore, three integration sites in three other genes, i.e APC, F9 and BTK, have been reported to be used twice independently for L1 EN-mediated retrotransposon insertions [24], [39]. Taken together with our findings in the NF1 gene, a tenth (6/66) of the well characterized sites that harbor disease causing retrotransposon insertions have been used multiple times with half of them identified in the current study. It remains to be elucidated which features make these sites particularly vulnerable to L1 EN-mediated insertions. The fact that some of these sites are embedded in a larger sequence context that appears to be a hotspot region for insertions may indicate that flanking sequences and possibly also the chromatin structure in these regions may play a role.
It has been suggested that Alu and L1 elements have a similar retrotransposition efficiency via-à-vis the molecular retrotransposition machinery [40]. However, as in the NF1 gene also in previously published disease causing retrotranspositional insertions a preponderance of Alu over L1 insertions can be observed. As a possible explanation Dewannieux et al. [3] propose that L1 elements harboring a disrupted ORF1, but retaining a functional ORF2, should still be competent for trans mobilization of Alu elements even if they are unable to promote their own transposition. In other words, there appear to be more potential “drivers” for Alu retrotransposition than there are for L1 retrotransposition [4] potentially explaining the preponderance of disease-causing Alu over L1 insertions [24].
18 novel Alu/L1 insertions substantially increase the number of known disease-causing mutations of this type
With this report we substantially increase the number of known disease causing Alu, L1 and simple poly(T) insertions. Hence, our data confirm and extend several observations deduced from 48 previously analyzed pathological insertions [24]. All inserted retrotransposons in our study contained a poly(A)/poly(T) tail with a size in the range from 60–178 bp and all but one were flanked by TSDs with a size in the range of 6–17 bp (see Table 1 and Table 2). All integration sites (Table 2) matched the previously reported consensus sequence of the L1 EN cleavage site [13], [41] and there was no preferential orientation of the inserts with respect to the NF1 coding sequence. All Alu insertions belong to the evolutionary youngest “Y” Alu family (see Figure S4). Three of them (MUI-2, UAB-R07118 and UAB-R869001 in Table 1) lacked 17–39 bp at their 5′ end when compared to the Alu consensus sequence (Figure S4). According to [42] these three elements would fall into the Group II Alu inserts which represent ∼8% of all Alu elements polymorphic with respect to presence or absence in the human genome. The poly(T) sequence inserted into the NF1 exon 25 (19b) may represent a severely truncated Group III Alu insertion [42]. Equally, this poly(T) stretch may result from a severely truncated L1 element or even a processed pseudogene. One AluY element inserted into intron 10 (8) was accompanied by a 71-bp deletion and lacked a TSD. Different possible mechanisms for the loss of genomic sequences in association with EN-dependent retroelement insertions have been discussed [24]. One suggested mechanism depicted in Figure 6A in [33] assumes that the second-strand nick at the top strand of the L1 target site is made a few bp to the “left” of the initial nick on the bottom strand rather than to the “right” causing the loss of a few bp at the insertion site. A model that would explain the larger deletions of several kb assumes that the reverse-transcribed cDNA strand of the retroelement is involved in double-strand break processing and invades a double-strand break to the “left” of the first-strand nick (see Figure 6B in [33]). Both models may theoretically apply to the 71-bp deletion associated with the AluY insertion into NF1 intron 10 (8). In both models the first-strand L1 EN-mediated nick would have occurred at position c.1186-85_1186-86 in intron 10 (8) (as tentatively given in Table 1 and Table 2). However, it is also possible that an EN-independent integration mechanism as previously observed in certain Chinese hamster ovary (CHO) cell models [41], [43] has led to the TSD-lacking and deletion-associated integration of this AluY element.
According to Kojima [44] L1 elements of the human genome can be classified into three categories: full-length, 5′-truncated and 5′-inverted, comprising respectively 25.5%, 43.8% and 30.7% of all L1 elements. Here we found an example of each category. The full-length element belonging to the youngest L1(Ta-1d) subset was a simple insertion in sense orientation with regard to the NF1 coding sequence flanked by a 12-bp TSD. The 5′-truncated element lacked the promoter region, the entire ORF1 and the first 2080 bp of the 3825-bp ORF2. Of note, this is only the second reported pathogenic insertion of an older and less active L1(pre-Ta) element [2]; the first was found in the F8 gene in a hemophilia A patient [21]. The 5′ inverted L1 element contained a poly(T) tail at the 5′ end but the analyzed sequence at the 3′ end was inserted in sense orientation and contained 1088 bp from the center of the 3825-bp ORF2 (Figure S6). This suggests that during the process of retrotransposition the orientation of the reverse transcription from the L1-RNA template that started at the sense strand of the NF1 gene switched and continued from the anti-sense strand. The presence of a 2-bp micro-homology that may have promoted strand switching due to secondary binding between the RNA-template and the 3′-end of the 5′-overhang at the antisense strand supports the model of twin priming [45] (depicted also in Figure 1 in [44]) as the underlying mechanism leading to the insertion of this 5′ inverted L1 element.
Splicing defects are the prevalent effect of L1 EN-mediated retroelement insertions
Only four of the Alu insertions identified here directly affect splice sites. Nevertheless, all L1 EN-mediated insertions in this study affected NF1 splicing in the patients. The predominant effect of the 13 exonic Alu, L1 and poly(T) insertions was exon skipping (6/13 cases) or use of a cryptic splice site -either a cryptic 5′ splice site upstream (3/13 cases) or a cryptic 3′ splice site downstream of the integration site (1/13 cases). In one of 13 cases both of these splice effects were observed in the same patient. In only two cases part of the Alu or L1 element were contained in the mRNA transcripts, inserted into exon 12 (10a) and 39 (30) respectively. Overall, we do not believe that ascertainment bias can explain why exon skipping and/or use of cryptic splice site are the main effects of the de novo Alu and L1 insertions in exonic sequences. Firstly, because we have no evidence that the long-range RT-PCR reactions used in the applied assays would miss Alu insertions in exons or introns that lead to full or partial exonisation of the inserted sequences in the mRNA transcripts. On the contrary, duplications of single or multiple exons which lead to a similar size increase of the RT-PCR products were readily detected by our assays [46]. Secondly, in agreement with our observations, exon skipping was also the reported effect of 5/7 L1 EN-mediated integrations for which RNA data were available in the literature (see Table 1 in [28]). Hence, following an exon definition model of splicing [47], our data indicate that the main effect of exonic Alu and L1 insertion is weakening of the exon definition resulting in altered splicing of the affected exon. Currently it is not fully evaluated by which mechanisms the inserted sequences reduce exon recognition by the splicing machinery. It has been proposed that inserted sequences may disrupt specific cis-acting exonic splice elements, such as exonic splicing enhancers (ESEs) (see [28] and references cited therein). However, it is unlikely this pertains to all or to the majority of exonic de novo insertions. Therefore, we favor the hypothesis that the insertion of a relatively large number of nucleotides (300–2000 bp) weakens exon definition simply by increasing the size of the affected exon and/or by disrupting the exonic structure of cis acting elements in a more general sense, e.g. by increasing the distance of exonic cis regulatory elements and the splice sites of the exon. In this respect it is of note that the use of cryptic 5′ - and 3′ - splice sites in exons 21 (16) and 25 (19b) is also observed in transcripts from mutant NF1 alleles that carry single nucleotide alterations destroying the respective natural 5′ - and 3′ - splice sites of these exons ([48] and Messiaen unpublished results). This observation may indicate that some of the inserted sequences weaken particularly the downstream or upstream splice site while the definition of the respective other splice site is unaffected which in certain instances favors use of a cryptic splice over skipping of the affected exon [48]–[49].
The integration of an inverted L1 element into intron 9 (7) caused a complex splicing effect, i.e. insertion of a cryptic exon embedded in the inserted L1 element and skipping of the preceding exon 9 (7). A similar splicing defect was described in a chronic granulomatous disease patient who carried a truncated L1 element in intron 5 of the CYBB gene [50]. As in the NF1 gene, the cryptic exon inserted in the transcripts of CYBB was not better defined by the splice sites compared to the flanking natural exons that were skipped in those transcripts containing the L1 derived cryptic exon. Hence, it remains to be explained why insertion of these L1 elements lead to exonisation of the cryptic exon on the account of the adjacent natural ones.
Concluding remarks
Our results clearly show that RNA-based mutation analysis strategies have the potential to detect disease-causing L1 and Alu insertions. In addition, the RNA-based comprehensive NF1 mutation detection approaches unambiguously identify and functionally characterize other classes of mutations usually missed by DNA-based mutation detection strategies, such as intronic alterations outside the canonical splice site dinucleotides (GT-AG) (including deep intronic mutations), as well as silent and missense mutations with an effect on splicing [48], [51]. Still this approach may underestimate L1 EN-mediated insertions. De novo retrotransposon insertions within the 3′ - or 5′-UTR can lead to reduced expression by the disruption of gene regulatory elements [52]–[53]. Furthermore, insertion of L1 elements within introns has been shown to reduce mRNA transcript levels. This phenomenon is related to RNA polymerase II elongation defects and/or premature polyadenylation caused by the L1 elements [54]–[55]. Thus, it is possible that insertions especially in the regulatory or intronic regions of the gene may still be missed by the here applied NF1 mutation analysis assay.
Material and Methods
Patients
All mutations reported were uncovered in samples from unrelated index patients sent for clinical NF1 testing to two centers, i.e. the Medical Genomics Laboratory at the University of Alabama in Birmingham (UAB) and the Division of Human Genetics, Medical University Innsbruck (MUI). Informed consent was obtained from all patients. This study was approved by the ethical committee from both institutions.
Identification of splicing defects by direct cDNA sequencing
In both laboratories the primary assay for comprehensive NF1 mutation analysis is a direct cDNA sequencing approach that is based on the amplification of the entire NF1 coding region in three (Birmingham, AL) or five (Innsbruck) overlapping RT-PCR fragments and subsequent sequencing of the entire PCR products with 18 (20) internal primers. Details on the RT-PCR reactions and primers can be found in [37], [56]. To avoid illegitimate splicing, known to lead to multiple aberrant splice variants that impede the detection of mutations in an RNA-based approach [51], [57], total RNA is extracted from phytohemagglutinin (PHA)-stimulated short-term lymphocyte cultures treated with 200 µg/ml puromycin for 4 h prior to cell harvest to prevent the nonsense-mediated RNA decay [51]. Details on cell culture, RNA-extraction and cDNA synthesis can be found in [37]. BigDye Terminator Cycle Sequencing chemistry was used for sequencing (Applied Biosystems, Foster City, CA). The sequencing reactions were subsequently run on an automated capillary sequencer and analyzed using the sequence analysis program SeqScape v2.5 (Applied Biosystems, Foster City, CA) and/or SequencePilot (JSI Medical Systems, Kippenheim, Germany). In some instances it was necessary to manually analyze (read) the aberrant sequences in order to determine all splicing defects deducible from multiple overlaying sequences (see Figure S1).
NF1 nucleotide numbering is based on GenBank reference sequence NM_000267.3 with the A of the ATG start codon being nucleotide position c.1. The integration sites are given according to the HGVS nomenclature after the first duplicated sequence regardless of whether the first nick occurred at the sense or the anti-sense strand and the thereof resulting orientation of the insertion with respect to the NF1 coding sequence. Exons are numbered according to the reference sequence with the widely known legacy numbering given in parenthesis.
Identification and confirmation of L1 EN-mediated insertions
Exons and flanking intronic sequences affected by splicing defects were analyzed by sequencing from genomic DNA and by multiplex ligation dependent probe amplification (MLPA) with the current SALSA-MLPA-kit P081-B1 and/or P082-B1 (MRC-Holland, Amsterdam, the Netherlands). When MLPA results were negative for a genomic deletion of the respective exon and no alteration was identified explaining the observed splicing defect, sequences were meticulously reanalyzed to uncover a possible Alu/L1 insertion within or in close vicinity of the exon (see Figure S2). To diminish allelic drop-out of the mutant allele due to the increased exon size by an Alu/L1 insertion, the PCR conditions were modified to allow for the amplification of larger sequences from gDNA of the patients. Primers and PCR conditions used to amplify the affected exons are listed in Table S1A and S1B. Mutant exons containing Alu elements were amplified using Takara Ex Taq (Takara Biotechnology Co. LTD, Madison, WI) or Taq DNA Polymerase (Invitrogen, Carlsbad, CA). To amplify the mutant alleles containing a truncated 1753-bp and a full-length 6-kb L1 element, respectively, Expand Long PCR Taq (Roche Diagnostics GmbH, Roche Applied Science, Mannheim, Germany) was used. To determine the precise sequence of the inserted Alu element as well as the duplicated sequence at the insertion site the PCR products showing a band of increased size were cloned according to the manufacturer's instructions into the TOPO-TA cloning vector pCR 4-TOPO (Invitrogen, Carlsbad, CA) and individually sequenced. Alternatively or in addition, primers were designed to specifically amplify only the mutant alleles containing the Alu insertions. These primers contained at their 5′-end the exonic sequences immediately upstream/downstream of the insertion site and at the 3′-end a few nucleotides of either the Alu or the poly(A/T) stretch. All primers are listed in Table S1A. PCR products generated with these primers used together with the regular exon primer at the opposite site of the exon were subsequently sequenced (see as an example Figure S3). Alignment of the identified Alu sequences with the consensus sequences of the different Alu families (as deposited in Repbase Giri [29]) was performed with the program ClustalW v.1.83 [58]. To amplify the much larger mutant alleles containing a truncated 1753-bp and a full-length 6-kb L1 element, the Expand Long PCR Taq kit (Roche Diagnostics GmbH, Roche Applied Science, Mannheim, Germany) specifically designed to amplify fragments up to around 20 kb, was used. To sequence the 5′-end of a full length L1 element integrated in exon 39 (30) a L1-specific reverse primer (L1-5_39r in Table S1) was used together with the exon 39 (30) forward primer (39f). Thereafter, the entire 6021-bp L1 sequence was characterized by sequencing of the PCR product generated with primers 39f and 39r (Figure 2) using L1-sequence specific internal primers (Table S1). Similarly, to amplify a mutant 1800-bp fragment containing a truncated L1 element in exon 23 (18), and thereafter sequence the 5′ and 3′ends of this mutant fragment, we used the primers 23f and 23r. In addition, sequence analysis was performed with a L1-specific primer, L1_Fam (Table S1), located 86 bp upstream of a 3-bp diagnostic site that distinguishes the young L1 subfamilies (Ta) and pre-(Ta) from the older ones [35]. To determine the sequence inserted into intron 9 (7) that lead, at the transcript level, to loss of exon 9 (7) and concomitant insertion of an L1-derived 130-bp sequence between exons 8 (6) and 10 (8) in patient UAB-R91409, a reverse and a forward primer (Line9_r and Line10_f in Table S1) were used in two PCR reactions together with the exon 9 (7) forward (9f) and the exon 10 (8) reverse (10r) primer, respectively. The resulting PCR products were sequenced in both directions.
Supporting Information
Zdroje
1. LanderESLintonLMBirrenBNusbaumCZodyMC 2001 Initial sequencing and analysis of the human genome. Nature 409 860 921
2. BrouhaBSchustakJBadgeRMLutz-PriggeSFarleyAH 2003 Hot L1s account for the bulk of retrotransposition in the human population. Proc Natl Acad Sci U S A 100 5280 5285
3. DewannieuxMEsnaultCHeidmannT 2003 LINE-mediated retrotransposition of marked Alu sequences. Nat Genet 35 41 48
4. CordauxRHedgesDJBatzerMA 2004 Retrotransposition of Alu elements: how many sources? Trends Genet 20 464 467
5. DeiningerPLBatzerMAHutchisonCA3rdEdgellMH 1992 Master genes in mammalian repetitive DNA amplification. Trends Genet 8 307 311
6. DanielsGRDeiningerPL 1991 Characterization of a third major SINE family of repetitive sequences in the galago genome. Nucleic acids research 19 1649 1656
7. SwergoldGD 1990 Identification, characterization, and cell specificity of a human LINE-1 promoter. Mol Cell Biol 10 6718 6729
8. DeiningerPLBatzerMA 2002 Mammalian retroelements. Genome research 12 1455 1465
9. DombroskiBAMathiasSLNanthakumarEScottAFKazazianHHJr 1991 Isolation of an active human transposable element. Science 254 1805 1808
10. ScottAFSchmeckpeperBJAbdelrazikMComeyCTO'HaraB 1987 Origin of the human L1 elements: proposed progenitor genes deduced from a consensus DNA sequence. Genomics 1 113 125
11. FengQMoranJVKazazianHHJrBoekeJD 1996 Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell 87 905 916
12. MathiasSLScottAFKazazianHHJrBoekeJDGabrielA 1991 Reverse transcriptase encoded by a human transposable element. Science 254 1808 1810
13. JurkaJ 1997 Sequence patterns indicate an enzymatic involvement in integration of mammalian retroposons. Proc Natl Acad Sci U S A 94 1872 1877
14. LuanDDKormanMHJakubczakJLEickbushTH 1993 Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell 72 595 605
15. KazazianHHJr 2004 Mobile elements: drivers of genome evolution. Science 303 1626 1632
16. BatzerMADeiningerPL 2002 Alu repeats and human genomic diversity. Nat Rev Genet 3 370 379
17. BeckCRCollierPMacfarlaneCMaligMKiddJM 2010 LINE-1 retrotransposition activity in human genomes. Cell 141 1159 1170
18. HuangCRSchneiderAMLuYNiranjanTShenP 2010 Mobile interspersed repeats are major structural variants in the human genome. Cell 141 1171 1182
19. IskowRCMcCabeMTMillsREToreneSPittardWS 2010 Natural mutagenesis of human genomes by endogenous retrotransposons. Cell 141 1253 1261
20. WitherspoonDJXingJZhangYWatkinsWSBatzerMA 2010 Mobile element scanning (ME-Scan) by targeted high-throughput sequencing. BMC Genomics 11 410
21. KazazianHHJrWongCYoussoufianHScottAFPhillipsDG 1988 Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man. Nature 332 164 166
22. BelancioVPHedgesDJDeiningerP 2008 Mammalian non-LTR retrotransposons: for better or worse, in sickness and in health. Genome research 18 343 358
23. GoodierJLKazazianHHJr 2008 Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell 135 23 35
24. ChenJMStensonPDCooperDNFerecC 2005 A systematic analysis of LINE-1 endonuclease-dependent retrotranspositional events causing human genetic disease. Hum Genet 117 411 427
25. WallaceMRAndersenLBSaulinoAMGregoryPEGloverTW 1991 A de novo Alu insertion results in neurofibromatosis type 1. Nature 353 864 866
26. KazazianHHJrMoranJV 1998 The impact of L1 retrotransposons on the human genome. Nat Genet 19 19 24
27. CallinanPABatzerMA 2006 Retrotransposable elements and human disease. Genome Dyn 1 104 115
28. ChenJMFerecCCooperDN 2006 LINE-1 endonuclease-dependent retrotranspositional events causing human genetic disease: mutation detection bias and multiple mechanisms of target gene disruption. J Biomed Biotechnol 2006 56182
29. JurkaJ 2000 Repbase update: a database and an electronic journal of repetitive elements. Trends Genet 16 418 420
30. BoissinotSChevretPFuranoAV 2000 L1 (LINE-1) retrotransposon evolution and amplification in recent human history. Mol Biol Evol 17 915 928
31. MyersJSVincentBJUdallHWatkinsWSMorrishTA 2002 A comprehensive analysis of recently integrated human Ta L1 elements. Am J Hum Genet 71 312 326
32. GilbertNLutzSMorrishTAMoranJV 2005 Multiple fates of L1 retrotransposition intermediates in cultured human cells. Mol Cell Biol 25 7780 7795
33. GilbertNLutz-PriggeSMoranJV 2002 Genomic deletions created upon LINE-1 retrotransposition. Cell 110 315 325
34. SymerDEConnellyCSzakSTCaputoEMCostGJ 2002 Human l1 retrotransposition is associated with genetic instability in vivo. Cell 110 327 338
35. SkowronskiJFanningTGSingerMF 1988 Unit-length line-1 transcripts in human teratocarcinoma cells. Mol Cell Biol 8 1385 1397
36. KazazianHHJr 1999 An estimated frequency of endogenous insertional mutations in humans. Nat Genet 22 130
37. MessiaenLWimmerK 2011 Mutation analysis of the NF1 gene by cDNA-based sequencing of the coding region Soares Gonçalves CunhaKGellerM Advances in Neurofibromatosis Research Nova Science Publishers
38. TeugelsEDe BrakeleerSGoelenGLissensWSermijnE 2005 De novo Alu element insertions targeted to a sequence common to the BRCA1 and BRCA2 genes. Hum Mutat 26 284
39. ConleyMEPartainJDNorlandSMShurtleffSAKazazianHHJr 2005 Two independent retrotransposon insertions at the same site within the coding region of BTK. Hum Mutat 25 324 325
40. CordauxRHedgesDJHerkeSWBatzerMA 2006 Estimating the retrotransposition rate of human Alu elements. Gene 373 134 137
41. MorrishTAGilbertNMyersJSVincentBJStamatoTD 2002 DNA repair mediated by endonuclease-independent LINE-1 retrotransposition. Nat Genet 31 159 165
42. ChenJMFerecCCooperDN 2007 Mechanism of Alu integration into the human genome. Genomic Med 1 9 17
43. MorrishTAGarcia-PerezJLStamatoTDTaccioliGESekiguchiJ 2007 Endonuclease-independent LINE-1 retrotransposition at mammalian telomeres. Nature 446 208 212
44. KojimaKK 2010 Different integration site structures between L1 protein-mediated retrotransposition in cis and retrotransposition in trans. Mob DNA 1 17
45. OstertagEMKazazianHHJr 2001 Twin priming: a proposed mechanism for the creation of inversions in L1 retrotransposition. Genome research 11 2059 2065
46. WimmerKYaoSClaesKKehrer-SawatzkiHTinschertS 2006 Spectrum of single - and multiexon NF1 copy number changes in a cohort of 1,100 unselected NF1 patients. Genes Chromosomes Cancer 45 265 276
47. BergetSM 1995 Exon recognition in vertebrate splicing. J Biol Chem 270 2411 2414
48. WimmerKRocaXBeiglbockHCallensTEtzlerJ 2007 Extensive in silico analysis of NF1 splicing defects uncovers determinants for splicing outcome upon 5′ splice-site disruption. Hum Mutat 28 599 612
49. KralovicovaJVorechovskyI 2007 Global control of aberrant splice-site activation by auxiliary splicing sequences: evidence for a gradient in exon and intron definition. Nucleic Acids Res 35 6399 6413
50. MeischlCBoerMAhlinARoosD 2000 A new exon created by intronic insertion of a rearranged LINE-1 element as the cause of chronic granulomatous disease. Eur J Hum Genet 8 697 703
51. MessiaenLMCallensTMortierGBeysenDVandenbrouckeI 2000 Exhaustive mutation analysis of the NF1 gene allows identification of 95% of mutations and reveals a high frequency of unusual splicing defects. Hum Mutat 15 541 555
52. KobayashiKNakahoriYMiyakeMMatsumuraKKondo-IidaE 1998 An ancient retrotransposal insertion causes Fukuyama-type congenital muscular dystrophy. Nature 394 388 392
53. YoshidaKNakamuraAYazakiMIkedaSTakedaS 1998 Insertional mutation by transposable element, L1, in the DMD gene results in X-linked dilated cardiomyopathy. Hum Mol Genet 7 1129 1132
54. HanJSSzakSTBoekeJD 2004 Transcriptional disruption by the L1 retrotransposon and implications for mammalian transcriptomes. Nature 429 268 274
55. Perepelitsa-BelancioVDeiningerP 2003 RNA truncation by premature polyadenylation attenuates human mobile element activity. Nat Genet 35 363 366
56. VandenbrouckeIvan DoornRCallensTCobbenJMStarinkTM 2004 Genetic and clinical mosaicism in a patient with neurofibromatosis type 1. Hum Genet 114 284 290
57. WimmerKEckartMRehderHFonatschC 2000 Illegitimate splicing of the NF1 gene in healthy individuals mimics mutation-induced splicing alterations in NF1 patients. Hum Genet 106 311 313
58. ThompsonJDHigginsDGGibsonTJ 1994 CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22 4673 4680
Štítky
Genetika Reprodukční medicína
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2011 Číslo 11
-
Všechny články tohoto čísla
- De Novo Origins of Human Genes
- Duplication Hotspots Are Associated with Late-Replicating Regions of the Genome
- De Novo Origin of Human Protein-Coding Genes
- Cyclin D/CDK4 and Cyclin E/CDK2 Induce Distinct Cell Cycle Re-Entry Programs in Differentiated Muscle Cells
- Short Day–Mediated Cessation of Growth Requires the Downregulation of AINTEGUMENTALIKE1 Transcription Factor in Hybrid Aspen
- Physiological IRE-1-XBP-1 and PEK-1 Signaling in Larval Development and Immunity
- Role of Pirh2 in Mediating the Regulation of p53 and c-Myc
- Signatures of Environmental Genetic Adaptation Pinpoint Pathogens as the Main Selective Pressure through Human Evolution
- FOXO Regulates Organ-Specific Phenotypic Plasticity In
- Heritable Epigenetic Variation among Maize Inbreds
- Foxn1 Regulates Lineage Progression in Cortical and Medullary Thymic Epithelial Cells But Is Dispensable for Medullary Sublineage Divergence
- Attenuation of the Sensing Capabilities of PhoQ in Transition to Obligate Insect–Bacterial Association
- A Novel Protein LZTFL1 Regulates Ciliary Trafficking of the BBSome and Smoothened
- Activation of Bmp2-Smad1 Signal and Its Regulation by Coordinated Alteration of H3K27 Trimethylation in -Induced Senescence
- Histone H3K56 Acetylation, CAF1, and Rtt106 Coordinate Nucleosome Assembly and Stability of Advancing Replication Forks
- The SUN Protein Mps3 Is Required for Spindle Pole Body Insertion into the Nuclear Membrane and Nuclear Envelope Homeostasis
- Evidence-Based Annotation of Gene Function in MR-1 Using Genome-Wide Fitness Profiling across 121 Conditions
- Effect of Host Species on the Distribution of Mutational Fitness Effects for an RNA Virus
- Pch2 Acts through Xrs2 and Tel1/ATM to Modulate Interhomolog Bias and Checkpoint Function during Meiosis
- SOX9 Governs Differentiation Stage-Specific Gene Expression in Growth Plate Chondrocytes via Direct Concomitant Transactivation and Repression
- from the Aphid : A Missing Link from Facultative to Obligate Insect Endosymbiont
- Recessive Antimorphic Alleles Overcome Functionally Redundant Loci to Reveal Function in Flowers and Meristems
- Over-Expression of DSCAM and COL6A2 Cooperatively Generates Congenital Heart Defects
- Consequences of Eukaryotic Enhancer Architecture for Gene Expression Dynamics, Development, and Fitness
- Distinct Genetic Architectures for Male and Female Inflorescence Traits of Maize
- Capture of MicroRNA–Bound mRNAs Identifies the Tumor Suppressor miR-34a as a Regulator of Growth Factor Signaling
- For Male , Sperm Activation Is a “Just-in-Time” Event
- PcG Complexes Set the Stage for Epigenetic Inheritance of Gene Silencing in Early S Phase before Replication
- The Gene Contains Hotspots for L1 Endonuclease-Dependent Insertion
- Relative Burden of Large CNVs on a Range of Neurodevelopmental Phenotypes
- Multiple Means to the Same End: The Genetic Basis of Acquired Stress Resistance in Yeast
- Genome-Wide Crossover Distribution in Meiosis Reveals Sex-Specific Patterns along Chromosomes
- TRY-5 Is a Sperm-Activating Protease in Seminal Fluid
- Homologs of Retinoblastoma-Associated Protein 46/48 Associate with a Histone Deacetylase to Act Redundantly in Chromatin Silencing
- Genetic Interaction Maps in Reveal Functional Crosstalk among Cell Envelope Biogenesis Pathways
- The ERI-6/7 Helicase Acts at the First Stage of an siRNA Amplification Pathway That Targets Recent Gene Duplications
- PBX1 Genomic Pioneer Function Drives ERα Signaling Underlying Progression in Breast Cancer
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Evidence-Based Annotation of Gene Function in MR-1 Using Genome-Wide Fitness Profiling across 121 Conditions
- De Novo Origins of Human Genes
- Capture of MicroRNA–Bound mRNAs Identifies the Tumor Suppressor miR-34a as a Regulator of Growth Factor Signaling
- TRY-5 Is a Sperm-Activating Protease in Seminal Fluid
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



