-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaEpigenetic Silencing of Spermatocyte-Specific and Neuronal Genes by SUMO Modification of the Transcription Factor Sp3
SUMO modification of transcription factors is linked to repression of transcription. The physiological significance of SUMO attachment to a particular transcriptional regulator, however, is largely unknown. We have employed the ubiquitously expressed murine transcription factor Sp3 to analyze the role of SUMOylation in vivo. We generated mice and mouse embryonic fibroblasts (MEFs) carrying a subtle point mutation in the SUMO attachment sequence of Sp3 (IKEE553D mutation). The E553D mutation impedes SUMOylation of Sp3 at K551 in vivo, without affecting Sp3 protein levels. Expression profiling revealed that spermatocyte-specific genes, such as Dmc1 and Dnahc8, and neuronal genes, including Paqr6, Rims3, and Robo3, are de-repressed in non-testicular and extra-neuronal mouse tissues and in mouse embryonic fibroblasts expressing the SUMOylation-deficient Sp3E553D mutant protein. Chromatin immunoprecipitation experiments show that transcriptional de-repression of these genes is accompanied by the loss of repressive heterochromatic marks such as H3K9 and H4K20 tri-methylation and impaired recruitment of repressive chromatin-modifying enzymes. Finally, analysis of the DNA methylation state of the Dmc1, Paqr6, and Rims3 promoters by bisulfite sequencing revealed that these genes are highly methylated in Sp3wt MEFs but are unmethylated in Sp3E553D MEFs linking SUMOylation of Sp3 to tissue-specific CpG methylation. Our results establish SUMO conjugation to Sp3 as a molecular beacon for the assembly of repression machineries to maintain tissue-specific transcriptional gene silencing.
Published in the journal: . PLoS Genet 6(11): e32767. doi:10.1371/journal.pgen.1001203
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1001203Summary
SUMO modification of transcription factors is linked to repression of transcription. The physiological significance of SUMO attachment to a particular transcriptional regulator, however, is largely unknown. We have employed the ubiquitously expressed murine transcription factor Sp3 to analyze the role of SUMOylation in vivo. We generated mice and mouse embryonic fibroblasts (MEFs) carrying a subtle point mutation in the SUMO attachment sequence of Sp3 (IKEE553D mutation). The E553D mutation impedes SUMOylation of Sp3 at K551 in vivo, without affecting Sp3 protein levels. Expression profiling revealed that spermatocyte-specific genes, such as Dmc1 and Dnahc8, and neuronal genes, including Paqr6, Rims3, and Robo3, are de-repressed in non-testicular and extra-neuronal mouse tissues and in mouse embryonic fibroblasts expressing the SUMOylation-deficient Sp3E553D mutant protein. Chromatin immunoprecipitation experiments show that transcriptional de-repression of these genes is accompanied by the loss of repressive heterochromatic marks such as H3K9 and H4K20 tri-methylation and impaired recruitment of repressive chromatin-modifying enzymes. Finally, analysis of the DNA methylation state of the Dmc1, Paqr6, and Rims3 promoters by bisulfite sequencing revealed that these genes are highly methylated in Sp3wt MEFs but are unmethylated in Sp3E553D MEFs linking SUMOylation of Sp3 to tissue-specific CpG methylation. Our results establish SUMO conjugation to Sp3 as a molecular beacon for the assembly of repression machineries to maintain tissue-specific transcriptional gene silencing.
Introduction
A plethora of proteins involved in regulating gene expression such as promoter-specific transcription factors, cofactors and chromatin-modifying enzymes are reversibly modified by the Small Ubiquitin-like MOdifier SUMO (reviewed in [1], [2]). With few exceptions, SUMO modification of transcriptional regulators correlates with repression of transcription [3]–[5].
The ubiquitously expressed transcription factor Sp3 represents a well-studied paradigm for regulation of activity by SUMOylation [6]–[8]. Sp3 belongs to the Sp (specificity protein) family of transcription factors that is implicated in the expression of a wide variety of genes including housekeeping, tissue-specific, developmentally and cell-cycle regulated genes [9]–[12]. A major feature of Sp3 is that, depending on promoter context, it can either activate or repress transcription in reporter gene assays [6], [7], [13]. Two glutamine-rich domains are known to exercise the activation function of Sp3 [13]; whereas the repressive activity of Sp3 is mediated by attachment of SUMO to lysine 551. K551 lies within the SUMO consensus motif IKEE located between the second Q-rich activation domain and the DNA-binding domain [6], [7] (Figure 1A and 1B). The functional complexity of Sp3 is further increased in vivo by the expression of four different isoforms that differ in their N-terminal extension [14]. All of these isoforms are SUMO-modified at K551, giving rise to a composite pattern of at least eight distinct protein species [14].
Fig. 1. Targeting of the mouse Sp3 SUMO attachment site. 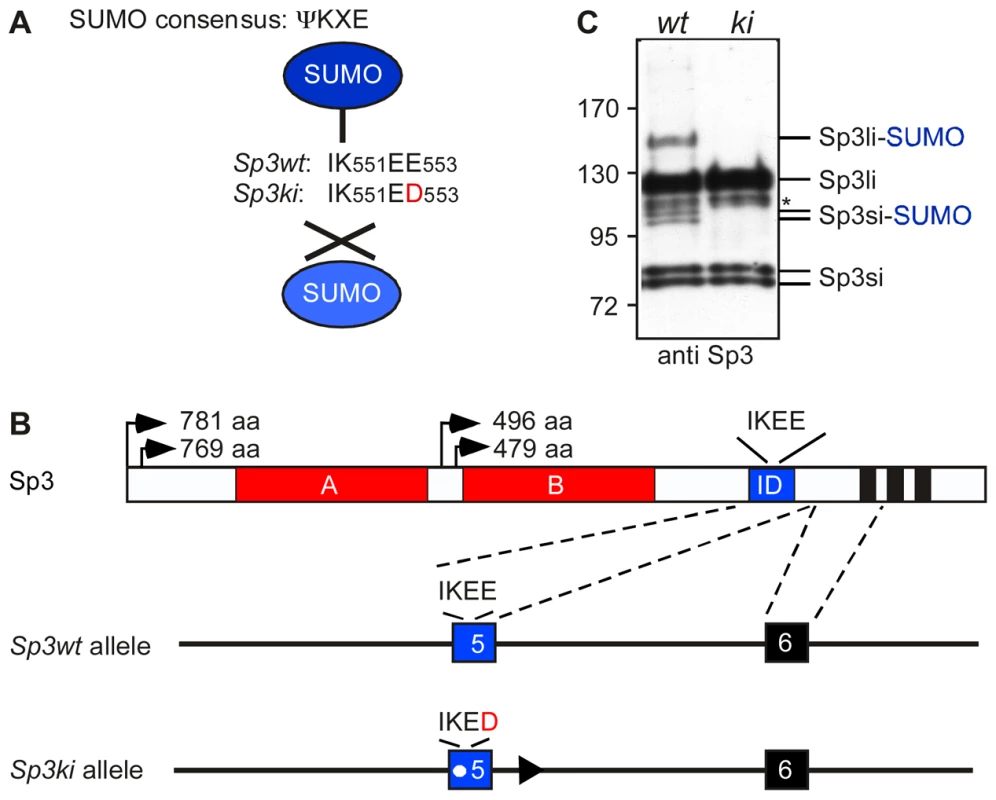
(A) Sp3 is posttranslationally modified at K551 within the SUMO consensus sequence ΨKXE. In Sp3ki/ki mice the IK551EE553 sequence is replaced by IK551ED553. (B) Schematic presentation of Sp3 protein structure and part of the Sp3 gene. The glutamine-rich activation domains A and B, the SUMO attachment site (IKEE) within the inhibitory domain (ID) and the zinc fingers (black bars) of the DNA-binding domain are indicated. Arrows depict the four translational start sites and the lengths of the resulting Sp3 isoforms are shown. In the mutated Sp3ki allele, wild type exon 5 is replaced by the mutant exon 5 carrying the E553D mutation. (C) Sp3ki/ki mice lack SUMO modification of Sp3. MEFs obtained from Sp3wt (wt) and Sp3ki/ki (ki) embryos at E13.5 were subjected to Western blot analysis with Sp3 antibodies. The asterisk denotes an aspecific band. Previous investigations of the molecular events associated with Sp3-SUMO-dependent repression have provided mechanistic clues underlying SUMO-dependent gene silencing [15], [16]. SUMO-modification can act as a signal for the recruitment of various chromatin-associated repression components including the chromatin remodeler Mi-2, the MBT-domain proteins L3MBTL1 and L3MBTL2, heterochromatin protein 1 (HP1) and the histone methyltransferases (HMTs) SETDB1/ESET and SUV4-20H, concomitant with the establishment of repressive histone modifications such as H3K9 and H4K20 tri-methylation [16].
Despite extensive studies on the repression function of SUMOylated Sp3 and other transcription factors, the significance of SUMO attachment for the expression of endogenous genes in vivo is still largely unknown. Here, we report the generation of mice and mouse embryonic fibroblasts (MEFs) with a point mutation in the SUMO attachment sequence of Sp3. Expression profiling revealed that SUMOylation of Sp3 is required for silencing of spermatocyte-specific genes such as Dmc1 and Dnahc8 in somatic cells, and neuronal genes including Paqr6, Rims3 and Robo3 in non-neuronal cells. Transcriptional de-repression of these genes in MEFs expressing the Sp3E553D mutant protein is accompanied by the loss of repressive heterochromatic marks such as H3K9 and H4K20 tri-methylation, impaired recruitment of repressive chromatin-modifying enzymes and loss of DNA methylation. Our results establish that SUMO-modification of Sp3 acts as a platform for the assembly of repression machineries to maintain tissue-specific transcriptional gene silencing.
Results
Generating mutant mice deficient for SUMO modification of Sp3
To investigate the in vivo function of Sp3 SUMOylation, we generated mice in which the SUMO attachment site IK551EE is mutated to IK551ED (Figure 1A). We chose glutamic acid residue E553 for mutation because K551 might also be a target for other posttranslational modifications such as methylation or acetylation. A vector carrying the Sp3E553D mutation and a floxed neomycin-resistance cassette was used for targeting of ES cells to generate heterozygous mutant mice (Figure 1B; Figure S1). The neomycin-resistance gene was subsequently removed by mating with appropriate Cre recombinase-expressing mice [17]. Mice carrying the Sp3E553D mutation will from hereon be referred to as Sp3 knockin (Sp3ki) mutant. Heterozygous Sp3wt/ki mice and homozygous Sp3ki/ki mice were fertile, born at the expected Mendelian frequency and exhibited no obvious phenotype (Table S1).
To ensure that the E553D mutation impaired SUMOylation of Sp3, we performed Western blotting of adult mouse tissues and MEFs derived from E13.5 embryos. SUMO-modification of Sp3 was readily detectable in Sp3wt and heterozygous Sp3wt/ki but not in homozygous Sp3ki/ki tissues and MEFs (Figure 1C; Figure S1). This result demonstrates that the glutamic acid residue within the SUMOylation consensus motif ΨKXE is absolutely essential for the attachment of SUMO to endogenous Sp3. We conclude that the E553D mutation carried by Sp3ki/ki mice impedes SUMOylation of Sp3 at K551 in vivo, without affecting Sp3 protein levels.
SUMOylated Sp3 represses spermatocyte-specific and neuronal genes
To identify genes that are regulated by SUMO-modified Sp3, we performed gene expression profiling with RNA extracted from primary Sp3wt and Sp3ki/ki MEFs derived from E13.5 littermates. This identified 68 genes that were upregulated and 7 genes that were downregulated by more than 2-fold in Sp3ki/ki MEFs (Table S2). Notably, top candidate genes that were upregulated in Sp3ki/ki MEFs encode developmentally-regulated meiotic and neuronal proteins. Dmc1 and Dnahc8 are expressed in meiotic spermatocytes and encode a RecA-like recombinase and a flagellar protein, respectively [18], [19]. Paqr6, Rims3 and Robo3 are expressed in the central nervous system [20]–[22]. The expression pattern of another upregulated gene (Villin-like, Vill) is largely unknown, although a low level of expression in early embryogenesis was reported [23].
To validate aberrant de-repression of these genes, we analyzed their expression in MEF cultures by quantitative RT-PCR. Dmc1, Dnahc8, Paqr6, Rims3, Robo3 and Vill mRNA levels were elevated in primary Sp3ki/ki MEFs as well as in immortalized Sp3ki/ki MEFs (Figure 2). We also analyzed expression of Dmc1, Dnahc8, Paqr6, Rims3, Robo3 and Vill in Sp3-deficient (Sp3-/-) MEFs obtained from E13.5 Sp3 knockout embryos [24]. Consistent with an Sp3-SUMO-dependent silencing function, all six genes were upregulated also in Sp3-/- MEFs as compared to corresponding Sp3wt MEFs derived from littermates (Figure 2). We also performed immunoblot analysis for Dmc1 and Dnahc8 but failed to detect these proteins probably due to their low expression levels. However, de-repression of the Dmc1 gene in Sp3ki/ki and Sp3-/- MEFs was verified with different amplimers spanning different exons (data not shown), thereby precluding the possibility that the qRT-PCR analyses detected an aberrant Dmc1 transcript.
Fig. 2. SUMOylation of Sp3 is required for silencing of spermatocyte-specific and neuronal genes in MEFs. 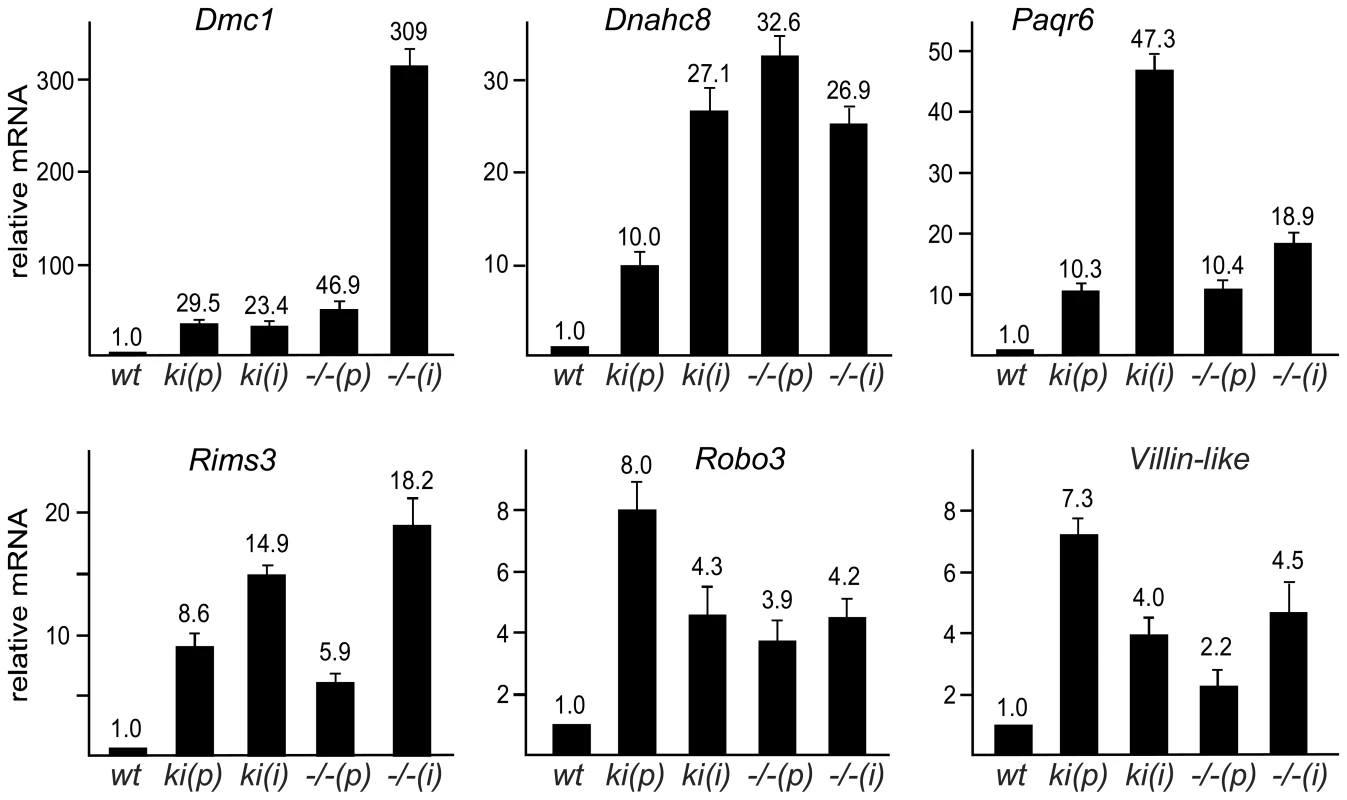
Quantitative real-time RT-PCR analysis of Dmc1, Dnahc8, Paqr6, Rims3, Robo3 and Vill expression in primary (p) and immortalized (i) Sp3wt (wt), Sp3ki/ki (ki) and Sp3-/- (−/−) MEFs. Normalized mRNA levels are presented relative to wild type MEFs arbitrarily set to 1. Data are expressed as mean +/−SD. De-repression of Dmc1, Dnahc8, Paqr6, Rims3, Robo3 and Vill in Sp3ki/ki and Sp3-/- MEF cultures suggested that SUMO modification of Sp3 might be essential for silencing these genes in tissues other than testis and brain, respectively. We analyzed RNA from various tissues of adult Sp3wt and Sp3ki/ki mice. As expected, Dmc1 and Dnahc8 were strongly expressed in testis of Sp3wt and Sp3ki/ki mice at similar levels but were not or only marginally expressed in other tissues such as brain, heart, intestine, kidney, liver, lung and spleen. In Sp3ki/ki mice, Dmc1 and Dnahc8 mRNA levels were significantly higher in all tissues (Figure 3). However, the amount of RNA in testis was still one to three orders of magnitude higher indicating that testis-specific activators further enhance expression of Dmc1 and Dnahc8 in spermatocytes.
Fig. 3. SUMOylation of Sp3 restricts tissue-specific gene expression in mouse organs. 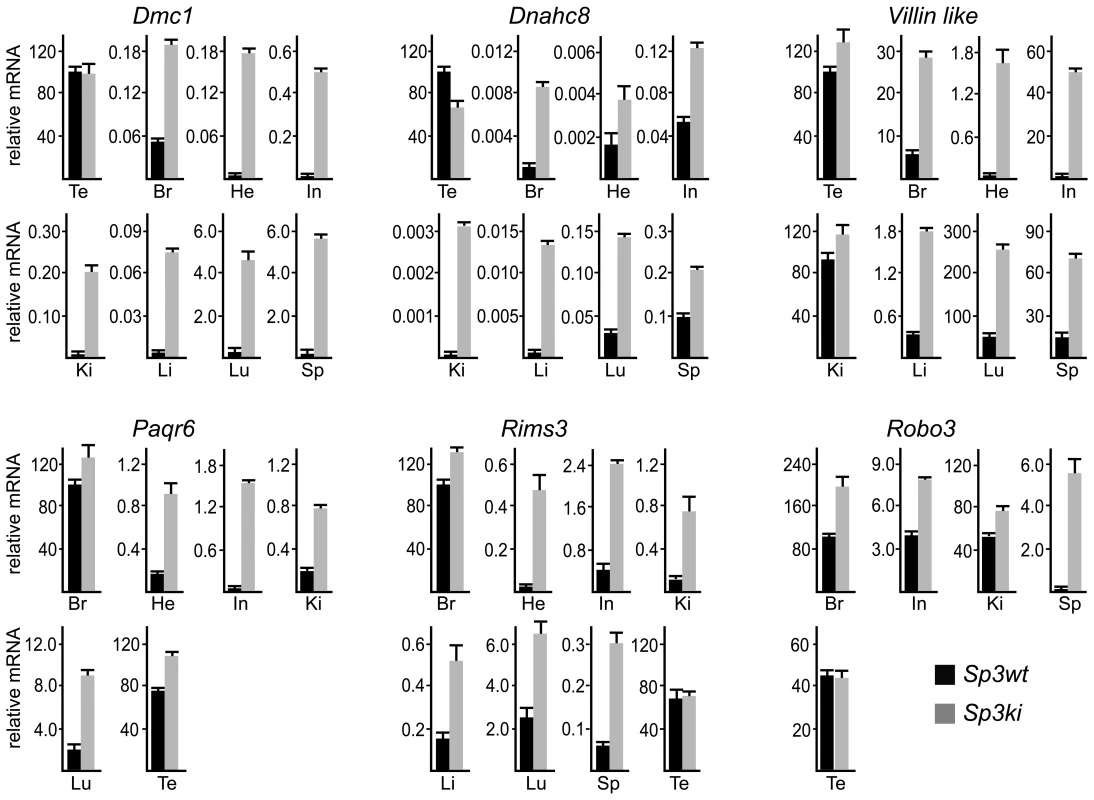
RNA from adult mouse organs was analyzed for Dmc1, Dnahc8, Villin-like, Paqr6, Rims3 and Robo3 expression by qRT-PCR. Expression levels were normalized to Gapdh mRNA and are presented relative to testis (Dmc1, Dnahc8 and Villin-like) or brain (Paqr6, Rims3 and Robo3) RNA arbitrarily set to 100. Abbreviations: Br, brain; Te, testis; He, heart; In, intestine; Ki, kidney; Li, liver, Lu, lung; Sp, spleen. Data are expressed as mean +/−SD. Previous reports have attributed silencing of several meiotic and male germ-line-specific genes in somatic cells such as Smc1ß and Stag3 to E2F6 [25], [26], a repressive member of the E2F family of transcription factors. We analyzed expression of Smc1ß and Stag3 in tissues of Sp3ki/ki mice as well. Both genes were only detectable in testis RNA preparations and were not de-repressed in non-testicular Sp3ki/ki tissues or Sp3ki/ki MEFs (data not shown). Vice versa, the Dmc1 gene is not de-repressed in E2F6-/- MEFs [27] although the Dmc1 promoter region is bound by E2F6 in vivo at a conserved binding site [27]. This observation suggests that different transcription factors and mechanisms are responsible for repressing spermatocyte-specific genes in somatic cells.
Consistent with published data, Paqr6, Rims3 and Robo3 mRNA levels were highest in mouse brain. Strikingly, all three genes were also highly expressed in testis and in the case of Robo3 also in kidney. In all other organs these mRNAs were either not detectable or expressed only at a very low level (Figure 3). Nevertheless, expression of Paqr6, Rims3 and Robo3 as well as Vill mRNA was significantly elevated in several organs of Sp3ki/ki mice (Figure 3). Taken together, these results demonstrate that SUMOylation of Sp3 is essential for silencing of a subset of spermatocyte-specific and neuronal genes in somatic and non-neuronal tissues, respectively, implying an important role of the SUMO moiety attached to Sp3 in establishing tissue-specific gene expression patterns.
Gene silencing of meiotic and neuronal genes is rescued by re-expression of SUMOylation-competent Sp3 in Sp3-/- MEFs
To substantiate the notion that SUMOylation of Sp3 is directly responsible for silencing testis - and neuronal-specific genes in MEFs, we re-expressed the short and long isoforms of Sp3 (Sp3si-wt and Sp3li-wt) in Sp3-/- MEFs by retroviral transduction (Figure 4A). As controls, we used the corresponding SUMOylation-deficient Sp3 mutants (Sp3si-K551D and Sp3li-K551R). Particularly, re-expression of the long isoform of Sp3 resulted in significantly reduced expression of the Dmc1, Dnahc8, Paqr6, Rims3, Robo3 and Vill genes. Repression of these genes by re-expression of the small isoforms of Sp3 was less pronounced (Figure 4B–4G). The weaker effects observed with the small isoforms of Sp3 could be due to their lower expression level (see Figure 4A). Potentially, simultaneous expression of all four wild type Sp3 isoforms would be necessary to restore repression completely. Nevertheless, in contrast to the wild type Sp3 isoforms, introduction of the SUMOylation-deficient Sp3 mutants failed to rescue gene silencing but instead further enhanced expression of these genes (Figure 4B–4G). These results show that reintroduction of Sp3 partially reverses de-repression of these genes in a SUMOylation-dependent manner.
Fig. 4. Sp3-SUMO–dependent rescue of gene silencing. 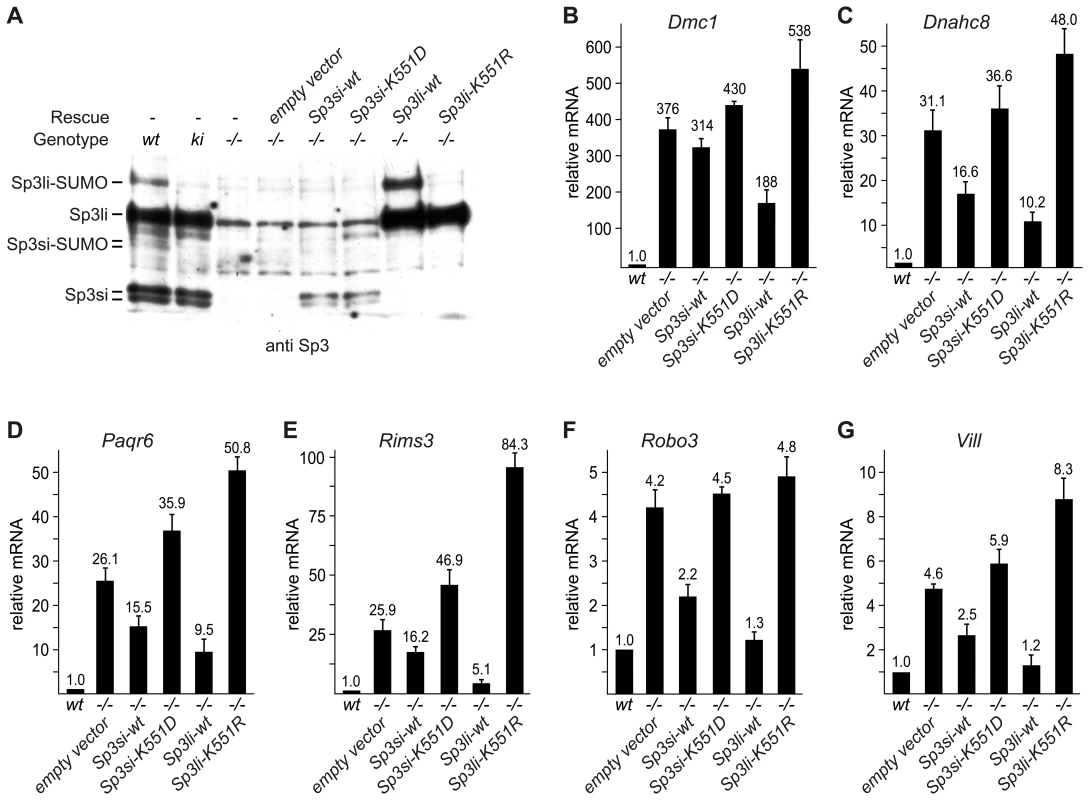
Sp3-/- MEFs were transduced with retroviral expression vectors for the SUMOylation-competent long and short Sp3 isoforms (Sp3li-wt and Sp3si-wt), and corresponding SUMOylation-deficient mutants (Sp3li-K551R and Sp3si-K551D), respectively. (A) Immunoblot analysis of the various MEF cultures. (B–G) Expression of Dmc1, Dnahc8, Paqr6, Robo3, Rims3 and Vill in rescue MEFs. Gapdh-normalized mRNA expression levels are presented relative to the expression in wild type MEFs arbitrarily set to 1. Data are expressed as mean +/−SD. Sp3 is bound to the promoters of repressed genes in vivo
To analyze whether Sp3 is bound to the promoters of genes that are repressed by Sp3-SUMO, we performed ChIP analyses. Because of the lack of precise promoter information for Dnahc8, Robo3 and Vill we focused on Dmc1, Paqr6 and Rims3. All three promoters contain several potential binding sites for Sp3. Antibodies to Sp3 precipitated all three promoters from Sp3wt and Sp3ki/ki MEF chromatin but not from Sp3-/- MEF chromatin (Figure 5), demonstrating that both wild type Sp3 and the SUMOylation-deficient Sp3E553D mutant were bound to the Dmc1, Paqr6 and Rims3 promoters.
Fig. 5. Sp3-SUMO–dependent association of heterochromatic histone modifications and repressive chromatin components. 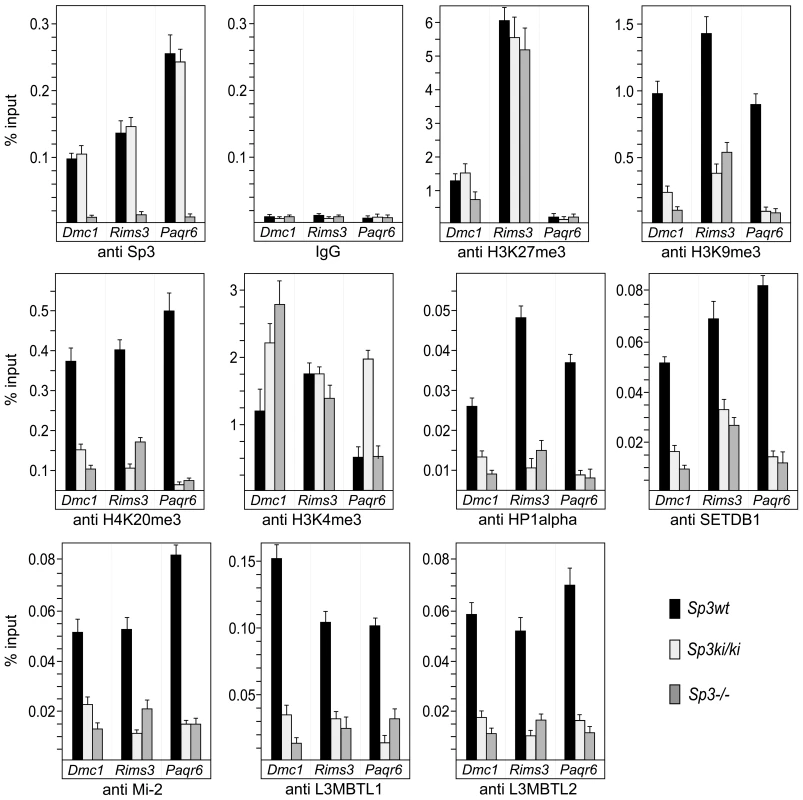
Sp3wt, Sp3ki/ki and Sp3-/- MEFs were subjected to ChIP analysis with the indicated antibodies. Precipitated DNA was amplified by qPCR with promoter-specific primers for Dmc1, Rims3 and Paqr6. The locations of the primers are indicated as filled arrowheads in the schematic presentation of the promoters in Figure 6. DNA recoveries are expressed as percentage of input (mean +/−SD). Repressive chromatin components are enriched on promoters silenced by Sp3-SUMO
Next, we analyzed the Dmc1, Paqr6 and Rims3 promoters for the presence of repressive histone modifications (Figure 5). In Sp3wt MEFs, the H3K27me3 mark is abundantly present at the Dmc1 and Rims3 promoters but not at the Paqr6 promoter. Moreover, this mark is not or only marginally reduced in the absence of SUMOylated Sp3 suggesting that H3K27me3 does not contribute to SUMO-dependent gene silencing of these three genes. In contrast, H3K9me3 and H4K20me3 marks are present at all three promoters in Sp3wt MEFs but are strongly reduced in Sp3ki/ki and in Sp3-/- MEFs. Consistently, HP1α, which binds H3K9me3, is present at the Dmc1, Paqr6 and Rims3 promoters in Sp3wt MEFs but not in Sp3ki/ki and Sp3-/- MEFs. Thus, the presence or absence of H3K9me3, H4K20me3 and HP1α at the Dmc1, Paqr6 and Rims3 promoters correlates strictly with the repressed or de-repressed state of these genes. We also analyzed for the presence of H3K4 trimethylation, an epigenetic mark characteristic for promoter-proximal nucleosomes of most active as well as inactive genes [28]. The co-occurrence of H3K9me3, H3K27me3 and H3K4me3 marks is a characteristic property of “bivalent” promoters of euchromatic genes in ES cells, believed to reflect a repressed but poised transcriptional state [29]. The H3K4me3 mark was abundantly present on the Dmc1, Paqr6 and Rims3 promoters in Sp3wt MEFs. In Sp3ki/ki MEFs we found an approximately 2-fold and 3-fold increase of H3K4 trimethylation on the Dmc1 promoter and on the Paqr6 promoter, respectively, but not on the Rims3 promoter (Figure 5). In Sp3-/- cells, higher H3K4me3 levels were detected on the Dmc1 promoter but not on the Rims3 and Paqr6 promoters. Thus, there is no strict correlation between the changes of the H3K4me3 mark and the expression state of the different target genes.
Our previous investigations revealed that the establishment of repressive nucleosomal signatures on a chromatinized Gal4-driven reporter gene by Gal4-Sp3-SUMO involves the recruitment of the histone methyltransferase SETDB1, the chromatin remodeler Mi-2, and the chromatin-compacting MBT-domain proteins L3MBTL1 and L3MBTL2 [16]. Therefore, we analyzed the Dmc1, Paqr6 and Rims3 promoters for the presence of these proteins. ChIP analysis revealed that all four proteins are abundantly present on the promoters in Sp3wt MEFs but strongly reduced in Sp3ki/ki and Sp3-/- MEFs (Figure 5). The recruitment of these chromatin-modifying proteins to the endogenous Dmc1, Paqr6 and Rims3 promoters is thus dependent on the presence of the Sp3 SUMO moiety. Taken together, these results support the conclusion that the posttranslational modification of Sp3 at K551 provokes a local repressive chromatin structure on a subset of spermatocyte - and neuronal-specific genes in somatic and non-neuronal cells, respectively.
Repression of many nervous system-specific genes in unrelated tissues has been attributed to the corepressor CoREST1. CoREST1 is recruited to promoters by the transcriptional repressor REST/NRSF [30] and, alternatively, by a REST/NRSF-independent mechanism that involves direct interaction between CoREST1 and a thus far unknown SUMO2/3-modified transcription factor [31]. To examine whether CoREST1 is also involved in silencing the Dmc1, Paqr6 and Rims3 promoters, we performed ChIP analysis for the presence of CoREST1. CoREST1 was to some extent detectable on the Rims3 promoter but not on the Dmc1 and Paqr6 promoters irrespectively of the Sp3 SUMOylation status (Figure S2). This finding indicates that SUMOylation of Sp3 represents an alternative, CoREST1-independent pathway mediating extra-neuronal repression.
SUMO modification of Sp3 is essential for DNA methylation of target genes
An EMBOS CpGPlot analysis (http://www.ebi.ac.uk/Tools/emboss/cpgplot/index.html) revealed that the Dmc1, Paqr6 and Rims3 promoters are embedded in CpG islands. CpG island methylation may contribute to silencing of these genes in MEFs and may be reversed in Sp3ki/ki and Sp3-/- MEFs. Therefore, we analyzed the methylation states of these promoters in Sp3wt, Sp3ki/ki and Sp3-/- MEFs. Bisulfite sequencing revealed that the proximal promoters and the first exons of the Dmc1, Paqr6 and Rims3 genes are highly methylated in Sp3wt MEFs. In contrast, CpG methylation is strongly reduced in Sp3ki/ki and in Sp3-/- MEFs (Figure 6). In summary, there is a very tight correlation between SUMO modification of Sp3, transcriptional repression, repressive histone modifications and DNA methylation of these promoters. Lack of SUMOylation of Sp3 results in their de-repression accompanied by the absence of repressive histone modifications and the absence of CpG methylation.
Fig. 6. SUMOylation-dependent CpG methylation of the Dmc1, Paqr6, and Rims3 genes. 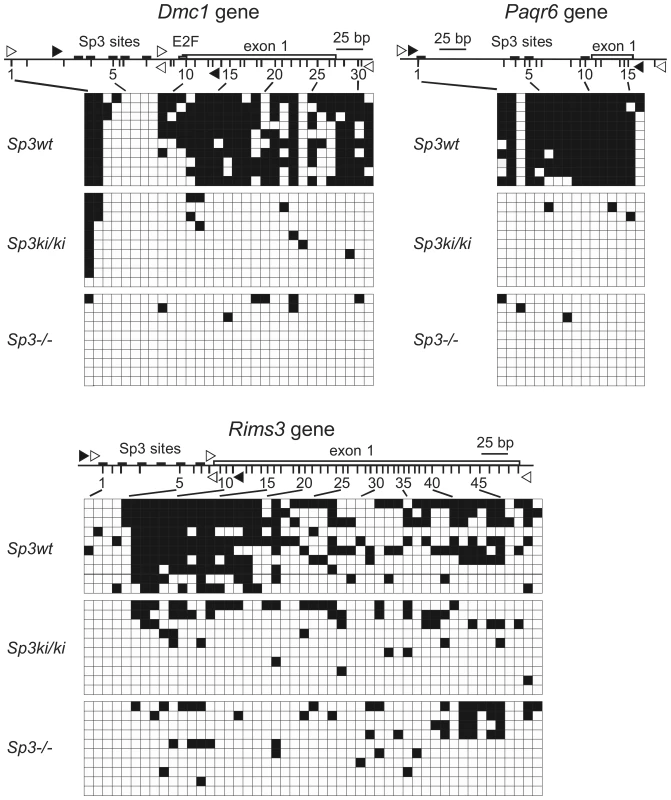
The methylation state of individual CpGs in Sp3wt, Sp3ki/ki and Sp3-/- MEFs was analyzed by bisulfite sequencing of ten clones. Non-methylated CpGs are indicated as open squares and methylated CpGs as filled squares. The promoters along with the position of Sp3 binding sites and the first exon are depicted schematically. Vertical lines depict the positions of CpG dinucleotides. Unfilled arrowheads indicate the locations of the primers used for PCR, and filled arrowheads indicate the locations of the primers used for the ChIP experiments shown in Figure 5. Discussion
The generation and analysis of mice with a subtle point mutation in the SUMO attachment site of the transcription factor Sp3 has revealed the relevance of SUMO modification of Sp3 for gene silencing in vivo. SUMO attachment to Sp3 serves as a molecular beacon for the recruitment of chromatin-modifying machineries that impose epigenetic silencing on a subset of spermatocyte-specific and neuronal genes. Our data are consistent with the bidirectional crosstalk between repressive histone modification and DNA methylation, that was established by demonstrating direct interactions between SETDB1 and the de novo methylase DNMT3A [32]. However, SETDB1 may also be recruited indirectly through interaction with the methyl-CpG binding protein MBD1 that forms a stable complex with SETDB1 [33]. We note that a functional SUMO interaction motif (SIM) is present in the histone methyltransferase SETDB1 [34] and that potential SIMs are also present in DNMTs. Thus, SUMO-modified Sp3 might recruit SETDB1 and DNMTs independently. Sp3-SUMO-mediated repression might involve a timely coordinated recruitment of chromatin remodelers, nucleosome compactors such as MBT domain proteins, HMTs and DNMTs. Alternatively, these different types of epigenetic players might be recruited simultaneously. The Sp3ki/ki and Sp3-/- MEFs described here provide essential tools for future experiments addressing these questions.
The lack of Sp3 SUMOylation in Sp3ki/ki mice causes aberrant expression of several spermatocyte-specific and neuronal genes in various somatic tissues. Although de-repression of these genes is best described by the loss of the repressive function of SUMOylated Sp3 in Sp3ki/ki mice, it is conceivable that the activation function of the Sp3E553D mutant protein may contribute directly or indirectly to the aberrant expression of these genes as well. However, expression of the spermatocyte-specific and neuronal genes in testis and brain, respectively, is not affected. This tissue-selectivity could be due to low-level Sp3 SUMOylation in spermatocytes and neurons. To investigate this, we performed Western blot analysis of testis and brain extracts. Consistent with the lack of repression in testis and brain, SUMOylated Sp3 species were barely detectable in these tissues (Figure S3). Future studies using purified spermatocytes and neurons might provide further insight on the cell type-specific SUMOylation state of Sp3. Interestingly, it has been reported that Sp3 expression in germ cells declines during the leptotene to pachytene transition whereas the related transcription factor Sp1 did not decline until the mid-pachytene phase of meiosis [35]. Dmc1 is expressed in leptotene to zygotene spermatocytes [19], and Dnahc8 from mid-pachytene to diplotene spermatocytes [18]. Thus, activation of the Dmc1 and Dnahc8 genes at these meiotic stages correlates with down-regulation of Sp3. Accordingly, one could image a scenario in which down-regulation of the Sp3 protein level facilitates Sp1-mediated activation of these two Sp3-SUMO target genes during spermatocyte development.
The promoters of the three Sp3-SUMO target genes that we analyzed in detail share several features. They contain multiple GC-boxes, lack a TATA box and are embedded in CpG islands. Such a promoter arrangement is reminiscent of housekeeping genes. In contrast to the three Sp3 target genes, housekeeping genes are ubiquitously expressed and remain unmethylated in all tissues. It is currently unclear why the three Sp3 target genes are expressed in such a highly tissue-specific manner. We have not detected obvious features in the spacing or orientation of Sp-binding sites that can account for this interesting difference. Further investigations comparing these two types of promoters are required to address this enigmatic point.
The aberrant expression of several testicular and neuronal genes in Sp3ki/ki mice apparently does not lead to obvious histological or behavioral abnormalities under standard mouse housing conditions indicating that essential functions of differentiated cell types are not grossly impaired. We note that the overall amounts of aberrantly produced mRNA transcripts in somatic extra-neuronal cells are low. This provides an explanation for the absence of clear anatomical and physiological anomalies in the Sp3ki/ki mice.
The phenotype of the Sp3ki/ki mice differs significantly from mice lacking the entire Sp3 protein. Sp3-deficient mice display skeletal, tooth, hematopoietic and heart defects at late embryonic development, and die immediately after birth due to respiratory failure [24], [36]–[38]. Given that SUMOylation-deficient Sp3 proteins are strong activators [13], [14] the defects observed in Sp3-/- mice have to be attributed largely to the activation function of Sp3.
Materials and Methods
Ethics statement
Research involving mice have been conducted according to the German Animal Protection Law (Tierschutzgesetz). The application for the experiments was reviewed and approved by the responsible local authorities (Regierungspräsidium Giessen, reference number V 54–19 c 20/15 cMR20/27).
Generation of Sp3ki/ki mice
A targeting vector containing the Sp3E553D mutation was constructed and transfected into ES cells (Text S1). For selection of ES cells, we used a floxed IRES-LacZ-neo-polyA cassette that integrates into intron 5 of the Sp3 gene by homologous recombination. A single clone out of >200 G418-resistant colonies showed the homologous recombination event. After karyotyping, the ES clone was injected into C57BL/6 blastocysts. Breeding of the chimeras revealed germ-line transmission of the targeted Sp3 allele. The IRES-lacZ-neo cassette was removed by mating heterozygous mice with mice expressing the Cre recombinase under control of the cytomegalovirus-immediate early enhancer-chicken beta-actin hybrid (CAG) promoter [17]. Offspring were genotyped by Southern blotting and PCR (Figure S1). The Sp3wt/ki heterozygous offspring were intercrossed and homozygous Sp3ki/ki mice were obtained.
Retroviral infections
Retroviral vectors for expression of the long and short isoforms of Sp3 and corresponding SUMOylation-deficient mutants were generated by cloning of appropriate wild type and mutant Sp3 cDNA fragments [14] into the pBABE-puro plasmid. Retroviral packaging in Phoenix cells and infection of immortalized Sp3-/- MEFs [24] were performed according to standard procedures. Transduced cells were selected for uptake of retrovirus with 2 µg/mL of puromycin.
Western blotting
Whole cell extract from MEFs and mouse tissues were prepared as described [7], separated on SDS-polyacrylamide gels, blotted on PVDF membranes and probed with anti Sp3 antibodies (Santa Cruz Biotechnology, sc-644). Secondary antibodies were visualized using the Immobilon Western HRP substrate (Millipore).
Expression profiling
Total RNA was prepared from freshly isolated Sp3wt and Sp3ki/ki mouse embryonic fibroblasts of E13.5 siblings using the RNeasy kit (Qiagen). Purified RNA was labeled with the two-color Quick-Amp Labeling kit (Agilent) and hybridized to a whole genome microarray 4x44K 60mer slide (G4122F) according to the manufacturer's instructions (Agilent). Microarray data were analyzed using Bioconductor [39]. The loess method implemented in the Bioconductor package marray was applied for normalization. Two biological replicates (male and female MEFs) were performed. Genes were considered as regulated when they had a fold change of ≥2, a logarithmic intensity value (base 2) of ≥5 in Sp3ki/ki, and when the expression level of replicates were similar. Similarity for two log2 transformed expression levels was determined ad hoc by the constraint max(1, |e1, e2| ×0.75,) > |e1 – e2|.
Data deposition
Microarray data were deposited at ArrayExpress (www.ebi.ac.uk/arrayexpress) under accession number E-MEXP-2755.
Quantitative real-time PCR
One microgram of total RNA prepared from MEFs and mouse organs was used for cDNA synthesis along with 0.5 µg of oligo(dT) primer and 200 U of M-MLV reverse transcriptase (Invitrogen). Quantitative RT-PCR was performed with 1 µL of 1∶20 diluted cDNA using gene-specific primers (Text S1). qPCRs were performed in quadruplicate using the Absolute SYBRGreen qPCR Mix (Abgene) on the Mx3000P real-time PCR system (Stratagene). Values were normalized to Gapdh and/or Sp1 mRNA content.
Chromatin immunoprecipitation
Chromatin immunoprecipitation was performed using the One Day ChIP kit (Diagenode) in accordance to the manufacturer's instructions. Primer sequences specific for Dmc1, Paqr6 and Rims3 promoter regions can be found in Text S1. Antibodies used for ChIP analysis are described in [16].
Bisulfite sequencing
For DNA methylation analysis, 2 µg of genomic DNA derived from immortalized Sp3wt, Sp3-/- and Sp3ki/ki MEFs were subjected to sodium bisulfite conversion of unmethylated cytosines using the EpiTect Bisulfite Kit (Qiagen) in accordance to the manufacturer's instructions. Converted DNA was subjected to PCR amplification using promoter-specific BamHI- and KpnI-tailed primers (Text S1) and the ImmoMix PCR reagent (Bioline). PCR products were cloned into the pcDNA3 vector and 10 clones were sequenced using the BGHrev primer.
Supporting Information
Zdroje
1. Geiss-FriedlanderR
MelchiorF
2007 Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol 8 947 956
2. SeelerJS
DejeanA
2003 Nuclear and unclear functions of SUMO. Nat Rev Mol Cell Biol 4 690 699
3. Garcia-DominguezM
ReyesJC
2009 SUMO association with repressor complexes, emerging routes for transcriptional control. Biochim Biophys Acta 1789 451 459
4. GillG
2005 Something about SUMO inhibits transcription. Curr Opin Genet Dev 15 536 541
5. HayRT
2005 SUMO: a history of modification. Mol Cell 18 1 12
6. RossS
BestJL
ZonLI
GillG
2002 SUMO-1 modification represses Sp3 transcriptional activation and modulates its subnuclear localization. Mol Cell 10 831 842
7. SapetschnigA
RischitorG
BraunH
DollA
SchergautM
2002 Transcription factor Sp3 is silenced through SUMO modification by PIAS1. EMBO J 21 5206 5215
8. ValinA
GillG
2007 Regulation of the dual-function transcription factor Sp3 by SUMO. Biochem Soc Trans 35 1393 1396
9. BouwmanP
PhilipsenS
2002 Regulation of the activity of Sp1-related transcription factors. Mol Cell Endocrinol 195 27 38
10. PhilipsenS
SuskeG
1999 A tale of three fingers: the family of mammalian Sp/XKLF transcription factors. Nucleic Acids Res 27 2991 3000
11. SuskeG
1999 The Sp-family of transcription factors. Gene 238 291 300
12. SuskeG
BrufordE
PhilipsenS
2005 Mammalian SP/KLF transcription factors: bring in the family. Genomics 85 551 556
13. DennigJ
BeatoM
SuskeG
1996 An inhibitor domain in Sp3 regulates its glutamine-rich activation domains. EMBO J 15 5659 5667
14. SapetschnigA
KochF
RischitorG
MennengaT
SuskeG
2004 Complexity of translationally controlled transcription factor Sp3 isoform expression. J Biol Chem 279 42095 42105
15. StielowB
SapetschnigA
KrügerI
KunertN
BrehmA
2008 Identification of SUMO-dependent chromatin-associated transcriptional repression components by a genome-wide RNA interference screen. Mol Cell 29 742 754
16. StielowB
SapetschnigA
WinkC
KrügerI
SuskeG
2008 SUMO-modified Sp3 represses transcription by provoking local heterochromatic gene silencing. EMBO Rep 9 899 906
17. SakaiK
MiyazakiJ
1997 A transgenic mouse line that retains Cre recombinase activity in mature oocytes irrespective of the cre transgene transmission. Biochem Biophys Res Commun 237 318 324
18. SamantSA
OgunkuaO
HuiL
FossellaJ
PilderSH
2002 The T complex distorter 2 candidate gene, Dnahc8, encodes at least two testis-specific axonemal dynein heavy chains that differ extensively at their amino and carboxyl termini. Dev Biol 250 24 43
19. YoshidaK
KondohG
MatsudaY
HabuT
NishimuneY
1998 The mouse RecA-like gene Dmc1 is required for homologous chromosome synapsis during meiosis. Mol Cell 1 707 718
20. CamurriL
MambetisaevaE
SundaresanV
2004 Rig-1 a new member of Robo family genes exhibits distinct pattern of expression during mouse development. Gene Expr Patterns 4 99 103
21. LiangF
ZhangB
TangJ
GuoJ
LiW
2007 RIM3gamma is a postsynaptic protein in the rat central nervous system. J Comp Neurol 503 501 510
22. TangYT
HuT
ArterburnM
BoyleB
BrightJM
2005 PAQR proteins: a novel membrane receptor family defined by an ancient 7-transmembrane pass motif. J Mol Evol 61 372 380
23. FuX
KampsMP
1997 E2a-Pbx1 induces aberrant expression of tissue-specific and developmentally regulated genes when expressed in NIH 3T3 fibroblasts. Mol Cell Biol 17 1503 1512
24. BouwmanP
GöllnerH
ElsässerHP
EckhoffG
KarisA
2000 Transcription factor Sp3 is essential for post-natal survival and late tooth development. EMBO J 19 655 661
25. PohlersM
TrussM
FredeU
ScholzA
StrehleM
2005 A role for E2F6 in the restriction of male-germ-cell-specific gene expression. Curr Biol 15 1051 1057
26. StorreJ
SchaferA
ReichertN
BarberoJL
HauserS
2005 Silencing of the meiotic genes SMC1beta and STAG3 in somatic cells by E2F6. J Biol Chem 280 41380 41386
27. KehoeSM
OkaM
HankowskiKE
ReichertN
GarciaS
2008 A conserved E2F6-binding element in murine meiosis-specific gene promoters. Biol Reprod 79 921 930
28. GuentherMG
LevineSS
BoyerLA
JaenischR
YoungRA
2007 A chromatin landmark and transcription initiation at most promoters in human cells. Cell 130 77 88
29. BilodeauS
KageyMH
FramptonGM
RahlPB
YoungRA
2009 SetDB1 contributes to repression of genes encoding developmental regulators and maintenance of ES cell state. Genes Dev 23 2484 2489
30. LunyakVV
BurgessR
PrefontaineGG
NelsonC
SzeSH
2002 Corepressor-dependent silencing of chromosomal regions encoding neuronal genes. Science 298 1747 1752
31. OuyangJ
ShiY
ValinA
XuanY
GillG
2009 Direct binding of CoREST1 to SUMO-2/3 contributes to gene-specific repression by the LSD1/CoREST1/HDAC complex. Mol Cell 34 145 154
32. LiH
RauchT
ChenZX
SzaboPE
RiggsAD
2006 The histone methyltransferase SETDB1 and the DNA methyltransferase DNMT3A interact directly and localize to promoters silenced in cancer cells. J Biol Chem 281 19489 19500
33. SarrafSA
StanchevaI
2004 Methyl-CpG binding protein MBD1 couples histone H3 methylation at lysine 9 by SETDB1 to DNA replication and chromatin assembly. Mol Cell 15 595 605
34. IvanovAV
PengH
YurchenkoV
YapKL
NegorevDG
2007 PHD Domain-Mediated E3 Ligase Activity Directs Intramolecular Sumoylation of an Adjacent Bromodomain Required for Gene Silencing. Mol Cell 28 823 837
35. MaW
HorvathGC
KistlerMK
KistlerWS
2008 Expression patterns of SP1 and SP3 during mouse spermatogenesis: SP1 down-regulation correlates with two successive promoter changes and translationally compromised transcripts. Biol Reprod 79 289 300
36. GöllnerH
DaniC
PhillipsB
PhilipsenS
SuskeG
2001 Impaired ossification in mice lacking the transcription factor Sp3. Mech Dev 106 77 83
37. Van LooPF
BouwmanP
LingKW
MiddendorpS
SuskeG
2003 Impaired hematopoiesis in mice lacking the transcription factor Sp3. Blood 102 858 866
38. Van LooPF
MahtabEA
WisseLJ
HouJ
GrosveldF
2007 Transcription factor Sp3 knockout mice display serious cardiac malformations. Mol Cell Biol 27 8571 8582
39. GentlemanRC
CareyVJ
BatesDM
BolstadB
DettlingM
2004 Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5 R80
Štítky
Genetika Reprodukční medicína
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2010 Číslo 11
-
Všechny články tohoto čísla
- Genome-Wide Association Meta-Analysis of Cortical Bone Mineral Density Unravels Allelic Heterogeneity at the Locus and Potential Pleiotropic Effects on Bone
- Beyond QTL Cloning
- An Evolutionary Framework for Association Testing in Resequencing Studies
- Genome-Wide Association Study Identifies Two Novel Regions at 11p15.5-p13 and 1p31 with Major Impact on Acute-Phase Serum Amyloid A
- The Functional Interplay between Protein Kinase CK2 and CCA1 Transcriptional Activity Is Essential for Clock Temperature Compensation in Arabidopsis
- Endogenous Viral Elements in Animal Genomes
- Analysis of the 10q11 Cancer Risk Locus Implicates and in Human Prostate Tumorigenesis
- DNA Methylation and Normal Chromosome Behavior in Neurospora Depend on Five Components of a Histone Methyltransferase Complex, DCDC
- Sarcomere Formation Occurs by the Assembly of Multiple Latent Protein Complexes
- Genetic Basis of Growth Adaptation of after Deletion of , a Major Metabolic Gene
- Nomadic Enhancers: Tissue-Specific -Regulatory Elements of Have Divergent Genomic Positions among Species
- The Parental Non-Equivalence of Imprinting Control Regions during Mammalian Development and Evolution
- CTCF-Dependent Chromatin Bias Constitutes Transient Epigenetic Memory of the Mother at the Imprinting Control Region in Prospermatogonia
- Systematic Dissection and Trajectory-Scanning Mutagenesis of the Molecular Interface That Ensures Specificity of Two-Component Signaling Pathways
- Nucleolin Is Required for DNA Methylation State and the Expression of rRNA Gene Variants in
- The Complex Genetic Architecture of the Metabolome
- ATM Limits Incorrect End Utilization during Non-Homologous End Joining of Multiple Chromosome Breaks
- Mutation Disrupts Synaptonemal Complex Formation, Recombination, and Chromosome Segregation in Mammalian Meiosis
- Mismatch Repair–Independent Increase in Spontaneous Mutagenesis in Yeast Lacking Non-Essential Subunits of DNA Polymerase ε
- The Kinesin-3 Motor UNC-104/KIF1A Is Degraded upon Loss of Specific Binding to Cargo
- Epigenetic Silencing of Spermatocyte-Specific and Neuronal Genes by SUMO Modification of the Transcription Factor Sp3
- A Coastal Cline in Sodium Accumulation in Is Driven by Natural Variation of the Sodium Transporter AtHKT1;1
- Cyclin B3 Is Required for Multiple Mitotic Processes Including Alleviation of a Spindle Checkpoint–Dependent Block in Anaphase Chromosome Segregation
- Altered DNA Methylation in Leukocytes with Trisomy 21
- Human-Specific Evolution and Adaptation Led to Major Qualitative Differences in the Variable Receptors of Human and Chimpanzee Natural Killer Cells
- Leptotene/Zygotene Chromosome Movement Via the SUN/KASH Protein Bridge in
- RACK-1 Acts with Rac GTPase Signaling and UNC-115/abLIM in Axon Pathfinding and Cell Migration
- Genome-Wide Effects of Long-Term Divergent Selection
- Endless Forms Most Viral
- Conflict between Noise and Plasticity in Yeast
- Essential Functions of the Histone Demethylase Lid
- The Transcriptional Regulator Rok Binds A+T-Rich DNA and Is Involved in Repression of a Mobile Genetic Element in
- The Cellular Robustness by Genetic Redundancy in Budding Yeast
- Localization of a Guanylyl Cyclase to Chemosensory Cilia Requires the Novel Ciliary MYND Domain Protein DAF-25
- A Buoyancy-Based Screen of Drosophila Larvae for Fat-Storage Mutants Reveals a Role for in Coupling Fat Storage to Nutrient Availability
- A Functional Genomics Approach Identifies Candidate Effectors from the Aphid Species (Green Peach Aphid)
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Genome-Wide Association Study Identifies Two Novel Regions at 11p15.5-p13 and 1p31 with Major Impact on Acute-Phase Serum Amyloid A
- Analysis of the 10q11 Cancer Risk Locus Implicates and in Human Prostate Tumorigenesis
- The Parental Non-Equivalence of Imprinting Control Regions during Mammalian Development and Evolution
- Genome-Wide Effects of Long-Term Divergent Selection
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



