-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaCTCF-Dependent Chromatin Bias Constitutes Transient Epigenetic Memory of the Mother at the Imprinting Control Region in Prospermatogonia
Genomic imprints—parental allele-specific DNA methylation marks at the differentially methylated regions (DMRs) of imprinted genes—are erased and reestablished in germ cells according to the individual's sex. Imprint establishment at paternally methylated germ line DMRs occurs in fetal male germ cells. In prospermatogonia, the two unmethylated alleles exhibit different rates of de novo methylation at the H19/Igf2 imprinting control region (ICR) depending on parental origin. We investigated the nature of this epigenetic memory using bisulfite sequencing and allele-specific ChIP–SNuPE assays. We found that the chromatin composition in fetal germ cells was biased at the ICR between the two alleles with the maternally inherited allele exhibiting more H3K4me3 and less H3K9me3 than the paternally inherited allele. We determined genetically that the chromatin bias, and also the delayed methylation establishment in the maternal allele, depended on functional CTCF insulator binding sites in the ICR. Our data suggest that, in primordial germ cells, maternally inherited allele-specific CTCF binding sets up allele-specific chromatin differences at the ICR. The erasure of these allele-specific chromatin marks is not complete before the process of de novo methylation imprint establishment begins. CTCF–dependent allele-specific chromatin composition imposes a maternal allele-specific delay on de novo methylation imprint establishment at the H19/Igf2 ICR in prospermatogonia.
Published in the journal: . PLoS Genet 6(11): e32767. doi:10.1371/journal.pgen.1001224
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1001224Summary
Genomic imprints—parental allele-specific DNA methylation marks at the differentially methylated regions (DMRs) of imprinted genes—are erased and reestablished in germ cells according to the individual's sex. Imprint establishment at paternally methylated germ line DMRs occurs in fetal male germ cells. In prospermatogonia, the two unmethylated alleles exhibit different rates of de novo methylation at the H19/Igf2 imprinting control region (ICR) depending on parental origin. We investigated the nature of this epigenetic memory using bisulfite sequencing and allele-specific ChIP–SNuPE assays. We found that the chromatin composition in fetal germ cells was biased at the ICR between the two alleles with the maternally inherited allele exhibiting more H3K4me3 and less H3K9me3 than the paternally inherited allele. We determined genetically that the chromatin bias, and also the delayed methylation establishment in the maternal allele, depended on functional CTCF insulator binding sites in the ICR. Our data suggest that, in primordial germ cells, maternally inherited allele-specific CTCF binding sets up allele-specific chromatin differences at the ICR. The erasure of these allele-specific chromatin marks is not complete before the process of de novo methylation imprint establishment begins. CTCF–dependent allele-specific chromatin composition imposes a maternal allele-specific delay on de novo methylation imprint establishment at the H19/Igf2 ICR in prospermatogonia.
Introduction
Imprinted genes are epigenetically modified during germ cell development, such that their expression in somatic cells depends on the parent of origin [1], [2]. Allele-specific differential DNA methylation is associated with most imprinted genes [3]. Male or female-specific methylation of the germ line differentially methylated regions (DMRs) is inherited from the gametes, survives the global wave of demethylation during early embryogenesis and is faithfully maintained in somatic cells during the life of the individual. Deletion studies showed that some DMRs are critical for allele-specific monoallelic expression of imprinted genes [4]–[8]. The importance of DNA methylation in the establishment and maintenance of genomic imprinting has been demonstrated in mice in which DNA methyltransferase genes have been inactivated [9]–[12].
The paternally expressed insulin-like growth factor 2 (Igf2) and maternally expressed H19 genes on mouse distal chromosome 7 [13] are coordinately expressed during embryonic development, due to shared tissue-specific enhancers (Figure 1A) [14], [15]. A paternally methylated germ line DMR between Igf2 and H19 [16]–[18] is responsible for monoallelic expression of both H19 and Igf2 [19]–[21], and therefore, is called an imprinting control region (ICR). The regulatory functions of the ICR depend on allele-specific DNA methylation. Inactivation of the H19 promoter takes place in post-implantation development on the paternal chromosome and it depends on ICR methylation [22]. The ICR functions as a methylation regulated enhancer blocker [23]–[27]: CTCF protein [28]–[30] binds in the unmethylated maternal allele and insulates between the Igf2 promoters and the shared enhancers. DNA methylation in the paternal allele inhibits CTCF binding, hence the ICR has no insulator activity, and the Igf2 promoters and the enhancers can interact. Targeted mutagenesis of the CTCF binding sites in the mouse results in a loss of enhancer-blocking activity and increased DNA methylation in the mutant maternal chromosome [31]–[33]. CTCF binding in the ICR is the major organizer of chromatin composition in the maternal allele along the entire imprinted domain [34]–[36]. CTCF recruits active histone tail modification marks to the ICR and to the H19 gene [34] and also recruits at a distance, Polycomb-mediated H3K27me3 repressive marks at the Igf2 promoter and at the Igf2 DMRs [34], [35].
Fig. 1. The imprint cycle at the H19/Igf2 ICR. 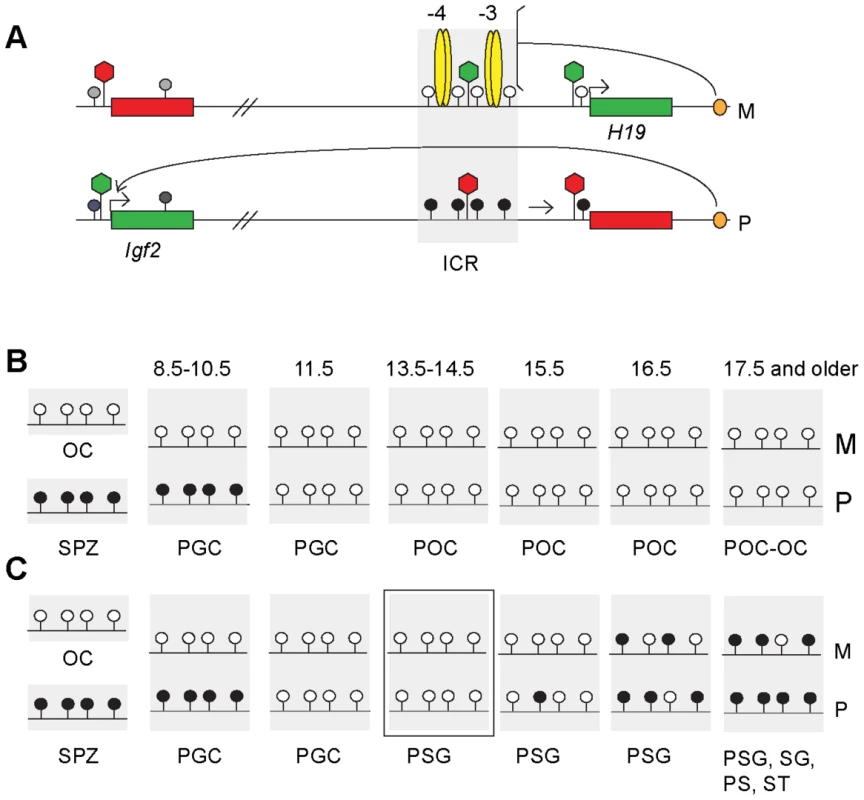
Schematic representation of epigenetic features at the H19/Igf2 imprinted domain based on publications referenced in the Introduction. (A) The H19/Igf2 imprinted domain in the soma. Maternal chromosome (M): unmethylated (white lollipops) ICR (shaded area) is inherited from the egg. CTCF protein (yellow ovals) at binding sites 1–2 and 3–4 at about −4 kb and −3 kb upstream of the H19 transcription start site imparts insulator activity (bracket) between the Igf2 promoters and the shared, downstream enhancers (orange oval). Paternal chromosome (P): methylated (black lollipops) ICR is inherited from the sperm, CTCF cannot bind, hence ICR has no insulator activity, Igf2 promoters and enhancers can interact. Early in postimplantation development, the H19 promoter is inactivated by an ICR-dependent mechanism (horizontal arrow). Active or repressive chromatin (green or red hexagon) is present at expressed or silent alleles of genes (green-red rectangles) and at respective alleles of the ICR. (B) Fate of the imprint in the female germ line. Methylation status of the ICR is depicted in the mature oocyte (OC), spermatozoon (SPZ), primordial germ cells (PGC) primary oocytes (POC) at gestational stages (in dpc). (C) Fate of the imprint in the male germ line. Methylation status is depicted in OC, SPZ and PGC as above and in prospermatogonia (PSG), spermatogonia (SG) pachytene spermatocytes (PS) and round spermatids (ST). The developmental stage under investigation is marked by a rectangle. CpG methylation at DMRs is reset during germ cell development: inherited gametic marks are erased in primordial germ cells (PGCs) followed by the establishment of new gametic marks in the female and male germ lines according to the individual's sex (Figure 1B and 1C). The umethylated versus methylated status of the H19/Igf2 ICR in oocytes versus spermatozoa constitutes the female and male gametic mark. Methylation of the paternal allele is erased in female and male germ cells by 13.5 days post coitum (dpc) [37]–[40] (Figure 1B and 1C). In the female germ line the ICR remains unmethylated during fetal and postnatal stages of oogenesis (M and P alleles in Figure 1B). In male germ cells, the ICR methylation imprint is laid down between 15.5–17.5 dpc, and is almost fully established by 18.5 dpc [31], [41]. The germ line-specific processes that target differential methylation to the ICR are unknown but are entirely separate from the later somatic ICR functions of chromatin insulation and H19 promoter silencing. CTCF binding is not required to establish an unmethylated ICR during oogenesis or a methylated ICR during spermatogenesis. The ICR that lacks functional CTCF binding sites is unmethylated in female fetal germ cells and ovulated oocytes but is methylated in perinatal spermatogonia [31], [32].
The timing of DNA methylation between the maternally and paternally inherited alleles (M and P alleles in Figure 1C) is different during spermatogenesis, methylation of the paternally inherited allele preceding that of the maternally inherited allele, implying that the two parental alleles can be distinguished from each other by the de novo DNA methylation machinery in the absence of DNA methylation [37], [39], [41], [42]. We sought to investigate the nature of this epigenetic memory in spermatogonia. We hypothesized that differences in CTCF protein binding and/or chromatin composition between the paternally or maternally inherited alleles are responsible for discriminating between the parental alleles in the male germ line. We based this hypothesis on previous observations: We have shown that migratory PGCs exhibit strict imprinted maternal allele-specific H19 expression at 8.5 dpc, and paternal allele-specific Igf2 expression at 10.5 dpc [43]. Expression of H19 and Igf2 becomes biallelic by the early post-migratory stage of 11.5 dpc [43], [44] and remains biallelic during fetal and postnatal stages of spermatogenesis [44]. Because parental allele-specific expression of both H19 and Igf2 depends on CTCF insulator binding in the maternally inherited ICR allele [31]–[33], CTCF binding in the ICR must be maternal allele-specific in migratory PGCs and biallelic or missing at later stages of spermatogenesis. It is not known if allele-specific chromatin differences exist in PGCs or if these become erased at the time when DNA methylation marks are erased at DMRs. CTCF binding, however, likely organizes the chromatin composition of the ICR in the maternal allele in PGCs, similarly to its role in somatic cells [34]. The allele-specific chromatin difference may also need to be erased in postmigratory spermatogonia, for example H3K4 methylation would be removed from the maternal allele, because H3K4 methylation is not permissive for de novo DNA methylation [45]. Erasure of chromatin marks may occur synchronously with the global dynamic changes of chromatin reorganization that take place in germ cells around mid-gestation [46]–[48]. If allele-specific chromatin marks are not fully erased in prospermatogonia after methylation imprint erasure is complete, they may influence the rate of de novo methylation. We can test this possibility directly and specifically by perturbing the chromatin bias of the ICR in prospermatogonia. After maternal transmission of the ICR CTCF site mutations [31], [34] we expect to find loss of allele-specific differences in chromatin composition and methylation establishment in prospermatogonia.
Using allele-specific chromatin immunoprecipitation single nucleotide primer extension (ChIP-SNuPE) assays we found that in normal prospermatogonia the chromatin composition was biased between the two alleles after complete erasure of CpG methylation. The CTCF site mutant maternal ICR allele, however, no longer exhibited those allele-specific chromatin differences and delayed methylation establishment. Our data suggest that CTCF dependent allele-specific chromatin composition gives de novo methylation imprint establishment an allele-specific bias at the H19/Igf2 ICR.
Results
To assess DNA methylation and chromatin in prospermatogonia and primary oocytes, we obtained high purity germ cell populations using the TgOG2 transgenic mouse line [43], in which the enhanced green fluorescent protein (EGFP) is expressed from the Pou5f1 promoter in gestational-stage germ cells. We have shown earlier using flow cytometry that in TgOG2 transgenic embryos the EGFP positive cell population is highly synonymous at the premigratory and early postmigratory stages with the populations staining positive for other PGC markers, alkaline phosphatase 2 and stage specific embryonic antigen (SSEA) [43]. Immunocytochemistry of the fetal germ cell populations before and after cell sorting using anti-DDX4 antibody confirmed that flow sorting resulted in a high level of enrichment (Figure S1). Additionally, bisulfite sequencing of the KvDMR1 [6], [49] correctly only detected unmethylated chromosomes in fetal germ cells at 13.5, 15.5 and 17.5 days post coitum (dpc) (Figure S2). This maternally methylated DMR is unmethylated in fetal germ cells at 12.5–13.5 dpc [46] and only becomes methylated postnatally, in growing oocytes [50]. Fully methylated chromosomes (methylated maternal allele) would indicate contamination from the somatic cells.
Bisulfite sequencing confirms a delay of imprint establishment at the ICR in the maternally inherited allele in prospermatogonia
In prospermatogonia, the paternally inherited ICR allele becomes methylated earlier than the maternally inherited allele in reciprocal crosses between C57BL/6J (B6) and JF1 [39], [41]. Similarly, when the ICR carries the B6 type allele in the maternal allele and the CAST/Ei type allele in the paternal allele, the B6 type maternal allele is delayed compared to the CAST/Ei type paternal allele in prospermatogonia between 14.5 and 18.5 dpc [37]. We tested the reciprocal situation when the CAST/Ei type ICR allele is inherited from the mother and the B6 type allele is inherited from the father. Females of FVB/NJ.CAST/Ei(N7), a distal chromosome 7 partial congenic strain for CAST/Ei (CS) [31] were mated with TgOG2 homozygous transgenic males [43] resulting in CS X OG2 fetuses. We isolated male and female germ cells from 13.5, 14.5, 15.5, 16.5 and 17.5 dpc gonads. We performed two or more independent bisulfite conversion reactions for each sample and sequenced at least twelve clones of each sample. A single nucleotide polymorphism in the CS strain was used to identify the parental alleles. We confirmed previous observations [37]–[40] that DNA methylation erasure is complete by 13.5–14.5 dpc at the ICR (Figure 2 and Figure S3). We found that primary oocytes exhibited no methylation of the ICR region between 13.5 and 16.5 dpc (Figure S3A) and prospermatogonia attained CpG methylation gradually (Figure 2) between 15.5 dpc and 17.5 dpc as expected [37], [39]. We confirmed that the maternal allele (CS type) was delayed compared to the paternal allele (B6 type) in CS X OG2 prospermatogonia (Figure 2) similar to the reciprocal B6 X CS situation [37]. Regardless of mouse strains used, there exists a time gap in methylation imprint establishment between the two chromosomes depending on the inheritance from the mother or father (M and P alleles in Figure 1) during spermatogenesis [37], [39], [41], [42]. Therefore, the two parental alleles must be distinguished from each other in 13.5–14.5 dpc prospermatogonia by epigenetic means other than DNA methylation.
Fig. 2. Methylation dynamics at the ICR in normal prospermatogonia. 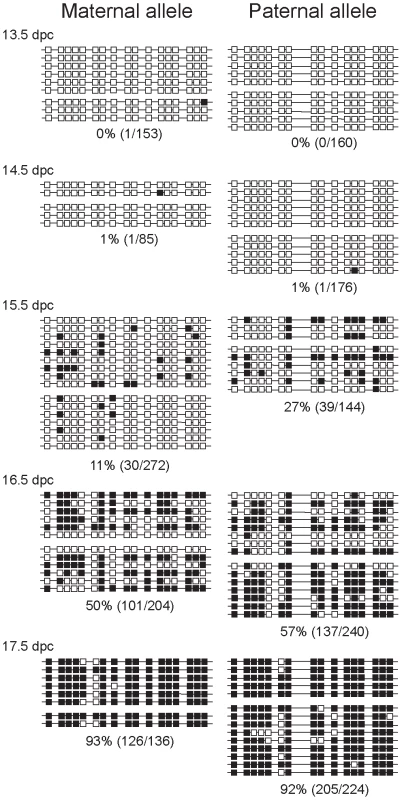
Bisulfite sequencing results are shown at fetal stages (in dpc). Prospermatogonia of CS X OG2 fetuses were analyzed. Unmethylated CpGs (white squares) and methylated CpGs (black squares) are shown along independent chromosomes (horizontal lines). Groups of chromosomes were derived from the same bisulfite reaction. CTCF sires 1 and 2 of the ICR are included in the analyzed region. CpG site 8 is polymorphic and is only present in the CS type allele. The percentage of methylated CpGs (methylated CpG/total CpG) at each developmental stage is indicated for each allele. CTCF site mutations abolish delayed methylation imprint establishment of the maternal ICR allele
We tested the hypothesis that functional CTCF binding sites in the maternally inherited H19/Igf2 ICR allele are responsible for the delayed methylation of the maternally inherited, compared to the paternally inherited allele in male germ cells. Female mice homozygous for CTCF site mutations (−/−) [31] were mated with TgOG2 homozygous transgenic males [43] (wild type ICR). In the resulting CTCFm X OG2 fetuses, the maternally inherited ICR allele was mutant, lacking functional binding sites. Germ cells were collected at 13.5, 14.5, 15.5 and 16.5 dpc. Bisulfite DNA sequencing was performed on agarose-embedded germ cells as described before [31] according to Olek et al. [51]. Nucleotide changes, introduced with the mutations aided discrimination between the mutant and wild type alleles. We found that due to the ICR CTCF site mutations the maternally inherited mutant allele did not lag behind the paternal allele in male germ cells (Figure 3). The increased rate of methylation in the CTCF site mutant ICR maternal allele (Figure 3) compared to the normal ICR allele (Figure 2) was statistically significant. At 15.5 days the p-value = 0.0014 and at 16.5 days the p-value = 0.0183 according to Fisher's exact test. This argues that intact CTCF protein binding sites in the ICR are required for the transient epigenetic memory that delays methylation of the maternally inherited allele during male fetal germ cell development. When the paternal ICR carried the CTCF site mutations in the control OG2 X CTCFm male germ cells (Figure S4), its rate of methylation was similar to the normal paternal allele in the CS X OG2 cross (Figure 2), indicating that simply having less CpG sites in the CTCF site mutant ICR is not sufficient to alter the rate of methylation. The control female germ cells did not attain methylation in the mutant allele (Figure S3B).
Fig. 3. Methylation dynamics at the ICR in ICR CTCF site mutant prospermatogonia. 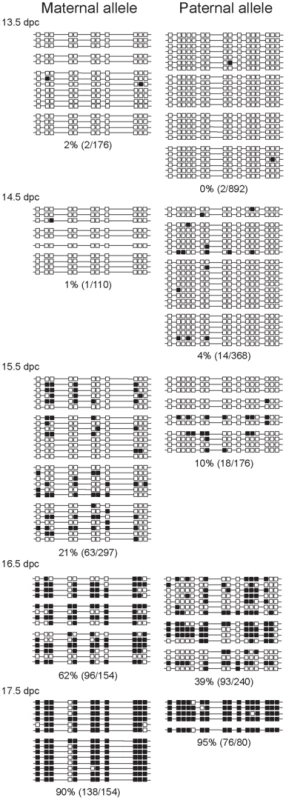
Bisulfite sequencing results of prospermatogonia from CTCFm X OG2 fetuses are shown. CpG sites 4–5 and 12–13 had been eliminated in the maternal allele by the CTCF site mutations. Other details are as in Figure 2. The mutant maternal allele was, unexpectedly, more prone to methylation than the wild-type paternal allele in the same cell. The wild type paternal and mutant maternal alleles are different in two respects, in the strain and in the presence or absence of the CTCF site mutations. The best comparison can be made when the CTCFm allele is compared between paternal and maternal inheritance. The methylation levels of these chromosomes, indeed, were very similar at 14.5 and 15.5 dpc (Figure 3 and Figure S4). We noted that the sum level of methylation in the two alleles did not change between the wild type and CTCF site mutant prospermatogonia, indicating perhaps that the two alleles are in competition for a methylation inducing factor that has limited concentration at 15.5–16.5 dpc.
Chromatin composition at the ICR is biased between parental alleles in 13.5 and 14.5 dpc prospermatogonia
We considered the possibility that differences in CTCF binding and chromatin composition between the paternally or maternally inherited alleles might be responsible for discriminating between the parental ICR alleles in the male germ line. If this is correct, we would expect in spermatogonia a slight bias in chromatin composition between the maternally and paternally inherited alleles at the ICR such that the paternally inherited allele would be more permissive to DNA methylation. We developed ChIP-SNuPE assays based on mass spectrometry Sequenom allelotyping [36], [52] to distinguish allele-specific incorporation of ddNTPs into the SNuPE primer based on differences in molecular mass at sites of single nucleotide polymorphisms (SNPs) between 129 or OG2 and CS mouse genomic sequences along the H19/Igf2 ICR [34]. The Sequenom assay used SNPs at two halves of the ICR at −4 kb and −3 kb distances from the H19 transcriptional start site. Both assays were rigorously quantitative, as shown by DNA mixing experiments (Figure S5A). The number of fetal germ cells is limiting for ChIP assays, we can obtain 100,000–300,000 germ cells per dissection. We decided to use 100,000 germ cells per ChIP. We validated the ChIP-SNuPE assays using 100,000 129 X CS mouse embryo fibroblasts (MEFs). We found that CTCF binding and active chromatin (H3K4me2 enrichment) was highly specific to the maternal allele in the ICR whereas repressive chromatin (H3K9me3) was highly specific to the paternal allele in MEFs (Figure S6A) as we previously reported using large number of the same 129 X CS MEF cells [34]. We found that the assay correctly measured 50% 129 and CS alleles in the input chromatin samples for MEFs (Figure S6A) and for CS X OG2 and CTCFm X OG2 fetal germ cells (Figure S6C).
We isolated male and control female germ cells from 13.5 and 14.5 gonads from the CS X OG2 mouse cross and performed ChIP-SNuPE assays using 100,000 germ cells per ChIP reaction. The control, nonspecific IgG-precipitated chromatin samples did not exhibit a clear pattern of allele-specific skewing (Figure S6B). The results did not show consistency between the −4 kb and −3 kb regions (A and B regions, respectively) or between the 13.5 and 14.5 dpc stages. Specific antibodies, on the other hand gave reproducible results using germ cell chromatin (Figure 4, Figure 5, Figure 6). CTCF binding was slightly biased toward the maternal ICR allele in male and female germ cells at 14.5 dpc (Figure 4). CTCF binding in the paternal allele would likely be inhibited by DNA methylation in PGCs similarly to somatic cells [23], [24], [27], but not in fetal prospermatogonia at 13.5–14.5 dpc in the lack of DNA methylation. The slight maternal bias is consistent with the possibility that allele-specific CTCF binding is not completely erased at 14.5 dpc, after DNA methylation erasure had been completed. The total level of CTCF binding at the ICR was very low in germ cells at 14.5 dpc compared to MEFs (Figure S7). This suggests that CTCF has been almost completely removed from both ICR alleles in germ cells by 14.5 dpc, consistent with biallelic Igf2 expression in the absence of insulation [31]–[33], [43]. The almost complete lack of CTCF binding, however is not due to the absence of CTCF from prospermatogonia at these stages. This would be expected based on that CTCF and CTCFL (BORIS) proteins exhibit mutually exclusive expression in adult male germ cells, round spermatids and spermatocytes, respectively [53] and that CTCFL is expressed in 14.5 dpc prospermatogonia [54]. It is not known whether CTCF is expressed in embryonic and fetal germ cells. We addressed this question by performing immunocytochemistry with anti-CTCF antibody using fetal germ cells (Figure S8). We found that CTCF staining in male and female germ cells was similar to that of control gonadal somatic cells at 12.5 dpc and 14.5 dpc. The mutually exclusive expression of CTCF and CTCFL, therefore, does not apply in germ cells at 14.5 dpc. CTCF may be inhibited to bind in the ICR at these stages because of changes in its covalent modifications [55], cofactors, or due to an RNA-dependent mechanism [56].
Fig. 4. Allele-specific bias in CTCF binding chromatin at the H19/Igf2 ICR in 14.5 dpc fetal germ cells. 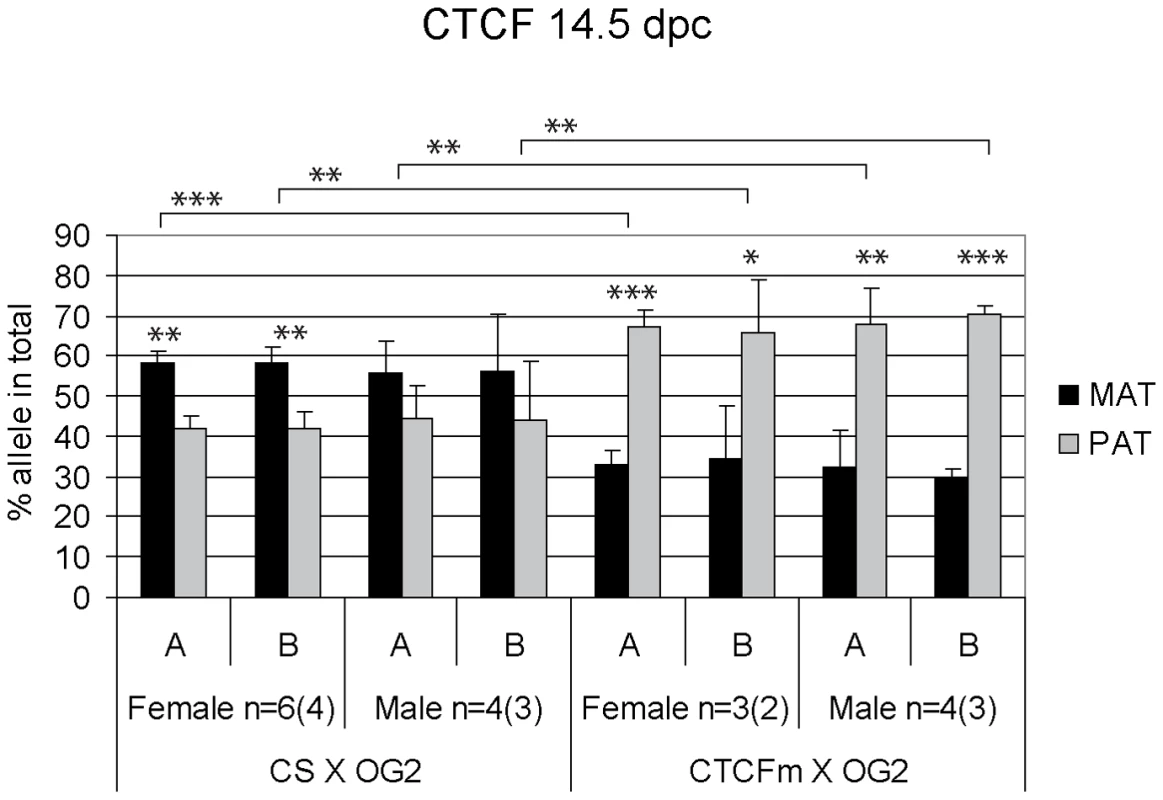
Female and male germ cell chromatin was precipitated from 14.5 dpc CS X OG2 and CTCFm X OG2 fetuses with the anti-CTCF antibody. Allele-specific enrichment in the immunoprecipitated chromatin was assessed at the H19/Igf2 ICR −4 kb and −3 kb regions (A and B, respectively) using ChIP-SNuPE assays. The number of ChIP reactions (n) is indicated and the number of independent germ cell pools/chromatin preparations is given in parentheses. Average maternal (MAT) and paternal (PAT) allele contributions are shown with standard deviations. Statistical significance of the difference between alleles and between wild type and mutant samples was evaluated using Student T-test (p values are shown by asterisks: <0.001***; <0.01**; <0.05*). Fig. 5. Allele-specific bias in H3K4me2 enrichment at the H19/Igf2 ICR in fetal germ cells. 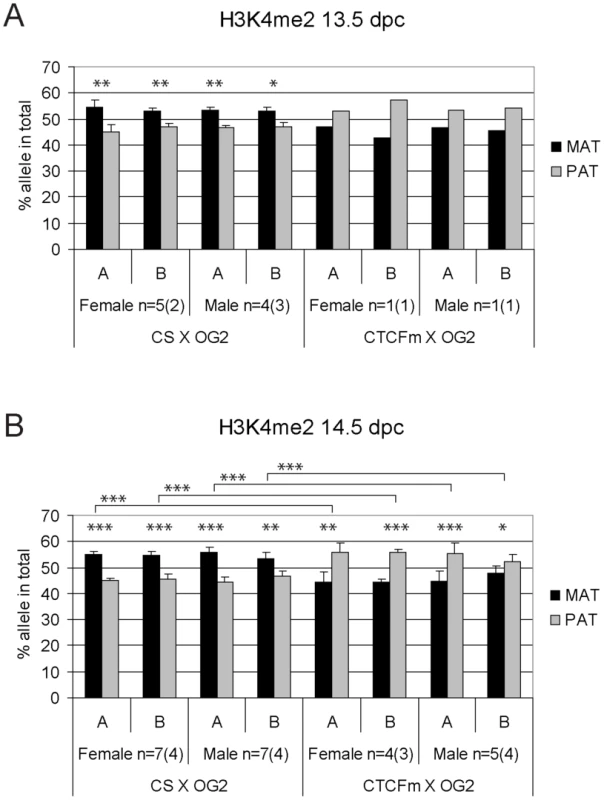
ChIP-SNuPE Sequenom assay results of H3K4me2-precipitated (A) 13.5 dpc and (B) 14.5 dpc fetal germ cell chromatin are shown. Other details are as in Figure 4. Fig. 6. Allele-specific bias in H3K9me3 enrichment at the H19/Igf2 ICR in fetal germ cells. 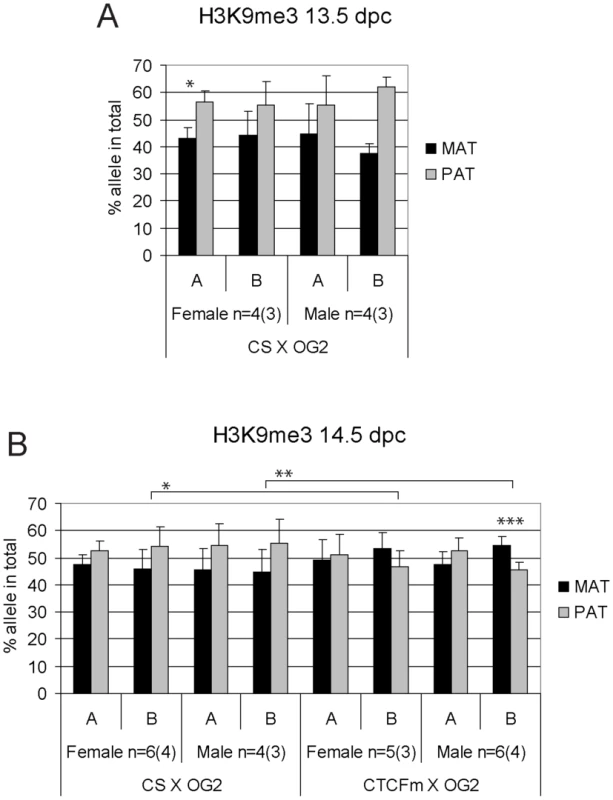
ChIP-SNuPE Sequenom assays results of H3K9me3-precipitated (A) 13.5 dpc and (B) 14.5 dpc fetal germ cell chromatin is shown. Other details are as in Figure 4. We found a slight (∼10%), but reproducible bias in the H3K4me2 levels toward the maternally inherited allele in male and female germ cell ChIP samples at 13.5 and 14.5 dpc (Figure 5). The bias was present in the ICR at −3 kb and −4 kb positions. H3K4me2 enrichment in germ cells was similar to the level found in MEFs (Figure S9), suggesting that the ICR had not been stripped of this mark at 13.5–14.5 dpc. H3K9me3 was reciprocally biased: the paternally inherited allele exhibited about 10% higher enrichment at 13.5 and 14.5 dpc (Figure 6). The allele-specificity of the bias for H3K4me2 and H3K9me3 in 13.5 dpc germ cells was in agreement with the somatic pattern (Figure S6A), being maternal and paternal specific, respectively, suggesting that it originates in premigratory PGCs (Figure 7A). The amplitude of the bias, however, was smaller than in the soma, consistent with the possibility that the chromatin differences are being erased in germ cells around mid-gestation and only the remnants of the allele-specific differences can be detected at 13.5–14.5 dpc. H3K9me3 levels at the ICR, however, were very low in germ cells at these stages (not shown), consistent with the possibility that similarly to CTCF but unlike H3K4me2 this mark is almost completely removed by 13.5 dpc.
Fig. 7. Model. 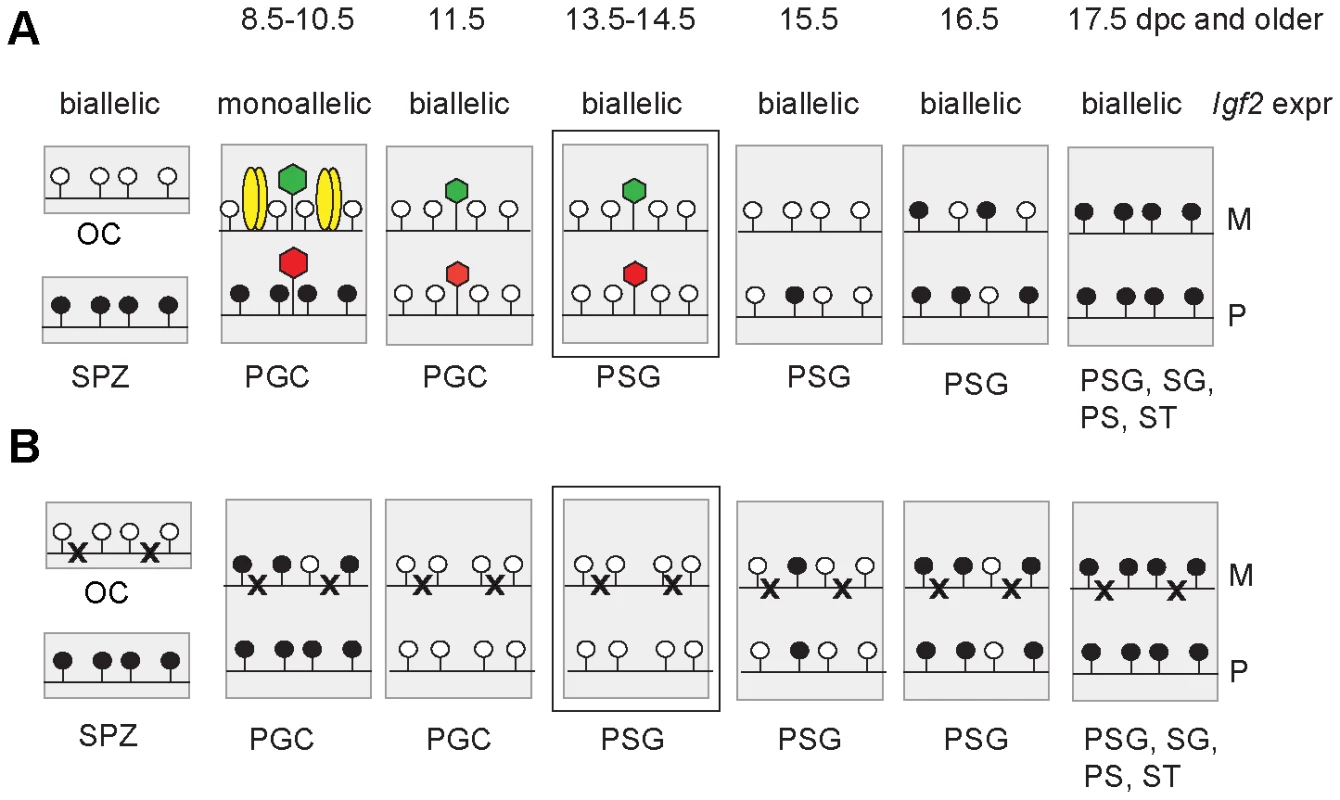
Functional CTCF sites are required for chromatin bias and delayed methylation of the maternally inherited ICR allele. Expected CTCF binding and chromatin composition is depicted in primordial germ cells (PGC). Observed chromatin bias is depicted in prospermatogonia (PSG). Other details are as Figure 1. The developmental stages are indicated above in dpc. (A) Imprint establishment of the ICR in the normal male germ line. Chromatin bias is observed in the normal ICR between the parental alleles in the absence of CpG methylation at 13.5–14.5 dpc. (B) Imprint establishment at the CTCF site mutant ICR in the male germ line. CTCF cannot bind in the maternal allele in PGCs because of the mutations (x) or in the paternal allele because of CpG methylation. The chromatin bias, found in normal cells, is no longer observed between parental alleles in the mutant cells at 13.5–14.5 dpc and the maternal allele's methylation is not delayed at 15.5–17.5 dpc. The allele-specific bias of H3K4me2 in 14.5 dpc germ cells was present with only trace amounts of CTCF binding (Figure S7) in the ICR. We concluded that the H3K4me2 histone mark could be a potential candidate that provides the epigenetic memory of the mother at the ICR in 13.5–14.5 dpc prospermatogonia in the absence of CpG methylation.
CTCF site mutations abolish parental allele–specific chromatin bias at the ICR in prospermatogonia
CTCF binding is essential for the maternal allele's chromatin composition along the H19/Igf2 imprinted domain in the soma [34]. We decided to analyze if CTCF binding site mutations abolish the enrichment bias of histone covalent modifications between the maternally and paternally inherited alleles in fetal male germ cells. Female mice homozygous for the CTCF site mutations [31] were mated with TgOG2/TgOG2 transgenic males [43]. In the resulting CTCFm X OG2 fetuses, the maternally inherited allele was mutant, lacking functional binding sites. Male and control female germ cells were collected at 13.5 and 14.5 dpc and ChIP was performed with 100,000 germ cells per reaction using the H3K4me2 and H3K9me3 antibodies. Allele-specific precipitation was assessed using ChIP-SNuPE Sequenom assays that can distinguish the CTCF site mutation sites from the normal allele at CTCF binding sites 1 and 3 (at −4 kb and −3 kb positions, respectively) in the ICR. Each assay was rigorously quantitative, as shown by DNA mixing experiments (Figure S5B).
Contrary to what we found in CS X OG2 fetal germ cells, CTCF did not exhibit a slight bias toward the maternally inherited allele but instead a strong bias toward the paternal allele in 14.5 dpc CTCFm X OG2 germ cells (Figure 4). The reduction of maternal-allele specificity is consistent with impaired binding of CTCF to the mutant sites in the maternal allele. The paternal allele-specificity is likely due to the potential of CTCF binding in the paternal allele in the lack of methylation at 13.5–14.5 dpc. Using gel shift competition assays [31] and in vivo ChIP analysis [34] we have shown previously that the CTCF site mutations completely abolished CTCF binding in the ICR sequences. The fact that we do not measure a complete lack of maternal allele-specific CTCF binding in 14.5 dpc CTCFm X OG2 germ cells is most likely due to the limitation of the assay at extremely low copy numbers (Figure S7).
H3K9me3 was slightly paternally biased at 13.5–14.5 dpc in CS X OG2 germ cells (Figure 6) but was not consistently biased in CTCFm X OG2 germ cells at 14.5 dpc (Figure 6B). We observed a switch from a slight maternal - to a slight paternal H3K4me2 bias at −4 kb and also at −3 kb along the ICR (Figure 5B) suggesting that intact CTCF binding sites are required for distinguishing the maternal allele by H3K4 dimethylation in male and female germ cells at 14.5 dpc.
Discussion
This is the first study addressing the chromatin composition of a DMR at any imprinted region in gestational stage germ cells. We tested the hypothesis whether the epigenetic memory of the father and the mother exists in fetal germ cells in the form of an allele-specific bias of chromatin composition after the erasure of the DNA methylation imprint at the H19/Igf2 ICR. We found that the chromatin composition was biased at the ICR between the two alleles in fetal germ cells and this bias depended on functional CTCF insulator binding sites in the ICR. The CTCF site mutant maternal ICR allele no longer exhibited delayed methylation establishment. Our data suggest that CTCF dependent allele-specific chromatin composition gives de novo methylation imprint establishment a maternal allele-specific delay at the H19/Igf2 ICR in prospermatogonia. A more general implication of our results is that the erasure of the allele-specific chromatin imprints is not fully synchronized with the erasure of CpG methylation at DMRs.
Erasure of allele-specific chromatin marks follows the erasure of DNA methylation
We hypothesized that chromatin differences exist between parental alleles of DMRs in PGCs at the time of monoallelic expression of imprinted genes and that these chromatin differences are erased in the germ line. It would be extremely challenging technically to assess allele-specific chromatin in migratory PGCs because of the very low germ cell numbers at those stages. We found, however, evidence that parental allele-specific chromatin bias exists in the H3K4me2 and H3K9me3 residues in postmigratory germ cells at the H19/Igf2 ICR at 13.5 and 14.5 dpc; thus the erasure of allele-specific chromatin lags behind the erasure of DNA methylation at the H19/Igf2 ICR (Figure 7). The erasure of allele-specific chromatin at the ICR, therefore, is not required for the erasure of DNA methylation imprint.
It will be interesting to investigate the mechanism of how allele-specific chromatin marks are erased at DMRs. It is important to note that fetal germ cells do not divide after 13.5 dpc: spermatogonia enter mitotic arrest whereas primary oocytes arrest at the diplotene phase of meiosis, therefore, a passive loss of chromatin marks at DMRs is possible only before 13.5 dpc. DMR chromatin erasure might be linked with global chromatin remodeling events [46]–[48] around mid-gestation. The mechanism of global chromatin remodeling in PGCs is not known but is speculated to be mediated by chromatin chaperons [46]. We found that the rate of erasure at the ICR was different for the H3K4me2 and H3K9me3 marks. H3K4me2 overall enrichment appeared to hold on longer whereas H3K9me3 was largely removed by 14.5 dpc. This difference suggests that chromatin mark erasure at DMRs likely occurs by specific chromatin modifying enzymes, such as histone demethylases and does not involve nucleosome removal. Overall H3K4me2 erasure is likely completed at a later stage during spermatogenesis, because H3K4 dimethylation is absent at the ICR in postnatal male germ cells spermatocytes, round spermatids and elongating spermatids [57].
CTCF sites are responsible for chromatin differences between alleles and for delayed methylation of the maternal ICR allele in prospermatogonia
We confirmed previous observations [37]–[40] that DNA methylation erasure at the ICR is complete by 13.5–14.5 dpc. If epigenetic memory existed of the mother or father in prospermatogonia that could distinguish the parental alleles at this time, it had to be distinct from CpG methylation. 5-hydroxy-methyl C (5hmC) emerges as a second covalent DNA modification with potential for epigenetic regulation [58], [59]. Because bisulfite sequencing recognizes not only 5mC but also 5hmC [60], our data are consistent with the absence of epigenetic memory of a parent in the form of both of these DNA covalent modifications at 13.5–14.5 dpc. Prospermatogonia attained CpG methylation at the ICR gradually between 15.5 dpc and 17.5 dpc with an allele-specific bias in the rate of methylation, confirming that there was epigenetic distinction between the parental alleles. Methylation of the maternal allele was slower than the paternal allele in normal spermatogonia, but not in CTCFm X OG2 spermatogonia where the ICR CTCF sites were mutant, arguing that functional CTCF sites are required in the maternal allele for its delayed methylation. We found maternally biased CTCF binding in the ICR at 13.5–14.5 dpc, consistent with the possibility that a bias in CTCF binding may provide the epigenetic memory of the mother. However, CTCF binding was only at trace levels suggesting that CTCF is not likely the factor that physically delays DNA methylation in the maternal allele at 15.5 dpc. Our data are in agreement with the model (Figure 7) that CTCF binding in the maternal allele organizes allele-specific chromatin differences at the ICR in PGCs and these chromatin marks are erased with a slower rate than the rate of DNA methylation erasure. The remnants of chromatin differences at 13.5–14.5 dpc may simply reflect their history and may not be responsible for the methylation bias. Alternatively, these marks may constitute the epigenetic memory that distinguishes the parental alleles for de novo methylation, commencing at 15.5 dpc. Indeed, in the absence of CTCF binding in the mutant ICR there was no maternal-allele-specific H3K4me2 bias and the methylation rate of the maternal allele was not delayed compared to the paternal allele in prospermatogonia, giving support to our model (Figure 7).
Chromatin difference constitutes the transient epigenetic memory of the parental alleles in prospermatogonia
With the erasure of genomic imprints around mid-gestation the female and male germ lines are preparing for the establishment of the new imprints according to the individual's sex. It will be important to find out how the chromatin composition provides clues to the methylation imprint establishment. The chromatin composition at the paternally methylated DMRs is expected to be permissive to de novo methylation in 15.5–18.5 dpc spermatogonia and refractory to de novo methylation in growing oocytes. Our results argue that the erasure of chromatin clues at the H3K4me2 and H3K9me3 residues overlaps with the initiation phase of de novo methylation imprint establishment at the ICR and the incomplete erasure of these allele-specific chromatin marks can affect the rate of the new methylation imprint establishment in prospermatogonia.
Histone covalent modifications could take active part in or influence DNA methylation imprint establishment in the germ line, based on studies describing the interplay between histone methylation and DNA methylation. Histone H3K9 methylation controls DNA methylation in Neurospora crassa [61], [62] and in Arabidopsis thaliana [63], [64]. Histone lysine methylation by Suv39h1 is required for DNA methylation at the pericentric heterochromatin in mice [65]. Our genetic system [31], [34] is uniquely suited for asking the question whether disturbing the bias in chromatin composition specifically at the H19/Igf2 locus would abolish the bias of methylation imprint establishment at the ICR in male fetal germ cells. H3K9me3 was biased toward the paternal ICR allele at 13.5 dpc, and H3K4me2 was biased towardF the maternal allele at 13.5–14.5 dpc in prospermatogonia. In the absence of paternal H3K9me3 bias in the 13.5 dpc CTCFm X OG2 prospermatogonia, the paternal allele's methylation rate was reduced, whereas in the lack of maternal H3K4m2 bias in 13.5–14.5 dpc prospermatogonia, the maternal allele's methylation rate increased. These findings suggest that chromatin composition differences between the parental alleles may influence the rate of their de novo methylation at the ICR.
Male and female germ cells behaved similarly with respect to the dynamics of the overall levels and the allele-specificity of H3K4m2 and H3K9me3 enrichment at the H19/Igf2 ICR at 13.5 and 14.5 dpc, yet methylation imprint establishment was affected only in male germ cells. The maintenance of the unmethylated state of the ICR in fetal female germ cells was not affected by the chromatin bias. It is likely that the chromatin composition provides clues to exclude or target the de novo DNA methyltransferase complex to DMRs. Because Dnmt3a and Dnmt3L are specifically expressed in male versus female fetal germ cells [66]–[69], these would be affected by allele-specifically biased chromatin in prospermatogonia but not in primary oocytes.
The level of H3K4me2 bias toward the maternal allele was about 10% and thus was similar to the average 15% maternal allele-specific bias in delay of DNA methylation at 15.5 dpc. The H3K4me2 bias between the parental alleles existed in the lack of DNA methylation and with only a trace amount of CTCF binding in the ICR. We concluded that the H3K4me2 histone mark could provide the epigenetic memory of the mother in prospermatogonia at 13.5–14.5 dpc that delays de novo CpG methylation in the maternal ICR allele. Significantly, H3K4 demethylase KDM1B is required at certain DMRs for the establishment of maternal methylation imprints in oocytes [45], indicating that methylated H3K4 is refractory to DNA de novo methylation. Additionally, the DNA de novo methylation cofactor, Dnmt3L [70] requires a DNA substrate in association with histones containing unmethylated H3K4 [71].
Two other paternally methylated DMRs, the Rasgrf1 DMR and the Dlk1/Gtl2 DMR (IG-DMR) also exhibit paternal allele-specific bias in de novo methylation imprint establishment [41]. The maternally methylated Snrpn, Zac1 and Peg1/Mest DMRs are methylated faster in the maternal allele in growing oocytes [50], [72]. Similarly to the H19/Igf2 ICR, allele-specific bias in chromatin composition of PGC origin may be responsible for providing epigenetic memory of the mother or father at these DMRs.
Materials and Methods
The experiments involving mice had been approved by the IACUC of the City of Hope. Housing and care of the animals has been consistent with the Public Health Service Policy, the NIH “Guide for the Care and Use of Laboratory Animals” and the Animal Welfare Act.
Purification of germ cells by flow cytometry
Male mice of the homozygous transgenic TgOG2 line [B6;CBA-Tg(Pou5f1-EGFP)2Mnn], which expresses the EGFP reporter gene specifically in germ cells [43] were mated to wild type females of FVB/NJ.CAST/Ei(N7) (CS), a distal chromosome 7 partial congenic strain [31] or to females carrying the H19/Igf2 ICR CTCF site mutations (CTCFm) where the mutatant allele was derived from the 129SI/ImJ strain [31]. Pregnant females were sacrificed and from the fetuses female or male gonads were isolated and dispersed according to Buehr and McLaren [73]. Isolates were placed into 0.15 ml of trypsin-EDTA, incubated for 20 min at 37C° then dissociated into a single cell suspension. A total of 0.3 ml of 25% (v/v) fetal bovine serum in medium M2 [74] was added before flow cytometry. Cell suspensions were analyzed and sorted on a MoFlo flow cytometer (Beckman Coulter, Fort Collins, CO). Data were acquired using 488 nm excitation from an Innova-306 Argon laser (Coherrent, Santa Clara, CA) at 500 mW. EGFP emission was measured through a 530DF30 filter (Omega Optical, Brattleboro, VT).
Bisulfite genomic sequencing
Fetal germ cells were flow-sorted, collected by centrifugation and embedded into agarose beads. Bisulfite sequencing of the ICR A region was done as before [31] according to Olek [51]. The average number of germ cells used per bisulfite reaction was 20,000. The range was between 1,200 and 27,000.
Chromatin Immunoprecipitation
Chromatin preparation from 129 X CS primary MEFs was done as described earlier [34]. Chromatin was prepared from flow-sorted fetal germ cells similarly with modifications. We used chromatin from 100,000 cells per ChIP estimated by the number of sorted EGFP+ cells. We formaldehyde-crosslinked the chromatin in suspension for 2 min, stopped crosslinking by adding glycine, washed the cell pellet in PBS and resuspended the cells in M2 for flow cytometry. After sorting we resuspended the germ cells in lysis buffer, snap froze the chromatin aliquots and kept them deep frozen until sufficient quantities were obtained for several immunoprecipitations. We thawed the chromatin aliquots, sheared the chromatin by sonication and performed ChIP with different antibodies. The following antibodies were used in the chromatin immunoprecipitation (ChIP) assays: anti CTCF, 07-729; anti-dimethyl-histone H3 (Lys4), 07-030; anti-trimethyl-histone H3 (Lys9), 17–625; were purchased from Millipore and nonspecific IgG, sc-2027; was from Santa Cruz Biotechnology. The chromatin immunoprecipitation was performed as described previously [34] with minor modifications. Pre-blocked A/G beads from Santa Cruz (Cat#sc-2003) were used.
Real-time PCR
Real-time PCR was performed to measure the region-specific overall ChIP enrichment levels at the H19-Igf2 ICR as described [34].
Analysis of allele-specific histone enrichment
To measure allele-specific chromatin differences we used the MALDI-TOF allelotyping analysis method from Sequenom [52] as we have done earlier [36]. Mass spectrometry was performed to quantify the extended SNuPE primers based on the differences in molecular mass between alleles. SNPs for the H19-Igf2 region were obtained by DNA sequencing of inbred 129S1 (129) and CAST/Ei (CS) at specific regions of interest as described [34] or were provided by the introduced mutations [31]. Polymerase chain reaction and extension primers for the normal ICR (forward, reverse and UEP, respectively) were: SNuPE-H19-4kb: 5′-ACGTTGGATGTTGCGCCAAACCTAAAGAGC-3′; 5′-ACGTTGGATGAGGTACTGAACTTGGGTGAC-3′; 5′-CATTTGTGAATTCCAATACC-3′; SNuPE-H19-3kb: 5′-ACGTTGGATGACACTTGTGTTTCTGGAGGG-3′; 5′-ACGTTGGATGATGCCTTCCTATAGTGAGCC-3′; 5′-AAGGGGTCCCTTTGGTC-3′. Polymerase chain reaction and extension primers for the CTCF site mutant ICR (forward, reverse and UEP, respectively) were: SNuPE-CTCFm1#2 : 5′-ACGTTGGATGCTTTAGGTTTGGCGCAATCG-3′; 5′-ACGTTGGATGCGTCTGCTGAATCAGTTGTG-3′; 5′-CGCAATCGATTTTGCTG-3′; SNuPE-CTCFm3#1 : 5′-ACGTTGGATGGCTGTTATGTGCAACAAGGG-3′; 5′-ACGTTGGATGTGGGCCACGATATATAGGAG-3′; 5′-AAGGGAACGGATGCTAC-3′.
Supporting Information
Zdroje
1. Ferguson-SmithAC
SuraniMA
2001 Imprinting and the epigenetic asymmetry between parental genomes. Science 293 1086 1089
2. ReikW
WalterJ
2001 Genomic imprinting: parental influence on the genome. Nat Rev Genet 2 21 32
3. MannJR
SzabóPE
ReedMR
Singer-SamJ
2000 Methylated DNA sequences in genomic imprinting. Crit Rev Eukaryot Gene Expr 10 241 257
4. WutzA
SmrzkaOW
SchweiferN
SchellanderK
WagnerEF
1997 Imprinted expression of the Igf2r gene depends on an intronic CpG island. Nature 389 745 749
5. ThorvaldsenJL
BartolomeiMS
2000 Molecular biology. Mothers setting boundaries. Science 288 2145 2146
6. FitzpatrickGV
SolowayPD
HigginsMJ
2002 Regional loss of imprinting and growth deficiency in mice with a targeted deletion of KvDMR1. Nat Genet 32 426 431
7. Ferguson-SmithA
LinSP
TsaiCE
YoungsonN
TevendaleM
2003 Genomic imprinting—insights from studies in mice. Semin Cell Dev Biol 14 43 49
8. WilliamsonCM
TurnerMD
BallST
NottinghamWT
GlenisterP
2006 Identification of an imprinting control region affecting the expression of all transcripts in the Gnas cluster. Nat Genet 38 350 355
9. HataK
OkanoM
LeiH
LiE
2002 Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development 129 1983 1993
10. LiE
BeardC
JaenischR
1993 Role for DNA methylation in genomic imprinting. Nature 366 362 365
11. Bourc'hisD
XuGL
LinCS
BollmanB
BestorTH
2001 Dnmt3L and the establishment of maternal genomic imprints. Science 294 2536 2539
12. KanedaM
OkanoM
HataK
SadoT
TsujimotoN
2004 Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature 429 900 903
13. BrannanCI
BartolomeiMS
1999 Mechanisms of genomic imprinting. Curr Opin Genet Dev 9 164 170
14. KafferCR
GrinbergA
PfeiferK
2001 Regulatory mechanisms at the mouse Igf2/H19 locus. Mol Cell Biol 21 8189 8196
15. LeightonPA
SaamJR
IngramRS
StewartCL
TilghmanSM
1995 An enhancer deletion affects both H19 and Igf2 expression. Genes Dev 9 2079 2089
16. BartolomeiMS
WebberAL
BrunkowME
TilghmanSM
1993 Epigenetic mechanisms underlying the imprinting of the mouse H19 gene. Genes Dev 7 1663 1673
17. TremblayKD
DuranKL
BartolomeiMS
1997 A 5′ 2-kilobase-pair region of the imprinted mouse H19 gene exhibits exclusive paternal methylation throughout development. Mol Cell Biol 17 4322 4329
18. TremblayKD
SaamJR
IngramRS
TilghmanSM
BartolomeiMS
1995 A paternal-specific methylation imprint marks the alleles of the mouse H19 gene. Nat Genet 9 407 413
19. LeightonPA
IngramRS
EggenschwilerJ
EfstratiadisA
TilghmanSM
1995 Disruption of imprinting caused by deletion of the H19 gene region in mice. Nature 375 34 39
20. RipocheMA
KressC
PoirierF
DandoloL
1997 Deletion of the H19 transcription unit reveals the existence of a putative imprinting control element. Genes Dev 11 1596 1604
21. ThorvaldsenJL
DuranKL
BartolomeiMS
1998 Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Genes Dev 12 3693 3702
22. SrivastavaM
HsiehS
GrinbergA
Williams-SimonsL
HuangSP
2000 H19 and Igf2 monoallelic expression is regulated in two distinct ways by a shared cis acting regulatory region upstream of H19. Genes Dev 14 1186 1195
23. BellAC
FelsenfeldG
2000 Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405 482 485
24. HarkAT
SchoenherrCJ
KatzDJ
IngramRS
LevorseJM
2000 CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 405 486 489
25. KafferCR
SrivastavaM
ParkKY
IvesE
HsiehS
2000 A transcriptional insulator at the imprinted H19/Igf2 locus. Genes Dev 14 1908 1919
26. KanduriC
PantV
LoukinovD
PugachevaE
QiCF
2000 Functional association of CTCF with the insulator upstream of the H19 gene is parent of origin-specific and methylation-sensitive. Curr Biol 10 853 856
27. SzabóP
TangSH
RentsendorjA
PfeiferGP
MannJR
2000 Maternal-specific footprints at putative CTCF sites in the H19 imprinting control region give evidence for insulator function. Curr Biol 10 607 610
28. BellAC
WestAG
FelsenfeldG
1999 The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell 98 387 396
29. MoonH
FilippovaG
LoukinovD
PugachevaE
ChenQ
2005 CTCF is conserved from Drosophila to humans and confers enhancer blocking of the Fab-8 insulator. EMBO Rep 6 165 170
30. FilippovaGN
FagerlieS
KlenovaEM
MyersC
DehnerY
1996 An exceptionally conserved transcriptional repressor, CTCF, employs different combinations of zinc fingers to bind diverged promoter sequences of avian and mammalian c-myc oncogenes. Mol Cell Biol 16 2802 2813
31. SzabóPE
TangSH
SilvaFJ
TsarkWM
MannJR
2004 Role of CTCF binding sites in the Igf2/H19 imprinting control region. Mol Cell Biol 24 4791 4800
32. SchoenherrCJ
LevorseJM
TilghmanSM
2003 CTCF maintains differential methylation at the Igf2/H19 locus. Nat Genet 33 66 69
33. PantV
MarianoP
KanduriC
MattssonA
LobanenkovV
2003 The nucleotides responsible for the direct physical contact between the chromatin insulator protein CTCF and the H19 imprinting control region manifest parent of origin-specific long-distance insulation and methylation-free domains. Genes Dev 17 586 590
34. HanL
LeeDH
SzabóPE
2008 CTCF is the master organizer of domain-wide allele-specific chromatin at the H19/Igf2 imprinted region. Mol Cell Biol 28 1124 1135
35. LiT
HuJF
QiuX
LingJ
ChenH
2008 CTCF regulates allelic expression of Igf2 by orchestrating a promoter-polycomb repressive complex 2 intrachromosomal loop. Mol Cell Biol 28 6473 6482
36. SinghP
HanL
RivasGE
LeeDH
NicholsonTB
2010 Allele-specific H3K79 Di - versus trimethylation distinguishes opposite parental alleles at imprinted regions. Mol Cell Biol 30 2693 2707
37. DavisTL
YangGJ
McCarreyJR
BartolomeiMS
2000 The H19 methylation imprint is erased and re-established differentially on the parental alleles during male germ cell development. Hum Mol Genet 9 2885 2894
38. LiJY
Lees-MurdockDJ
XuGL
WalshCP
2004 Timing of establishment of paternal methylation imprints in the mouse. Genomics 84 952 960
39. UedaT
AbeK
MiuraA
YuzurihaM
ZubairM
2000 The paternal methylation imprint of the mouse H19 locus is acquired in the gonocyte stage during foetal testis development. Genes Cells 5 649 659
40. HajkovaP
ErhardtS
LaneN
HaafT
El-MaarriO
2002 Epigenetic reprogramming in mouse primordial germ cells. Mech Dev 117 15 23
41. KatoY
KanedaM
HataK
KumakiK
HisanoM
2007 Role of the Dnmt3 family in de novo methylation of imprinted and repetitive sequences during male germ cell development in the mouse. Hum Mol Genet 16 2272 2280
42. DavisTL
TraslerJM
MossSB
YangGJ
BartolomeiMS
1999 Acquisition of the H19 methylation imprint occurs differentially on the parental alleles during spermatogenesis. Genomics 58 18 28
43. SzabóPE
HubnerK
ScholerH
MannJR
2002 Allele-specific expression of imprinted genes in mouse migratory primordial germ cells. Mech Dev 115 157 160
44. SzabóPE
MannJR
1995 Biallelic expression of imprinted genes in the mouse germ line: implications for erasure, establishment, and mechanisms of genomic imprinting. Genes Dev 9 1857 1868
45. CicconeDN
SuH
HeviS
GayF
LeiH
2009 KDM1B is a histone H3K4 demethylase required to establish maternal genomic imprints. Nature 461 415 418
46. HajkovaP
AncelinK
WaldmannT
LacosteN
LangeUC
2008 Chromatin dynamics during epigenetic reprogramming in the mouse germ line. Nature 452 877 881
47. SekiY
HayashiK
ItohK
MizugakiM
SaitouM
2005 Extensive and orderly reprogramming of genome-wide chromatin modifications associated with specification and early development of germ cells in mice. Dev Biol 278 440 458
48. SekiY
YamajiM
YabutaY
SanoM
ShigetaM
2007 Cellular dynamics associated with the genome-wide epigenetic reprogramming in migrating primordial germ cells in mice. Development 134 2627 2638
49. YatsukiH
JohK
HigashimotoK
SoejimaH
AraiY
2002 Domain regulation of imprinting cluster in Kip2/Lit1 subdomain on mouse chromosome 7F4/F5: large-scale DNA methylation analysis reveals that DMR-Lit1 is a putative imprinting control region. Genome Res 12 1860 1870
50. HiuraH
ObataY
KomiyamaJ
ShiraiM
KonoT
2006 Oocyte growth-dependent progression of maternal imprinting in mice. Genes Cells 11 353 361
51. OlekA
OswaldJ
WalterJ
1996 A modified and improved method for bisulphite based cytosine methylation analysis. Nucleic Acids Res 24 5064 5066
52. JurinkeC
DenissenkoMF
OethP
EhrichM
van den BoomD
2005 A single nucleotide polymorphism based approach for the identification and characterization of gene expression modulation using MassARRAY. Mutat Res 573 83 95
53. LoukinovDI
PugachevaE
VatolinS
PackSD
MoonH
2002 BORIS, a novel male germ-line-specific protein associated with epigenetic reprogramming events, shares the same 11-zinc-finger domain with CTCF, the insulator protein involved in reading imprinting marks in the soma. Proc Natl Acad Sci U S A 99 6806 6811
54. JelinicP
StehleJC
ShawP
2006 The testis-specific factor CTCFL cooperates with the protein methyltransferase PRMT7 in H19 imprinting control region methylation. PLoS Biol 4 e355 doi:10.1371/journal.pbio.0040355
55. FarrarD
RaiS
ChernukhinI
JagodicM
ItoY
2010 Mutational analysis of the poly(ADP-ribosyl)ation sites of the transcription factor CTCF provides an insight into the mechanism of its regulation by poly(ADP-ribosyl)ation. Mol Cell Biol 30 1199 1216
56. LefevreP
WithamJ
LacroixCE
CockerillPN
BoniferC
2008 The LPS-induced transcriptional upregulation of the chicken lysozyme locus involves CTCF eviction and noncoding RNA transcription. Mol Cell 32 129 139
57. DelavalK
GovinJ
CerqueiraF
RousseauxS
KhochbinS
2007 Differential histone modifications mark mouse imprinting control regions during spermatogenesis. Embo J 26 720 729
58. KriaucionisS
HeintzN
2009 The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 324 929 930
59. TahilianiM
KohKP
ShenY
PastorWA
BandukwalaH
2009 Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324 930 935
60. JinSG
KadamS
PfeiferGP
2010 Examination of the specificity of DNA methylation profiling techniques toward 5-methylcytosine and 5-hydroxymethylcytosine. Nucleic Acids Res 38 e125
61. TamaruH
SelkerEU
2001 A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature 414 277 283
62. TamaruH
ZhangX
McMillenD
SinghPB
NakayamaJ
2003 Trimethylated lysine 9 of histone H3 is a mark for DNA methylation in Neurospora crassa. Nat Genet 34 75 79
63. JacksonJP
JohnsonL
JasencakovaZ
ZhangX
PerezBurgosL
2004 Dimethylation of histone H3 lysine 9 is a critical mark for DNA methylation and gene silencing in Arabidopsis thaliana. Chromosoma 112 308 315
64. JacksonJP
LindrothAM
CaoX
JacobsenSE
2002 Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature 416 556 560
65. LehnertzB
UedaY
DerijckAA
BraunschweigU
Perez-BurgosL
2003 Suv39h-mediated histone h3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr Biol 13 1192 1200
66. La SalleS
MertineitC
TaketoT
MoensPB
BestorTH
2004 Windows for sex-specific methylation marked by DNA methyltransferase expression profiles in mouse germ cells. Dev Biol 268 403 415
67. Lees-MurdockDJ
ShovlinTC
GardinerT
De FeliciM
WalshCP
2005 DNA methyltransferase expression in the mouse germ line during periods of de novo methylation. Dev Dyn 232 992 1002
68. LefevreC
MannJR
2008 RNA expression microarray analysis in mouse prospermatogonia: identification of candidate epigenetic modifiers. Dev Dyn 237 1082 1089
69. SakaiY
SuetakeI
ShinozakiF
YamashinaS
TajimaS
2004 Co-expression of de novo DNA methyltransferases Dnmt3a2 and Dnmt3L in gonocytes of mouse embryos. Gene Expr Patterns 5 231 237
70. ChedinF
LieberMR
HsiehCL
2002 The DNA methyltransferase-like protein DNMT3L stimulates de novo methylation by Dnmt3a. Proc Natl Acad Sci U S A 99 16916 16921
71. OtaniJ
NankumoT
AritaK
InamotoS
AriyoshiM
2009 Structural basis for recognition of H3K4 methylation status by the DNA methyltransferase 3A ATRX-DNMT3-DNMT3L domain. EMBO Rep 10 1235 1241
72. LuciferoD
MannMR
BartolomeiMS
TraslerJM
2004 Gene-specific timing and epigenetic memory in oocyte imprinting. Hum Mol Genet 13 839 849
73. BuehrM
McLarenA
1993 Isolation and culture of primordial germ cells. Methods Enzymol 225 58 77
74. WoodMJ
WittinghamDG
RallWF
1987 The low temperature preservation of mouse oocytes and embryos, Mammalian Development: A Practical Approach (ed. M. Monk) pp 255–280.;
MonkM
Oxford IRL Press 255 280
Štítky
Genetika Reprodukční medicína
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2010 Číslo 11
-
Všechny články tohoto čísla
- Genome-Wide Association Meta-Analysis of Cortical Bone Mineral Density Unravels Allelic Heterogeneity at the Locus and Potential Pleiotropic Effects on Bone
- Beyond QTL Cloning
- An Evolutionary Framework for Association Testing in Resequencing Studies
- Genome-Wide Association Study Identifies Two Novel Regions at 11p15.5-p13 and 1p31 with Major Impact on Acute-Phase Serum Amyloid A
- The Functional Interplay between Protein Kinase CK2 and CCA1 Transcriptional Activity Is Essential for Clock Temperature Compensation in Arabidopsis
- Endogenous Viral Elements in Animal Genomes
- Analysis of the 10q11 Cancer Risk Locus Implicates and in Human Prostate Tumorigenesis
- DNA Methylation and Normal Chromosome Behavior in Neurospora Depend on Five Components of a Histone Methyltransferase Complex, DCDC
- Sarcomere Formation Occurs by the Assembly of Multiple Latent Protein Complexes
- Genetic Basis of Growth Adaptation of after Deletion of , a Major Metabolic Gene
- Nomadic Enhancers: Tissue-Specific -Regulatory Elements of Have Divergent Genomic Positions among Species
- The Parental Non-Equivalence of Imprinting Control Regions during Mammalian Development and Evolution
- CTCF-Dependent Chromatin Bias Constitutes Transient Epigenetic Memory of the Mother at the Imprinting Control Region in Prospermatogonia
- Systematic Dissection and Trajectory-Scanning Mutagenesis of the Molecular Interface That Ensures Specificity of Two-Component Signaling Pathways
- Nucleolin Is Required for DNA Methylation State and the Expression of rRNA Gene Variants in
- The Complex Genetic Architecture of the Metabolome
- ATM Limits Incorrect End Utilization during Non-Homologous End Joining of Multiple Chromosome Breaks
- Mutation Disrupts Synaptonemal Complex Formation, Recombination, and Chromosome Segregation in Mammalian Meiosis
- Mismatch Repair–Independent Increase in Spontaneous Mutagenesis in Yeast Lacking Non-Essential Subunits of DNA Polymerase ε
- The Kinesin-3 Motor UNC-104/KIF1A Is Degraded upon Loss of Specific Binding to Cargo
- Epigenetic Silencing of Spermatocyte-Specific and Neuronal Genes by SUMO Modification of the Transcription Factor Sp3
- A Coastal Cline in Sodium Accumulation in Is Driven by Natural Variation of the Sodium Transporter AtHKT1;1
- Cyclin B3 Is Required for Multiple Mitotic Processes Including Alleviation of a Spindle Checkpoint–Dependent Block in Anaphase Chromosome Segregation
- Altered DNA Methylation in Leukocytes with Trisomy 21
- Human-Specific Evolution and Adaptation Led to Major Qualitative Differences in the Variable Receptors of Human and Chimpanzee Natural Killer Cells
- Leptotene/Zygotene Chromosome Movement Via the SUN/KASH Protein Bridge in
- RACK-1 Acts with Rac GTPase Signaling and UNC-115/abLIM in Axon Pathfinding and Cell Migration
- Genome-Wide Effects of Long-Term Divergent Selection
- Endless Forms Most Viral
- Conflict between Noise and Plasticity in Yeast
- Essential Functions of the Histone Demethylase Lid
- The Transcriptional Regulator Rok Binds A+T-Rich DNA and Is Involved in Repression of a Mobile Genetic Element in
- The Cellular Robustness by Genetic Redundancy in Budding Yeast
- Localization of a Guanylyl Cyclase to Chemosensory Cilia Requires the Novel Ciliary MYND Domain Protein DAF-25
- A Buoyancy-Based Screen of Drosophila Larvae for Fat-Storage Mutants Reveals a Role for in Coupling Fat Storage to Nutrient Availability
- A Functional Genomics Approach Identifies Candidate Effectors from the Aphid Species (Green Peach Aphid)
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Genome-Wide Association Study Identifies Two Novel Regions at 11p15.5-p13 and 1p31 with Major Impact on Acute-Phase Serum Amyloid A
- Analysis of the 10q11 Cancer Risk Locus Implicates and in Human Prostate Tumorigenesis
- The Parental Non-Equivalence of Imprinting Control Regions during Mammalian Development and Evolution
- Genome-Wide Effects of Long-Term Divergent Selection
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



