-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaAnalysis of the 10q11 Cancer Risk Locus Implicates and in Human Prostate Tumorigenesis
Genome-wide association studies (GWAS) have established a variant, rs10993994, on chromosome 10q11 as being associated with prostate cancer risk. Since the variant is located outside of a protein-coding region, the target genes driving tumorigenesis are not readily apparent. Two genes nearest to this variant, MSMB and NCOA4, are strong candidates for mediating the effects of rs109939934. In a cohort of 180 individuals, we demonstrate that the rs10993994 risk allele is associated with decreased expression of two MSMB isoforms in histologically normal and malignant prostate tissue. In addition, the risk allele is associated with increased expression of five NCOA4 isoforms in histologically normal prostate tissue only. No consistent association with either gene is observed in breast or colon tissue. In conjunction with these findings, suppression of MSMB expression or NCOA4 overexpression promotes anchorage-independent growth of prostate epithelial cells, but not growth of breast epithelial cells. These data suggest that germline variation at chromosome 10q11 contributes to prostate cancer risk by influencing expression of at least two genes. More broadly, the findings demonstrate that disease risk alleles may influence multiple genes, and associations between genotype and expression may only be observed in the context of specific tissue and disease states.
Published in the journal: . PLoS Genet 6(11): e32767. doi:10.1371/journal.pgen.1001204
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1001204Summary
Genome-wide association studies (GWAS) have established a variant, rs10993994, on chromosome 10q11 as being associated with prostate cancer risk. Since the variant is located outside of a protein-coding region, the target genes driving tumorigenesis are not readily apparent. Two genes nearest to this variant, MSMB and NCOA4, are strong candidates for mediating the effects of rs109939934. In a cohort of 180 individuals, we demonstrate that the rs10993994 risk allele is associated with decreased expression of two MSMB isoforms in histologically normal and malignant prostate tissue. In addition, the risk allele is associated with increased expression of five NCOA4 isoforms in histologically normal prostate tissue only. No consistent association with either gene is observed in breast or colon tissue. In conjunction with these findings, suppression of MSMB expression or NCOA4 overexpression promotes anchorage-independent growth of prostate epithelial cells, but not growth of breast epithelial cells. These data suggest that germline variation at chromosome 10q11 contributes to prostate cancer risk by influencing expression of at least two genes. More broadly, the findings demonstrate that disease risk alleles may influence multiple genes, and associations between genotype and expression may only be observed in the context of specific tissue and disease states.
Introduction
Variation at rs10993994 on chromosome 10q11 is associated with prostate cancer risk [1]–[5]. The risk polymorphism is located at the telomeric end of a 50 kilobase (kb) linkage disequilibrium block and is within 60 base pairs (bp) of the transcription start site of beta-microseminoprotein (MSMB). MSMB has been characterized as a tumor suppressor [6], and lower levels of its product, PSP94, are associated with more aggressive forms of prostate cancer [7]. MSMB has therefore been a target of recent investigation into the mechanism of chromosome 10q-associated risk. Reporter assays demonstrate that plasmids carrying the rs10993994 risk allele (T) significantly decrease luciferase activity compared with the wild-type allele (C) [8], [9]. In addition, in 19 cancer cell lines of various tissue types expressing MSMB, those carrying the TT genotype have decreased MSMB expression relative to those carrying a C allele [9]. However, no study has definitively linked MSMB expression to risk allele status in human prostate tissue. A second gene, nuclear receptor co-activator 4 (NCOA4, also known as ARA70), is a ligand-dependent androgen receptor co-activator [10], [11] and is within 16 kb telomeric of rs10993994. Given its proximity to the risk variant and its activity in the prostate gland [12], NCOA4 has also been considered a candidate gene involved in the mechanism of disease risk [8].
Gene expression is a heritable trait [13]–[16] and represents a powerful avenue for connecting risk variants with their target genes. Studies have demonstrated that variation at intergenic or intronic disease-associated loci can act through gene regulatory mechanisms [17]–[20]. Because regulatory elements can interact with many genes [21], and since both MSMB and NCOA4 are strong candidates for prostate cancer risk, we evaluated the relationship between risk allele status and transcript abundance of these genes across both normal and tumor prostate tissues. We also tested the functional consequences of altering the expression levels of these candidate genes in immortalized prostate epithelial cells.
Results
A total of 180 individuals of European ancestry were genotyped for the rs10993994 polymorphism, and MSMB and NCOA4 mRNA levels were quantified in tissue isolated from radical prostatectomy surgical specimens. Samples were derived from two cohorts - the Gelb Center at Dana-Farber Cancer Institute (DFCI) (N = 121 – histologically normal and tumor prostate tissue) and the Physicians’ Health Study (PHS) [22] (N = 59 – prostate tumor tissue only). In the DFCI cohort, transcript levels were measured in both normal and tumor prostate tissue using a quantitative competitive PCR strategy (Methods). Two MSMB and five NCOA4 isoforms annotated in the Ensembl database (build 52) were evaluated (Figure 1A). Each isoform of MSMB and NCOA4 was expressed in both normal and tumor prostatic tissue. Expression levels of MSMB and NCOA4 transcripts were significantly higher in normal compared with tumor tissue (p<0.0001), consistent with previously published reports [23]–[25]. In the PHS cohort, only tumor tissue was isolated from radical prostatectomy specimens. For this cohort the probes used to measure expression captured both MSMB isoforms and all but one of the NCOA4 isoforms.
Fig. 1. RNA expression of MSMB and NCOA4 in normal and tumor prostate tissue by rs10994994 genotype. 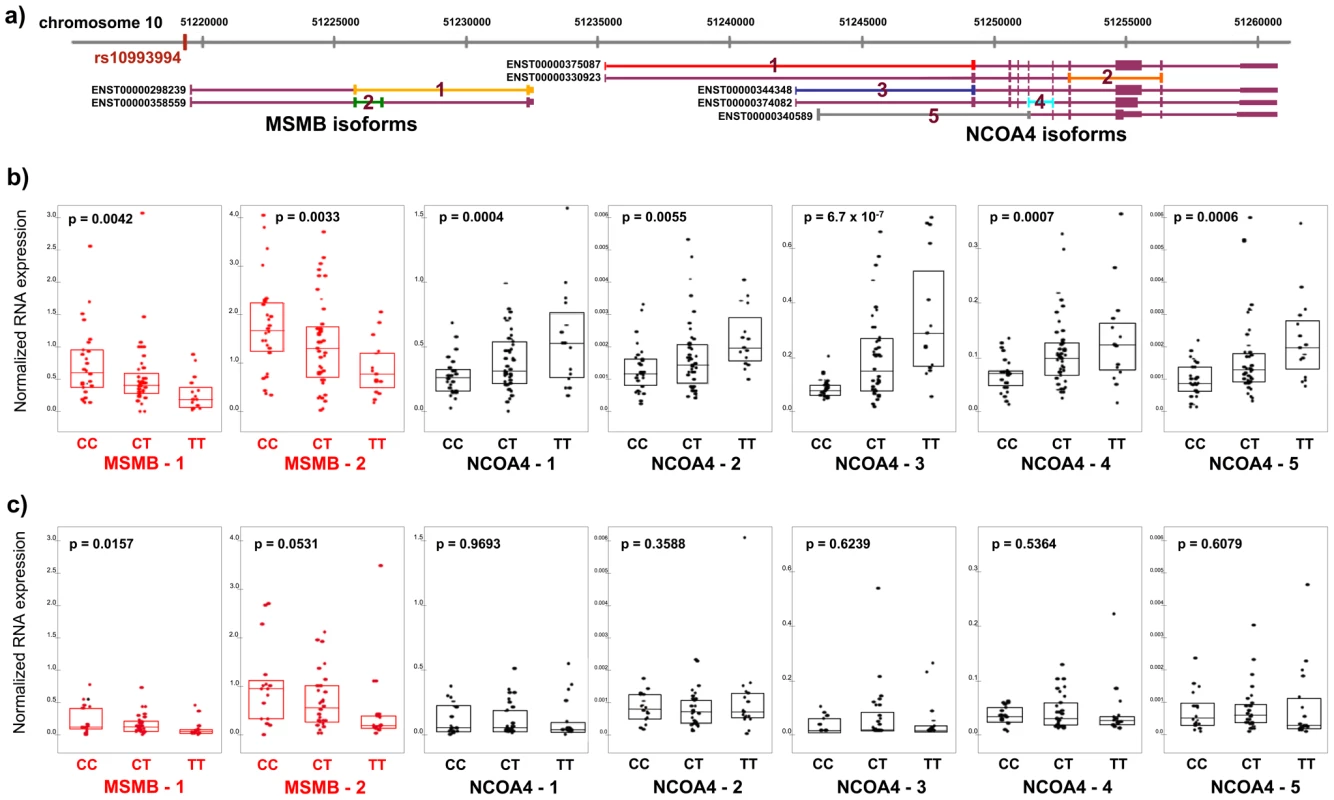
A. Chromosome 10q11 with isoforms of MSMB and NCOA4 (Ensembl build 52). Primers for competitive PCR were designed to cross exon-exon boundaries depicted by colored lines 1–2 (MSMB) and 1–5 (NCOA4). B. Expression in histologically normal prostate tissue (n = 84). Each point represents absolute RNA expression for one individual, normalized to three housekeeping genes. The top and bottom of the boxes within each graph represent the upper and lower quartiles for expression at each genotype. The band inside each box marks the median value. P-value for each graph denotes the significance for association between expression and genotype. C. Expression in prostate tumor tissue in the Dana-Farber Cancer Institute series (n = 61). The T (risk) allele is significantly associated with transcript levels of both MSMB and NCOA4 in histologically normal prostate tissue (N = 84). While the T allele is associated with decreased expression of both MSMB isoforms (p-value range, 0.0033–0.0042), it is associated with increased expression of all five isoforms of NCOA4 (p-value range, 6.7×10−7–0.0055) (Figure 1B). In tumor tissue, MSMB retains its association with risk allele status (p-value range, 0.016–0.053, Figure 1C, Figure S1). NCOA4 expression levels, however, are no longer associated with genotype in tumor tissue (p>0.30, Figure 1C, Figure S1). The associations are specific to the genes in this region. Expression levels of TIMM23, the next closest gene to the risk locus and 1.4 kb telomeric to NCOA4, are not correlated with genotype status in either normal or tumor tissue. (p>0.30, Figure S2).
Because rs10993994 is not a risk allele in colon or breast cancers, we reasoned that the association between genotype and disease-relevant genes may be specific to prostate tissue and not observed in other tissue types. If an association between genotype and expression of MSMB or NCOA4 were observed in tissue other than prostate, then that gene may be less likely to be involved in prostate cancer risk. MSMB and NCOA4 expression levels were measured in histologically normal colon (N = 72) and breast (N = 56) tissue samples. While breast tissue expresses both genes, colon tissue only expresses NCOA4. Unlike prostate tissue, neither breast nor colon tissue demonstrates convincing or consistent associations with genotype across isoforms (Figure S3).
To evaluate the functional implications of the genetic findings, we tested the effect of increasing NCOA4 and suppressing MSMB expression levels in immortalized prostate epithelial cells (LHSAR) [26]. Specifically, we assessed the ability of NCOA4 and MSMB to promote anchorage-independent growth, a phenotype strongly associated with cell transformation [27]–[30]. Suppression of MSMB expression in LHSAR cells led to a significant increase in anchorage-independent colony growth (p-values 0.0023–0.0001; Figure 2A, Figure S4). Overexpression of NCOA4 in LHSAR cells also resulted in robust anchorage-independent colony growth (p-value 0.0074; Figure 2B, Figure S4). To assess whether these alterations were specific for prostate epithelial cells, similar functional studies were performed in immortalized human mammary epithelial cells [31]. In contrast to what was observed in prostate epithelial cells, manipulating expression levels of MSMB or NCOA4 did not result in any consistently significant change in the anchorage-independent growth in mammary epithelial cells (Figure S5). Together, these observations implicate a role for both NCOA4 and MSMB in the transformation of prostate epithelial cells.
Fig. 2. Suppressing MSMB or overexpressing NCOA4 is associated with increased anchorage-independent growth of prostate epithelial cells. 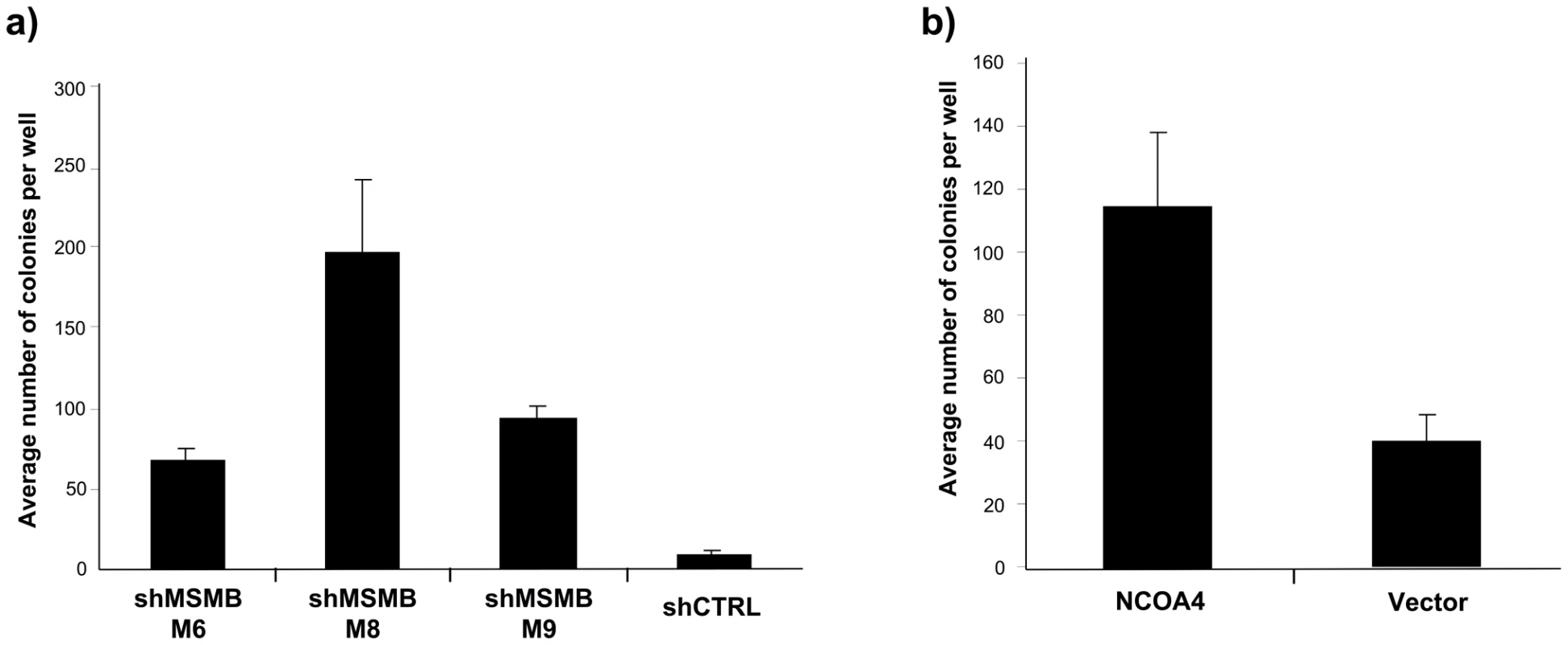
A. Effects of suppressing MSMB with three independent shRNAs in LHSAR cells (p-values 0.0001; M6, 0.0023; M8, and 0.0001; M9). The increase in anchorage-independent growth inversely correlates with the degree of MSMB suppression (Figure S4). B. Anchorage-independent growth of LHSAR cells overexpressing NCOA4 and a control vector (p = 0.0074). Discussion
Genetic data presented here demonstrate that the chromosome 10q11 prostate cancer risk locus is associated with decreased levels of MSMB and increased levels of NCOA4 RNA expression. Strikingly, the functional data fully corroborate the genetic data. When MSMB is knocked down or NCOA4 is overexpressed in immortalized prostate epithelial cells, the cells become anchorage independent. Our data suggest that both MSMB and NCOA4 mediate prostate tumorigenesis, and this study is the first, to our knowledge, to implicate these genes in actual human prostate tissue. As expected in a cohort comprised of subjects who underwent radical prostatectomy, a large majority of individuals included in the analysis were diagnosed with low - or intermediate-risk prostate cancer. Despite a relatively homogenous cohort, the results presented here are likely generalizable to most prostate cancer cases since rs10993994 appears to confer risk for all levels of prostate cancer aggressiveness [1], [3], [32], [33], [34].
Similar to the 10q11 risk allele, other disease risk loci have been shown to affect expression of more than one gene [17], [34]. A variant associated with systemic lupus erythematous at chromosome 8p23, for example, is associated with increased expression of one gene (BLK) and decreased expression of another (C8orf13) in B cell lines [34]. As more genes underlying complex traits are discovered, it may be that certain risk alleles mediate their effects through multiple genes, or alternatively, that two risk variants in tight linkage disequilibrium influence separate genes. The findings at 10q11 highlight the importance of considering multiple genes when analyzing GWAS results.
The findings at 10q11 also underscore the importance of evaluating risk loci in a tissue-specific context [35]. It is hypothesized that a fraction of non-protein coding risk alleles will alter disease risk by regulating gene activity, and these variants may exert their effects in a specific genetic and epigenetic context [21], [36]. In the present study, the 10q11 risk variant is associated with transcript levels of MSMB and NCOA4 in primary prostate tissue. In contrast, no convincing or consistent association is observed in colon or breast tissue. Similarly, alteration of MSMB and NCOA4 expression significantly affects anchorage-independence of prostate but not breast epithelial cells. These findings may have implications for future studies attempting to connect risk alleles with target gene(s). Evaluation of GWAS findings will focus, in part, on identifying the genes targeted by risk alleles, as these are the genes likely to drive the trait under study. Our findings suggest that this type of analysis should include evaluation of the tissue at risk for disease, although it is entirely plausible that variants associated with a particular disease may manifest their effects in tissues other than target tissue.
Notably, associations between rs10993994 genotype and expression of MSMB and NCOA4 are observed in histologically normal prostate tissue, whereas in tumor tissue an association is detected with only MSMB (albeit at an attenuated level relative to the normal tissue). It is conceivable that increased expression of NCOA4 is associated with tumor initiation, as reflected by its association with risk in solely normal tissue, while decreased expression of MSMB is associated with both tumor initiation and maintenance or progression. More studies, however, will be necessary before a general principle emerges.
Cellular context also appears to be an important determinant in the functional analysis of candidate risk genes. This is illustrated by comparing data in the present study to previous work involving NCOA4. The characteristics of two NCOA4 isoforms, alpha and beta, have been studied in functional assays. Upregulation of NCOA4beta increases anchorage-independent growth [11], while overexpression of NCOA4alpha inhibits growth in LNCaP cells [37]. Functional analysis of immortalized prostate epithelial cells presented here demonstrates increased colony growth in the setting of an overexpressed alpha isoform (Figure 2). Distinctions between the cell lines used in these studies may account for these divergent results. LNCaP cells are derived from metastatic prostate lesions. Immortalized prostate epithelial cells, on the other hand, are not tumorigenic and differentiate in the presence of androgen [26], suggesting that these cells are more closely related to normal prostate epithelial cells.
The data presented here suggest that tissue type (in this case, prostate versus non-prostate tissue) and cellular states (i.e., normal versus tumor) are likely important factors in the evaluation of complex trait loci. Chromatin context and differential use of gene regulatory elements across tissues and disease states may be the basis for expression effects specific to normal prostate tissue [36]. It can be difficult to accurately model these effects outside of the particular genetic and epigenetic context of specific tissues. Luciferase reporter assays, for example, are often utilized to define a relationship between a polymorphism and a gene. Reporter assays cannot, however, detect situations where a risk variant is associated with opposing transcriptional effects on two loci, as observed with rs10993994. An alternative explanation for the effects on the two transcripts is that two separate variants in linkage disequilibrium are responsible for the different transcriptional effects.
In contrast to Mendelian diseases, where resequencing protein-coding regions often reveals the causal gene, common complex trait alleles often occur outside of protein-coding regions. As is the case at 10q11 and other loci [38], [39], these alleles may be associated with expression of nearby and/or distal candidate genes. There are also situations in which associations with strong candidate genes, however, cannot be established by measuring steady-state expression at a single point in time [19]. In order to better understand the gene(s) involved in complex trait pathogenesis, experiments will need to integrate and to account for the genetic and epigenetic contexts of the particular tissue type and cellular state.
Materials and Methods
Ethics statement
This study was conducted according to the principles expressed in the Declaration of Helsinki. The study was approved by the Institutional Review Board of Dana-Farber Cancer Institute. All patients provided written informed consent for the collection of samples and subsequent analysis.
Cohorts and RNA isolation
A total of 180 patients treated with radical prostatectomy (RP) for prostate cancer and 92 patients treated for colon cancer consented to provide tissue. Additionally, histologically normal breast tissue from 56 from women undergoing cosmetic reduction surgery was analyzed in the present study.
Fresh frozen radical prostatectomy specimens were available from 121 subjects at the Dana-Farber Cancer Institute (DFCI) and Brigham and Women's Hospital (Boston, MA) and were reviewed by a pathologist (J.C. or R.F.). Over 95% of the patients in the cohort were diagnosed with Gleason 6 or Gleason 7 disease and median PSA was 5.1 ng/ml. Areas of tumor consisted of >60% tumor cells and areas of benign tissue consisted of >80% non-neoplastic epithelial cells at least 5 mm away from any tumor focus. Biopsy cores of fresh frozen tissue were processed for RNA extraction using a modified Qiagen Allprep DNA/RNA protocol.
Archival FFPE blocks were available for 59 men with prostate cancer enrolled in the Physicians’ Health Study (PHS) [22], [40]. These men were diagnosed with prostate cancer between 1983 and 2003 and treated by radical prostatectomy. RNA were extracted from paraffin-embedded tumor tissue as described previously [40]. Areas of tumor consisted of >90% tumor cells.
Fresh frozen colorectal cancer tissue samples were reviewed by a pathologist (J.C.) and areas of benign tissue were selected where 80% of cells consisted of non-neoplastic epithelium. RNA was extracted using a modified Qiagen Allprep DNA/RNA protocol. Fresh frozen normal breast tissue samples were reviewed to identify tissue blocks containing >40% normal epithelial cells. RNA was extracted using a modified Qiagen RNeasy protocol.
Ethnicity was self-reported by most, but not all, subjects. Subjects in the DFCI cohort of unknown ancestry were genotyped for 59 ancestry-informative SNPs in order to ascertain ethnicity. The marker set primarily captured ancestral differences between European and African ancestries (D. Reich, personal communication). Five samples found to be from subjects of African ancestry were excluded from analysis.
The human tissues analyzed in this study were from patients treated at Brigham and Women’s Hospital, Dana-Farber Cancer Institute or Vall d'Hebron University Hospital in Barcelona, Spain, all of whom provided informed consent. The study was approved by the Institutional Review Board at Dana-Farber Cancer Institute.
Expression analysis
cDNA was prepared for expression analysis using Invitrogen SuperScript III Reverse Transcription kit. DFCI prostate samples, colon samples and breast samples were analyzed via competitive RT-PCR using Sequenom iPLEX matrix-assisted laser desorption/ionization (MALDI)-time of flight mass spectrometry technology. Expression levels of two MSMB isoforms and five NCOA4 isoforms were measured. These splice variants represent all isoforms reported in Ensembl genebuild 52. RNA expression of TIMM23 and three housekeeping genes (ACTB, MYL6 and RPL13A) were also measured. Primer, probe and competitor oligo sequences are available upon request. Reactions were performed in quadruplicate using 8 serial dilutions of competitor, and the EC50 was calculated using QGE Analyzer software (Sequenom). The PHS subgroup was analyzed using Illumina cDNA-mediated Annealing, Selection, Extension and Ligation (DASL) expression assay (Illumina).
Expression data analysis
A gene expression normalization factor was calculated using the geometric mean of expression level of the three housekeeping genes. Linear regression was used to assess whether expression levels increased (or decreased) as the number of T-alleles of rs10993994 increased; for prostate tissue, differences between prostate tumor and normal tissue levels were also assessed and random effects linear regression was used to account for within-sample correlation of tumor/normal pairs.
Genotyping
Genotyping of DNA from each subject was carried out using Sequenom iPLEX matrix-assisted laser desorption/ionization (MALDI)-time of flight mass spectrometry technology.
Cell culture
LHSAR: Prostate epithelial cells (PrECs) immortalized with hTERT, SV40 Large T and small t antigens and overexpressing androgen receptor were grown in PREGM (Lonza CC-3166). HMLE: Human mammary epithelial cells immortalized with hTERT, Large T and small t antigens were grown in MEGM (Lonza CC-3150). All growth media were supplemented with 100 ug/ml Penicillin/Streptomycin.
Soft agar colony formation assay
The bottom layer contained 0.6% agar (Sigma A5431) in DMEM and 8% FBS. The top layer contained 0.3% agar in PREGM or MEGM for LHSAR or HMLE respectively. Fifteen thousand cells were seeded in the top agar layer in triplicate wells of a 6 well plate. Colonies were counted from 2 to 6 weeks post-seeding. Image of each well was taken at a 6× magnification and analyzed with Image J software. Colonies that were 50 sq. pixels or larger were counted.
Quantitative RT-PCR
Qiagen RNeasy kit was used for RNA extraction. Reverse transcription was carried out with Clonetech Advantage RT-to-PCR kit while the quantitative PCR was carried out using SYBR Green Master Mix (Applied Biosystems). Two sets of NCOA4 and one set of MSMB primers were used:
-
NCOA4 exon 3-4 - Forward CAGCAGCTCTACTCGTTATTGG
Reverse TCTCCAGGCACACAGAGACT
-
NCOA4 exon 5-6 - Forward CTCTCAAAACCATTCAAATTCCT
Reverse CTCTGGCATGGAGATACAGC
-
MSMB exon 1-2 - Forward GCTTATCACAATGAATGTTCTCCT
Reverse AATCTCCTGGAACTCCCTCA
Expression constructs
NCOA4 expression constructs were received from the human ORFeome V5.1 and cloned into pWZL-Neo retroviral expression vector. Retrovirus production, infection and selection were carried out as described previously [41].
RNA interference
Short hairpins in pLKO.1 lentiviral constructs were received from the RNAi Consortium (TRC). Lentivirus production and infection were carried out as described previously [42].
-
Hairpin shMSMB M6 TRC Clone ID TRCN0000147237
Target sequence GTTCTGTCAGTGAATGGATAA
-
Hairpin shMSMB M8 TRC Clone ID TRCN0000146396
Target sequence CACCTTCGTGACTTTATGCAA
-
Hairpin shMSMB M9 TRC Clone ID TRCN0000146343
Target sequence CAAAGGAAACAAACACCCAAT
Supporting Information
Zdroje
1. EelesRA
Kote-JaraiZ
GilesGG
OlamaAA
GuyM
2008 Multiple newly identified loci associated with prostate cancer susceptibility. Nat Genet 40 316 321
2. Kote-JaraiZ
EastonDF
StanfordJL
OstranderEA
SchleutkerJ
2008 Multiple novel prostate cancer predisposition loci confirmed by an international study: the PRACTICAL Consortium. Cancer Epidemiol Biomarkers Prev 17 2052 2061
3. ThomasG
JacobsKB
YeagerM
KraftP
WacholderS
2008 Multiple loci identified in a genome-wide association study of prostate cancer. Nat Genet 40 310 315
4. WatersKM
Le MarchandL
KolonelLN
MonroeKR
StramDO
2009 Generalizability of associations from prostate cancer genome-wide association studies in multiple populations. Cancer Epidemiol Biomarkers Prev 18 1285 1289
5. ZhengSL
SunJ
WiklundF
GaoZ
StattinP
2009 Genetic variants and family history predict prostate cancer similar to prostate-specific antigen. Clin Cancer Res 15 1105 1111
6. ReevesJR
DuludeH
PanchalC
DaigneaultL
RamnaniDM
2006 Prognostic value of prostate secretory protein of 94 amino acids and its binding protein after radical prostatectomy. Clin Cancer Res 12 6018 6022
7. ShukeirN
ArakelianA
KadhimS
GardeS
RabbaniSA
2003 Prostate secretory protein PSP-94 decreases tumor growth and hypercalcemia of malignancy in a syngenic in vivo model of prostate cancer. Cancer Res 63 2072 2078
8. ChangBL
CramerSD
WiklundF
IsaacsSD
StevensVL
2009 Fine mapping association study and functional analysis implicate a SNP in MSMB at 10q11 as a causal variant for prostate cancer risk. Hum Mol Genet 18 1368 1375
9. LouH
YeagerM
LiH
BosquetJG
HayesRB
2009 Fine mapping and functional analysis of a common variant in MSMB on chromosome 10q11.2 associated with prostate cancer susceptibility. Proc Natl Acad Sci U S A 106 7933 7938
10. NiuY
YehS
MiyamotoH
LiG
AltuwaijriS
2008 Tissue prostate-specific antigen facilitates refractory prostate tumor progression via enhancing ARA70-regulated androgen receptor transactivation. Cancer Res 68 7110 7119
11. PengY
LiCX
ChenF
WangZ
LigrM
2008 Stimulation of prostate cancer cellular proliferation and invasion by the androgen receptor co-activator ARA70. Amer J Pathol 172 225 235
12. HuYC
YehS
YehSD
SampsonER
HuangJ
2004 Functional domain and motif analyses of androgen receptor coregulator ARA70 and its differential expression in prostate cancer. J Biol Chem 279 33438 33446
13. CheungVG
ConlinLK
WeberTM
ArcaroM
JenKY
2003 Natural variation in human gene expression assessed in lymphoblastoid cells. Nat Genet 33 422 425
14. SchadtEE
MonksSA
DrakeTA
LusisAJ
CheN
2003 Genetics of gene expression surveyed in maize, mouse and man. Nature 422 297 302
15. MorleyM
MolonyCM
WeberTM
DevlinJL
EwensKG
2004 Genetic analysis of genome-wide variation in human gene expression. Nature 430 743 747
16. JiaZ
XuS
2007 Mapping quantitative trait loci for expression abundance. Genetics 176 611 623
17. MoffattMF
KabeschM
LiangL
DixonAL
StrachanD
2007 Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature 448 470 473
18. MeyerKB
MaiaAT
O'ReillyM
TeschendorffAE
ChinSF
2008 Allele-specific up-regulation of FGFR2 increases susceptibility to breast cancer. PLoS Biol 6 e108 doi:10.1371/journal.pbio.0060108
19. PomerantzMM
AhmadiyehN
JiaL
HermanP
VerziMP
2009 The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. Nat Genet 41 882 884
20. TuupanenS
TurunenM
LehtonenR
HallikasO
VanharantaS
2009 The common colorectal cancer predisposition SNP rs6983267 at chromosome 8q24 confers potential to enhanced Wnt signaling. Nat Genet 41 885 890
21. BirneyE
StamatoyannopoulosJA
DuttaA
GuigoR
GingerasTR
2007 Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447 799 816
22. GannPH
HennekensCH
StampferMJ
1995 A prospective evaluation of plasma prostate-specific antigen for detection of prostatic cancer. Jama 273 289 294
23. LiP
YuX
GeK
MelamedJ
RoederRG
2002 Heterogeneous expression and functions of androgen receptor co-factors in primary prostate cancer. Amer J Pathol 161 1467 1474
24. VanajaDK
ChevilleJC
IturriaSJ
YoungCY
2003 Transcriptional silencing of zinc finger protein 185 identified by expression profiling is associated with prostate cancer progression. Cancer Res 63 3877 3882
25. LapointeJ
LiC
HigginsJP
van de RijnM
BairE
2004 Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proc Natl Acad Sci U S A 101 811 816
26. BergerR
FebboPG
MajumderPK
ZhaoJJ
MukherjeeS
2004 Androgen-induced differentiation and tumorigenicity of human prostate epithelial cells. Cancer Res 64 8867 8875
27. FreedmanVH
ShinSI
1974 Cellular tumorigenicity in nude mice: correlation with cell growth in semi-solid medium. Cell 3 355 359
28. ShinSI
FreedmanVH
RisserR
PollackR
1975 Tumorigenicity of virus-transformed cells in nude mice is correlated specifically with anchorage independent growth in vitro. Proc Natl Acad Sci U S A 72 4435 4439
29. BouckN
di MayorcaG
1976 Somatic mutation as the basis for malignant transformation of BHK cells by chemical carcinogens. Nature 264 722 727
30. SpandidosDA
SiminovitchL
1977 Transfer of anchorage independence by isolated metaphase chromosomes in hamster cells. Cell 12 675 682
31. ElenbaasB
SpirioL
KoernerF
FlemingMD
ZimonjicDB
2001 Human breast cancer cells generated by oncogenic transformation of primary mammary epithelial cells. Genes Dev 15 50 65
32. CampNJ
FarnhamJM
WongJ
ChristensenGB
ThomasA
2009 Replication of the 10q11 and Xp11 prostate cancer risk variants: results from a Utah pedigree-based study. Cancer Epidemiol Biomarkers Prev 18 1290 1294
33. FitzgeraldLM
KwonEM
KoopmeinersJS
SalinasCA
StanfordJL
2009 Analysis of recently identified prostate cancer susceptibility loci in a population-based study: associations with family history and clinical features. Clin Cancer Res 15 3231 3237
34. HomG
GrahamRR
ModrekB
TaylorKE
OrtmannW
2008 Association of systemic lupus erythematosus with C8orf13-BLK and ITGAM-ITGAX. N Engl J Med 358 900 909
35. EmilssonV
ThorleifssonG
ZhangB
LeonardsonAS
ZinkF
2008 Genetics of gene expression and its effect on disease. Nature 452 423 428
36. DimasAS
DeutschS
StrangerBE
MontgomerySB
BorelC
2009 Common regulatory variation impacts gene expression in a cell type-dependent manner. Science 325 1246 1250
37. LigrM
LiY
ZouX
DanielsG
MelamedJ
2010 Tumor suppressor function of androgen receptor coactivator ARA70 alpha in prostate cancer. Am J Pathol 176 1891 900
38. LibioulleC
LouisE
HansoulS
SandorC
FarnirF
2007 Novel Crohn disease locus identified by genome-wide association maps to a gene desert on 5p13.1 and modulates expression of PTGER4. PLoS Genet 3 e58 doi:10.1371/journal.pgen.0030058
39. CooksonW
LiangL
AbecasisG
MoffattM
LathropM
2009 Mapping complex disease traits with global gene expression. Nat Rev Genet 10 184 194
40. SetlurSR
MertzKD
HoshidaY
DemichelisF
LupienM
2008 Estrogen-dependent signaling in a molecularly distinct subclass of aggressive prostate cancer. J Natl Cancer Inst 100 815 825
41. BoehmJS
HessionMT
BulmerSE
HahnWC
2005 Transformation of human and murine fibroblasts without viral oncoproteins. Mol Cell Biol 25 6464 6474
42. MoffatJ
GruenebergDA
YangX
KimSY
KloepferAM
2006 A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell 124 1283 1298
Štítky
Genetika Reprodukční medicína
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2010 Číslo 11
-
Všechny články tohoto čísla
- Genome-Wide Association Meta-Analysis of Cortical Bone Mineral Density Unravels Allelic Heterogeneity at the Locus and Potential Pleiotropic Effects on Bone
- Beyond QTL Cloning
- An Evolutionary Framework for Association Testing in Resequencing Studies
- Genome-Wide Association Study Identifies Two Novel Regions at 11p15.5-p13 and 1p31 with Major Impact on Acute-Phase Serum Amyloid A
- The Functional Interplay between Protein Kinase CK2 and CCA1 Transcriptional Activity Is Essential for Clock Temperature Compensation in Arabidopsis
- Endogenous Viral Elements in Animal Genomes
- Analysis of the 10q11 Cancer Risk Locus Implicates and in Human Prostate Tumorigenesis
- DNA Methylation and Normal Chromosome Behavior in Neurospora Depend on Five Components of a Histone Methyltransferase Complex, DCDC
- Sarcomere Formation Occurs by the Assembly of Multiple Latent Protein Complexes
- Genetic Basis of Growth Adaptation of after Deletion of , a Major Metabolic Gene
- Nomadic Enhancers: Tissue-Specific -Regulatory Elements of Have Divergent Genomic Positions among Species
- The Parental Non-Equivalence of Imprinting Control Regions during Mammalian Development and Evolution
- CTCF-Dependent Chromatin Bias Constitutes Transient Epigenetic Memory of the Mother at the Imprinting Control Region in Prospermatogonia
- Systematic Dissection and Trajectory-Scanning Mutagenesis of the Molecular Interface That Ensures Specificity of Two-Component Signaling Pathways
- Nucleolin Is Required for DNA Methylation State and the Expression of rRNA Gene Variants in
- The Complex Genetic Architecture of the Metabolome
- ATM Limits Incorrect End Utilization during Non-Homologous End Joining of Multiple Chromosome Breaks
- Mutation Disrupts Synaptonemal Complex Formation, Recombination, and Chromosome Segregation in Mammalian Meiosis
- Mismatch Repair–Independent Increase in Spontaneous Mutagenesis in Yeast Lacking Non-Essential Subunits of DNA Polymerase ε
- The Kinesin-3 Motor UNC-104/KIF1A Is Degraded upon Loss of Specific Binding to Cargo
- Epigenetic Silencing of Spermatocyte-Specific and Neuronal Genes by SUMO Modification of the Transcription Factor Sp3
- A Coastal Cline in Sodium Accumulation in Is Driven by Natural Variation of the Sodium Transporter AtHKT1;1
- Cyclin B3 Is Required for Multiple Mitotic Processes Including Alleviation of a Spindle Checkpoint–Dependent Block in Anaphase Chromosome Segregation
- Altered DNA Methylation in Leukocytes with Trisomy 21
- Human-Specific Evolution and Adaptation Led to Major Qualitative Differences in the Variable Receptors of Human and Chimpanzee Natural Killer Cells
- Leptotene/Zygotene Chromosome Movement Via the SUN/KASH Protein Bridge in
- RACK-1 Acts with Rac GTPase Signaling and UNC-115/abLIM in Axon Pathfinding and Cell Migration
- Genome-Wide Effects of Long-Term Divergent Selection
- Endless Forms Most Viral
- Conflict between Noise and Plasticity in Yeast
- Essential Functions of the Histone Demethylase Lid
- The Transcriptional Regulator Rok Binds A+T-Rich DNA and Is Involved in Repression of a Mobile Genetic Element in
- The Cellular Robustness by Genetic Redundancy in Budding Yeast
- Localization of a Guanylyl Cyclase to Chemosensory Cilia Requires the Novel Ciliary MYND Domain Protein DAF-25
- A Buoyancy-Based Screen of Drosophila Larvae for Fat-Storage Mutants Reveals a Role for in Coupling Fat Storage to Nutrient Availability
- A Functional Genomics Approach Identifies Candidate Effectors from the Aphid Species (Green Peach Aphid)
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Genome-Wide Association Study Identifies Two Novel Regions at 11p15.5-p13 and 1p31 with Major Impact on Acute-Phase Serum Amyloid A
- Analysis of the 10q11 Cancer Risk Locus Implicates and in Human Prostate Tumorigenesis
- The Parental Non-Equivalence of Imprinting Control Regions during Mammalian Development and Evolution
- Genome-Wide Effects of Long-Term Divergent Selection
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



