-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaGranulocytes Impose a Tight Bottleneck upon the Gut Luminal Pathogen Population during Typhimurium Colitis
Salmonella Typhimurium can colonize the human intestine and cause severe diarrhea. In recent years, it has become clear that this pathogen profits from inflammatory changes in the intestinal lumen, as the inflamed gut helps Salmonella to out-compete the resident microbiota. Granulocytes transmigrating into the gut lumen were found to “foster” luminal Salmonella growth by providing nutrients (used by Salmonella, not the microbiota) and by releasing growth inhibitors affecting the microbiota, but not the pathogen. In this study, we extend this “fostering” concept by showing that gut luminal Salmonella Typhimurium population is itself surprisingly vulnerable to the host's inflammatory response. Indeed, inflammation reduces the size of the gut luminal Salmonella population by as much as 105-fold at day 2 post infection. Thus, triggering of mucosal inflammation is in fact a double-edged sword by providing S. Typhimurium with a relative growth advantage against the microbiota in the gut lumen and by killing 99.999% of the gut luminal pathogen population at day 2. However, the pathogen population can recover and grow up again during the subsequent days. This changes the current view: Inflammation is not simply “beneficial” for the pathogen in the gut lumen. Instead, pathogen growth in the inflamed gut must be considered as an equilibrium between inflammation-inflicted killing and fostering growth of the surviving bacteria.
Published in the journal: . PLoS Pathog 10(12): e32767. doi:10.1371/journal.ppat.1004557
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1004557Summary
Salmonella Typhimurium can colonize the human intestine and cause severe diarrhea. In recent years, it has become clear that this pathogen profits from inflammatory changes in the intestinal lumen, as the inflamed gut helps Salmonella to out-compete the resident microbiota. Granulocytes transmigrating into the gut lumen were found to “foster” luminal Salmonella growth by providing nutrients (used by Salmonella, not the microbiota) and by releasing growth inhibitors affecting the microbiota, but not the pathogen. In this study, we extend this “fostering” concept by showing that gut luminal Salmonella Typhimurium population is itself surprisingly vulnerable to the host's inflammatory response. Indeed, inflammation reduces the size of the gut luminal Salmonella population by as much as 105-fold at day 2 post infection. Thus, triggering of mucosal inflammation is in fact a double-edged sword by providing S. Typhimurium with a relative growth advantage against the microbiota in the gut lumen and by killing 99.999% of the gut luminal pathogen population at day 2. However, the pathogen population can recover and grow up again during the subsequent days. This changes the current view: Inflammation is not simply “beneficial” for the pathogen in the gut lumen. Instead, pathogen growth in the inflamed gut must be considered as an equilibrium between inflammation-inflicted killing and fostering growth of the surviving bacteria.
Introduction
Acute infections constitute highly complex interactions between pathogens and their hosts. The complexity arises from dynamic changes in pathogen gene expression, pathogen growth, barriers limiting the initial colonization and host defenses which limit further pathogen growth and survival during the course of an infection. Identifying the relevant interactions and how they affect the progression of the disease is of great value for understanding the infection process and may reveal new targets for prevention or therapy.
Mixed inoculation provides a powerful approach to decipher pathogen-host interactions [1]. In such experiments, genetic markers carried by some members of the pathogen population are used to follow how the pathogen population disseminates, grows or is killed during the course of an infection [2]–[13]. This can reveal “barriers”, which limit the infection. Barriers can be of varying nature, including chemical barriers (i.e. antimicrobial peptides, stomach acid), physical obstacles (e.g. the mucus layer separating gut luminal bacteria from the epithelial surface [14]) or immune responses killing the pathogen. Such barriers can impose “bottlenecks” onto the pathogen population which can be detected as loss of marker diversity. Thus, barriers are important characteristics of an infection process as they indicate how hosts can interfere with pathogen colonization and survival.
We employed a mixed inoculum approach to study Salmonella enterica subspecies 1 serovar Typhimurium (termed S. Tm hereafter) growth in the inflamed gut using the well-established streptomycin mouse model for Salmonella colitis [15]. In this model, the resident microbiota is transiently suppressed by a single dose of streptomycin [16]. This bypasses the initial phase of the natural infection where the pathogen has to competitively grow in the face of an intact, dense microbiota [17] and allows us to focus on the next stage where the pathogen triggers disease and grows in the inflamed gut [15], which is still incompletely understood.
S. Tm mainly elicits mucosal inflammation via virulence factors encoded in genomic islands, i.e. the SPI-1 and SPI-2 type III secretion systems [18], [19]. The inflammatory response has been shown to foster efficient colonization of the host's gut lumen by the pathogen, as the milieu in the inflamed gut can help the pathogen to outcompete and/or suppress the resident microbiota [20]. Some of the molecular mechanisms have been identified [15]. This includes the elicitation of antimicrobial peptides which kill some of the microbiota (but not S. Tm; [21]), the limitation of iron and zinc uptake by microbiota species (but not S. Tm; [22], [23]) and the provision of terminal electron acceptors fuelling S. Tm growth by anaerobic respiration [24], [25]. These findings have established S. Tm as a pathogen subverting gut luminal inflammation to efficiently colonize this niche. However, it was previously unclear whether the inflammatory response also inflicts an additional barrier limiting pathogen colonization of the gut. This would seem reasonable as inflammation is generally mounted to fight infection, i.e. in infected host tissues [26], [27]. Indeed, some studies have observed transient, 10-fold reductions of gut luminal pathogen loads at day 2 p.i. [20] and Reg3β, an antimicrobial peptide released by the inflamed mucosa, was found to kill fast-growing S. Tm cells [28].
In this study, we have employed a mixed inoculum approach to identify barriers limiting gut luminal colonization by S. Tm. Using S. Tm strains chromosomally tagged with bar coded sequences (WITS, wild-type isogenic tagged strains; [5], [6], [29]) we identified a pronounced bottleneck in the gut luminal pathogen population and oligo-clonal expansion post crisis. The barrier was attributable to pathogen-induced inflammation, in particular granulocytes, a prominent phagocytic cell type infiltrating the infected mucosa and the gut lumen. Our data extend the current paradigm by establishing that not only the tissue-infiltrating bacteria, but also the gut luminal pathogen population, can be temporarily restricted by the host's inflammatory response.
Results
WITS infections reveal neutral genetic diversity loss by S. Tm in the cecum lumen and feces of streptomycin pretreated mice
To identify barriers limiting gut luminal pathogen colonization, we monitored the S. Tm community composition by using seven wild-type isogenic tagged S. Tm strains (WITS), which are phenotypically identical (S1 Figure). Each WITS harbors a 40 nucleotide “bar code” between two pseudogenes which allow quantification of their relative proportion by rtqPCR [5], [6], [30]. In case of perturbations, the equal proportions of the WITS-tagged subpopulations will shift towards an uneven population structure in which some WITS are dominant while others disappear (i.e. loss of diversity; reduced “evenness”). Dilution of the tagged strains by untagged S. TmWT probes the degree of diversity loss (S1C Figure).
First, we screened for optimal assay conditions by titrating the fraction of tagged WITS strains in the inoculum. A “1∶7” inoculum was prepared by mixing seven different WITS at an equal ratio (1∶1∶1∶1∶1∶1∶1; S1 Figure). The “1∶70”, 1∶700” and “1∶7000” inocula were generated by diluting the original WITS mixture with an untagged, isogenic wild type strain (1∶10, 1∶100 and 1∶1000, respectively). Streptomycin pretreated C57BL/6 mice were infected with the indicated inocula (5×107 cfu total, by gavage; without sodium bicarbonate-mediated neutralization of stomach acid, 2 independent experiments using a total of 5 or 6 mice per group) and we monitored pathogen loads in the stool by plating fecal pellets (day 1 p.i.), the cecal content, the mesenteric lymph nodes (mLN), the livers and the spleens (day 4 p.i.). All animals featured high pathogen loads in the feces at day 1 and in the cecum lumen at day 4 p.i. (≈109 cfu/g; Fig. 1A–D, black bars). Furthermore, we observed efficient colonization of the mLN, spleen and liver and all mice displayed profound cecum inflammation by day 4 p.i. (Fig. 1E). This is well in line with previous work [16], [19] and verified that the tags do not interfere with the infection process.
Fig. 1. Systematic dilution experiments of the tagged S. TmWT strains allow assessing the level of WITS diversity loss. 
Infections were performed by diluting the tagged S. TmWT WITS with the untagged isogenic strain to a final proportion of (A) 1∶7, (B) 1∶70, (C) 1∶700, (D) 1∶7000 of each strain. The population composition was monitored in fecal samples at day 1 post infection (day 1 p.i.) and in cecal content at day 4 post infection (day 4 p.i.) of each mouse (individual mouse identifiers were plotted on the x-axis). The cfu/g fecal or cecal content for the whole population and each single tagged strain were plotted. Systemic spread (cfu per organ in mLN, spleen and liver) and cecal pathology on day 4 p.i. (E) confirmed a typical S. TmWT-induced course of disease. Grey lines depict the detection limit for plating. Blue numbers indicate the median of the evenness indices of single mice. To detect diversity loss and identify optimal assay conditions, we quantified each WITS in the inoculum, the feces (day 1 p.i.) and the cecum content (day 4 p.i.). To analyze the WITS-diversity in a quantitative fashion, we have used the “evenness index”, an indicator commonly used to describe differences in the distribution of goods or money in a society ( = Gini-index [31]; see also S1 Figure). In our experiments, this index had a value close to 1 in the inoculum (0.913, median). Slight deviations (i.e. values from 0.9 to 1) are most likely attributable to technical errors (e.g. pipetting errors, PCR bias, cfu determination, DNA recovery, enrichment culture, etc.), not loss of genetic diversity in the biological system (S2 Figure).
At day 1 p.i., all WITS were detectable in the feces, no matter which inoculum was applied, and evenness indices ranged from 0.914–0.950 (1∶7 inoculum; median = 0.939) to 0.833–0.893 (1∶7000 inoculum; median = 0.868; Fig. 1A–D). This high degree of evenness indicated that the population did not encounter detectable bottlenecks, “selective sweeps” [32] or other effects limiting the genetic diversity during the transit through the gastrointestinal tract or the initial colonization of the host's large intestinal lumen by day 1 p.i.
In stark contrast, we detected dramatically reduced evenness indices of cecum luminal S. Tm populations at day 4 p.i., in particular at the highest WITS-dilutions (1∶7000; median = 0.033; Table 1). In the 5 mice, 18 of the 35 WITS were lost from the cecum lumen at day 4, and one animal did not retain any detectable WITS (mouse #G752; Fig. 1D). The identity of the dominant WITS and the WITS lost from the population differed from animal to animal and from experiment to experiment (Fig. 1A–D, see color code). These observations provided additional evidence that all WITS had equivalent fitness in our model and suggested that the diversity is reduced by a stochastic process.
Tab. 1. Statistics on population evenness. 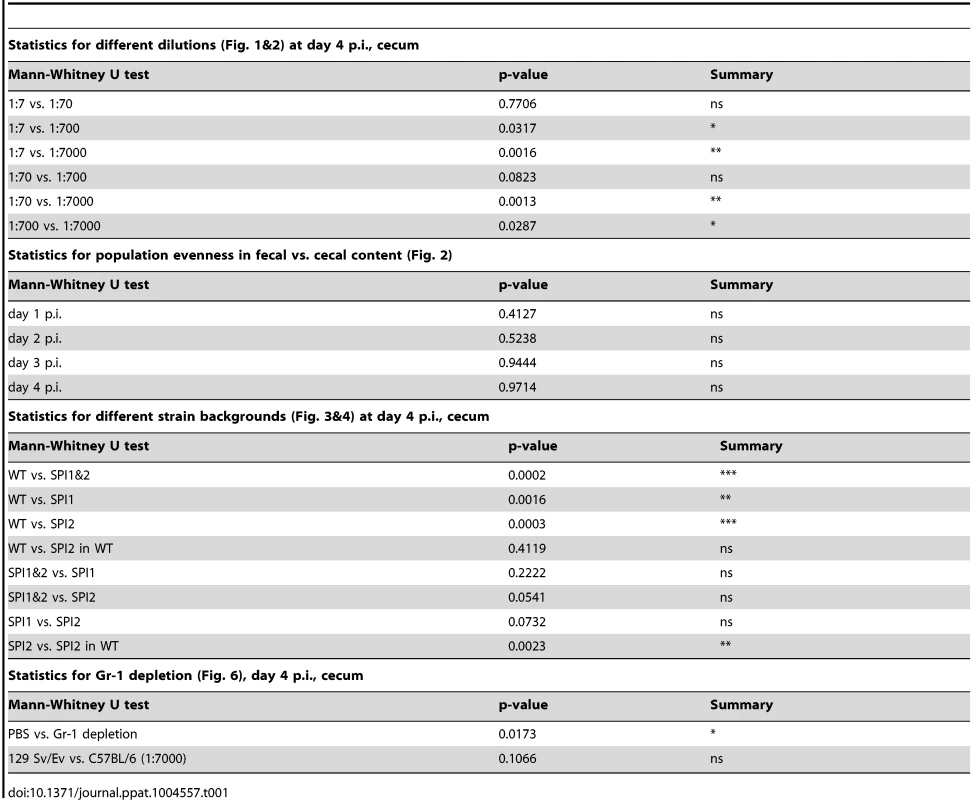
Formally, the reduced evenness observed at higher WITS-dilutions could be explained either by bottlenecks in the pathogen population or by selective sweeps (e.g. up-growth of fitter S. Tm mutants [33]). The latter was ruled out in competitive infections with re-isolated clones. These control experiments verified that (in general), the dominant WITS-isolates retained the fitness of the original strain (S3 Figure). Thus, selective sweeps [32] of beneficial mutations cannot explain the loss of WITS-diversity in the experiment shown in Fig. 1. Therefore, these data provided a first hint that a bottleneck might limit the diversity of the gut luminal pathogen population between days 1–4 post infection.
Taken together, our data suggested that a population-bottleneck explains the reduced evenness index at day 4 p.i. Equivalent observations were made in an Nramp-positive mouse line (129SvEv; see below), indicating that the barrier reducing the evenness may be a general feature, independent of the mouse line used. The gut luminal S. Tm population should encounter this bottleneck after day one and before day 4 p.i. The 1∶7000 inoculum seemed well suited for further analysis of this phenotype.
The gut luminal pathogen population encounters a pronounced bottleneck at day 2 p.i
Next, we analyzed the time course of changes in the WITS genetic diversity. We employed a “1∶7000” WITS inoculum, as the data above had indicated that this retains high evenness indices for stool samples by day 1 and should allow sensitive detection of changes during the subsequent days. The 1∶7000 inoculum was prepared as described above and we infected streptomycin pretreated C57BL/6 mice (5×107 cfu total, by gavage; 2 independent experiments using a total of 5 mice per group). Total pathogen loads and the WITS in the inoculum, the stool, the cecal content, the mLN, the spleen and the liver were quantified by plating and rtqPCR at day 1, 2, 3 or 4 post infection (Materials and Methods). In line with earlier results, S. Tm efficiently colonized the feces and the cecum contents within the first day (108–109 cfu/g; Fig. 2A,B, black bars), elicited pronounced cecum tissue contraction and inflammation (Fig. 2D,E) and the pathogen spread to the mesenteric lymph nodes, the spleen and the liver (S4 Figure, black bars). This verified that the infection had proceeded as expected.
Fig. 2. WITS infections reveal severe WITS diversity loss ( = reduction in population evenness) during oral infection of C57BL/6 mice starting at day 2 p.i. 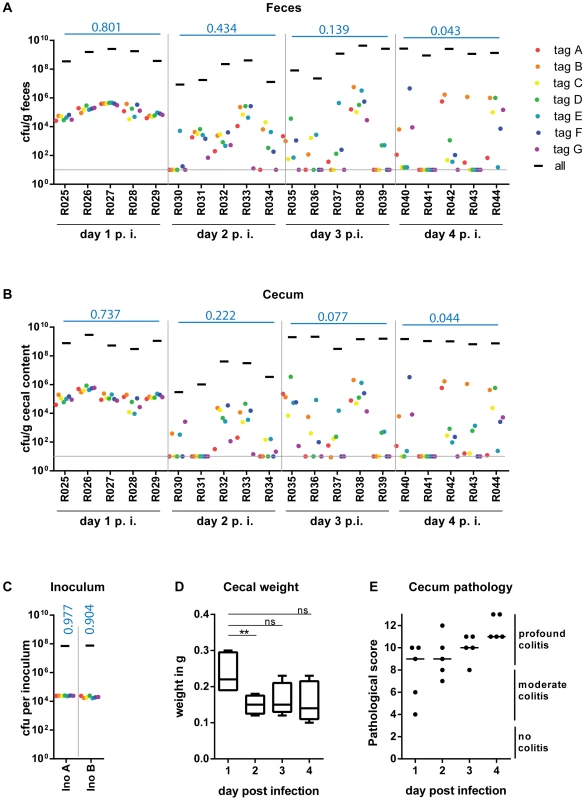
Streptomycin-pretreated C57BL/6 mice were gavaged with a mixture of 7 WITS strains in equal ratios, which were further diluted with untagged S. TmWT to a final proportion of 1∶7000. The cfu/g fecal (A) or cecal (B) content of the whole population and the individually tagged strains were monitor daily until day 4 post infection (day 4 p. i.) of each mouse (individual mouse identifiers were plotted on the x-axis). Grey lines depict the detection limit for plating (10 cfu/g). (C) rtqPCR analysis and selective plating of the inoculum used for infection confirmed the even distribution of the seven tagged strain, and the 1000-fold overrepresentation of the untagged S. TmWT strain. (D) To analyze the whole cecal population, the whole cecal content was retrieved. As the cecum shrinks in the course of S. TmWT infection [16], we monitored the total cecal weight over time. (E) At day 4 p.i. all animals were highly inflamed as assessed by a cecal pathology score. Blue numbers indicate the median of the evenness indices of single mice. At day 1 p.i., the bacterial populations in the feces and in the cecum content featured evenness indices almost as high as the inoculum (medianfeces = 0.801; mediancecum content = 0.737; each WITS present; Fig. 2 ABC). This was in stark contrast to the S. Tm populations in the mLN, spleens and livers, which harbored no WITS at all (S4 Figure; evenness index below detection). In line with earlier work, this indicated that systemic dissemination is partly restricted by a pronounced barrier and that these organs are populated by local growth of a few “founder” bacteria ([6], [29]; not analyzed further in this study). In contrast, the gut luminal population retained all WITS and the high evenness indices indicated that S. Tm did not encounter detectable bottlenecks during the transit through the stomach and gastrointestinal tract or the initial colonization of the gut lumen. Based on the 1∶7000 composition of the inoculum, we applied a simple binomial selection model to estimate that the luminal S. Tm population size never drops below 2×104 cfu during this initial day of infection (Table 2; Materials and Methods). Please note that this is an estimate of the lower boundary (detectable with WITS-dilutions of 1∶7000 and using 10 mice) and that the actual S. Tm population size might in fact be much larger than this.
Tab. 2. Bottleneck estimates. 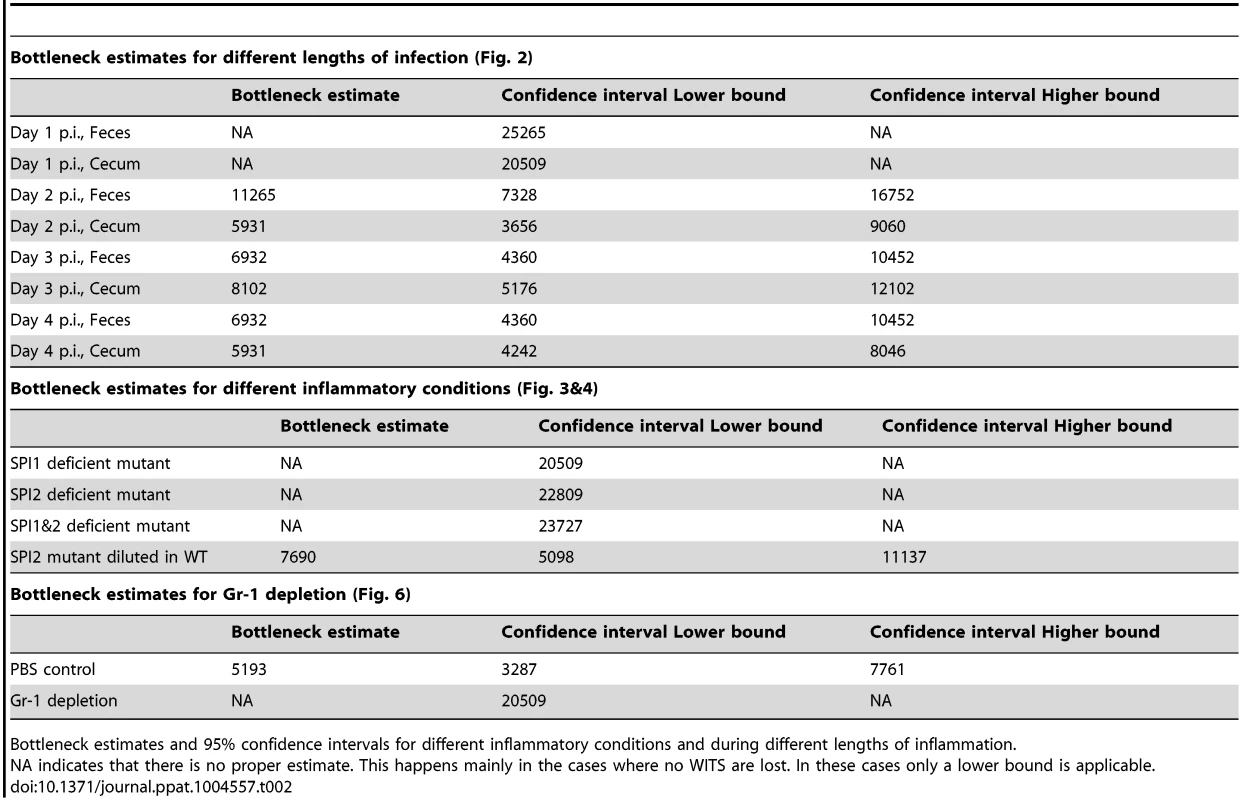
Bottleneck estimates and 95% confidence intervals for different inflammatory conditions and during different lengths of inflammation. Reduced evenness indices were observed in the cecum lumen and the feces by day 2 p.i. and evenness further declined until day 4 p.i. (medianfeces = 0.434→0.043; mediancecum content = 0.222→0.044; Fig. 2 A, B). By day 4 p.i., most mice had “lost” one or more WITS from the cecal contents and the feces (detection limit: 10 cfu/g). This indicated that some type of barrier may restrict the gut luminal S. Tm population from day 2 on.
A simple estimate of the number of bacteria that can cross this barrier can be done from the data on the presence or absence of WITS at day 4 after inoculation at a dilution of 1∶7000 (see Materials and Methods). In combination with the 1∶7000 dilution experiment shown in Fig. 1D, we had a total of 10 animals infected for 4 days. In these mice, 30 of the 70 WITS were lost from the cecum lumen at day 4 (Fig. 1D, Fig. 2B). Assuming that each WITS succeeds in crossing the barrier according to a binomial process with probability 1/7000, we estimated a bottleneck size of 5931 bacteria with a 95% confidence interval ranging from 4242 to 8046 bacteria (Materials and Methods).
Within each animal, the WITS distribution patterns of the cecal content and the feces were strikingly similar, in particular at days 3 and 4 p.i. (R2 = 0.854 or 0.961, respectively). Therefore, these populations are linked and fecal samples can be used to monitor the cecum luminal pathogen population at days 3 and 4 p.i. Conceptually, this suggests that the cecum lumen may produce (or seed) the pathogen population shed in the feces.
In conclusion, these data established that the gut luminal S. Tm population is restricted by a significant barrier by day 2 p.i. and that this bottleneck can be detected via the reduced evenness index of the WITS-diversity. It is interesting to note that the cecum luminal pathogen density transiently dropped at day 2 p.i. and was as low as 105–106 cfu/g in two of the mice (Fig. 2B, mouse #R030 and R031, black bars). Thus, a transient reduction of the total gut luminal S. Tm population size (a “population bottleneck”) may contribute to the barrier restricting the gut luminal S. Tm population. The nature of this barrier remained to be determined.
A SPI-1 SPI-2 double mutant does not encounter a bottleneck in the gut lumen
Next, we analyzed if mucosal inflammation contributes to the gut luminal population bottleneck. This seemed plausible, as inflammatory responses can (at least in tissues) eliminate pathogens via an elaborated arsenal of antimicrobial mechanisms [26]. Gut luminal colonization without the elicitation of mucosal inflammation can be achieved by using “avirulent” S. Tm mutants deficient in the SPI-1 and SPI-2 type III secretion systems [18], [19]. To this end, we constructed “avirulent” WITS by P22 phage transduction of the chromosomal tags into an invGssaV mutant (WITSSPI-1 & SPI-2; Materials and Methods) and prepared a “1∶7000” inoculum by mixing with an untagged S. TmSPI-1 & SPI-2 strain as described above. Streptomycin pretreated C57BL/6 mice were infected (5×107 cfu total, by gavage) and we analyzed pathogen loads in the stool, the WITS diversity in stool samples, gut pathology and pathogen loads at systemic sites (Fig. 3A). As expected, the S. Tm mutant yielded only minimal colonization of the spleen and the liver, reduced pathogen loads in the mLN, reduced total gut luminal loads by day 4 p.i. and no signs of overt mucosal inflammation by day 4 p.i. (pathoscore 2.5+/−1). The WITS evenness index in the stool was high at day 1 p.i. and only slightly reduced by day 4 p.i. (median = 0.799→0.676; Table 1). Conceivably, the slightly reduced evenness index detected at day 4 p.i. might be attributable (at least in part) to a reduced total S. Tm population size, as loads in the stool reached very high levels at day 1 p.i. (109–1010 cfu/g), but decreased by about 10-fold to 108–109 cfu/g by day 4 p.i. (Fig. 3A, black bars). This is in line with earlier work which established that, in the absence of gut inflammation, the re-growing microbiota can outcompete avirulent S. Tm mutants lacking functional SPI-1 and SPI-2 type III secretion systems [20]. Nevertheless, the WITS-diversity was only slightly reduced by day 4 p.i., suggesting that the tight bottleneck observed in wild type S. Typhimurium infections is attributable to mucosal inflammation.
Fig. 3. WITS diversity loss is inflammation-dependent. 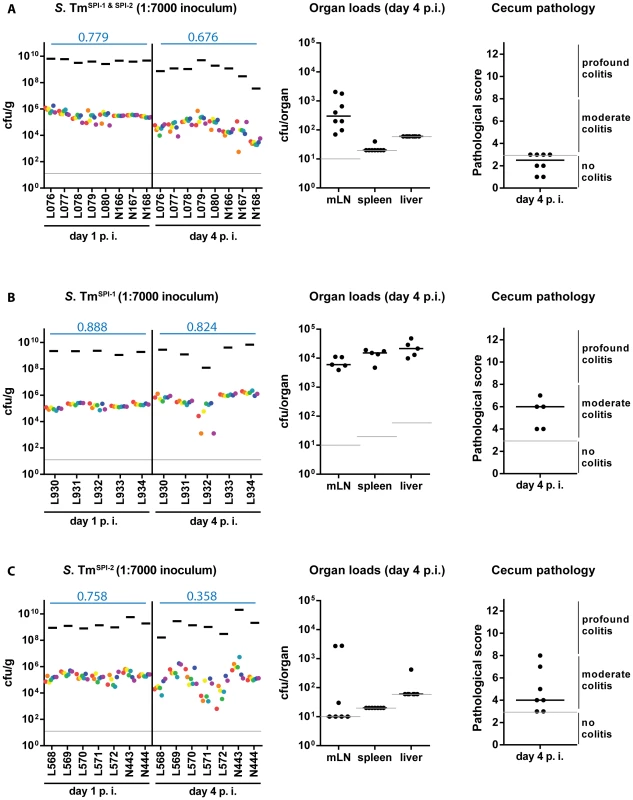
Streptomycin-pretreated C57BL/6 mice were orally infected with a WITS cocktails in the dilution 1∶7000 in different strain backgrounds: (A) S. TmSPI-1 & SPI-2, (B) S. TmSPI-1 and (C) S. TmSPI-2 and WITS compositions throughout the infection process were monitored by rtqPCR. For each conditions, organ loads and cecum pathology at day 4 p. i. were monitored. Grey lines depict the detection limit for plating (10 cfu/g for feces, cecal content and mLN, 20 cfu/organ for spleen and 60 cfu/organ for liver). Blue numbers indicate the median of the evenness indices of single mice. SPI-1 is more important than SPI-2 for eliciting WITS-diversity loss
Next, we tested the individual contributions of SPI-1 and SPI-2 to WITS-diversity loss. For this we used S. Tm mutants lacking either a functional SPI-1 TTSS (S. TmSPI-1, ΔinvG; strong defect in eliciting gut inflammation at day 2 p.i.) or a functional SPI-2 TTSS (S. TmSPI-2, ssaV::cat; slight defect in eliciting gut inflammation at day 2 p.i.), which do efficiently colonize the gut lumen of streptomycin pretreated mice, but elicit reduced levels of mucosal inflammation on day 2 p.i. [16], [18], [19], the time when the gut luminal bottleneck is observed. The respective WITS were constructed by P22 phage transduction thus yielding WITSSPI-1 (ΔinvG background; [6], [34], [35]) and WITSSPI-2 (ssaV::cat background; [6],[35] Materials and Methods) and mice were infected with “1∶7000” inocula and analyzed as described for Fig. 1.
As expected, the SPI-1 mutant efficiently colonized the spleen, the liver and the mLN and elicited moderate mucosal inflammation by day 4 p.i. (Fig. 3B). In line with earlier data, the mucosa featured a “patchy” distribution of inflammatory foci typical for the “SPI-2 mediated” disease [19], [36] which was even at day 4 p.i. significantly less pronounced than the inflammation elicited by wild type S. Tm (compare Fig. 1E, 2E with Fig. 3B). The stool loads remained high (109–1010 cfu/g) throughout the experiment and we observed at most a slight decrease of the evenness index (median = 0.888→0.824; Table 1).
In line with the key role of SPI-2 in systemic infection [19], [37], the SPI-2 mutant did not efficiently colonize the spleen, liver and the mLN, but elicited mucosal inflammation (Fig. 3C). Please note that the “SPI-1 mediated” mucosal inflammation elicited by this strain is much more pronounced than the “SPI-2 mediated” inflammation at day 2 p.i. [19]. In the present experiment, we analyzed the gut inflammation at a much later time point, i.e. day 4 p.i. Here, inflammation was much milder than the pathology elicited by wild type S. Tm (compare Fig. 1E, 2E with Fig. 3C; [6], [19]). Strikingly, the WITSSPI-2 population featured a moderate drop of the evenness index between days 1 and 4 p.i. (median = 0.758→0.358; Table 1). These data are in line with the notion that mucosal inflammation (i.e. the degree of inflammation at day 2 p.i.) may drive the WITS-diversity loss and suggest that the grade of inflammation may dictate (at least in part) barrier efficiency.
Mixed infection experiments verify that mucosal inflammation is required for WITS-diversity loss
To study the effect of mucosal inflammation on strains incapable of eliciting disease one can perform a modified version of our mixed inoculum experiments. The inflammation elicited by wild type S. Tm will affect all bacteria present in the gut lumen (i.e. wild type and mutant [20], [38]–[42]). Thus, we performed a variant of the assay described above to verify that the bottleneck is indeed affected by the grade of the inflammatory response (not genetic predisposition of the S. Tm strain). In particular, we wanted to assess the evenness index of WITSSPI-2 in the face of full blown mucosal inflammation elicited by wild type S. Tm. This was of particular interest, as WITSSPI-2 can colonize the gut lumen, but fails to efficiently grow in the mucosal lamina propria and at systemic sites [19]. Thus, using WITSSPI-2 would help to exclude possible re-seeding from such sites and therefore allow focusing on the gut luminal pathogen population. To this end, we mixed WITSSPI-2 with an excess of wild type S. Tm [6], [38]. Streptomycin pretreated C57BL/6 mice were infected for 1 or 4 days with a “1∶7000” inoculum composed of WITSSPI-2 and untagged S. TmWT and we analyzed the infection as in Fig. 1. Pathogen organ loads (i.e. WITSSPI-2 plus untagged S. TmWT) and cecum pathological scoring confirmed an overall “wild type”-like intestinal disease progression during this mixed infection experiment (Fig. 4A,B, compare to Fig. 2E; black bars). Remarkably, in this experiment, the WITSSPI-2 evenness index dropped much further than in infections performed with WITSSPI-2 alone by day 4 p.i. (median = 0.06; Fig. 4A, compare to Fig. 3C). In fact, the evenness index at day 4 p.i. was strikingly similar to that of wild type WITS (median = 0.033 and 0.044; see Fig. 1D and 2A,B; Table 1). We therefore conclude that the bottleneck is determined by the degree of inflammation, not by the genetic background of the tagged subpopulation.
Fig. 4. Severity of inflammation, not genetic background of the tagged strains, determines WITS diversity loss. 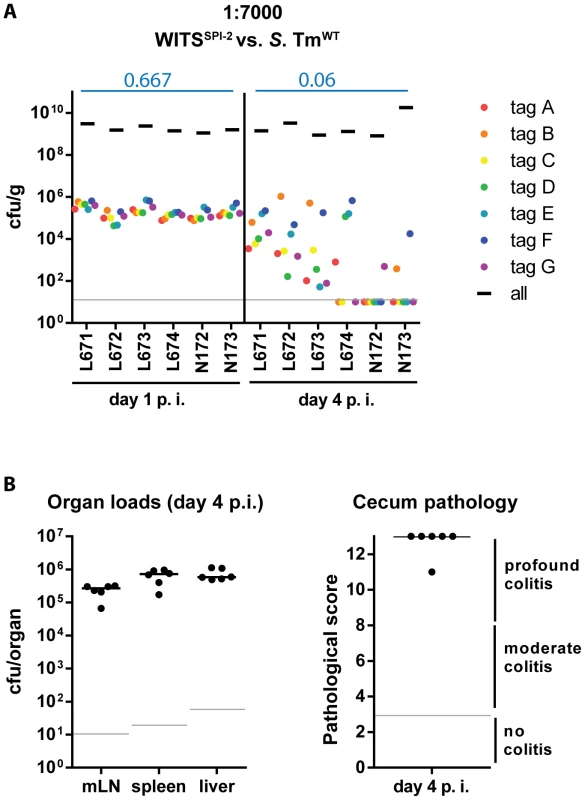
(A) To exclude a strain-intrinsic property of the SPI-2 WITS to be responsible for the decrease in WITS diversity loss observed in the S. TmSPI-2 background, we tested a chimeric situation in which inflammation was caused by S. TmWT but WITS-tags were in a SPI-2 deficient background. Experiments were performed using a WITS cocktail in a dilution of 1∶7000. Grey lines depict the detection limit for plating (10 cfu/g). Blue numbers indicate the median of the evenness indices of single mice. (B) Organ loads and cecum pathology at day 4 p.i. confirmed the “WT-like” severity of the infection. The size of the cecum luminal S. Tm population is diminished at day 2 post infection
Reduced WITS-evenness can be explained by a drastic reduction in the cecal S. Tm population size and a subsequent (oligo-clonal) regrowth of the population. In fact, transient reductions of the cecal and fecal population densities at day 2 p.i. have been observed repeatedly (though never followed up in detail), over the past decade ([20]; Fig. 2A,B). To specifically analyze the population size at this critical time point, we monitored changes in cecum luminal S. Tm population size by sacrificing S. TmWT-infected mice at day 1 and at day 2 p. i. and by determining the respective pathogen loads. In line with earlier data, the mice featured moderate colitis by day 1 and pronounced colitis by day 2 p.i. (Fig. 5B). The pathogen densities in the cecum lumen were high at day 1 p.i. (≈109 cfu/g), but dropped significantly by day 2 (median = 3×107 cfu/g, Fig. 5A). Strikingly, the gut luminal pathogen density featured pronounced animal-to-animal variation. In some animals, the pathogen density dropped down to 3×105 cfu/g. Without further information, these data cannot tell whether the dip in the gut luminal pathogen density is more pronounced in some animals than in others. Alternatively, equivalent dips may occur in all mice however with slightly different kinetics. Nevertheless, the data suggests that a transient dip in the cecum luminal population size may contribute to the drop in WITS evenness at least in some of the mice.
Fig. 5. Decrease in cecal population size between day 1 and day 2 p.i. only occurs during S. TmWT induced inflammation, but not during inflammation in a SPI-2 deficient background. 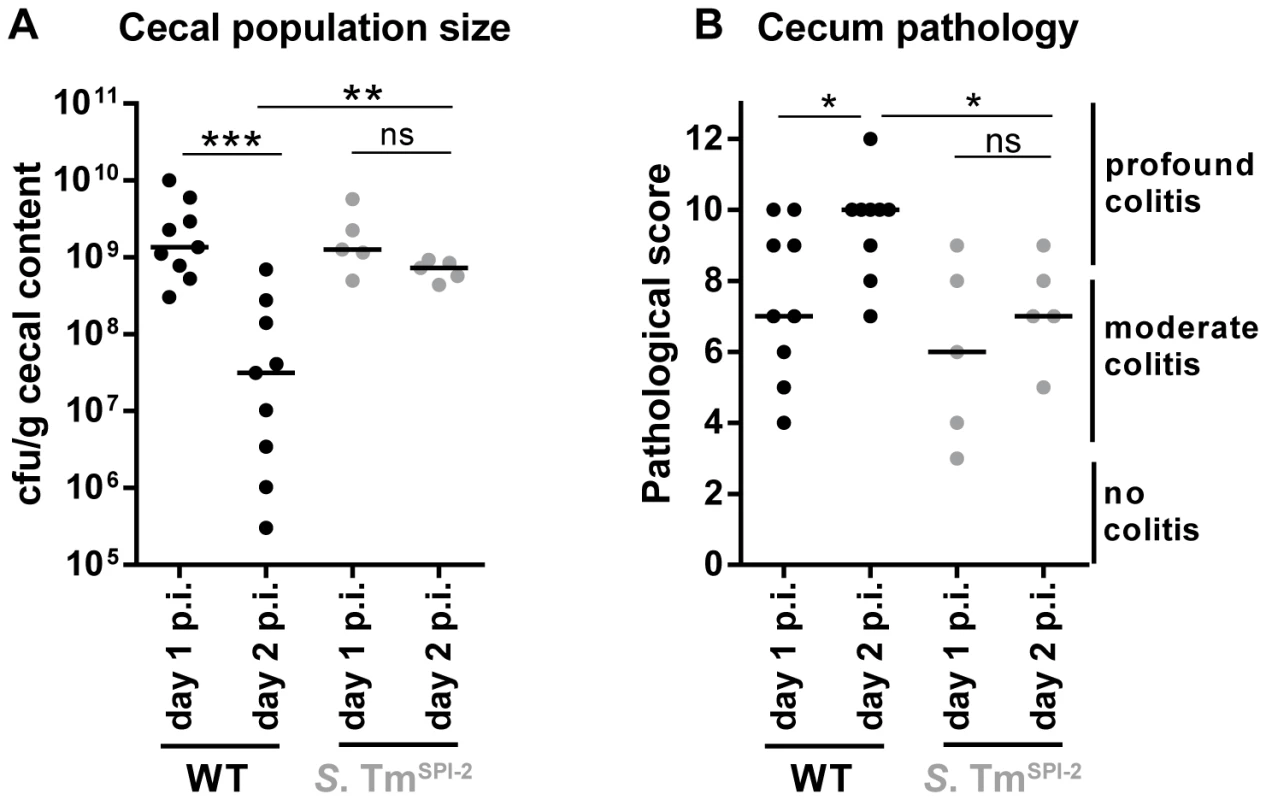
Streptomycin-pretreated C57BL/6 mice were orally infected with S. TmWT or S. TmSPI-2, respectively. Mice were sacrificed at day 1 or day 2 p.i. and bacterial population size (A) and cecum pathology at the day of sacrifice (B) were plotted. ns = not significant (P≥0.05), * P<0.05, ** P<0.01, *** P<0.001; Mann-Whitney U test. If pathogen-triggered inflammation was responsible for the dip at day 2 p.i., cecal S. Tm densities should remain higher in cases of reduced mucosal inflammation. To test this hypothesis, we infected mice with S. TmSPI-2 and compared cecal population sizes at day 1 and day 2 p. i. Indeed, under these conditions cecal S. Tm loads remained equally high from day 1 to day 2 (Fig. 5).
Taken together, our observations suggest that S. TmWT triggered inflammation can inflict a transient population bottleneck at day 2 p. i. and that this contributes at least in part to the reduced evenness index, observed in our WITS experiments.
Depletion of Gr1+-cells alleviates the bottleneck
Thus far, our data suggested that the grade of the mucosal inflammation is a key determinant of the bottleneck at day 2 p.i. However, the mechanisms explaining this barrier to infection remained to be identified. One hallmark of S. Tm induced inflammation is the tissue infiltration by granulocytes, their transmigration into the gut lumen and the formation of characteristic crypt abscesses [15]. As granulocytes can attack bacteria by bactericidal mechanisms such as phagocytosis, release of antimicrobial substances and formation of neutrophil extracellular traps (NETs) [43], they might contribute to the gut luminal bottleneck. However, to the best of our knowledge, it has not been assessed previously, whether granulocytes can have bactericidal activity upon transmigration into the gut lumen.
To test if granulocyte-mediated killing contributes to the gut luminal bottleneck, we employed a cell depletion strategy. Initially, we depleted granulocytes during S. TmWT infection using an α-Gr1 antibody (100 µg i.p., once per day; Materials and Methods; Fig. 6A) which binds to Ly6G on neutrophilic granulocytes (PMN) and to Ly6C, a marker expressed on neutrophils and on dendritic cells, some subpopulations of lymphocytes and some monocytes [44]. For this experiment, we used 129SvEv mice (Nramp/slc11α1+/+) as their monocytes can control S. Tm growth at systemic sites much more efficiently than those from slc11α1−/− C57BL/6 mice [45], [46]. This allowed infecting Gr1+-depleted mice for 4 days without compromising the viability of the animal (Fig. 6A).
Fig. 6. Depletion of Gr1+- cells reduces WITS diversity loss and prevents the cecal population crash between day 1 and day 2 p.i. 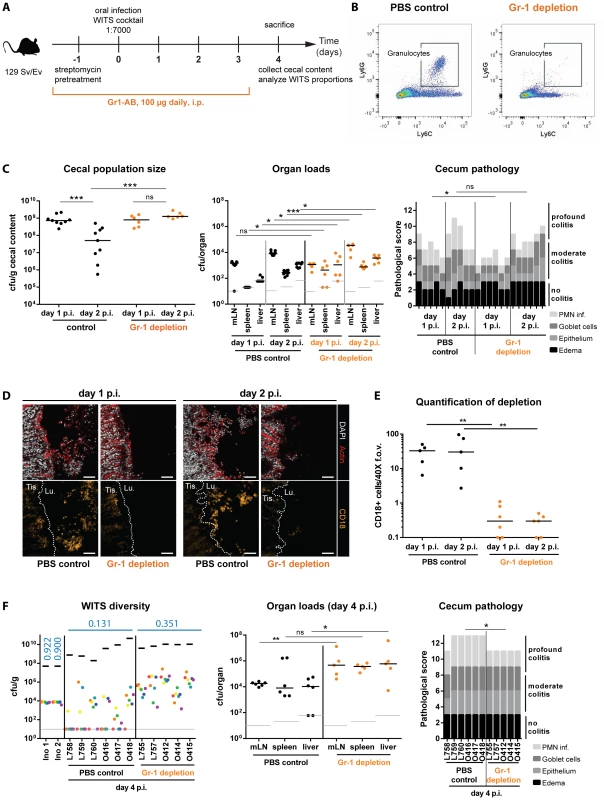
(A) Schematic overview of the experimental setup. Starting with streptomycin-pretreatment one day prior to infection with the S. TmWT WITS cocktail in a dilution of 1∶7000, Gr1+-cells (mostly neutrophilic granulocytes) were depleted by daily i.p. injection of 100 µg of anti-mouse-Gr1-antibody. The control group was injected with an equal volume of PBS. (B) Successful reduction in Ly6C+ Ly6G+ cells in the murine blood was verified daily using a flow cytometry assay. A representative blot is given in panel B. (C) The cecal S. Tm population size was monitored in the Gr1+-cell depleted mice and the control group and organ loads and cecum pathology of respective animals were assessed. (D) CD18 immunofluorescent staining of cecal section from Gr1+-cell depleted mice and PBS treated control groups confirmed reduction of granulocytes in the course of the experiment. Scale bars: 50 µm (E) Quantification of luminal granulocytes from tissue sections. (F) At day 4 p.i. animals were sacrificed and the population composition in cecal contents were analyzed by rtqPCR. Organ pathogen loads and cecum pathology at day 4 p.i. indicate systemic pathogen spread and high intestinal inflammation. ns = not significant (P≥0.05), * P<0.05, ** P<0.01, *** P<0.001, Mann-Whitney U test. Grey lines indicate detection limits. Blue numbers indicate the median of the evenness indices of single mice. First we analyzed the effect of granulocyte depletion on the cecum luminal pathogen density at day 2 p.i. Gr1+-cell-depleted mice, and PBS-treated control animals were pretreated with streptomycin and infected for 1 or 2 days with S. TmWT. The efficiency of granulocyte depletion was verified by flow cytometry (CD45+, Ly6G+, Ly6C+ cells; Fig. 6B) of murine blood. The detailed effect of Gr-1 neutralization on inflammatory myeloids in the cecal mucosa of 129SvEv mice revealed that the procedure depleted not only granulocytes, but also other phagocyte populations (S5 Figure, S1 Text). Pathogen loads in the mLN, the spleens and the livers did rise in both groups by 10–100 fold between day 1 and day 2 p.i. We detected (at most) slightly elevated tissue loads in the Gr1+-depleted mice, while both groups featured equivalent S. TmWT loads in the cecal lumen at day 1 p.i. (Fig. 6C). Strikingly, the cecum luminal pathogen loads remained high at day 2 p.i. in the Gr1+-depleted mice, but plummeted in the PBS controls. This went along with reduced gut luminal granulocyte (CD18+) numbers in the Gr1+-depleted mice (Fig. 6D,E) and suggested that granulocytes may indeed affect the bottleneck.
A second experiment was performed to assess effects on the WITS evenness index. Gr1+-depleted mice, and PBS-treated control animals were pretreated with streptomycin and infected for 4 days with a “1∶7000” inoculum of S. TmWT WITS as described for Fig. 2. In line with earlier work, the Gr1+-depleted mice featured increased organ loads in the mLN, the spleens and the livers [47]. This verified that granulocytes indeed significantly limit systemic infection. In contrast, cecum luminal total S. TmWT densities did not differ between both groups (Fig. 6F, left panel, black bars). Strikingly, the WITS evenness index remained much higher in the Gr1+-depleted mice, as compared to the PBS controls (Fig. 6F, Table 1). In a complementary, yet more specific strategy, we applied a combination of α-Ly6G and α-G-CSF antibodies to selectively reduce granulocytes but spare monocytes from depletion [48], [49]. Again, the depleted animals featured higher WITS evenness indices than the control group treated with the respective isotype controls of both antibodies (S6 Figure, S1 Text). These data supported our hypothesis that Gr1+ cells (mainly granulocytes) form a major bottleneck for the gut luminal S. Tm population by day 2 p.i. However, these results raised the question whether this outcome is a direct effect of the bactericidal activity of granulocytes (e.g. NO/ROS production) or an indirect effect (e.g. reduction of granulocyte-released cytokines or antimicrobial proteins). First, we examined WITS diversity loss in Nos2-deficient mice to clarify the requirement of inflammation-mediated NO production in reducing luminal S. Tm. As Nos2 was shown to be dispensable for controlling S. Tm during the initial days of infection [50], we hypothesized that the Nos2-deficient mice should restrict gut luminal S. Typhimurium loads as efficiently as the wild type animals. In line with this, the WITS evenness indices in Nos2−/− mice and the littermate controls did not differ at day 3 p.i. (S7A Figure, S2 Table) suggesting that NO is not the predominant cause for the WITS diversity loss.
Next, we tested the contribution of oxidative burst in the reduction of WITS evenness. As NADPH-oxidase is a key antimicrobial factor of granulocytes [43] at systemic sites and in the intestinal tissue [51] and as these cells do transmigrate in large numbers into the lumen of the inflamed gut, we speculated that Cybb−/− mice might be incapable of restricting gut luminal pathogen loads in the inflamed gut. Strikingly, WITS evenness in the NADPH-oxidase deficient (Cybb−/−) mice at day 3 p.i. was increased compared to a heterozygous control group (S8A Figure, S2 Table) pointing to a possible involvement of direct ROS-mediated killing to the bottleneck effect. However, interpretation of WITS analysis in the Cybb−/− mice is limited by a high S. Tm colonization at systemic sites up to levels where WITS-tagged strains are found systemically (S8B Figure). As possible re-seeding events of these strains into the gut lumen may compromise the unequivocal interpretation of our WITS-diversity analysis, these results should be taken with caution.
Taken together, the gut luminal S. Tm population bottleneck is mostly caused by granulocytes, possibly via a direct ROS-mediated killing mechanism. However, our data do not exclude the contribution of additional factors to this bottleneck effect.
Discussion
To identify barriers limiting S. Tm in the gut lumen, we have performed mixed inoculation experiments. During the first day, the pathogen density rose to ≈109 cfu/g in the large intestinal lumen without encompassing obvious bottlenecks. In the case of wild type S. Tm infection, cecum colonization was accompanied by pronounced mucosal inflammation which arises during the first day (8–12 h p.i.; [15], [52]) and lasts for four days (or longer; [46], [53]). By day 2 p.i., the inflammatory response dramatically reduces the gut luminal pathogen density. This inflammation-inflicted, granulocyte-dependent bottleneck transiently restricts the gut luminal S. Tm population to about 6000 cfu. By days 3 and 4, the pathogen density rises again to ≈109 cfu/g of gut content. These data establish that mucosal inflammation can represent a barrier limiting the gut luminal colonization by the pathogen.
Our findings significantly extend earlier work on S. Tm growth in the lumen of the inflamed gut. That earlier work had indicated that the pathogen subverts the inflammatory milieu in the gut lumen for its own advantage, i.e. for outcompeting the resident microbiota (reviewed in [15], [54], [55]). Granulocytes infiltrating the inflamed mucosa and transmigrating into the gut lumen are thought to contribute in several different ways i.e., by i) providing/regenerating terminal electron acceptors (tetrathionate, nitrate) used by S. Tm, thereby fuelling anaerobic respiration and fast growth of the pathogen (not the commensals, which are mainly fermenters [24], [25]); by ii) releasing lipocalin-2, an antimicrobial peptide blocking siderophore-mediated iron acquisition by E. coli. Interestingly, S. Tm is endowed with the iroBCDEN variant of such system which can bypass the lipocalin-2-mediated restriction [23]; by iii) releasing calprotectin which sequesters Zn2+, an essential micronutrient, from the gut lumen. This restricts microbiota growth, while S. Tm can bypass this restriction using a high affinity zinc transporter (znuABC; [22]). Furthermore, S. Tm is endowed with enzymes inactivating PMN-derived reactive oxygen and nitrogen compounds [56]–[58]. Overall, these earlier findings indicated that granulocyte activity (and mucosa inflammation in general) foster pathogen growth in the intestine and prevent growth of the resident microbiota. However, the quantitative contribution of inflammation-mediated fuelling versus restriction of S. Tm growth in the gut lumen had remained unclear. Our findings demonstrate that the gut luminal S. Tm population is highly vulnerable to the inflammatory response in particular at day 2 p.i. We conclude that the size of the pathogen population in the inflamed gut is controlled by a fine balance between inflammation-fuelled pathogen growth (and competition against resident microbiota) and inflammation/granulocyte-inflicted losses to the pathogen population.
How many S. Tm cells survive the bottleneck in the gut lumen? Our best estimate is that 6000+/−2000 bacteria survive this dip at day 2. This value is obtained from a simple binomial selection model fitted to the data on the detectability (presence/absence) of WITS at day 4 for the 1∶7000 dilution. This model assumes that there are no fitness differences between WITS and that the population bottleneck is effective for only a short time. The classical cfu-plating revealed reduced, but highly variable pathogen population sizes at day 2 p.i. (median = 2×106 cfu/g). In a few mice, we detected as few as 3×105 cfu/g of cecum content by day 2 p.i. This corresponds to a total population size of 15000 bacterial cells, considering that the infected cecum (tissue+lumen) has a total volume of no more than 200 µl, and that the cecal luminal volume might range at approx. 50 µl at this stage. The high variance of the cfu-data measured at day 2 p.i. and the similarity of the evenness-indices across animals suggest that the pathogen population may be reduced for no more than a few hours before net re-growth occurs. Using 24 h sampling intervals, classical cfu-plating would therefore miss the low in most mice, in particular if the time-course would differ from animal to animal. Therefore, we conclude that the bottleneck allows the survival of just 6000 bacteria. Thus, the mucosal inflammation is a strikingly efficient host defense, reducing the total cecum luminal population by about 105–106 fold (108 cfu per cecum at day 1 p.i. to 6000 bacteria by day 2). This is almost as efficient as chemical disinfectants (defined as reducing bacterial survival by ≥106-fold; [59]) and far more effective than many other barriers, e.g. the IFNγ or inflammasome mediated restriction of systemic spread (≈10-fold restriction by day 2 p.i.; [60], [61]) or the gut luminal growth defect inflicted by adaptive sIgA response directed against the LPS O-sidechain (≈10-fold; [53], [62]) limiting S. Tm loads at different stages of an infection. In conclusion, the mucosal inflammation severely reduces the gut luminal S. Tm population by day 2 p.i. However, this is not quite sufficient to yield sterilizing protection and to prevent subsequent pathogen-regrowth.
In this work, the importance of granulocytes in controlling S. Tm within the gut lumen is re-emphasized and extended. Already in the early studies of streptomycin pretreated mice, neutrophil infiltration was detected in the cecal mucosa by 10 h p.i. [63]. Similarly, granulocytes are prominent in the infected mucosa of cows and man [64], [65], they transmigrate into the gut lumen and form the archetypical “crypt abscesses”, i.e. densely packed clusters of granulocytes located within the intestinal crypts [66]–[69]. By extension, these observations suggest that granulocyte-mediated growth and restriction might be of general importance also in other hosts including the human patient.
During the first day of infection, granulocytes might also serve as a host cell permissive for intracellular growth (or at least survival) of the pathogen. In the streptomycin mouse model, gut luminal neutrophils harbored significant numbers of S. Tm by 20 hours p.i. and most of these bacterial cells were viable, as judged from the expression of iroBCDE - and ttss-2 driven GFP reporters [70]. Other studies have also indicated that S. Tm can survive in murine neutrophils [70]–[72]. The lack of a bottleneck during the first day of infection would be in line with this. Our data suggest that the barrier detected by day 2 p.i. is attributable to some quantitative or qualitative change in the inflammatory response and/or the granulocyte activity, e.g. by modulating NADPH-oxidase dependent defenses. Identifying the underlying mechanism controlling the granulocyte-inflicted bottleneck will be of interest for future work and might identify strategies for bolstering the gut luminal barrier. Still, our data suggest that pronounced granulocyte migration into the gut lumen can have detrimental effects on the S. Tm population, at least by day 2 p.i. Furthermore, our data provide first hints that ROS might play a central role in decimating luminal S. Tm, despite of the low oxygen-availability in the gut lumen and the fact that S. Tm expresses three catalases and three periplasmic Cu,Zn superoxide dismutase enzymes [73]. Thus, the interaction of S. Tm with granulocytes in the gut lumen is clearly important, but more complex than previously anticipated.
In summary, investigating mixed inocula allows precious insights into colonization strategies and host defenses limiting infection. The population bottlenecks identified by such approaches may represent attractive points for therapeutic interventions, as pathogen populations are already reduced to a minimum.
Materials and Methods
Bacterial strains
The wild type strain SB300 is a clone of S. Tm SL1344 and the SPI-1 and SPI-2 deficient mutant strains (S. TmWT, S. TmSPI-1, S. TmSPI-2), as well as their isogenic tagged strains have been described previously [5], [6], [34], [35], [74]. WITS-tags were introduced into S. TmSPI-1 & SPI-2 (SL1344 invGssaV; [17] and into S. TmSPI-2 (SL1344 ssaV::cat; [35]) by P22 phage transduction and subsequent selection on kanamycin. The presence of the correct WITS-tag was confirmed by PCR using tag-specific primers (see S1 Table).
Mouse lines
Wild type C57BL/6 mice were bred and kept under specified pathogen free (SPF) conditions in individually ventilated cages at the Rodent Center RCHCI (ETH Zurich). They harbor a complex microbiota and are called “conventional” mice. For reliable and efficient infection with S. Tm, streptomycin pretreatment of these mice was performed in order to overcome colonization resistance in all experiments described herein [16]. 129 Sv/Ev mice are Nramp1(+/+) (Slc11α1(+/+)) mice which can control systemic S. Tm infection with low bacterial loads at systemic sites but develop acute S. Tm colitis upon streptomycin pretreatment [46]. 129 Sv/Ev mice are specified pathogen free animals with a complex microbiota. Cybb−/− (B6.129S-Cybbtm1Din/J; C57BL/6 background) and Nos2−/− (B6.129P2-Nos2tm1Lau/J; C57BL/6 background) were bred at the Rodent Center HCI (ETH Zurich, Switzerland). Both mouse lines have been described previously [75], [76].
Animal infection experiments
Animal infection experiments were performed in 8 to 12 week old mice as described previously [16]: C57BL/6 mice or 129 Sv/Ev mice were pretreated with 20 mg streptomycin 24 hours prior to infection. For infection, bacteria were grown for 12 h in 0.3 M NaCl supplemented LB medium containing the appropriate antibiotic(s), diluted 1∶20 and sub-cultured for 4 h in the same medium without supplementation of antibiotics. Tagged and un-tagged strains were mixed as indicated, washed twice with PBS and mice were infected with 5×107 bacteria by gavage. Each animal was kept in a single cage to avoid transmission between mice. Animals were sacrificed at day 4 p.i. by cervical dislocation. Freshly collected fecal pellets or whole cecal content were harvested, homogenized in 500 µl PBS with steel balls in a Tissue Lyser (Qiagen) for plating to determine the total population size. 250 µl of homogenized feces or cecal content were used to inoculate an over night culture in LB supplemented with 50 µg/ml kanamycin to select for tagged strains. Bacteria from 1 ml of this over night culture were harvested and frozen for genomic DNA extraction. HE-staining of cryo-embedded tissues and subsequent pathoscoring for granulocyte infiltration was performed as described previously [16]. To test for selection of beneficial mutations which might explain the variation in the subpopulations at day 4 p.i., we re-isolated dominant WITS-tagged bacteria from cryo-embedded tissues by plating on kanamycin. Untagged strains were isolated in the same way, but were selected for growth on streptomycin and remained kanamycin sensitive. Competitive infection experiments to test for increased fitness of re-isolated strains were performed by infecting streptomycin-pretreated mice with equal amounts of S. TmWT or a WITS-tagged S. TmWT strain and the isolates. By selective plating, population sizes of both were determined and a competitive index was calculated by dividing the ratio of the isolate to the background strains by the ratio of both strains in the inoculum.
WITS quantification
Genomic DNA from enrichment cultures was isolated with the QIAamp DNA Mini Kit (Qiagen, Cat. NO. 51306). rtqPCR analysis with FastStart Universal SYBR Green Master (Roche) was performed using primers and temperature profiles as described previously [5]. The population size of each tagged strain was calculated by multiplying the number of kanamycin resistant cfu/g recovered S. Tm with the ratio of WITS determined by rtqPCR.
Depletion of Gr-1+-cells
The monoclonal anti-Gr1 antibody NIMP-R14 is highly specific for the murine epitopes Ly6G and Ly6C and was shown to selectively deplete mouse neutrophils [77]. For depletion of Gr-1+-cells during S. TmWT infection, we injected 100 µg of anti-Gr1 i.p. on a daily basis, starting 24 h prior to infection. Granulocyte depletion was assessed daily by analysis of peripheral blood (and lamina propria after sacrificing the mice) in a flow cytometry assay using the following staining antibodies: anti-CD11b-PECy7 (clone M1/70), anti-Ly6C-FITC (clone AL-21), anti-Ly6G-450V (clone 1A8) and anti-CD45.2-APC (clone 104). NIMP-R14 hybridoma was a kind gift of Dr. Nancy Hogg (Cancer Research UK, London, U.K.) and antibodies for flow cytometry were purchased from BioLegend.
Immunofluorescence microsopy of fixed tissue
Cecal tissues were recovered, fixed for 5 h in 4% paraformaldehyde/4% sucrose, saturated in PBS/20% sucrose (overnight, 4°C), embedded in OCT medium (Sakura, Torrance, CA), snap-frozen in liquid nitrogen and stored at −80°C. Cryosections (20 µm) were air-dried, rehydrated with PBS and permeabilized with PBS/0.5% Triton X-100. Unspecific binding was blocked with PBS/10% Normal Goat Serum. The tissue sections were immunostained using rat-anti-mouse-CD18 (clone M18/2, Biolegend, (1∶300)). Goat-anti-rat-Cy3 (112-165-167, Jackson, 1∶200) was used as a secondary antibody. DNA was stained with DAPI (SIGMA Aldrich) and F-actin was stained using AlexaFluor647-conjugated phalloidin (Molecular probes). Sections were subsequently mounted with Mowiol (Calbiochem). Imaging was performed using a Zeiss Axiovert 200 m microscope equipped with 10×–100× objectives, a spinning disc confocal laser unit (Visitron) and parallel Evolve 512 EMCCD cameras (Photometrics). CD18+ cells were enumerated in 40× field of vision in several non-consecutive sections per mouse in a blind fashion and the average number of CD18+ cells were compared between experimental groups.
Statistical analysis
The exact Mann-Whitney U test was performed using the software Graphpad Prism Version 6.0 for Windows (GraphPad Software, La Jolla California USA, www.graphpad.com). P values of less than 0.05 (two-tailed) were considered as statistically significant. * P<0.05, ** P<0.01, *** P<0.001.
Evenness indices were calculated as described [31]. In cases where none of the seven WITS tagged strains were detectable any more, no evenness index was applicable and the corresponding mice were ignored for determination of the median evenness and the comparison between evenness indices in Table 1.
It should be noted that the 1∶7 inoculum also yielded a somewhat reduced evenness by day 4 p.i. (median = 0.528). This cannot be explained by technical error when using our simple population model and may suggest that a second, so far unidentified process may affect the gut luminal S. Tm population. However, this effect was much smaller than the WITS-diversity loss observed with the 1∶7000 inoculum. In the present paper, we have decided to focus on the latter phenomenon.
Estimation of the bottleneck size
We estimated the bottleneck size using the data on WITS loss after their inoculation at the lowest dilution of 1/7000. Let w be the number of WITS, which, in our experiments, was always w = 7. Further, let m be the number of mice in a given treatment group. Then we have a total of mw WITS in that treatment group. Lastly, let x denote the number of WITS that cannot be recovered across all mice. In the control group for example, 30 of the 70 WITS in the 10 mice were undetectable at day 4, and we thus have w = 7, m = 10, x = 30.
The procedure to estimate the bottleneck size is based on the simplest possible model of this process: binomial selection. We assume that during the bottleneck, B bacterial cells are selected. B is the bottleneck size, i.e. the number of bacteria that are able to traverse the bottleneck. The probability of selecting a certain WITS is determined by its dilution, d = 1/7000. Not selecting this WITS happens with a probability of p0 = (1−d)B. We can construct a log-likelihood for loosing x out of mw WITS by bottlenecking: ℓ = xlnp0+(mw−x)ln(1−p0). Maximizing this log-likelihood yields the maximum likelihood estimates for the bottleneck size listed in Table 2. To calculate confidence intervals for the estimates we used the profile likelihood [78]. If all or no WITS are lost in a treatment group, only upper or lower bounds of the bottleneck size can be calculated, respectively.
Ethical statement
All animal experiments were reviewed and approved by the Kantonales Veterinäramt, Zürich (license 223/2010) and are subject to the Swiss animal protection law (TschG).
Supporting Information
Zdroje
1. MastroeniP, GrantA, RestifO, MaskellD (2009) A dynamic view of the spread and intracellular distribution of Salmonella enterica. Nat Rev Microbiol 7 : 73–80.
2. BaccamP, BeaucheminC, MackenCA, HaydenFG, PerelsonAS (2006) Kinetics of influenza A virus infection in humans. J Virol 80 : 7590–7599.
3. BarnesPD, BergmanMA, MecsasJ, IsbergRR (2006) Yersinia pseudotuberculosis disseminates directly from a replicating bacterial pool in the intestine. J Exp Med 203 : 1591–1601.
4. BullRA, LucianiF, McElroyK, GaudieriS, PhamST, et al. (2011) Sequential bottlenecks drive viral evolution in early acute hepatitis C virus infection. PLoS Pathog 7: e1002243.
5. GrantAJ, RestifO, McKinleyTJ, SheppardM, MaskellDJ, et al. (2008) Modelling within-host spatiotemporal dynamics of invasive bacterial disease. PLoS Biol 6: e74.
6. KaiserP, SlackE, GrantAJ, HardtWD, RegoesRR (2013) Lymph node colonization dynamics after oral salmonella typhimurium infection in mice. PLoS Pathog 9: e1003532.
7. LayMD, PetravicJ, GordonSN, EngramJ, SilvestriG, et al. (2009) Is the gut the major source of virus in early simian immunodeficiency virus infection? J Virol 83 : 7517–7523.
8. LevinBR, LipsitchM, BonhoefferS (1999) Population biology, evolution, and infectious disease: convergence and synthesis. Science 283 : 806–809.
9. LimCH, VoedischS, WahlB, RoufSF, GeffersR, et al. (2014) Independent bottlenecks characterize colonization of systemic compartments and gut lymphoid tissue by salmonella. PLoS Pathog 10: e1004270.
10. MargolisE, LevinBR (2007) Within-host evolution for the invasiveness of commensal bacteria: an experimental study of bacteremias resulting from Haemophilus influenzae nasal carriage. J Infect Dis 196 : 1068–1075.
11. MeynellGG (1957) The applicability of the hypothesis of independent action to fatal infections in mice given Salmonella typhimurium by mouth. J Gen Microbiol 16 : 396–404.
12. MeynellGG, StockerBA (1957) Some hypotheses on the aetiology of fatal infections in partially resistant hosts and their application to mice challenged with Salmonella paratyphi-B or Salmonella typhimurium by intraperitoneal injection. J Gen Microbiol 16 : 38–58.
13. MoxonER, MurphyPA (1978) Haemophilus influenzae bacteremia and meningitis resulting from survival of a single organism. Proc Natl Acad Sci U S A 75 : 1534–1536.
14. JohanssonME, SjovallH, HanssonGC (2013) The gastrointestinal mucus system in health and disease. Nat Rev Gastroenterol Hepatol 10 : 352–361.
15. KaiserP, DiardM, StecherB, HardtWD (2012) The streptomycin mouse model for Salmonella diarrhea: functional analysis of the microbiota, the pathogen's virulence factors, and the host's mucosal immune response. Immunol Rev 245 : 56–83.
16. BarthelM, HapfelmeierS, Quintanilla-MartinezL, KremerM, RohdeM, et al. (2003) Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect Immun 71 : 2839–2858.
17. MaierL, VyasR, CordovaCD, LindsayH, SchmidtTS, et al. (2013) Microbiota-derived hydrogen fuels salmonella typhimurium invasion of the gut ecosystem. Cell Host Microbe 14 : 641–651.
18. CoburnB, LiY, OwenD, VallanceBA, FinlayBB (2005) Salmonella enterica serovar Typhimurium pathogenicity island 2 is necessary for complete virulence in a mouse model of infectious enterocolitis. Infect Immun 73 : 3219–3227.
19. HapfelmeierS, StecherB, BarthelM, KremerM, MullerAJ, et al. (2005) The Salmonella pathogenicity island (SPI)-2 and SPI-1 type III secretion systems allow Salmonella serovar typhimurium to trigger colitis via MyD88-dependent and MyD88-independent mechanisms. J Immunol 174 : 1675–1685.
20. StecherB, RobbianiR, WalkerAW, WestendorfAM, BarthelM, et al. (2007) Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol 5 : 2177–2189.
21. StelterC, KappeliR, KonigC, KrahA, HardtWD, et al. (2011) Salmonella-induced mucosal lectin RegIIIbeta kills competing gut microbiota. PLoS One 6: e20749.
22. LiuJZ, JellbauerS, PoeAJ, TonV, PesciaroliM, et al. (2012) Zinc sequestration by the neutrophil protein calprotectin enhances Salmonella growth in the inflamed gut. Cell Host Microbe 11 : 227–239.
23. RaffatelluM, GeorgeMD, AkiyamaY, HornsbyMJ, NuccioSP, et al. (2009) Lipocalin-2 resistance confers an advantage to Salmonella enterica serotype Typhimurium for growth and survival in the inflamed intestine. Cell Host Microbe 5 : 476–486.
24. LopezCA, WinterSE, Rivera-ChavezF, XavierMN, PoonV, et al. (2012) Phage-mediated acquisition of a type III secreted effector protein boosts growth of salmonella by nitrate respiration. MBio 3.
25. WinterSE, ThiennimitrP, WinterMG, ButlerBP, HusebyDL, et al. (2010) Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467 : 426–429.
26. MedzhitovR (2007) Recognition of microorganisms and activation of the immune response. Nature 449 : 819–826.
27. BardoelBW, KennyEF, SollbergerG, ZychlinskyA (2014) The Balancing Act of Neutrophils. Cell Host Microbe 15 : 526–536.
28. MikiT, HardtWD (2013) Outer membrane permeabilization is an essential step in the killing of gram-negative bacteria by the lectin RegIIIbeta. PLoS One 8: e69901.
29. KaiserP, RegoesRR, DolowschiakT, WotzkaSY, LengefeldJ, et al. (2014) Cecum lymph node dendritic cells harbor slow-growing bacteria phenotypically tolerant to antibiotic treatment. PLoS Biol 12: e1001793.
30. PeriaswamyB, MaierL, VishwakarmaV, SlackE, KremerM, et al. (2012) Live attenuated S. Typhimurium vaccine with improved safety in immuno-compromised mice. PLoS One 7: e45433.
31. Gini C (1912) Variabilità e Mutuabilità. Contributo allo Studio delle Distribuzioni e delle Relazioni Statistiche. C Cuppini, Bologna.
32. SmithJM, HaighJ (2007) The hitch-hiking effect of a favourable gene. Genet Res 89 : 391–403.
33. Barroso-BatistaJ, SousaA, LourencoM, BergmanML, SobralD, et al. (2014) The first steps of adaptation of Escherichia coli to the gut are dominated by soft sweeps. PLoS Genet 10: e1004182.
34. KanigaK, BossioJC, GalanJE (1994) The Salmonella typhimurium invasion genes invF and invG encode homologues of the AraC and PulD family of proteins. Mol Microbiol 13 : 555–568.
35. SuarM, PeriaswamyB, SonghetP, MisselwitzB, MullerA, et al. (2009) Accelerated type III secretion system 2-dependent enteropathogenesis by a Salmonella enterica serovar enteritidis PT4/6 strain. Infect Immun 77 : 3569–3577.
36. HapfelmeierS, MullerAJ, StecherB, KaiserP, BarthelM, et al. (2008) Microbe sampling by mucosal dendritic cells is a discrete, MyD88-independent step in DeltainvG S. Typhimurium colitis. J Exp Med 205 : 437–450.
37. HenselM, SheaJE, GleesonC, JonesMD, DaltonE, et al. (1995) Simultaneous identification of bacterial virulence genes by negative selection. Science 269 : 400–403.
38. AckermannM, StecherB, FreedNE, SonghetP, HardtWD, et al. (2008) Self-destructive cooperation mediated by phenotypic noise. Nature 454 : 987–990.
39. DiardM, GarciaV, MaierL, Remus-EmsermannMN, RegoesRR, et al. (2013) Stabilization of cooperative virulence by the expression of an avirulent phenotype. Nature 494 : 353–356.
40. IlgK, EndtK, MisselwitzB, StecherB, AebiM, et al. (2009) O-antigen-negative Salmonella enterica serovar Typhimurium is attenuated in intestinal colonization but elicits colitis in streptomycin-treated mice. Infect Immun 77 : 2568–2575.
41. StecherB, BarthelM, SchlumbergerMC, HaberliL, RabschW, et al. (2008) Motility allows S. Typhimurium to benefit from the mucosal defence. Cell Microbiol 10 : 1166–1180.
42. StecherB, DenzlerR, MaierL, BernetF, SandersMJ, et al. (2012) Gut inflammation can boost horizontal gene transfer between pathogenic and commensal Enterobacteriaceae. Proc Natl Acad Sci U S A 109 : 1269–1274.
43. AmulicB, CazaletC, HayesGL, MetzlerKD, ZychlinskyA (2012) Neutrophil function: from mechanisms to disease. Annu Rev Immunol 30 : 459–489.
44. DaleyJM, ThomayAA, ConnollyMD, ReichnerJS, AlbinaJE (2008) Use of Ly6G-specific monoclonal antibody to deplete neutrophils in mice. J Leukoc Biol 83 : 64–70.
45. MonackDM, BouleyDM, FalkowS (2004) Salmonella typhimurium persists within macrophages in the mesenteric lymph nodes of chronically infected Nramp1+/+ mice and can be reactivated by IFNgamma neutralization. J Exp Med 199 : 231–241.
46. StecherB, PaesoldG, BarthelM, KremerM, JantschJ, et al. (2006) Chronic Salmonella enterica serovar Typhimurium-induced colitis and cholangitis in streptomycin-pretreated Nramp1+/+ mice. Infect Immun 74 : 5047–5057.
47. ConlanJW (1996) Neutrophils prevent extracellular colonization of the liver microvasculature by Salmonella typhimurium. Infect Immun 64 : 1043–1047.
48. Trautwein-WeidnerK, GladiatorA, NurS, DiethelmP, LeibundGut-LandmannS (2014) IL-17-mediated antifungal defense in the oral mucosa is independent of neutrophils. Mucosal Immunol
49. WengnerAM, PitchfordSC, FurzeRC, RankinSM (2008) The coordinated action of G-CSF and ELR+CXC chemokines in neutrophil mobilization during acute inflammation. Blood 111 : 42–49.
50. MastroeniP, Vazquez-TorresA, FangFC, XuY, KhanS, et al. (2000) Antimicrobial actions of the NADPH phagocyte oxidase and inducible nitric oxide synthase in experimental salmonellosis. II. Effects on microbial proliferation and host survival in vivo. J Exp Med 192 : 237–248.
51. FelmyB, SonghetP, SlackEM, MullerAJ, KremerM, et al. (2013) NADPH oxidase deficient mice develop colitis and bacteremia upon infection with normally avirulent, TTSS-1 - and TTSS-2-deficient Salmonella Typhimurium. PLoS One 8: e77204.
52. SellinME, MullerAA, FelmyB, DolowschiakT, DiardM, et al. (2014) Epithelium-intrinsic NAIP/NLRC4 inflammasome drives infected enterocyte expulsion to restrict Salmonella replication in the intestinal mucosa. Cell Host Microbe 16 : 237–248.
53. EndtK, StecherB, ChaffronS, SlackE, TchitchekN, et al. (2010) The microbiota mediates pathogen clearance from the gut lumen after non-typhoidal Salmonella diarrhea. PLoS Pathog 6: e1001097.
54. StecherB, MaierL, HardtWD (2013) ‘Blooming’ in the gut: how dysbiosis might contribute to pathogen evolution. Nat Rev Microbiol 11 : 277–284.
55. ThiennimitrP, WinterSE, BaumlerAJ (2012) Salmonella, the host and its microbiota. Curr Opin Microbiol 15 : 108–114.
56. KimB, RichardsSM, GunnJS, SlauchJM (2010) Protecting against antimicrobial effectors in the phagosome allows SodCII to contribute to virulence in Salmonella enterica serovar Typhimurium. J Bacteriol 192 : 2140–2149.
57. ProstLR, SanowarS, MillerSI (2007) Salmonella sensing of anti-microbial mechanisms to promote survival within macrophages. Immunol Rev 219 : 55–65.
58. Vazquez-TorresA, FangFC (2001) Salmonella evasion of the NADPH phagocyte oxidase. Microbes Infect 3 : 1313–1320.
59. DGHM (2002) List of disinfectants - German Society of Hygiene and Microbiology. mhp Verlag GmbH
60. MullerAJ, HoffmannC, GalleM, Van Den BroekeA, HeikenwalderM, et al. (2009) The S. Typhimurium effector SopE induces caspase-1 activation in stromal cells to initiate gut inflammation. Cell Host Microbe 6 : 125–136.
61. SonghetP, BarthelM, StecherB, MullerAJ, KremerM, et al. (2011) Stromal IFN-gammaR-signaling modulates goblet cell function during Salmonella Typhimurium infection. PLoS One 6: e22459.
62. EndtK, MaierL, KappeliR, BarthelM, MisselwitzB, et al. (2012) Peroral ciprofloxacin therapy impairs the generation of a protective immune response in a mouse model for Salmonella enterica serovar Typhimurium diarrhea, while parenteral ceftriaxone therapy does not. Antimicrob Agents Chemother 56 : 2295–2304.
63. StecherB, HapfelmeierS, MullerC, KremerM, StallmachT, et al. (2004) Flagella and chemotaxis are required for efficient induction of Salmonella enterica serovar Typhimurium colitis in streptomycin-pretreated mice. Infect Immun 72 : 4138–4150.
64. TsolisRM, AdamsLG, FichtTA, BaumlerAJ (1999) Contribution of Salmonella typhimurium virulence factors to diarrheal disease in calves. Infect Immun 67 : 4879–4885.
65. WallisTS, GalyovEE (2000) Molecular basis of Salmonella-induced enteritis. Mol Microbiol 36 : 997–1005.
66. DayDW, MandalBK, MorsonBC (1978) The rectal biopsy appearances in Salmonella colitis. Histopathology 2 : 117–131.
67. McGovernVJ, SlavutinLJ (1979) Pathology of salmonella colitis. Am J Surg Pathol 3 : 483–490.
68. SachdevHP, ChadhaV, MalhotraV, VergheseA, PuriRK (1993) Rectal histopathology in endemic Shigella and Salmonella diarrhea. J Pediatr Gastroenterol Nutr 16 : 33–38.
69. BoydJF (1985) Pathology of the alimentary tract in Salmonella typhimurium food poisoning. Gut 26 : 935–944.
70. LoetscherY, WieserA, LengefeldJ, KaiserP, SchubertS, et al. (2012) Salmonella transiently reside in luminal neutrophils in the inflamed gut. PLoS One 7: e34812.
71. GeddesK, CruzF, HeffronF (2007) Analysis of cells targeted by Salmonella type III secretion in vivo. PLoS Pathog 3: e196.
72. RydstromA, WickMJ (2007) Monocyte recruitment, activation, and function in the gut-associated lymphoid tissue during oral Salmonella infection. J Immunol 178 : 5789–5801.
73. CraigM, SlauchJM (2009) Phagocytic superoxide specifically damages an extracytoplasmic target to inhibit or kill Salmonella. PLoS One 4: e4975.
74. HoisethSK, StockerBA (1981) Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature 291 : 238–239.
75. LaubachVE, SheselyEG, SmithiesO, ShermanPA (1995) Mice lacking inducible nitric oxide synthase are not resistant to lipopolysaccharide-induced death. Proc Natl Acad Sci U S A 92 : 10688–10692.
76. PollockJD, WilliamsDA, GiffordMA, LiLL, DuX, et al. (1995) Mouse model of X-linked chronic granulomatous disease, an inherited defect in phagocyte superoxide production. Nat Genet 9 : 202–209.
77. Tacchini-CottierF, ZweifelC, BelkaidY, MukankundiyeC, VaseiM, et al. (2000) An immunomodulatory function for neutrophils during the induction of a CD4+ Th2 response in BALB/c mice infected with Leishmania major. J Immunol 165 : 2628–2636.
78. Pawitan Y (2001) In all likelihood: Oxford University Press.
79. AnderssonDI, HughesD (1996) Muller's ratchet decreases fitness of a DNA-based microbe. Proc Natl Acad Sci U S A 93 : 906–907.
80. Vazquez-TorresA, Jones-CarsonJ, BaumlerAJ, FalkowS, ValdiviaR, et al. (1999) Extraintestinal dissemination of Salmonella by CD18-expressing phagocytes. Nature 401 : 804–808.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Selective Susceptibility of Human Skin Antigen Presenting Cells to Productive Dengue Virus InfectionČlánek P47 Mice Are Compromised in Expansion and Activation of CD8 T Cells and Susceptible to InfectionČlánek Molecular Evolution of Broadly Neutralizing Llama Antibodies to the CD4-Binding Site of HIV-1
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2014 Číslo 12- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Microbial Programming of Systemic Innate Immunity and Resistance to Infection
- Unique Features of HIV-1 Spread through T Cell Virological Synapses
- Measles Immune Suppression: Functional Impairment or Numbers Game?
- Cellular Mechanisms of Alpha Herpesvirus Egress: Live Cell Fluorescence Microscopy of Pseudorabies Virus Exocytosis
- Rubella Virus: First Calcium-Requiring Viral Fusion Protein
- Plasma Membrane-Located Purine Nucleotide Transport Proteins Are Key Components for Host Exploitation by Microsporidian Intracellular Parasites
- Selective Susceptibility of Human Skin Antigen Presenting Cells to Productive Dengue Virus Infection
- Loss of Dynamin-Related Protein 2B Reveals Separation of Innate Immune Signaling Pathways
- Intraspecies Competition for Niches in the Distal Gut Dictate Transmission during Persistent Infection
- Unveiling the Intracellular Survival Gene Kit of Trypanosomatid Parasites
- Extreme Divergence of Tropism for the Stem-Cell-Niche in the Testis
- HTLV-1 Tax-Mediated Inhibition of FOXO3a Activity Is Critical for the Persistence of Terminally Differentiated CD4 T Cells
- P47 Mice Are Compromised in Expansion and Activation of CD8 T Cells and Susceptible to Infection
- Hypercytotoxicity and Rapid Loss of NKp44 Innate Lymphoid Cells during Acute SIV Infection
- Molecular Evolution of Broadly Neutralizing Llama Antibodies to the CD4-Binding Site of HIV-1
- Crystal Structure of Calcium Binding Protein-5 from and Its Involvement in Initiation of Phagocytosis of Human Erythrocytes
- Chronic Parasitic Infection Maintains High Frequencies of Short-Lived Ly6CCD4 Effector T Cells That Are Required for Protection against Re-infection
- Specific Dysregulation of IFNγ Production by Natural Killer Cells Confers Susceptibility to Viral Infection
- HSV-2-Driven Increase in the Expression of αβ Correlates with Increased Susceptibility to Vaginal SHIV Infection
- Murine Anti-vaccinia Virus D8 Antibodies Target Different Epitopes and Differ in Their Ability to Block D8 Binding to CS-E
- Brothers in Arms: Th17 and Treg Responses in Immunity
- Granulocytes Impose a Tight Bottleneck upon the Gut Luminal Pathogen Population during Typhimurium Colitis
- A Negative Feedback Modulator of Antigen Processing Evolved from a Frameshift in the Cowpox Virus Genome
- Discovery of Replicating Circular RNAs by RNA-Seq and Computational Algorithms
- The Non-receptor Tyrosine Kinase Tec Controls Assembly and Activity of the Noncanonical Caspase-8 Inflammasome
- Targeted Changes of the Cell Wall Proteome Influence Ability to Form Single- and Multi-strain Biofilms
- Apoplastic Venom Allergen-like Proteins of Cyst Nematodes Modulate the Activation of Basal Plant Innate Immunity by Cell Surface Receptors
- The Toll-Dorsal Pathway Is Required for Resistance to Viral Oral Infection in
- Anti-α4 Antibody Treatment Blocks Virus Traffic to the Brain and Gut Early, and Stabilizes CNS Injury Late in Infection
- Initiation of ART during Early Acute HIV Infection Preserves Mucosal Th17 Function and Reverses HIV-Related Immune Activation
- Microbial Urease in Health and Disease
- Emergence of MERS-CoV in the Middle East: Origins, Transmission, Treatment, and Perspectives
- Blocking Junctional Adhesion Molecule C Enhances Dendritic Cell Migration and Boosts the Immune Responses against
- IL-28B is a Key Regulator of B- and T-Cell Vaccine Responses against Influenza
- A Natural Genetic Variant of Granzyme B Confers Lethality to a Common Viral Infection
- Neutral Sphingomyelinase in Physiological and Measles Virus Induced T Cell Suppression
- Differential PfEMP1 Expression Is Associated with Cerebral Malaria Pathology
- The Role of the NADPH Oxidase NOX2 in Prion Pathogenesis
- Rapid Evolution of Virus Sequences in Intrinsically Disordered Protein Regions
- The Central Role of cAMP in Regulating Merozoite Invasion of Human Erythrocytes
- Expression of Suppressor of Cytokine Signaling 1 (SOCS1) Impairs Viral Clearance and Exacerbates Lung Injury during Influenza Infection
- Cellular Oxidative Stress Response Controls the Antiviral and Apoptotic Programs in Dengue Virus-Infected Dendritic Cells
- SUMOylation by the E3 Ligase TbSIZ1/PIAS1 Positively Regulates VSG Expression in
- Monocyte Recruitment to the Dermis and Differentiation to Dendritic Cells Increases the Targets for Dengue Virus Replication
- Oral Streptococci Utilize a Siglec-Like Domain of Serine-Rich Repeat Adhesins to Preferentially Target Platelet Sialoglycans in Human Blood
- SV40 Utilizes ATM Kinase Activity to Prevent Non-homologous End Joining of Broken Viral DNA Replication Products
- Amphipathic α-Helices in Apolipoproteins Are Crucial to the Formation of Infectious Hepatitis C Virus Particles
- Proteomic Analysis of the Acidocalcisome, an Organelle Conserved from Bacteria to Human Cells
- Experimental Cerebral Malaria Pathogenesis—Hemodynamics at the Blood Brain Barrier
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Plasma Membrane-Located Purine Nucleotide Transport Proteins Are Key Components for Host Exploitation by Microsporidian Intracellular Parasites
- Rubella Virus: First Calcium-Requiring Viral Fusion Protein
- Emergence of MERS-CoV in the Middle East: Origins, Transmission, Treatment, and Perspectives
- Unique Features of HIV-1 Spread through T Cell Virological Synapses
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



