-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaBiofilm Development on by Is Facilitated by Quorum Sensing-Dependent Repression of Type III Secretion
Yersinia pseudotuberculosis forms biofilms on Caenorhabditis elegans which block nematode feeding. This genetically amenable host-pathogen model has important implications for biofilm development on living, motile surfaces. Here we show that Y. pseudotuberculosis biofilm development on C. elegans is governed by N-acylhomoserine lactone (AHL)-mediated quorum sensing (QS) since (i) AHLs are produced in nematode associated biofilms and (ii) Y. pseudotuberculosis strains expressing an AHL-degrading enzyme or in which the AHL synthase (ypsI and ytbI) or response regulator (ypsR and ytbR) genes have been mutated, are attenuated. Although biofilm formation is also attenuated in Y. pseudotuberculosis strains carrying mutations in the QS-controlled motility regulator genes, flhDC and fliA, and the flagellin export gene, flhA, flagella are not required since fliC mutants form normal biofilms. However, in contrast to the parent and fliC mutant, Yop virulon proteins are up-regulated in flhDC, fliA and flhA mutants in a temperature and calcium independent manner. Similar observations were found for the Y. pseudotuberculosis QS mutants, indicating that the Yop virulon is repressed by QS via the master motility regulator, flhDC. By curing the pYV virulence plasmid from the ypsI/ytbI mutant, by growing YpIII under conditions permissive for type III needle formation but not Yop secretion and by mutating the type III secretion apparatus gene, yscJ, we show that biofilm formation can be restored in flhDC and ypsI/ytbI mutants. These data demonstrate that type III secretion blocks biofilm formation and is reciprocally regulated with motility via QS.
Published in the journal: . PLoS Pathog 7(1): e32767. doi:10.1371/journal.ppat.1001250
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1001250Summary
Yersinia pseudotuberculosis forms biofilms on Caenorhabditis elegans which block nematode feeding. This genetically amenable host-pathogen model has important implications for biofilm development on living, motile surfaces. Here we show that Y. pseudotuberculosis biofilm development on C. elegans is governed by N-acylhomoserine lactone (AHL)-mediated quorum sensing (QS) since (i) AHLs are produced in nematode associated biofilms and (ii) Y. pseudotuberculosis strains expressing an AHL-degrading enzyme or in which the AHL synthase (ypsI and ytbI) or response regulator (ypsR and ytbR) genes have been mutated, are attenuated. Although biofilm formation is also attenuated in Y. pseudotuberculosis strains carrying mutations in the QS-controlled motility regulator genes, flhDC and fliA, and the flagellin export gene, flhA, flagella are not required since fliC mutants form normal biofilms. However, in contrast to the parent and fliC mutant, Yop virulon proteins are up-regulated in flhDC, fliA and flhA mutants in a temperature and calcium independent manner. Similar observations were found for the Y. pseudotuberculosis QS mutants, indicating that the Yop virulon is repressed by QS via the master motility regulator, flhDC. By curing the pYV virulence plasmid from the ypsI/ytbI mutant, by growing YpIII under conditions permissive for type III needle formation but not Yop secretion and by mutating the type III secretion apparatus gene, yscJ, we show that biofilm formation can be restored in flhDC and ypsI/ytbI mutants. These data demonstrate that type III secretion blocks biofilm formation and is reciprocally regulated with motility via QS.
Introduction
The human pathogenic Yersiniae (Yersinia pseudotuberculosis, Yersinia enterocolitica and Yersinia pestis) share a high degree of DNA identity, but cause distinct diseases ranging from enterocolitis (Y. enterocolitica and Y. pseudotuberculosis) to pneumonic, bubonic or septicaemic plague (Y. pestis). Essential for the virulence of all pathogenic Yersiniae, is the ∼70-kb pYV virulence plasmid, which encodes the Yop virulon. This consists of a type III secretion system which enables Yersinia to inject multiple Yop effector proteins directly into the cytosol of eukaryotic cells and so subvert host cell signalling pathways (for reviews see [1]–[3]. Yop virulon genes are tightly regulated by environmental conditions and in particular, temperature (only expressing at 37°C) and Ca2+ concentration (reviewed in [4]).
Y. pestis and Y. pseudotuberculosis are capable of forming biofilms around the anterior and along the surface of the nematode Caenorhabditis elegans [5], [6]. However, biofilm formation is strain-dependent and a study of over 40 different Y. pseudotuberculosis strains showed that some formed biofilms on C. elegans but not on abiotic polystyrene surfaces and vice versa [6]. No relationship was observed between strains forming biofilms on C. elegans and those that formed biofilms on polystyrene surfaces. These findings suggest that biofilm development on the living surface of C. elegans is different from that on an abiotic surface such as polystyrene.
Y. pestis is transferred between mammalian hosts by a flea borne vector that feeds on blood. The hmsHFRS operon is key to the colonisation and blockage of the flea proventriculus which results from the accumulation of biofilm [7]–[9] and hmsHFRS mutants of both Y. pestis and Y. pseudotuberculosis fail to form biofilms on C. elegans. Since C. elegans has been thoroughly studied at the genetic level and orthologous genes frequently studied in human health and disease, the C. elegans/Yersinia model can be used to identify genetic features of both the pathogen and the host that contribute to biofilm-mediated interactions between bacteria and invertebrates. These in turn have interesting implications for both the Yersinia/flea and human biofilm-centred infections. Although there are some limitations, the importance of C. elegans as a model organism for investigating prokaryotic/eukaryote interactions should not be overlooked given that nematodes are the most abundant animals on the Earth [10].
Although Y. pseudotuberculosis does not readily colonise fleas, biofilm formation may alternatively be involved in the prevention of predatory feeding as has been noted for other soil bacteria [11]. Whether the bacteria-invertebrate biofilm relationship is bacterially driven or is a two way interactive process between the bacteria and nematode is not fully understood. It has however been postulated that nematodes accumulate the bacterially derived extracellular matrix (ECM) passively by virtue of their movement through a lawn of bacteria [12] and there is evidence to show that biofilms do not accumulate on the surface of non-motile C. elegans. This implies that a prerequisite for biofilm formation is nematode translocation which provides the necessary contact between bacteria and nematode [12]. However, Y. pseudotuberculosis is unable to form biofilms on a number of motile C. elegans mutants such as srf-2, srf-3 and srf-5 [6] and bah-1, bah-2 and bah-3. Conversely many natural strains of Y. pseudotuberculosis fail to form biofilms on C. elegans as do a number of Y. pseudotuberculosis strains with mutations in lipopolysaccharide biosynthesis, signal transduction and hms genes [6]. Such findings imply the existence of an adaptive interaction between the nematode and the bacterium rather than simply the passive adherence of bacterially derived ECM [6].
Bacteria possess multiple integrated sensory systems that govern adaptation to environmental challenges including the local cell population density. Such population-dependent adaptive behaviour often takes the form of perception and processing of chemical information and is termed quorum sensing (QS). For many Gram negative bacteria this involves the use of self-generated diffusible signal molecules such as the N-acyl homoserine lactones (AHLs). These are usually synthesised and sensed via members of the LuxI AHL synthase and LuxR response regulator protein families respectively. QS enables bacteria to determine, by monitoring the concentration of a signal molecule, when the number of individuals in the population are sufficient (a quorum) to make a collective ‘decision’ to alter their behaviour in response to environmental challenges [13]–[16]. Such behavioural decisions impact on bacterial motility, secondary metabolism, virulence, and biofilm development [17].
Y. pseudotuberculosis produces four major AHLs via a QS system consisting of two genetic loci termed ypsR/ypsI and ytbR/ytbI which control cell aggregation/flocculation and swimming motility [18], [19]. This system is organized hierarchically with YpsR and its cognate AHLs regulating ytbR and ytbI as well as ypsR and ypsI. The YpsR/YpsI and YtbR/YtbI QS system in turn fine tunes swimming motility by governing the expression of two key regulators of the motility cascade, namely flhDC and fliA [19]. AHL-dependent QS also controls motility in Y. enterocolitica [20] Y. pestis produces a similar range of AHLs to Y. pseudotuberculosis [21] and retains an analogous QS system [22], [23]. However the relationship between QS and regulators of the motility cascade such as flhDC or fliA may be different in Y. pestis when compared with Y. pseudotuberculosis or Y. enterocolitica because Y. pestis is non-motile because of a frame-shift mutation in the motility master regulator flhD [24].
There is considerable evidence to show that AHL-dependent QS plays a significant role during the biofilm mode of growth on an abiotic surface since AHL production has been detected in glass and metal surface associated biofilms produced by bacteria such as Pseudomonas aeruginosa [25] and Aeromonas hydrophila [26]. Furthermore, in a variety of bacteria, QS controls the target genes required for different stages of biofilm development from adherence and aggregation to maturation and dispersal (for review see [27]). In addition QS determines the physiological response of biofilm communities to antimicrobial agents and host defences [28], [29].
In the present paper we sought to determine whether biofilm formation by Y. pseudotuberculosis on a living motile surface i.e. on C. elegans is an interactive, QS-dependent process. The results obtained revealed that QS in Y. pseudotuberculosis reciprocally regulates the C. elegans biofilm phenotype with type III secretion via the major motility regulators flhDC and fliA. Consequently the induction of type III secretion attenuates biofilm formation on C. elegans which can be restored in a QS mutant either by curing the pYV virulence plasmid from the ypsI/ytbI mutant, by growing YpIII under conditions permissive for type III needle formation but not Yop secretion or by mutating the type III secretion apparatus gene, yscJ, a key component of the type III injectisome.
Results
Y. pseudotuberculosis produces AHLs when growing as a biofilm on the surface of C. elegans
When C. elegans is infected with Y. pseudotuberculosis YpIII harboring the gfp-plasmid pSB2020 and examined by confocal microscopy, the bacterial microcolonies fluoresce green and are embedded in an ECM which fluoresces red (yellow when both bacteria and matrix are combined) (Figure 1A) when labelled with WGA-R consistent with the presence of bacterially generated N-acetyl-D-glucosamine [12]. An orthogonal image of Figure 1A showing the depth of the biofilm in x and y planes can be seen in Figure S1A. After 48 h incubation the biofilms on C. elegans became highly resistant to WGA-R labelling and only stained red on the outer surface while the inner mass remained green (compare Figure 1A with Figure 1B). In common with bacterial biofilms formed on abiotic surfaces [30], the Yersinia biofilm on C. elegans also contains extracellular DNA as revealed by DAPI staining (Figure 2A).
Fig. 1. Y. pseudotuberculosis QS mutants are attenuated for biofilm formation on C. elegans. 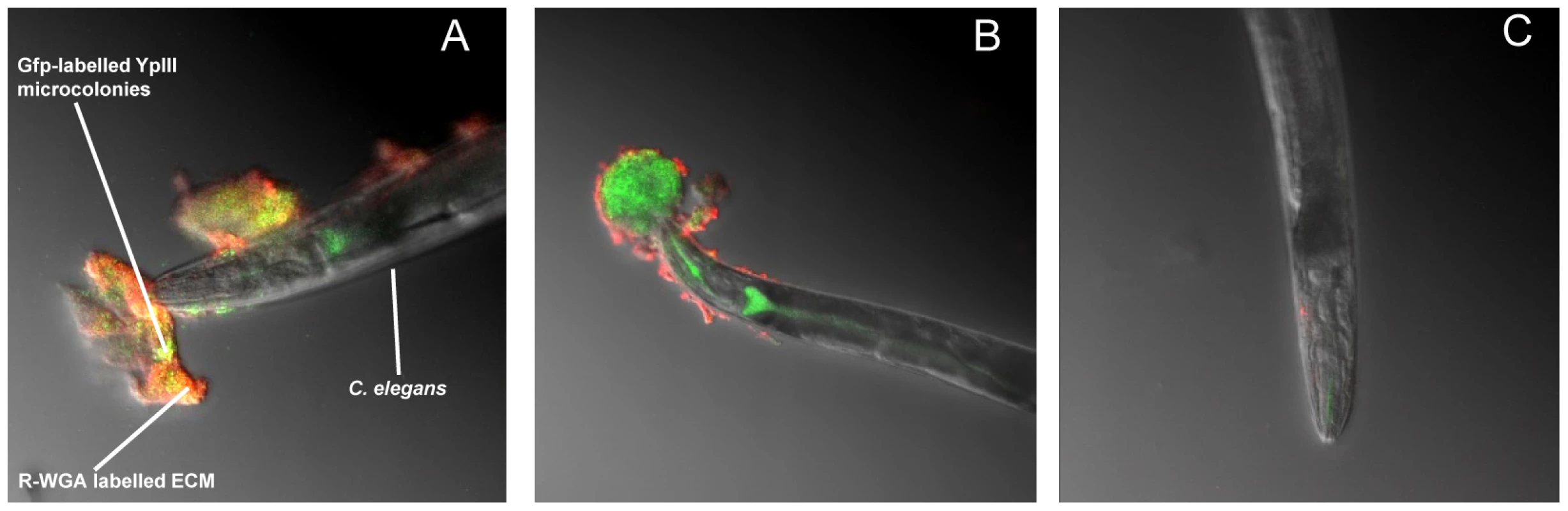
(A) Confocal image showing C. elegans heavily infected with Y. pseudotuberculosis YpIII embedded in a biofilm ECM which surrounds the anterior end of C. elegans and is spreading to other areas of the worm surface. Green, Gfp-labelled Y. pseudotuberculosis red, WGA-R binding to the ECM yellow, red and green overlay. (B) Confocal image of a Y. pseudotuberculosis YpIII biofilm on C. elegans after 48 h in which only the outer surface of the ECM stains with WGA-R which no longer penetrates deep into the biofilm. (C) C. elegans infected with the Y. pseudotuberculosis ypsI/ytbI double mutant. Fig. 2. Y. pseudotuberculosis biofilm ECM on C. elegans contains extracellular DNA. 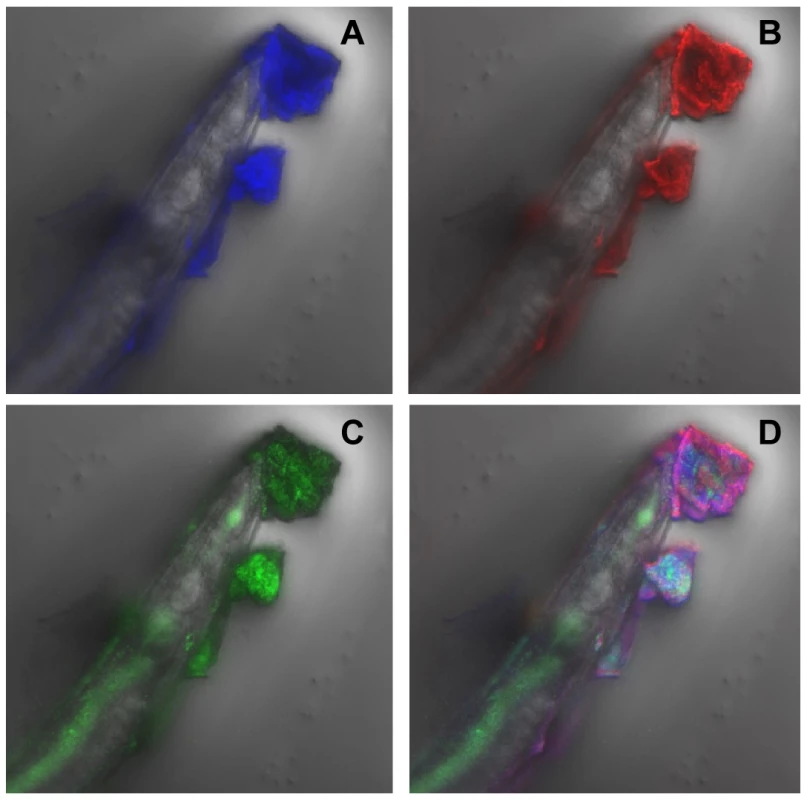
(A) The ECM fluoresces blue when stained with DAPI consistent with the presence of extracellular DNA. (B), (C) and (D) show the same image labelled with WGA-R (B; red), Gfp-labelled YpIII (C; green) and an overlay image (D) of the three fluorescent labels. To determine qualitatively whether AHLs are produced in the biofilms which accumulate on the surface of C. elegans, the biofilm matrix from heavily infected nematodes grown in the presence of Y. pseudotuberculosis for 24 h was extracted into dichloromethane and the extracts analysed using the AHL bioreporter C. violaceum CV026 in a well plate overlay assay [31]. As negative controls, AHL extractions were also carried out on nematodes which had been grown on E. coli OP50 and from the cell pellet of an overnight Y. pseudotuberculosis culture. Culture supernatant from the latter served as a positive control. Figure 3A (i) shows a purple halo of violacein around the agar well which contained the concentrated nematode extract taken from worms infected with parent Y. pseudotuberculosis. A similar result was obtained for the positive control (Figure 3A iv) while no violacein was observed around the negative control wells. Taken together these data indicate that AHLs are produced by Y. pseudotuberculosis growing as biofilms on the surface of C. elegans.
Fig. 3. AHLs are produced in Y. pseudotuberculosis YpIII biofilms on C. elegans. 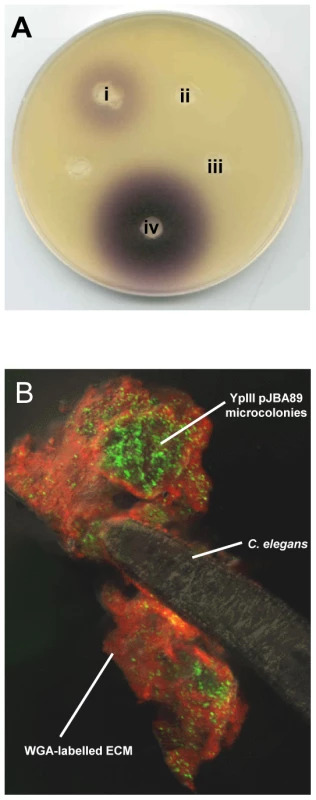
(A) C. violaceum AHL plate assay showing that AHLs are present in a Y. pseudotuberculosis biofilm growing on C. elegans. (i) Y. pseudotuberculosis YpIII biofilm extract harvested from C. elegans; (ii) extract from nematodes grown on E. coli OP50 (iii) cell pellet extract from an overnight liquid culture of Y. pseudotuberculosis and (iv) extract of an overnight liquid culture of Y. pseudotuberculosis YpIII. The AHL levels collected from the biofilm appear to be present at lower levels than in the culture supernatant. (B) Confocal image showing Y. pseudotuberculosis YpIII transformed with the AHL reporter, pJBA89 fluorescing green in response to AHLs in the biofilm. Red and yellow represent WGA-R stain of the ECM and the overlay of red and green respectively. To confirm that AHLs are synthesised in situ in the biofilms, Y. pseudotuberculosis was transformed with the gfp-biosensor, pJBA89 which fluoresces green in the presence of AHLs [32]. When infected with Y. pseudotuberculosis pJBA89 the characteristic biofilms which form on the surface of C. elegans after 24 h show green fluorescent Y. pseudotuberculosis pJAB89 embedded in the red WGA-R labelled biofilm matrix (Figure 3B) which were indistinguishable from those presented Figure 1A.
Quorum sensing regulates biofilm development on the surface of C. elegans
Since AHLs were detected in the biofilms formed on C. elegans, we used two approaches to determine whether QS was required for biofilm development on the nematode surface. Firstly, we exploited the lactonase, AiiA which hydrolyses the ester bond within the AHL homoserine lactone moiety generating the corresponding, inactive, N-acylhomoserine compound [33]. When aiiA is introduced into Y. pseudotuberculosis on the pSU18 derivative pSA236, the AHLs produced are hydrolysed, so generating an AHL-negative phenotype [19]. By comparing the parent YpIII strain with YpIII transformed with either the pSU18 control vector or pSA236, we evaluated the contribution of AHL-dependent QS to biofilm development. For these experiments, a biofilm severity incidence was calculated for the infected C. elegans population after 24 h incubation. Each nematode was assigned a score between 0 and 3 related to the severity of biofilm accumulation (examples of scores 0 and 3 can be taken from the biofilms shown in Figure 1A and C; and scored of 1 and 2 from Figure S1 B and C respectively). These assays revealed that Y. pseudotuberculosis and Y. pseudotuberculosis pSU18 had biofilm severity indices of 77.3% and 62.0% respectively. When C. elegans infected Y. pseudotuberculosis pSU18 were compared to nematodes infected with Y. pseudotuberculosis pSA236 the biofilm severity incidence was reduced to 38.7% (p = <0.05 and n = 3 respectively) (Figure 4A).
Fig. 4. QS controls Y. pseudotuberculosis biofilm formation on C. elegans. 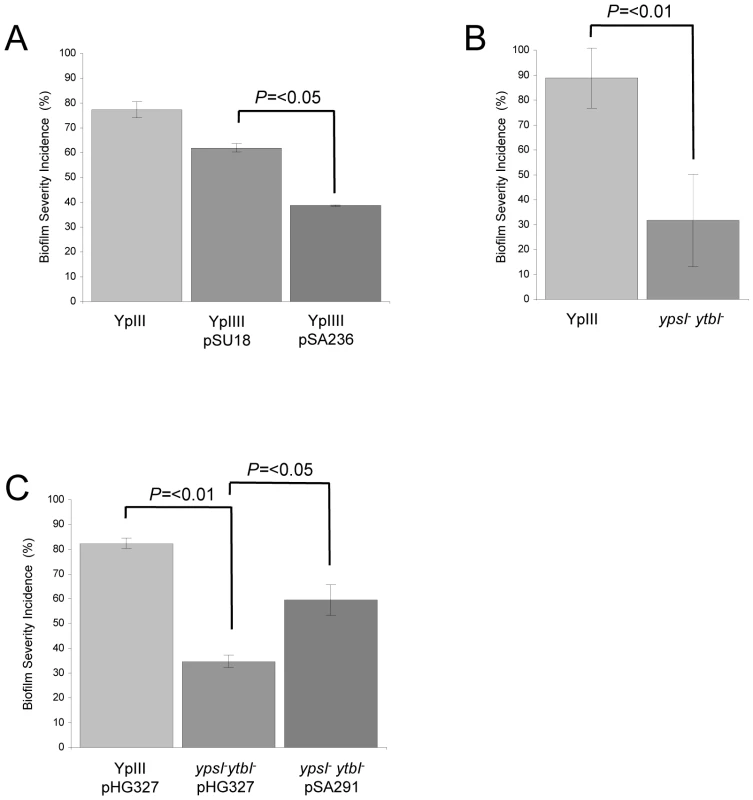
Biofilm severity as a measurement of biofilm formation by Y. pseudotuberculosis YpIII, transformed with the vector pSU18 or expressing the AHL lactonase AiiA on plasmid pSA236 (A) and for the ypsI/ytbI mutant (B) and complemented ypsI/ytbI mutant (C). Secondly we carried out C. elegans infection assays using Y. pseudotuberculosis YpIII QS mutants transformed with the constitutive gfp-plasmid, pSB2020. These included an AHL negative mutant in which both AHL synthase genes (ypsI and ytbI) have been disrupted and a second double mutant in which the two QS response regulators, ypsR and ytbR have been disrupted [18], [19]. When compared with the parent Y. pseudotuberculosis YpIII strain (Figure 1A), biofilm development was severely delayed in the ypsI/ytbI double mutant formed little or no biofilm (compare Figure 1A and 1C). Similar results were obtained for the ypsR/ytbR double mutant (data not shown). In addition, nematodes grown on YpIII, in contrast to those grown on E. coli OP50, exhibit exaggerated body bends (Figure 5), are unable translocate within 1.5 h and by 5 h become moribund. In contrast, C. elegans infected with either the ypsI/ytbI mutant or the ypsR/ytbR mutant translocate normally and make tracks in the agar which are identical to those presented in Figure 5A and only began to show signs of aberrant movement 3–4 h post infection. After 96 h growth, both the ypsI/ytbI and ypsR/ytbR mutants formed severe biofilms on the nematodes. In addition, we calculated a biofilm severity incidence for each yersinia strain. Figure 4B shows that after 24 h there is an ∼3 fold reduction in the amount of biofilm on nematodes infected by the ypsI/ytbI double mutant compared with the parent (32% compared with 89%; p = <0.01 n = 4). Similar results were obtained for the ypsR/ytbR double mutant (data not shown). Genetic complementation of the ypsI/ytbI mutation with pSA291 (Figure 4C) partially restored the biofilm severity incidence to that of the parent strain (Parent pHG327 (82%) compared with the ypsI/ytbI mutant pHG327 (35%) (p = 0.001 n = 3) and ypsI/ytbI mutant pSA291 (60%) compared with ypsI/ytbI mutant pHG327 (35%) (p = <0.05 n = 3)).
Fig. 5. Aberrant translocation of C. elegans on Y. pseudotuberculosis. 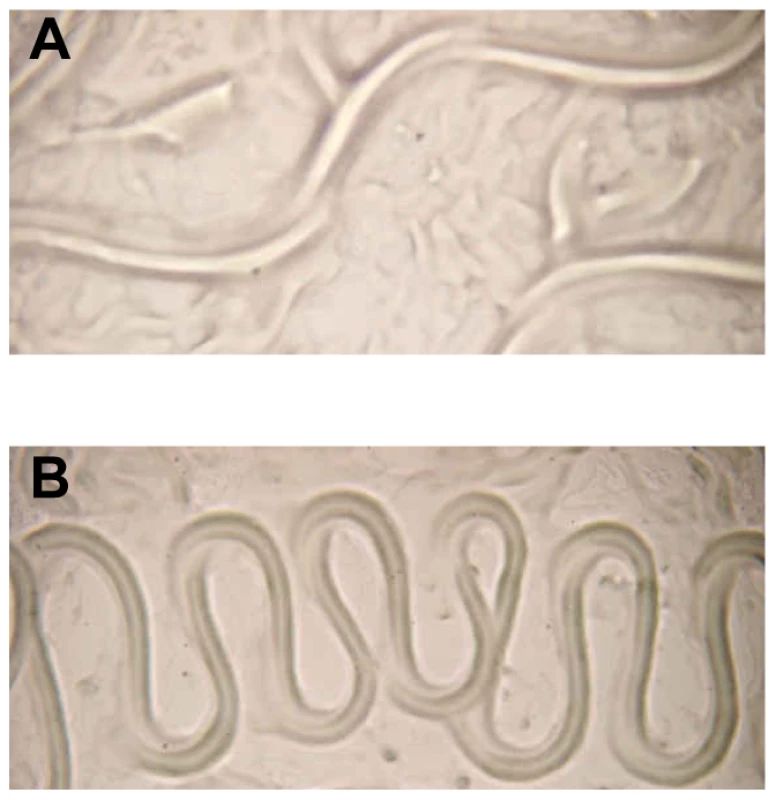
(A) E. coli OP50 and (B) Y. pseudotuberculosis YpIII. Worms infected with either the ypsI/ytbI or ypsR/ytbR mutants translocate normally and make tracks in the agar similar to those seen in (A) and only begin to show signs of aberrant movement comparable with (B), 3–4 h post infection. These data demonstrate that the loss of AHL synthesis either via enzyme-mediated inactivation or by mutagenesis of the AHL synthases results in the attenuation of biofilm formation on C. elegans. Consequently QS is pivotal to the timing and severity of biofilm development on C. elegans.
Flagellar-mediated motility is not required for biofilm development on C. elegans
Since the ypsR/ypsI and ytbR/ytbI loci are both involved in the regulation of motility via flhDC and fliA which code for the motility master regulator and flagellar specific sigma factor respectively [19], we sought to determine whether these downstream regulators contribute to the Yersinia/C. elegans biofilm phenotype. Figure 6A shows that the flhDC mutant was impaired in its ability to form biofilms on the surface of C. elegans (biofilm severity incidence for the parent of 57.5% compared to 24.7% for the flhDC mutant (p = <0.05, n = 3)) and genetic complementation of flhDC using pSA220 increased the biofilm severity to 61.6% when compared with the flhDC mutant (p = <0.02, n = 3). Figure 6B shows that the biofilm severity incidence for the fliA mutant was also reduced when compared with the parent (p = <0.05, n = 3). Since both regulators control swimming motility and as flhDC and fliA mutants are non-motile, these data suggested that biofilm formation may depend on flagellar-mediated motility. To explore this possibility, we first constructed a flagellin-negative strain by mutating the flagellin structural gene, fliC. This non-motile mutant formed biofilms on nematodes which were indistinguishable from the parent Y. pseudotuberculosis strain (Figure 6C and data not shown). Consequently, flagellar-mediated motility is not a necessary pre-requisite for biofilm formation on C. elegans. However, in Y. enterocolitica, the flagellar type III secretion apparatus may also secrete non-flagellar proteins termed ‘Fops’ (for Flagellar outer proteins) such as the phospholipase, YplA [34]. Since flagellin structural mutants still secrete Fops, we constructed a flhA mutant since this gene codes for a structural component of the flagellar protein export apparatus [35] and flhA mutants have been reported not to secrete Fops [34]. In common with the flhDC and fliA mutants and when compared to the parent, the flhA mutant exhibited attenuated biofilm formation (Figure 6B) (p = <0.05, n = 3), a finding which implies a possible role for a secreted protein(s).
Fig. 6. Y. pseudotuberculosis strains with mutations in flhDC, fliA or flhA but not fliC are attenuated for biofilm formation. 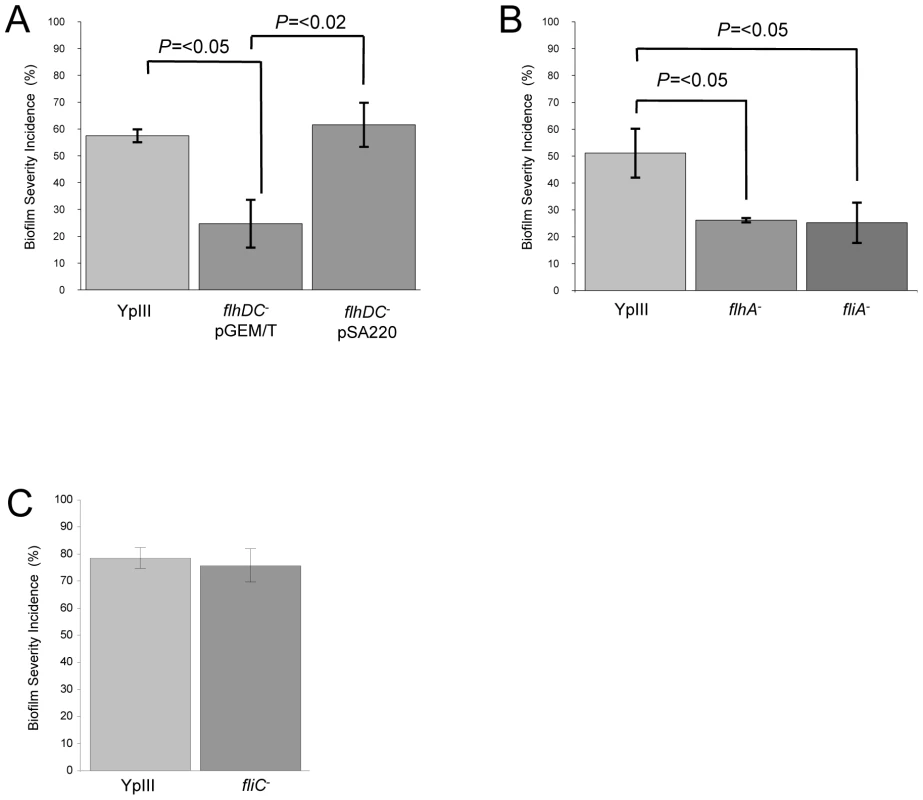
Biofilm severity indices are shown for flhDC and the complemented flhDC mutant (A), flhA and fliA (B) and fliC (C). To determine whether any secreted proteins could be involved in biofilm development on C. elegans, we first examined the extracellular protein profiles of the Y. pseudotuberculosis flhDC, fliA, flhA and fliC mutants grown overnight in LBmops at 30°C. Figure 7 shows that compared with the parent strain and fliC mutant, numerous proteins are up-regulated in each of the other motility mutants. MALDI-TOF MS analysis identified three of the major protein bands as YopM/H (41/51 kDa, these two proteins often co-migrate and could not be distinguished by MALDITOF sequencing), LcrV (37 kDa) and YopN (32 kDa) all of which are encoded on the pYV virulence plasmid and secreted by the Ysc-Yop type III secretion system. Two further up-regulated proteins were identified as KatY and GroEL which are not related to the Yop virulon (Figure 7).
Fig. 7. SDS-PAGE protein profiles of cell free supernatants prepared from Y. pseudotuberculosis YpIII parent, flhDC, fliA, flhA and fliC mutants grown at 30°C. 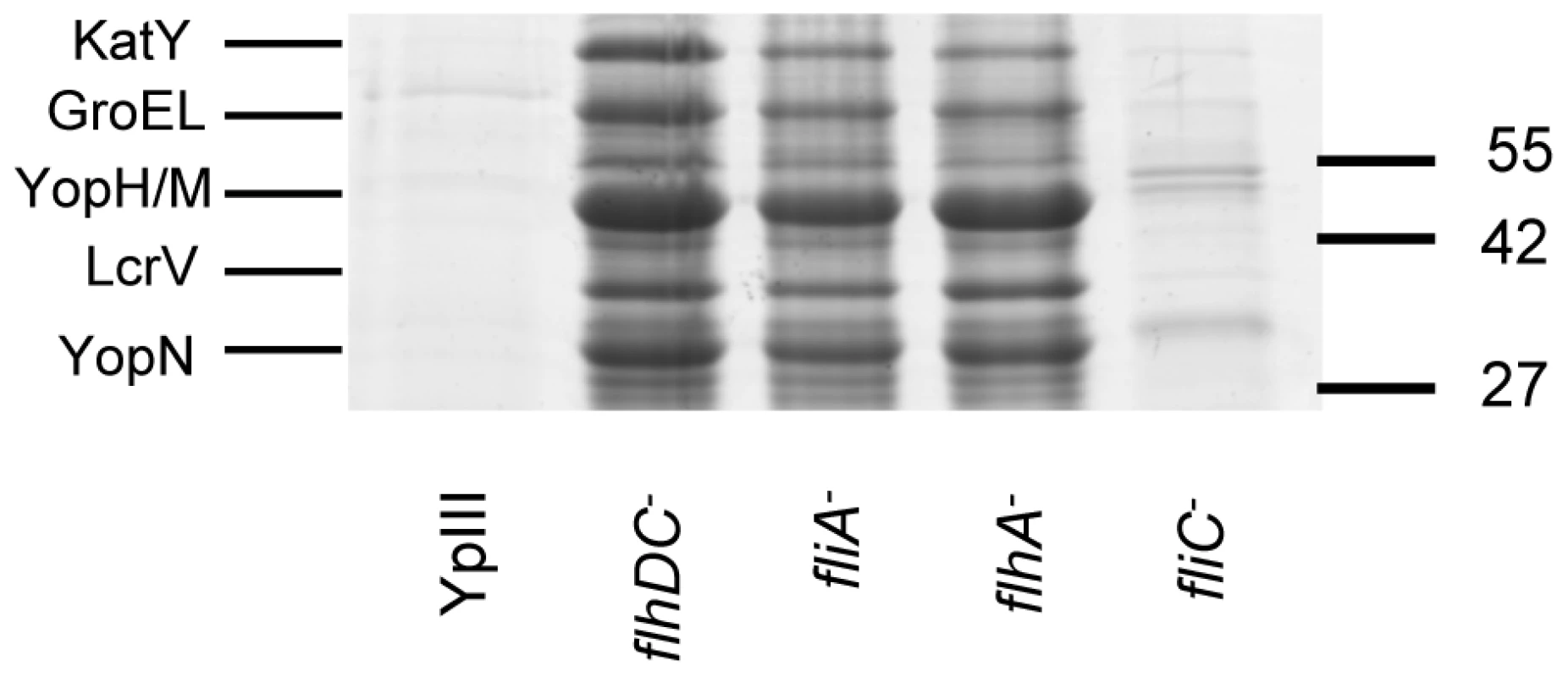
The up-regulated proteins YopN, YopM/H, LcrV, KatY and GroEL were identified by MALDI-TOF MS. Molecular masses of the marker proteins are in kDa. Quorum sensing represses type III secretion in Y. pseudotuberculosis
In contrast to the YpIII parent strain which only secretes Yops at 37°C in the absence of Ca2+ both flhDC and fliA mutants clearly secrete Yops at 30°C in the presence of Ca2+. Since both of these motility regulators are controlled by QS in Y. pseudotuberculosis [19], these data suggested that elements of the Yop virulon are also likely to be QS-controlled. Figure 8 shows that when grown in LBMops at 30°C overnight, at least 4 extracellular proteins are up-regulated in the ypsI/ytbI and ypsR/ytbR double mutants compared with the parent strain. The same proteins are also up-regulated in the ypsR, ytbR and ytbI single mutants whereas the ypsI mutant exhibits the same profile as the parent strain. MALDI-TOF MS analysis identified the proteins as YopM/YopH, FliC, LcrV and YopN. These proteins were also present in supernatants from the same mutants after growth at 37°C in LBMops but absent from the parent and ypsI mutant (Figure S2). In contrast, Yop proteins were absent from the supernatants of all of the strains grown at 22°C although two proteins, the flagellar capping protein (FliD; 48.6 KDa) and flagellin (FliC; 45 KDa) were up-regulated (data not shown).
Fig. 8. SDS-PAGE protein profiles of the Y. pseudotuberculosis parent and the QS mutants prepared from cell-free supernatants grown at 30°C. 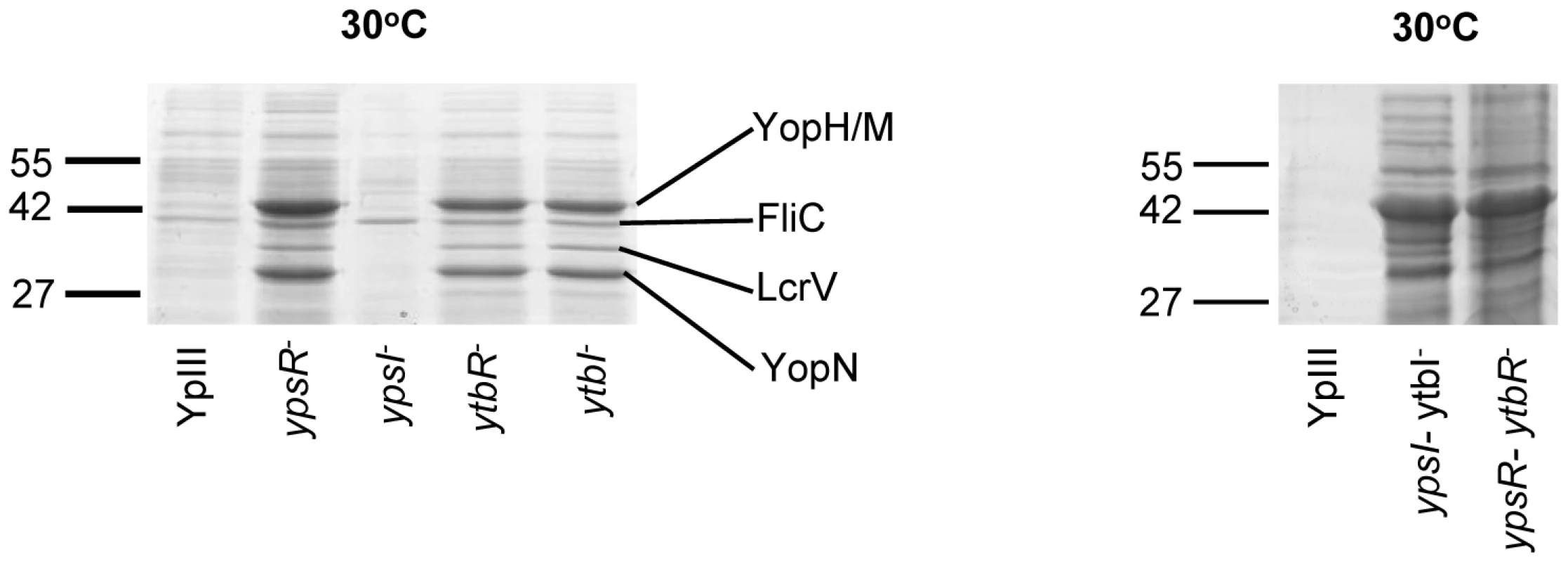
YopN, YopH/M, LcrV, and FliC were identified by MALDI-TOF MS. Molecular masses of the marker proteins are in kDa. The pYV plasmid inhibits biofilm formation by the Y. pseudotuberculosis ypsI/ytbI and flhDC mutants
The attenuation of biofilm formation on C. elegans observed for both the motility and QS mutants in conjunction with the elevated secretion of Yop virulon proteins at non-permissive temperatures raised the possibility that induction of type III secretion blocks biofilm development. Consequently, we predicted that biofilm formation would be restored in Y. pseudotuberculosis ypsI/ytbI and flhDC mutants cured of the pYV plasmid. To explore this hypothesis, we cured the pYV plasmid from the parent, ypsI/ytbI and flhDC mutants by repeated selection on CRMOX agar plates. The presence or absence of the pYV plasmid had no effect on the ability of the Y. pseudotuberculosis YpIII parent strain to form a biofilm on C. elegans (Figure 9A). However when similar experiments were performed using the ypsI/ytbI double mutant (Figure 9A and compare with Figure 4B) or flhDC (data not shown) cured of pYV, biofilm formation on C. elegans was restored to parental strain levels when compared with the biofilm levels observed on the ypsI/ytbI pYV+ double mutant (p = <0.01, n = 3). These data suggest that under these conditions, AHL-mediated QS represses the expression of a pYV gene(s) which would otherwise prevent biofilm formation.
Fig. 9. Impact of pYV and type III secretion on biofilm formation by Y. pseudotuberculosis YpIII on C. elegans. 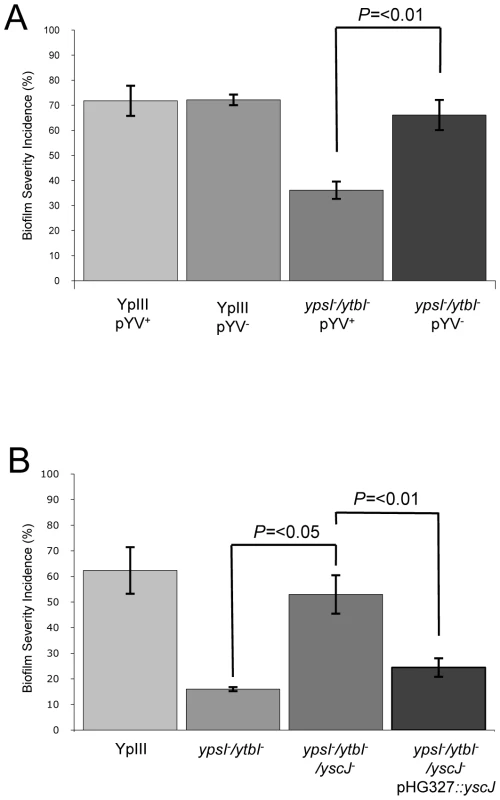
Biofilm severity indices are shown for (A) YpIII and the ypsI/ytbI mutant with or without pYV and (B) YpIII and the ypsI/ytbI mutant compared with the ypsI/ytbI/yscJ triple mutant and the triple mutant complemented with a plasmid borne copy of yscJ. To gain further evidence in support of a biofilm inhibitory role for pYV Yop virulon component(s), C. elegans was infected with the parent Y. pseudotuberculosis grown in LBmops MOX, conditions which promote Yop secretion (i.e. 37°C in the absence of Ca2+) rather than in LBmops at 30°C in which Yops will not be secreted. These seed cultures were then transferred to NGM plates containing MgCl2 and sodium oxalate to chelate Ca2+. Under such pre-conditions, the type III system is induced and no biofilms were formed on C. elegans (data not shown) providing additional support that induction of the Yop virulon prevents biofilm formation on C. elegans.
Biofilm formation on C. elegans is inhibited by induction of the type III injectisome
To demonstrate unequivocally that the inhibition of biofilm formation on C. elegans observed for the Y. pseudotuberculosis ypsI/ytbI mutant depends on the induction of functional type III secretion system rather than other genes present on the pYV plasmid, we modified the ypsI/ytbI mutant by mutating yscJ. This gene codes for a key component of the Ysc injectisome required for the assembly of a functional type III secretion apparatus [36]. Cell free culture supernatants taken from the ypsI/ytbI/yscJ triple mutant grown in LBMops at 30°C were examined by SDS-PAGE. This confirmed that, in contrast to the ypsI/ytbI mutant, Yop proteins were no longer secreted (data not shown). Yop secretion in the triple mutant grown under these conditions could however be restored by complementation with a plasmid-borne copy of yscJ (pHG::yscJ; data not shown). In the C. elegans biofilm assays, the biofilm severity index of the ypsI/ytbI/yscJ triple mutant was ∼4-fold higher than that of the ypsI/ytb double mutant (p = <0.05, n = 3) and comparable with that of the parent strain (Figure 9B). When the triple mutant was compared to its complemented counterpart containing a functional copy of yscJ (on plasmid pHG::yscJ) biofilm severity was reduced ∼two-fold (p = <0.01, n = 3) back to levels comparable with the ypsI/ytbI double mutant (Figure 9B). These results are consistent with a role for the type III injectisome in preventing biofilm development on C. elegans and demonstrate that either the type III needle or the secreted Yop proteins or both prevent biofilm development on C. elegans. To attempt to differentiate between these three possibilities, we grew the Y. pseudotuberculosis YpIII parent strain at 37°C in the presence of calcium which results in type III needle assembly but not Yop secretion [37]. This is because Ca2+ prevents Yop effector secretion even in the presence of a fully formed injectisome. YpIII was then subcultured onto NGM medium supplemented with calcium. When pre-cultured under these conditions and used to infect C. elegans at 22°C, Y. pseudotuberculosis YpIII failed to form a biofilm on C. elegans. The infected worms were indistinguishable from that shown in Figure 1C suggesting that the type III needle rather than the Yop effectors was responsible for preventing biofilm development.
Discussion
On abiotic surfaces, bacterial biofilm formation is generally considered as a step-wise process initiating from individual cells adhering to a substratum leading to microcolony formation, biofilm maturation and finally dispersal to new sites [38]–[42]. Although the nature and development of biofilms formed on biotic surfaces have not been as thoroughly investigated, biofilm development by Y. pseudotuberculosis on C. elegans involves attachment and maturation stages and the ECM contains both carbohydrate and extracellular DNA. Whether the DNA present in the biofilm is bacterial or nematode-derived has yet to be established. However, the WGA-stained carbohydrate present in the ECM appears to be bacterially-derived since it is present in the lawns of Y. pseudotuberculosis prior to the addition of nematodes which are not labelled by WGA [43]. The WGA-stained ECM carbohydrate could be either peptidoglycan which contains N-acetyl glucosamine in the sugar backbone [44] or polymeric N-acetyl-D-glucosamine or both. Y. pestis strains with mutations in the hmsHFRS locus, which is responsible for the biosynthesis of a poly β-1,6-N-acetyl-D-glucosamine-like polysaccharide [45], are defective for biofilm accumulation on C. elegans implying that this exopolysaccharide plays an essential role. An intact hmsHFRS is also required for biofilm formation on C. elegans by both Y. pseudotuberculosis and Xenorhabdus nematophila [46].
Apart from the hmsHFRS genes, other yersinia genes currently known to be required for biofilm formation on C. elegans include two genes involved in LPS biosynthesis, two genes of unknown function and a potential hybrid two component regulatory protein [6]. Both RcsA (a phosphorelay accessory protein which functions in concert with the response regulator, RcsB) and PhoP negatively regulate the formation of Y. pseudotuberculosis biofilms on nematodes [47] while the action of PhoP appears to be mediated at least in part by the down-regulation of HmsT [48]. This is interesting since HmsT is a cyclic diguanylate (c-di-GMP) synthase and c-di-GMP metabolism plays an important role in biofilm formation in many different bacteria including Y. pestis [49], [50].
Depending on the organism, QS may be involved in the early attachment or later maturation stages of biofilm development on abiotic surfaces [27]. In pathogens such as P. aeruginosa, QS is responsible for controlling the expression of key components of the biofilm extracellular matrix including exopolysaccharides and extracellular DNA release as well as the refractory nature of biofilms to host defences and antimicrobials [27]. The contribution of QS to yersinia biofilm development on C. elegans has not previously been investigated although for Y. pseudotuberculosis, QS controls cell aggregation (a type of suspended biofilm) in liquid culture [18]. A Y. pestis strain with combined mutations in ypsR/ypsI, ytbR/ytbI and luxS formed a similar biofilm on glass cover slips to the parental strain which could not be distinguished by crystal violet or Congo red staining although a very mild defect was observed using confocal microscopy [51]. Here, for Y. pseudotuberculosis YpIII, we have shown that AHL-dependent QS is functional in biofilms formed on C. elegans by demonstrating (i) the presence of AHL signal molecules within the nematode-associated biofilm matrix and (ii) that YpIII strains in which AHL biosynthesis is abrogated either by expressing an AHL-inactivating enzyme in situ or by mutating the AHL synthases (YpsI and YtbI) are attenuated for biofilm formation. Because Y. pseudotuberculosis YpIII does not form biofilms on polystyrene surfaces [6], these data indicate that the QS-dependent pathway for biofilm formation on C. elegans is different from that on abiotic surfaces. While QS signals have previously been identified in pseudomonas and aeromonas biofilms on abiotic surfaces [25], [26] to our knowledge they have not previously been detected directly in biofilms growing on a living, biotic surface. AHLs have however been shown to be produced in the tissues of mice infected with Y. enterocolitica [52] although no evidence was presented for biofilm formation in this acute experimental infection model.
In Y. pseudotuberculosis, YpsRI and YtbRI form a QS hierarchy in which ypsR is auto-regulated and also controls the expression of ypsI, ytbI and ytbR; YtbR also regulates ytbI expression [19]. In common with the ypsI/ytbI double synthase mutant, the ypsR/ytbR double response regulator mutant was also attenuated for biofilm development on C. elegans. The ypsR/ytbR mutant however produces a similar AHL profile to that of the parent strain [19] and therefore AHL production per se is not required for biofilm formation. The intermediate level biofilms formed by the single ypsR, ytbR and ytbI mutants (data not shown) reflect the interdependent nature of the Y. pseudotuberculosis QS system while the lack of biofilm attenuation observed for the ypsI mutant suggested that the AHLs synthesized via YtbI are primarily responsible for the biofilm phenotype observed.
A number of Gram-negative bacterial species rely on flagellar-mediated motility for specific stages of biofilm formation [38]. For example, in E. coli, mutations which lead to either the loss of flagella or flagella function (which include fliC or flhD) are unable to form mature biofilms indicating that the presence of functional flagella is a pre-requisite for biofilm development in a PVC attachment model [53]. Similarly, non-motile yet flagellate P. aeruginosa PA01 flgK mutants and Erwinia carotovora fliC or motA mutants cannot form biofilms on PVC surfaces [54], [55]. Furthermore, in Y. enterocolitica, mutations that abolish the structure or rotation of the flagellar greatly reduced biofilm formation in PVC microplate assays [56]. Thus, given the links between biofilm formation, flagella-mediated motility and the regulation of the two key motility regulators, flhDC and fliA by QS in Y. pseudotuberculosis [19], we investigated the contribution of motility to biofilm formation on C. elegans. Surprisingly, a Y. pseudotuberculosis fliC mutant formed similar biofilms to the parent strain indicating that on the nematode, the presence of flagellar is not a pre-requisite for biofilm formation. This provides further evidence to suggest that the biofilm developmental pathway on the living nematode surface is distinct from that occurring on an abiotic surface. Since flagellins are potent inducers of the innate immune response and are often considered as flags revealing the presence of bacteria [57], it may therefore be advantageous for Yersinia to repress their expression during growth on living surfaces.
Despite the lack of biofilm attenuation for the fliC mutant, non-motile strains with mutations in flhA, a structural component of the flagellar export apparatus as well as the motility cascade regulators, flhDC and fliA were significantly attenuated. Since QS governs the expression of key motility regulators [19] these data suggested that biofilm formation on C. elegans by Y. pseudotuberculosis was linked to QS via the motility cascade. As Y. pestis has a frameshift mutation in flhD, biofilm formation on C. elegans in Y. pestis may well be governed differently to Y. pseudotuberculosis.
Y. enterocolitica secretes FOP proteins such as YplA via the flagellar type III secretion apparatus [34]. Consequently, we considered it possible that the loss of Y. pseudotuberculosis FOP proteins by mutation of the motility genes may have been responsible for biofilm attenuation. However, SDS-PAGE analysis of the extracellular protein profile of these strains did not reveal any novel FOP proteins but rather the presence of several proteins associated with the Yop virulon and type III secretion. In particular, LcrV which is associated with the tip of the injectisome and with pore formation across the host cell membrane, YopN, a plug considered to limit Yop effector translocation through the needle and YopH, a phosphotyrosine phosphatase effector protein which inhibits phagocytosis (reviewed by [2]). Our findings are consistent with observations made by [58] that deletion of flhDC resulted in the up-regulation of the yop regulon in Y. enterocolitica as a consequence of FlhDC-mediated repression of the Yop virulon regulator gene, virF.
Since QS in Y. pseudotuberculosis regulates flhDC and fliA [19] we also examined cell free supernatants of strains with mutations in the ypsRI and ytbRI loci for the up-regulation of Yop virulon proteins. Apart from the single ypsI mutant, which exhibited the parental phenotype, each of the QS mutants exhibited the same protein profile on SDS-PAGE as the flhDC and fliA mutants when grown at 30°C in the presence of Ca2+. Since both injectisome and Yop effector proteins were up-regulated, these data suggest that QS represses the Yop virulon via the actions of FlhDC on virF. In addition, it is clear that mutation of QS results in the loss of both the temperature and Ca2+ dependence characteristic of type III secretion in Yersinia. Thus in Y. pseudotuberculosis, QS positively regulates motility but negatively controls type III secretion indicating that both phenotypes are population dependent. This would suggest that in the planktonic phase at high population densities in the presence of eukaryotic target cells, Yop secretion would be shut down in favour of bacterial migration to new sites where a fall in QS signal concentrations would stimulate the resumption of Yop secretion.
With respect to the biofilm phenotype of the QS and motility mutants, the de-repression of type III secretion at temperatures below 37°C suggested that type III secretion blocked biofilm formation on C. elegans. Since the Yop virulon genes are located entirely on the pYV plasmid, we examined the biofilm phenotype of the plasmid-cured parent, ypsI/ytbI and flhDC mutants respectively. The loss of pYV from the parent Y. pseudotuberculosis strain had no impact on biofilm formation an observation which is fully in agreement with Joshua et al., (2003) [6] who examined both YpIII and a range of Y. pseudotuberculosis strains with or without the virulence plasmid. However the attenuation of biofilm formation observed for both the ypsI/ytbI and flhDC mutants could be overcome by curing pYV, a finding which implied that QS represses the expression of pYV encoded gene(s) which block biofilm formation in the presence of Ca2+ and at 22°C, the temperature at which the C. elegans assays are carried out. Additional support for these observations was obtained when seed cultures of the Y. pseudotuberculosis parent strain were grown under conditions permissive for Yop release (37°C in the absence of Ca2+) and then transferred onto Ca2+-free modified NGM plates at 22°C whereupon biofilms did not form on C. elegans.
To rule out the possibility that other genes located on the pYV plasmid were responsible for the biofilm phenotype rather than the presence of a functional type III secretion system, we introduced a yscJ mutation into the ypsI/ytbI double mutant. The newly generated triple mutant resulted in the loss of type III secretion at 30°C in the presence of Ca2+ and the restoration of biofilm formation on C. elegans. This strongly implies that the presence of an intact injectisome blocks biofilm formation on C. elegans. However, these data alone could not determine whether the reduction in biofilm was due to the presence of an intact injectisome, extracellular Yops or both. Evidence to suggest that the type III injectisome rather than the Yop effectors were responsible for attenuating biofilm formation on C. elegans was obtained by first conditioning seed cultures of Y. pseudotuberculosis at 37°C in Ca2+ containing media prior to carrying out biofilm assays. We reasoned that the conditioned Y. pseudotuberculosis cells would possess intact injectisomes but would not release Yops [59]–[63]. Furthermore, the presence of Ca2+ in the NGM agar would continue to suppress Yop secretion during the biofilm assays. When biofilm assays were performed using pre-conditioned Y. pseudotuberculosis cells biofilm formation was suppressed. These data appear to preclude a requirement for extracellular Yops in order for biofilm formation to take place. The simplest explanation is that the presence of the fully formed needle acts as a physical barrier which blocks the interaction between a key, chromosomally encoded bacterial surface component and the nematode surface. This would also be consistent with the loss of biofilm formation which results from the mutation of a number of C. elegans surface-determining genes [64], [65]. However, at this stage we cannot rule out the possibility that contact between the injectisome and C. elegans results in the repression of as yet unidentified genes required for biofilm formation.
Materials and Methods
Strains and growth conditions
The Y. pseudotuberculosis, Escherichia coli and C. elegans strains and the plasmids used in this study are listed in Table S1 and Table S2 respectively. To aid visualisation of Y. pseudotuberculosis in biofilm assays, the bacterial cells were transformed with pSB2020 [66] which constitutively expresses gfp3. To determine whether biofilm formation on C. elegans could be attenuated by AHL hydrolysis, Y. pseudotuberculosis YpIII was also transformed with the lactonase gene, aiiA on pSA236 as described before [19]. Except where stated, bacterial cultures were routinely grown with shaking at 200 rpm in L broth Lennox [67] or on agar plates containing the appropriate antibiotics buffered to pH 6.8 with Mops (3-N-morpholino) propanesulphonic acid (YLBmops) to reduce alkaline hydrolysis of AHLs during bacterial growth [68]. To promote yop expression at 37°C some experiments were performed in YLBmops supplemented with MgCl2 (20 mM) and sodium oxalate (20 mM) as previously described [58]. Where required, pYV was cured from Y. pseudotuberculosis by the repeated sub-culture of white colonies onto Congo red-magnesium oxalate (CRMOX) plates [69].
The C. elegans wild-type (N2 Bristol) strain was obtained from the Caenorhabditis Genetics Centre (University of Minnesota, St. Paul, MN) and maintained on modified NGM plates [70] lacking MgCl2, seeded with E. coli OP50 unless otherwise stated. For Yop induction assays NGM was supplemented with MgCl2 (20 mM) and sodium oxalate (20 mM) but CaCl2 was omitted.
Y. pseudotuberculosis/C. elegans biofilm assay
NGM plates were seeded with 1 ml of the appropriate Y. pseudotuberculosis strain grown overnight at 30°C unless otherwise stated. For some C. elegans biofilm experiments, NGM agar plates were modified by the addition of sodium oxalate (20 mM) and MgCl2 (20 mM) to promote Yop secretion. For the assays in which biofilm severity incidence was calculated, Y. pseudotuberculosis were spread evenly over the agar surface, dried to remove excess liquid and 20–30 young adult C. elegans were aseptically transferred to the seeded plates. After incubation for 22°C for 24 h (unless otherwise stated), the worms were examined under low magnification using a Nikon SMZ1000 microscope and biofilm accumulation was classed as level 0 if no biofilm formed (e.g. Figure 1C); level 1 indicating a small accumulation of biofilm around the anterior end of the worm (e.g. Figure S1B); level 2 denoted larger accumulations of biofilm around the anterior end of the worm with some pockets of biofilm spreading back from the head (e.g. Figure S1C); level 3 by large accumulations of biofilm around the anterior end of the worm which extended to other parts of the nematode body surface (e.g. Figures 1A and 2B). Confocal images of C. elegans were taken using a Zeiss LSM700 inverted microscope. Replicate Z-stacks were taken at 5 µm intervals. The Zeiss Zen software package was used for image analysis. The level of biofilm accumulation on C. elegans was denoted as the biofilm severity incidence and was calculated according to the method of Tarr [71]: Biofilm severity incidence = {[∑(level X number of samples in this level)]/(highest level X total sample numbers)} X 100%. All assays in which the level of biofilm severity was assessed were carried out double blind, with at least three or four replicates and each experiment was performed more than once. The error bars shown on figures 4, 6 and 9 represent the standard deviation from the mean and when necessary independent two-sample t-tests were performed with values for p and n given in the text and on histograms where appropriate. For some experiments the presence of the N-acetyl-D-glucosamine in the ECM of Y. pseudotuberculosis biofilms was demonstrated using a wheat germ agglutinin (WGA)-rhodamine (WGA-R) conjugate as described by [12]. Extracellular DNA present in the biofilms was stained with DAPI following the method of Vilain et al., [72] in which low concentrations of DAPI are demonstrated to label the extracellular biofilm matrix without penetrating the bacterial cell and staining the intracellular DNA.
To determine whether biofilm formation was attenuated when worms were infected with Y. pseudotuberculosis containing the AHL lactonase AiiA, aiiA was excised from pSA302 [19] as an EcoRI fragment and then sub-cloned into the chloramphenicol resistant vector pSU18 [73] to give pSA236 which was transformed into Y. pseudotuberculosis. pSU18 was transformed into Y. pseudotuberculosis to act as a vector control.
DNA manipulations
Plasmids were isolated using the Promega Wizard system, agarose gel electrophoresis and standard methods for the preparation of competent cells, DNA ligation and electroporation were performed as previously described [20]. For the purification of DNA fragments from agarose gels, Qiaquick DNA purification columns were used (Qiagen Ltd). Restriction endonucleases, DNA ligase and other DNA modification enzymes were used according to the manufacturers' instructions (Promega).
AHL detection
C. elegans infected with Y. pseudotuberculosis were removed from 40 NGM plates in M9 wash solution [74]. The worms were washed, the pellet extracted into dichloromethane, reconstituted into 20 µl of acetonitrile and analysed using a well plate overlay assay using the C. violaceum CV026 biosensor which reports the presence of AHLs by producing the purple pigment violacein [31]. To detect AHLs produced in situ in biofilms on the surface of C. elegans, Y. pseudotuberculosis and the isogenic ypsI/ytbI double mutant were each transformed with the AHL biosensor, pJBA89 [32] which expresses gfp in the presence of AHLs. Infected worms were examined using fluorescent microscopy for the presence of green fluorescent bacteria within the biofilm matrix.
Construction of Y. pseudotuberculosis mutants
Y. pseudotuberculosis YpIII strains with deletions in fliA, flhA, fliC and yscJ were constructed as follows. The fliA mutant was constructed using a modified method of [75]. The primers used for mutant construction are listed in Table S3. Briefly, primer pairs fliA1up-F/fliA1up-R and fliA1down-F/fliA1down-R were used to amplify 510 and 511bp fragments of the up - and downstream regions of fliA (positions 2069236 to 2069746 and 2070363 to 2070874 in the published Y. pseudotuberculosis IP 32953 genome sequence [76]). Primer fliA1up-R and fliAdown-F also contained 25 and 22 bp respectively of sequence homologous to the first 25 bp and last 22 bp of kanamycin from pUC4K [77]. The kanamycin cassette was amplified from pUC4K (Pharmacia) using primer km-F and km-R under the following PCR conditions: 95°C for 5 min followed by 30 cycles of 95°C for 30 s, 56°C for 30 s and 74°C for 1 min and ending with 74°C for 5 min. The second and third step PCR conditions were as follows: 95°C for 5 min followed by 30 cycles of 95°C for 30 s, 60°C for 30 s and 74°C for 2 min and ending with 74°C for 5 min. The strategy for constructing the flhA and yscJ mutants was similar to that of fliA. For flhA, primer pairs flhA1up-F/flhA1up-R and flhA1down-F/flhA1down-R were used to amplify the up - and downstream regions of flhA (positions 2017164 to 2017699 and 2019587 to 2020179 on the published IP 32953 Y. pseudotuberculosis genome sequence) whereas for yscJ, primer pairs YscJaFor/YscJupR-Tet and YscJdownF-tet/YscJbRev were used to amplify the up - and downstream regions of yscJ (positions 59172 to 59743 and 60344 to 61135) on the published Y. pseudotuberculosis IP 32953 pYV virulence plasmid sequence. For flhA, primer flhA1up-R and flhA1down-F each contained 19 bp of sequence homologous to the first 19 bp or last 19 bp of the kanamycin cassette from pUC4K whereas for yscJ, YscJupR-Tet and YscJdownF-tet contained 21 bp or 22 bp of sequence homologous to a tetracycline cassette which was amplified as a 1191 bp product from pBlue-tet (a source of the tetracycline cassette initially amplified from pBR322 using primers Tet1 and Tet2 [19] and cloned into pBluescript as an xhoI fragment). All PCR conditions were the same as those for the construction of the fliA mutant.
To complement yscJ, primers YscJF-XbaI and YscJR-SalI were used to amplify an 842 bp product from Y. pseudotuberculosis (positions 59686 to 59703 on the IP32953 published sequence) which, after cloning into pBluescript and sequencing was excised as a KpnI and PstI fragment and sub-cloned into the low copy number vector pHG327 [78]. The resulting plasmid, pHG::yscJ was transformed into the Y. pseudotuberculosis ypsI/ytbI double mutant.
Colony PCR was used to amplify a fliC homologue from Y. pseudotuberculosis using the primers DC1 and DC2 and cloned into pGEMT/easy (Promega) to give pfliC. Sequencing revealed the 1,515 bp fragment to have an open reading frame of 1,110 bp and predicted protein product of 396 amino acids that shared significant amino acid similarity to several FliC homologues and was subsequently termed fliCYp (Genbank accession number AY244555). To construct a fliC mutant 616 bp was removed from pfliC using Csp45I and replaced with a kanamycin cassette from pUC4K (Pharmacia) as a blunt end fragment. The resulting construct was cloned into pDM4 as a SphI-SpeI fragment (pDM fliC-Km) and stably integrated into the chromosome of Y. pseudotuberculosis as previously described [18], [19].
To complement the Y. pseudotuberculosis YpIII flhDC mutant [19] flhDC was amplified by PCR (primers FlhDF and FlhCR), cloned into pGEMT/Easy (Promega) and the resulting pGEM::flhDC construct, pSA220 was transformed into the Y. pseudotuberculosis flhDC mutant. The flhDC, flhA, fliA and fliC mutants were examined for motility using swim agar plate assays and microscopy and the presence of flagella proteins was determined by SDS-PAGE once isolated from 24 h overnight liquid cultures grown at 22°C as previously described [20], [79].
SDS-PAGE and protein sequencing
Proteins present in 10 ml of cell-free supernatant taken from Y. pseudotuberculosis QS and motility mutants grown to the same OD600 (overnight in YLB at 22°C, 28°C and 37°C) were concentrated by trichloroacetic acid precipitation, subjected to SDS-PAGE and the relevant bands excised. After in-gel tryptic digestion, the resulting peptides were identified by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF)-MS sequencing as previously described [20].
Supporting Information
Zdroje
1. NavarroL
AltoNM
DixonJE
2005 Functions of the Yersinia effector proteins in inhibiting host immune responses. Curr Opin Microbiol 8 21 27
2. CornelisGR
2002 Yersinia type III secretion: send in the effectors. J Cell Biol 158 401 408
3. GalanJE
Wolf-WatzH
2006 Protein delivery into eukaryotic cells by type III secretion machines. Nature 444 567 573
4. RamamurthiKS
SchneewindO
2002 Type III protein secretion in Yersinia species. Annu Rev Cell Dev Biol 18 107 133
5. DarbyC
HsuJW
GhoriN
FalkowS
2002 Caenorhabditis elegans - Plague bacteria biofilm blocks food intake. Nature 417 243 244
6. JoshuaGWP
KarlyshevAV
SmithMP
IsherwoodKE
TitballRW
2003 A Caenorhabditis elegans model of Yersinia infection: biofilm formation on a biotic surface. Microbiology-Sgm 149 3221 3229
7. HinnebuschBJ
PerryRD
SchwanTG
1996 Role of the Yersinia pestis hemin storage (hms) locus in the transmission of plague by fleas. Science 273 367 370
8. StoodleyP
SauerK
DaviesDG
CostertonJW
2002 Biofilms as complex differentiated communities. Annu Rev Microbiol 56 187 209
9. JarrettCO
DeakE
IsherwoodKE
OystonPC
FischerER
2004 Transmission of Yersinia pestis from an infectious biofilm in the flea vector. J Infect Dis 190 783 792
10. PlattHM
1994 Forward in Phylogenetic systematics of free-living nematodes.
LorenzenS
London The Ray Society i ii
11. MatzC
KjellebergS
2005 Off the hook - how bacteria survive protozoan grazing. Trends Microbiol 13 302 307
12. TanL
DarbyC
2004 A movable surface: Formation of Yersinia sp biofilms on motile Caenorhabditis elegans. J Bacteriol 186 5087 5092
13. SalmondGPC
BycroftBW
StewartGSAB
WilliamsP
1995 The bacterial enigma-cracking the code of cell-cell communication. Mol Microbiol 16 615 624
14. WilliamsP
CámaraM
HardmanA
SwiftS
MiltonD
2000 Quorum sensing and the population-dependent control of virulence. Philos T Roy Soc B 355 667 680
15. SwiftS
DownieJA
WhiteheadNA
BarnardAML
SalmondGPC
WilliamsP
2001 Quorum sensing as a population-density-dependent determinant of bacterial physiology. Adv Microb Phys 45 199 270
16. CámaraM
WilliamsP
HardmanA
2002 Controlling infection by tuning in and turning down the volume of bacterial small-talk. Lancet Infect Dis 2 667 676
17. WilliamsP
WinzerK
ChanWC
CámaraM
2007 Look who's talking: communication and quorum sensing in the bacterial world. Philos T Roy Soc B 362 1119 1134
18. AtkinsonS
ThroupJP
StewartGSAB
WilliamsP
1999 A hierarchical quorum sensing system in Yersinia pseudotuberculosis is involved in the regulation of motility and clumping. Mol Microbiol 33 1267 1277
19. AtkinsonS
ChangCY
PatrickHL
BuckleyCMF
WangY
2008 Functional interplay between the Yersinia pseudotuberculosis YpsRI and YtbRI quorum sensing systems modulates swimming motility by controlling expression of flhDC and fliA. Mol Microbiol 69 137 151
20. AtkinsonS
ChangCY
SockettRE
CámaraM
WilliamsP
2006 Quorum sensing in Yersinia enterocolitica controls swimming and swarming motility. J Bacteriol 188 1451 1461
21. KirwanJP
GouldTA
SchweizerHP
BeardenSW
MurphyRC
2006 Quorum-sensing signal synthesis by the Yersinia pestis acyl homoserine lactone synthase YspI. J Bacteriol 188 784 788
22. SwiftS
IsherwoodKE
AtkinsonS
OystonP
StewartGSAB
1999 Quorum sensing in Aeromonas and Yersinia.
EnglandR
HobbsG
BaintonNJ
RobertsDM
Microbial Signalling and Communication Cambridge, UK Cambridge University Press 85 104
23. IsherwoodEK
2001 Quorum sensing in Yersinia pestis. PhD thesis The University of Nottingham
24. YoungGM
2004 Flagella:Organelles for motility and protein secretion.
CarnielE
HinnebuschBJ
Yersinia molecular and cellular biology Wymondham Horizon bioscience 243 256
25. CharltonTS
de NysR
NettingA
KumarN
HentzerM
2000 A novel and sensitive method for the quantification of N-3-oxoacyl homoserine lactones using gas chromatography-mass spectrometry: application to a model bacterial biofilm. Environ Microbiol 2 530 541
26. LynchMJ
SwiftS
KirkeDF
KeevilCW
DoddCER
2002 The regulation of biofilm development by quorum sensing in Aeromonas hydrophila. Environ Microbiol 4 18 28
27. AtkinsonS
CámaraM
WilliamsP
2007 N-Acylhomoserine lactones, quorum sensing and biofilm development in Gram-negative bacteria.
KjellbergS
GivskovM
The biofilm mode of life. Mechanisms and adaptations Wymondham Horizon bioscience 95 122
28. BjarnsholtT
JensenPO
BurmolleM
HentzerM
HaagensenJAJ
2005 Pseudomonas aeruginosa tolerance to tobramycin, hydrogen peroxide and polymorphonuclear leukocytes is quorum-sensing dependent. Microbiology-Sgm 151 373 383
29. JensenPO
BjarnsholtT
PhippsR
RasmussenTB
CalumH
2007 Rapid necrotic killing of polymorphonuclear leukocytes is caused by quorum sensing-controlled production of rhamnolipid by Pseudomonas aeruginosa. Microbiology-Sgm 153 1329 1338
30. Allesen-HolmM
BarkenKB
YangL
KlausenM
WebbJS
2006 A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol Microbiol 59 1114 1128
31. McCleanKH
WinsonMK
FishL
TaylorA
ChhabraSR
1997 Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology-Uk 143 3703 3711
32. AndersenJB
HeydornA
HentzerM
EberlL
GeisenbergerO
2001 gfp-based N-acyl homoserine-lactone sensor systems for detection of bacterial communication. Appl Environ Microbiol 67 575 585
33. RocheDM
ByersJT
SmithDS
GlansdorpFG
SpringDR
2004 Communications blackout? Do N-acylhomoserine lactone-degrading enzymes have any role in quorum sensing? Microbiology-Sgm 150 2023 2028
34. YoungGM
SchmielDH
MillerVL
1999 A new pathway for the secretion of virulence factors by bacteria: The flagellar export apparatus functions as a protein-secretion system. Proc Natl Acad Sci U S A 96 6456 6461
35. Saijo-HamanoY
ImadaK
MinaminoT
KiharaM
ShimadaM
2010 Structure of the cytoplasmic domain of FlhA and implication for flagellar type III protein export. Mol Microbiol 76 260 268
36. Silva-HerzogE
FerracciF
JacksonMW
JosephSS
PlanoGV
2008 Membrane localization and topology of the Yersinia pestis YscJ lipoprotein. Microbiology-Sgm 154 593 607
37. MarenneMN
MotaLJ
CornelisGR
2004 The pYV plasmid and the Ysc-Yop Type III secretion system.
CarnielE
HinnebuschBJ
Yersinia molecular and cellular biology Wymondham Horizon Bioscience 319 348
38. O'TooleG
KaplanHB
KolterR
2000 Biofilm formation as microbial development. Annu Rev Microbiol 54 49 79
39. KjellebergS
MolinS
2002 Is there a role for quorum sensing signals in bacterial biofilms? Curr Opin Microbiol 5 254 258
40. SauerK
CamperAK
EhrlichGD
CostertonJW
DaviesDG
2002 Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J Bacteriol 184 1140 1154
41. KlausenM
Aes-JorgensenA
MolinS
Tolker-NielsenT
2003 Involvement of bacterial migration in the development of complex multicellular structures in Pseudomonas aeruginosa biofilms. Mol Microbiol 50 61 68
42. WebbJS
ThompsonLS
JamesS
CharltonT
Tolker-NielsenT
2003 Cell death in Pseudomonas aeruginosa biofilm development. J Bacteriol 185 4585 4592
43. TanL
DarbyC
2006 Yersinia pestis YrbH is a multifunctional protein required for both 3-deoxy-D-manno-oct-2-ulosonic acid biosynthesis and biofilm formation. Mol Microbiol 61 861 870
44. SizemoreRK
CaldwellJJ
KendrickAS
1990 Alternate Gram staining technique using a fluorescent lectin. Appl Environ Microbiol 56 2245 2247
45. BobrovAG
KirillinaO
FormanS
MackD
PerryRD
2008 Insights into Yersinia pestis biofilm development: topology and co-interaction of Hms inner membrane proteins involved in exopolysaccharide production. Environmental Microbiology 10 1419 1432
46. DraceK
DarbyC
2008 The hmsHFRS operon of Xenorhabdus nematophila is required for biofilm attachment to Caenorhabditis elegans. Appl Environ Microbiol 74 4509 4515
47. SunYC
HinnebuschBJ
DarbyC
2008 Experimental evidence for negative selection in the evolution of a Yersinia pestis pseudogene. Proc Natl Acad Sci U S A 105 8097 8101
48. SunYC
KoumoutsiA
DarbyC
2009 The response regulator PhoP negatively regulates Yersinia pseudotuberculosis and Yersinia pestis biofilms. FEMS Microbiol Lett 290 85 90
49. KirillinaO
FetherstonJD
BobrovAG
AbneyJ
PerryRD
2004 HmsP, a putative phosphodiesterase, and HmsT, a putative diguanylate cyclase, control Hms-dependent biofilm formation in Yersinia pestis. Mol Microbiol 54 75 88
50. SimmR
FetherstonJD
KaderA
RomlingU
PerryRD
2005 Phenotypic convergence mediated by GGDEF-domain-containing proteins. J Bacteriol 187 6816 6823
51. BobrovAG
KirillinaO
PerryRD
2007 Regulation of biofilm formation in Yersinia pestis. Adv Exp Med Biol 603 201 210
52. JacobiCA
BachA
EberlL
SteidleA
HeesemannJ
2003 Detection of N-(3-oxohexanoyl)-L-homoserine lactone in mice infected with Yersinia enterocolitica serotype O8. Infect Immun 71 6624 6626
53. PrattLA
KolterR
1998 Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol Microbiol 30 285 293
54. O'TooleGA
KolterR
1998 Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol 30 295 304
55. HossainMM
TsuyumuS
2006 Flagella-mediated motility is required for biofilm formation by Erwinia carotovora subsp. carotovora. J Gen Plant Pathol 72 34 39
56. KimTJ
YoungBM
YoungGM
2008 Effect of flagellar mutations on Yersinia enterocolitica biofilm formation. Appl Environ Microbiol 74 5466 5474
57. Gomez-GomezL
BollerT
2002 Flagellin perception: a paradigm for innate immunity. Trends in Plant Sci 7 251 256
58. BlevesS
MarenneMN
DetryG
CornelisGR
2002 Up-regulation of the Yersinia enterocolitica yop regulon by deletion of the flagellum master operon flhDC. J Bacteriol 184 3214 3223
59. T
WattiauP
BrasseurR
RuysschaertJM
CornelisG
1990 Secretion of Yop Proteins by Yersiniae. Infect Immun 58 2840 2849
60. BrubakerRR
SurgallaMJ
1964 Effect of Ca2+ and Mg2+ on lysis growth and production of virulence antigens. J Infect Dis 114 13 25
61. BolinI
Wolf-WatzH
1984 Molecular cloning of the temperature inducible outer membrane protein-1 of Yersinia pseudotuberculosis. Infect Immun 43 72 78
62. BolinI
PortnoyDA
WatzHW
1985 Expression of the Temperature inducible outer membrane proteins of Yersiniae. Infect Immun 48 234 240
63. ForsbergA
ViitanenAM
SkurnikM
Wolf-WatzH
1991 The surface located YopN protein is involved in calcium signal transduction in Yersinia pseudotuberculosis. Mol Microbiol 5 977 986
64. DraceK
McLaughlinS
DarbyC
2009 Caenorhabditis elegans BAH-1 is a DUF23 protein expressed in seam cells and required for microbial biofilm binding to the cuticle. Plos One 4 e6741
65. DarbyC
ChakrabortiA
PolitzSM
DanielsCC
TanL
2007 Caenorhabditis elegans mutants resistant to attachment of Yersinia biofilms. Genetics 176 221 230
66. QaziSNA
ReesCED
MellitsKH
HillPJ
2001 Development of gfp vectors for expression in Listeria monocytogenes and other low G+C Gram-positive bacteria. Microb Ecol 41 301 309
67. LennoxES
1955 Transduction of linked genetic characters of the host by bacteriophage P1. Virology 1 190 206
68. YatesEA
PhilippB
BuckleyC
AtkinsonS
ChhabraSR
2002 N-acylhomoserine lactones undergo lactonolysis in a pH-, temperature-, and acyl chain length-dependent manner during growth of Yersinia pseudotuberculosis and Pseudomonas aeruginosa. Infect Immun 70 5635 5646
69. RileyG
TomaS
1989 Detection of pathogenic Yersinia enterocolitica by using congo red-magnesium oxalate agar medium. J Clin Microbiol 27 213 214
70. LewisJA
FlemingTJ
1995 Caenorhabditis elegans: Modern biological analysis of an organism. New York Academic Press 3 39
71. TarrSAJ
1972 The assesment of disease incidence and crop loss. London The Macmillan Press 430 454 In: Principles of plant pathology.
72. VilainS
PretoriusJM
TheronJ
BrozelVS
2009 DNA as an adhesin: Bacillus cereus requires extracellular DNA to form biofilms. Appl Environ Microbiol 75 2861 2868
73. BartolomeB
JubeteY
MartinezE
DelacruzF
1991 Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene 102 75 78
74. BrennerS
1974 Genetics of Caenorhabditis elegans. Genetics 77 71 94
75. DerbiseA
LesicB
DacheuxD
GhigoJM
CarnielE
2003 A rapid and simple method for inactivating chromosomal genes in Yersinia. FEMS Immunol Med Microbiol 38 113 116
76. ChainPSG
CarnielE
LarimerFW
LamerdinJ
StoutlandPO
2004 Insights into the evolution of Yersinia pestis through whole genome comparison with Yersinia pseudotuberculosis. Proc Natl Acad Sci U S A 101 13826 13831
77. YanischperronC
VieiraJ
MessingJ
1985 Improved M13 phage cloning vectors and host strains - nucleotide-sequences of the M13, Mp18 and pUC19 vectors. Gene 33 103 119
78. StewartGSAB
LubinskyminkS
JacksonCG
CasselA
KuhnJ
1986 pHG165-A pBR322 Copy Number Derivative of pUC8 for Cloning and Expression. Plasmid 15 172 181
79. SockettRE
1998 Characterising Flagella and Motile Behaviour.
WilliamsP
SalmondG
KetleyJM
Methods in microbiology: methods for studying pathogenic bacteria London Academic Press 227 237
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2011 Číslo 1- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Salivary Gland NK Cells Are Phenotypically and Functionally Unique
- Genetic Epidemiology of Tuberculosis Susceptibility: Impact of Study Design
- Early Target Cells of Measles Virus after Aerosol Infection of Non-Human Primates
- Multiple Plant Surface Signals are Sensed by Different Mechanisms in the Rice Blast Fungus for Appressorium Formation
- Biofilm Development on by Is Facilitated by Quorum Sensing-Dependent Repression of Type III Secretion
- Distinct Patterns of IFITM-Mediated Restriction of Filoviruses, SARS Coronavirus, and Influenza A Virus
- Molecular Basis of Increased Serum Resistance among Pulmonary Isolates of Non-typeable
- A Helminth Immunomodulator Exploits Host Signaling Events to Regulate Cytokine Production in Macrophages
- HCMV Spread and Cell Tropism are Determined by Distinct Virus Populations
- Characteristics of the Earliest Cross-Neutralizing Antibody Response to HIV-1
- Induction of a Peptide with Activity against a Broad Spectrum of Pathogens in the Salivary Gland, following Infection with Dengue Virus
- Structural Basis for the Recognition of Cellular mRNA Export Factor REF by Herpes Viral Proteins HSV-1 ICP27 and HVS ORF57
- Identification and Characterization of the Host Protein DNAJC14 as a Broadly Active Flavivirus Replication Modulator
- Dual-Use Research and Technological Diffusion: Reconsidering the Bioterrorism Threat Spectrum
- Pathogenesis of the 1918 Pandemic Influenza Virus
- A Cardinal Role for Cathepsin D in Co-Ordinating the Host-Mediated Apoptosis of Macrophages and Killing of Pneumococci
- Critical Role of IRF-5 in the Development of T helper 1 responses to infection
- The Pel Polysaccharide Can Serve a Structural and Protective Role in the Biofilm Matrix of
- Selective C-Rel Activation via Malt1 Controls Anti-Fungal T-17 Immunity by Dectin-1 and Dectin-2
- Imaging Single Retrovirus Entry through Alternative Receptor Isoforms and Intermediates of Virus-Endosome Fusion
- Aerosols Transmit Prions to Immunocompetent and Immunodeficient Mice
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Dual-Use Research and Technological Diffusion: Reconsidering the Bioterrorism Threat Spectrum
- Pathogenesis of the 1918 Pandemic Influenza Virus
- Critical Role of IRF-5 in the Development of T helper 1 responses to infection
- A Cardinal Role for Cathepsin D in Co-Ordinating the Host-Mediated Apoptosis of Macrophages and Killing of Pneumococci
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



