-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaInsect Resistance to Toxin Cry2Ab Is Conferred by Mutations in an ABC Transporter Subfamily A Protein
Transgenic crops expressing the insecticidal protein Cry2Ab from Bacillus thuringiensis (Bt) are used worldwide to suppress damage by lepidopteran pests, often used in combination with Cry1Ac toxin to delay resistance evolution. Until now, the Cry2Ab mode of action and the mechanism of resistance were unknown, with field-isolated Cry2Ab resistant Helicoverpa armigera showing no cross-resistance to Cry1Ac. In this study, biphasic linkage analysis of a Cry2Ab-resistant H. armigera family followed by EPIC marker mapping and candidate gene sequencing identified three independent INDEL mutations in an ATP-Binding Cassette transporter subfamily A gene (ABCA2). A deletion mutation was identified in the same gene of resistant H. punctigera. All four mutations are predicted to truncate the ABCA2 protein. This is the first molecular genetic characterization of insect resistance to the Cry2Ab toxin, and detection of diverse Cry2Ab resistance alleles will contribute to understanding the micro-evolutionary processes that underpinned lepidopteran Bt-resistance.
Published in the journal: . PLoS Genet 11(11): e32767. doi:10.1371/journal.pgen.1005534
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005534Summary
Transgenic crops expressing the insecticidal protein Cry2Ab from Bacillus thuringiensis (Bt) are used worldwide to suppress damage by lepidopteran pests, often used in combination with Cry1Ac toxin to delay resistance evolution. Until now, the Cry2Ab mode of action and the mechanism of resistance were unknown, with field-isolated Cry2Ab resistant Helicoverpa armigera showing no cross-resistance to Cry1Ac. In this study, biphasic linkage analysis of a Cry2Ab-resistant H. armigera family followed by EPIC marker mapping and candidate gene sequencing identified three independent INDEL mutations in an ATP-Binding Cassette transporter subfamily A gene (ABCA2). A deletion mutation was identified in the same gene of resistant H. punctigera. All four mutations are predicted to truncate the ABCA2 protein. This is the first molecular genetic characterization of insect resistance to the Cry2Ab toxin, and detection of diverse Cry2Ab resistance alleles will contribute to understanding the micro-evolutionary processes that underpinned lepidopteran Bt-resistance.
Introduction
In recent decades, agriculture has increasingly come to rely on toxins encoded by the Gram-positive bacterium Bacillus thuringiensis (Bt) for the production of insect–resistant transgenic crops. Narrow spectrum insecticides such as the protoxin crystals produced by Bt during sporulation are highly specific for certain insect groups including the Lepidoptera, Diptera and Coleoptera (e.g., [1]). Bt sprays have been used for many years, gaining widespread acceptance in pest management due to their relative target-specificity and their safety for humans, most other organisms, and the environment. However, the increasing cultivation of Bt transgenic crops poses a significant risk with various field populations of major lepidopteran pests reported to have developed resistance [2–4], threatening the sustainability of this strategy for crop protection. Indeed a major reason for the uptake of Bt was the evolution of resistance to chemical insecticides such as organochlorides, synthetic pyrethroids, and organophosphates in pests such as the cotton bollworm Helicoverpa armigera. This species is one of the most damaging and economically important lepidopteran pests known worldwide in a variety of crops, and one of four major pests in the genus Helicoverpa, the others being H. punctigera, H. zea and H. assulta [5–8].
For control of lepidopteran pests, genes encoding members of the Cry1A family were the first to be used in transgenic crops. However resistance alleles to the Cry1Ac toxin have been reported in field-collected H. punctigera from Australia [9,10], and H. armigera from China [11–13]. Resistance to Cry1Ac has also been reported in Indian populations of Pectinophora gossypiella [14] and H. zea from the New World [15–17]. Genetic studies on the field-derived strains have provided critical insight into the mode of action of this toxin, by identifying key receptors present on the surface of midgut epithelial cells (e.g., [18–23]; see also [24–26]). This binding of the activated toxins to specific receptors is crucial for formation of pores in the affected cells, leading eventually to the death of the larvae.
Resistance to the Cry1Ac toxin in the Lepidoptera was first shown to be associated with mutation of a gene encoding a 12-cadherin domain protein. Deletions of different lengths were observed in various regions of the gene in, e.g., Heliothis virescens [27], P. gossypiella [28] and H. armigera [11–13,24], as well as insertions by transposable elements [12,29,30]. Down regulation of cis-mediated transcription of the trypsin gene HaTryR allele due to mutations at the promoter region, mis-splicing of the ABCC2 gene, and a deletion mutation of the Aminopeptidase N (APN) gene have also been demonstrated to lead to resistance to Cry1Ac in H. armigera [31–33]. Recently, mutations in the ABCC2 gene belonging to Family C of the ATP-Binding Cassette (ABC) transporter family have also been shown to confer resistance to Cry1Ab and Cry1Ac in H. virescens [34], Plutella xylostella [35], Bombyx mori [36], Spodoptera exigua [37], and H. armigera [32]. Heterologous expression of ABCC2 from resistant and susceptible B. mori has shown that it aids in pore formation [38], and modification or deletion of ABCC2 is hypothesized to block the final step in the toxin's mode of action [39].
To prolong the efficacy of individual Bt toxins as transgenic control agents, multiple Bt genes, encoding different toxins with different modes of action, have been incorporated into plants. The second Bt gene adopted in many countries in transgenic plants has been a member of the Cry2A family, Cry2Ab. Cry1A resistance due to mutations in the cadherin or ABCC2 genes is not known to confer cross-resistance to Cry2Ab [22,34]. However, Cry2Ab resistance has now been reported in various lepidopteran pests (e.g., P. gossypiella [40]; H. zea [41]). In Australia, field-derived Cry2Ab resistance alleles in H. armigera and H. punctigera were first isolated in the summer of 2002/2003 and 2004/2005 respectively, and were used to establish the homozygous resistant lines SP15 [42] and Hp4-13 respectively [43]. All Cry2Ab resistant H. armigera and H. punctigera alleles isolated from Australia to-date have been shown to be recessive [9,21,42,43]. Isolates were captured using the "F2 screen" method [44] with a discriminating dose of Cry2Ab toxin, and confirmed as allelic by complementation tests [83]. These isolates are being used in F1 tests which involved crossing them to a field-collected insect (of unknown genotype), and screening the F1 offspring for resistance [45–47]. These F1 screens have estimated the frequency of Cry2Ab-resistance-conferring alleles in H punctigera and H. armigera field populations to range between 0.010 and 0.047, and 0.015 and 0.044, respectively, with no significant linear trend over time (from 2007 to 2014) [48–50].
The F1 and F2 screen techniques do not directly reveal the molecular identity of such resistance alleles, and the molecular basis of the Cry2Ab resistance is likely due to specific target-site alterations located within the midgut [21]. Whether the H. armigera and H. punctigera Cry2Ab resistance genes are homologous is not known, although parallel evolution in orthologous ABCC2 genes leading to Cry1Ac resistance in different species has been reported (e.g., [34,35]). A third generation of transgenic cotton (Bollgard III (BGIII)) expressing three Bt toxin genes (Cry1Ac, Cry2Ab and Vip3A) will soon become available in Australia. In light of these developments and whilst the Australian industry adopts a pre-emptive strategy to manage resistance to Bt, several key assumptions of this strategy are theoretically sound but empirically untested. Marker-assisted detection of the resistance alleles in insect populations will therefore not only enable a more efficient monitoring effort but will also enable assumptions about the ecology of resistance to be rigorously examined.
In this paper we report on the identification of the Cry2Ab resistance gene in H. armigera using linkage mapping and a chromosome walk with the assistance of exon-primed intron-crossing (EPIC)-PCR markers. This gene, which is expressed in the midgut, encodes an ABC transporter in the A subfamily—ABCA2—and is the likely site of mutations conferring resistance to Cry2Ab. By screening additional lab-isolated resistant lines derived from field-collected materials, we show that resistance to the Cry2Ab toxin in H. armigera occurred through independent evolutionary events involving different mutations, all of which were located in different exons of the same ABCA2 gene in both species. This work therefore provides the first insight into the detailed mode of action of a Cry2A toxin, which is conserved across different lepidopteran species, and is of considerable significance for the management of Bt resistance globally.
Results
Identification of a linkage group carrying the Cry2Ab resistance gene
The absence of crossing-over in female Lepidoptera makes it possible to map a recessive trait such as the Cry2Ab resistance in SP15 to a linkage group using biphasic linkage analysis with AFLPs as genetic markers [18]. Progeny from a female-informative backcross family were bioassayed with a discriminating dosage of Cry2Ab; 161 AFLPs segregating in this family were grouped into 31 independently assorting linkage groups. Linkage to Cry2Ab resistance was tested by comparing bioassayed survivors with untreated control progeny. Only AFLP linkage group (LG) 8 showed a significant association with resistance (χ2 = 19.44, P < 0.001; Fig 1); all 29 treated survivors were homozygous for the SP15 homolog of this linkage group.
Fig. 1. Tests of the association of AFLP linkage groups with Cry2Ab1 resistance in a bioassay of backcross progeny. 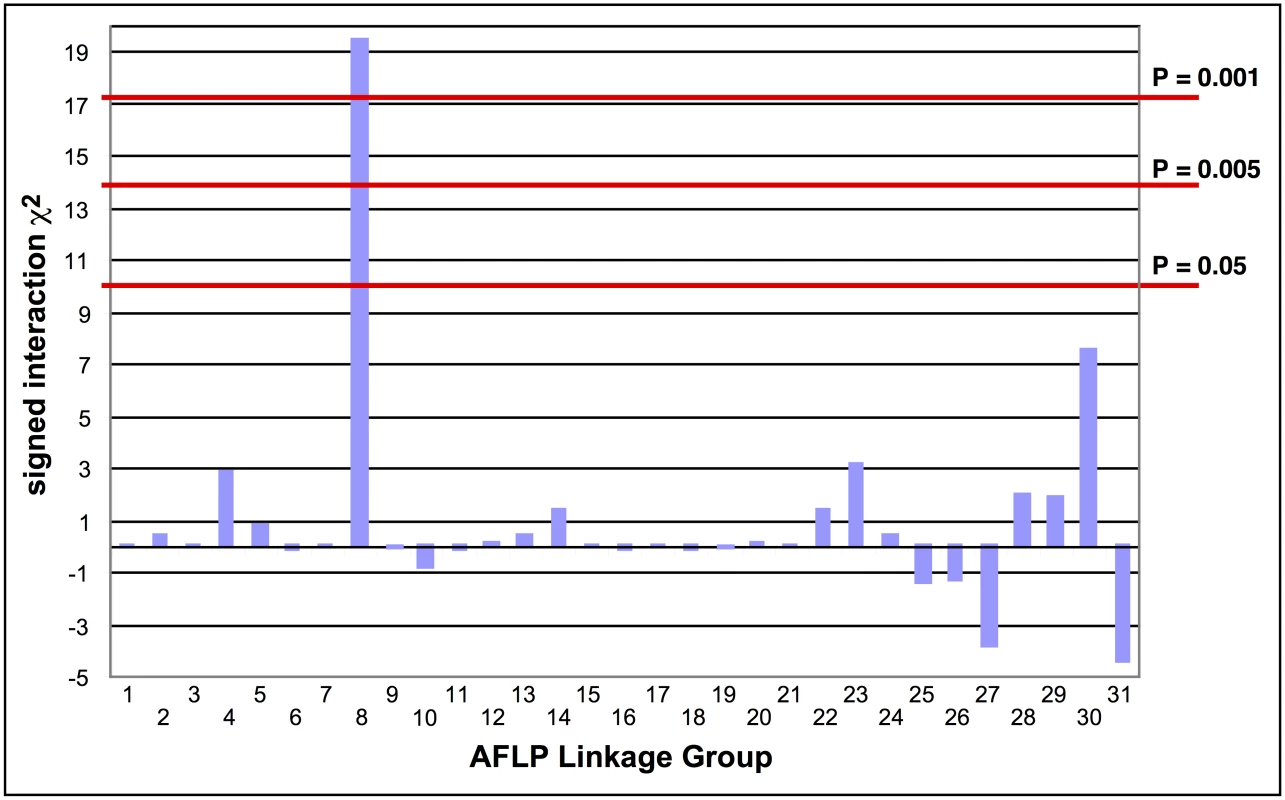
The signed interaction χ2 value shows the positive or negative association of the AFLP linkage group from the resistant grandparent with resistance in the progeny. Horizontal lines depict the Bonferroni-corrected probability values of P = 0.05, P = 0.005, and P = 0.001. Only AFLP linkage group 8 shows a significant positive association with resistance. Southern blot analysis of RFLPs in an unrelated Cry2Ab-susceptible H. armigera family showed that one AFLP from LG8 was linked to ribosomal protein gene RpL22. RpL22 in B. mori is located on chromosome 17 (BmChr17, KAIKObase [51]). Using specific probes for additional ribosomal protein genes, the Cry2Ab-resistance-associated linkage group in H. armigera was also shown to carry genes for RpL38 and RpS24, confirming homology with BmChr17 [52]. This assignment excludes a number of previously-identified genes as candidates for Cry2Ab resistance. Chromosomes harbouring homologs of previously-identified Cry1Ac resistance mutations in H. virescens include BmChr06 with the 12-cadherin-domain protein [27], BmChr15 with the ABCC2 protein [34], and BmChr21 with the BtR-5 gene [53]. Moreover, genes for previously identified Cry1Ac binding proteins map to chromosomes other than BmChr17: several aminopeptidase genes are located on BmChr09 [54], a membrane-bound alkaline phosphatase gene maps to BmChr03 [23], and the P252 glycoprotein gene is on BmChr25 [55,56]. Although different levels of cross-resistance between Cry1A and Cry2A toxins have been reported in H. armigera from China [57,58], in H. virescens [59], H. zea and P. gossypiella ([40], see also [60]); independent segregation of BmChr17 relative to all of these other chromosomes is nevertheless consistent with the absence of cross-resistance between Cry1Ac and Cry2Ab in both the SP15 H. armigera [42] and Hp4-13 H. punctigera [9,10] lines, thereby supporting the notion that the two toxins have different modes of action. However, we found that the ortholog of the bre-5 glycosyltransferase gene in a mutant of the nematode C. elegans resistant to the Cry4B toxin [19] is located on BmChr17 (Fig 2). This gene was therefore further investigated as a candidate gene for Cry2Ab resistance.
Fig. 2. Linkage map of B. mori Chromosome 17 (BmChr17), showing syntenic genes used in mapping of H. armigera. 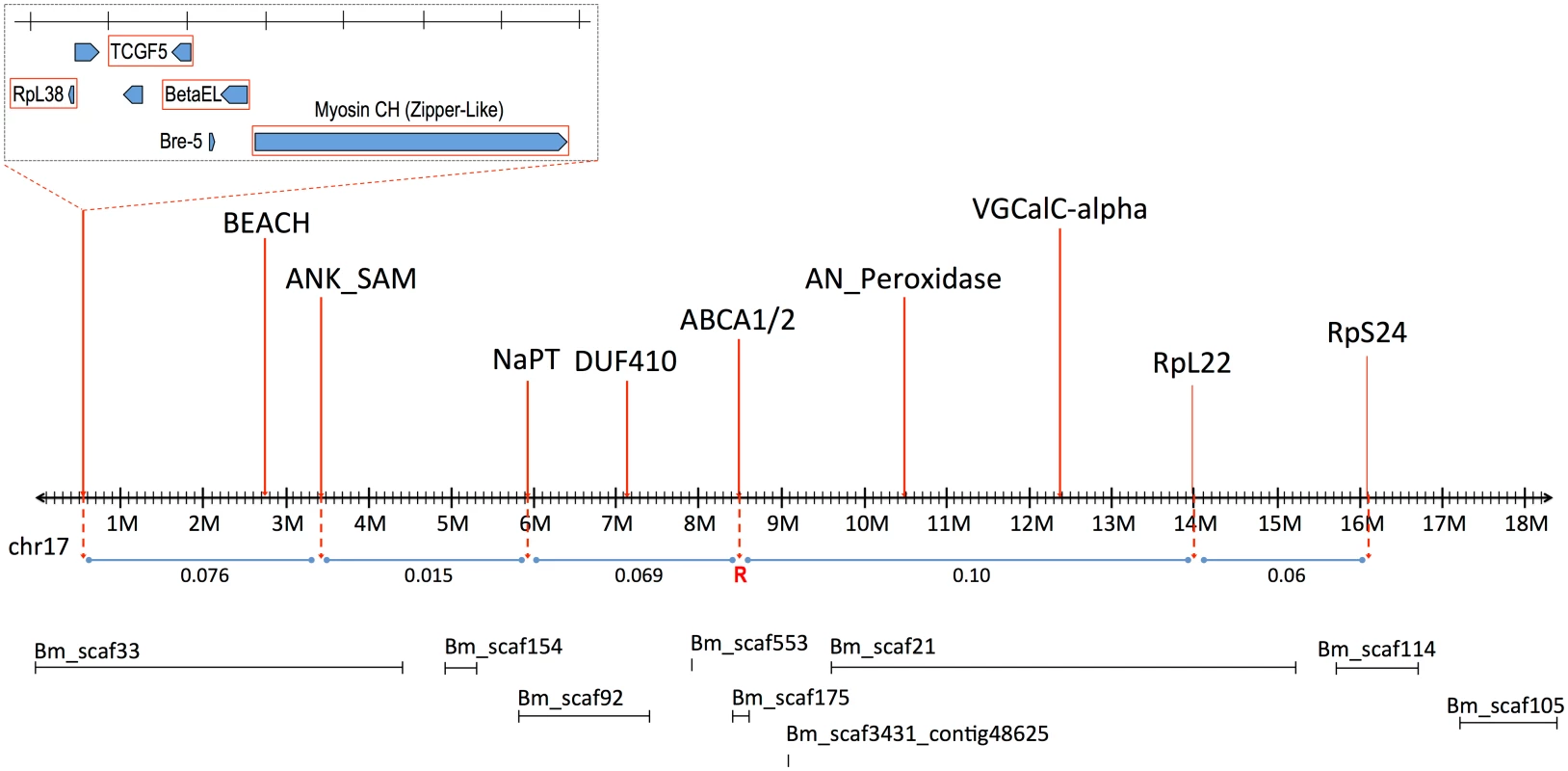
Approximate recombination rates between genes are provided in relation to the putative Cry2Ab resistance gene, indicated by ‘R’. The B. mori scaffolds on BmChr17 are also provided as obtained from KAIKObase <http://sgp.dna.affrc.go.jp/KAIKObase/>. Additional linkage mapping in a male-informative backcross and two F2 families was performed to further localize the resistance locus. A preliminary map based on 72 progeny from these families gave the gene order and recombination values as follows:
The order and spacing of the three marker loci was similar to that in B. mori on BmChr17. However, the large fraction of recombinants between the resistance locus and bre-5 ruled out the latter as a candidate (Fig 2).
Linkage mapping
The Cry2Ab resistance gene was further localised within BmChr17 using recombinational mapping in backcrosses with F1 males. For this work, markers were developed from H. armigera orthologs for genes mapped along BmChr17. Sequences allowing design of EPIC-PCR primers for the H. armigera orthologs for these genes were obtained from transcriptome sequencing of midgut RNA extracted from larvae of the GR susceptible colony.
Recombinational analysis of selected markers in H. armigera showed the linkage order of these markers to be the same as in B. mori (Fig 2), greatly assisting the subsequent analysis which employed the B. mori genome as a reference framework. Analysis of recombination rates between 3 of these markers (RpL38, Zip2, VGCal-A) and the Cry2Ab resistance allele placed it between Bre-5 (BGIBMGA005534) and RpL22 (BGIBMGA006986, at nt 14152986). The markers for the voltage-gated channel protein gene (orthologous to BGIBMGA007009, starting at 12341859 on BmChr17 –see S1 Table) further restricted the area containing the resistance gene. The target area could however be more narrowly defined, since the gene is ~10cM from BGIBMGA005534 and ~16cM from BGIBMGA006986; these genes are located at ~3Mbp and ~10Mbp respectively on the BmChr17 sequence (see S1 Table). In fine scale analysis of this region, markers for the genes BEACH, ANK_SAM, NaPT, DUF410, and AN_Peroxidase all showed recombination with the resistance trait. Of these, the marker for DUF410 (BGIBMGA007299, located at 7124451 on BmChr17) most closely restricted the target region on the proximal side, corresponding to less than 3Mbp of BmChr17, and containing fewer than 30 genes (S1 Table).
Two ABC transporter A subfamily genes are located adjacently between nts 8466000–8564000 on BmChr17. The first of these, termed BmABCA1, is well-predicted as BGIBMGA007221, while the other, BmABCA2, includes the partial predictions BGIBMGA007218 and BGIBMGA007217 (see [61,62] for analysis of the original uncorrected gene models). The sequence of HaABCA1, the H. armigera ortholog to the BGIBMGA007221 BmABCA1 gene, was obtained from RNAseq libraries, from total larvae of the susceptible GR colony. The EPIC-PCR marker for HaABCA1 (ABCA1; S2 Table) gave a genotype profile consistent with tight linkage to the Bt Cry2Ab resistance allele; the F2 bioassayed offspring (homozygous allele size of 272bp / 272bp) was identical to the SP15 grandmother (272bp / 272bp) in 100% of all samples tested (final n = 72). The GR grandfather was heterozygous with alleles 264bp / 282bp, leading to the F1 male being heterozygous with allele sizes 272bp / 282bp. The F2 control (n = 20) gave the expected 50 : 50 ratio with n = 11 being 272bp / 272bp homozygous and n = 9 heterozygous (272bp / 282bp). This tight linkage between the HaABCA1 gene and resistance made it a candidate for being the target of the resistance mutation.
Identification of resistant alleles
To assess whether HaABCA1 was the real location of resistance mutations, we checked whether this gene is expressed in the midgut. No evidence for significant midgut expression of ABCA1 was found in either B. mori (Bm-MDB, B. mori Microarray Database [63]; see S1 Table) or H. armigera, making it unlikely that this gene is actually involved in resistance. Initially detected in total larval and pupal transcripts, its expression was more specifically evident in larval foregut, hindgut, trachea and haemocytes. However the adjacent ABCA2 gene was significantly expressed in the midgut of both Bombyx and H. armigera; of the genes in this region of BmChr17, it is among the most highly expressed in the midgut [63] (see S1 Table). The full-length transcript of the H. armigera ortholog HaABCA2 (Fig 3) encodes a protein of 1,742 amino acids with 67.28% identity to the BmABCA2 gene in B. mori (S1 Fig).
Fig. 3. Predicted protein sequence of HaABC2. 
Feature predictions include 12 transmembrane helices (TM I to XII), two ATP-binding domains ATP1 and ATP2, two transporter motifs TpM1 and TpM2, and predicted glycosylation sites (highlighted in blue, and indicated by ‘Y’). Amino acid differences between H. armigera and H. punctigera are indicated by ‘.’ mutation sites are indicated by red and blue arrows for H. armigera and H. punctigera respectively—Ha2Ab-R01 (amino acid position 964); Ha2Ab-R02 (amino acid position 1,043); Ha2Ab-R03 (amino acid position 1,368); and Hp2Ab-R04 (amino acid position 1,504). We therefore further explored whether any changes were evident in the transcripts of the ABCA2 gene in midgut RNA from larvae of the resistant line. The ABCA2 candidate gene cDNA was fully sequenced from a SP15 resistant individual that identified a 73bp deletion at exon 16 (‘ G CTA GGA GTT CTG CGT TAC GTC ATG TCT TTA TCA CCA ACC ATT AGA ACT AGG TGG TTG TCG TTG GAA GAA GGG’ from nucleotides 2,889–2,961) / 8bp (‘C GGT TAA G’) insertion mutation (= allele 1, Ha2Ab-R01) (Fig 4a and 4b) of the coding sequence which resulted in the replacement of leucine (L) by glycine (G) at position 964, followed by a stop codon downstream from the mutation site at position 965. The 8bp ‘C GGT TAA G’ nucleotides matched completely to part of intron 16 (intron 16 nucleotide positions 56 to 62) of the SP15 H. armigera line. From the sequence of this mutated cDNA, we designed primers to screen additional independently isolated field resistant lines from 2005 (line 5–405); 2006 (lines 6–364 and 6–798), 2009 (line 9–4802), 2010 (line 10–485), and 2012 (line 12–2169), four of which (i.e., 5–405, 9–4802, 10–485, 12–2169) showed the same 73bp deletion/8bp insertion mutation as identified in the SP15 individual. A second resistance allele (= Ha2Ab-R02) with a 5bp (‘ACA AG’) deletion mutation at nucleotides 3,127–3,131 of the coding sequence was identified in a homozygous individual from the Cry2Ab resistant lines 6–364. A heterozygous resistant individual from line 6–798 was identified to possess one Ha2Ab-R02 allele, and a third resistance allele (= Ha2Ab-R03) at nucleotides 4,104–4,108 that represented a 5bp (GAATA) nucleotide duplication (Fig 4b), similar to the target site duplication (TSD) signature that is widespread in the H. armigera genome due to transposable element transposition activities (e.g., see [64]). cDNA sequencing of both the 6–364 and 6–798 lines identified the presence of the Ha2Ab-R02 allele as homozygous in the 6–364 line, and also both Ha2Ab-R02 and Ha2AB-R03 alleles at exons 18 and 24 respectively in line 6–798, thereby confirming that this resistant line was heterozygous for the ABCA2 gene (see S2 Fig). All three mutations identified to date in the Cry2Ab resistant lines are located at the 3’ region of the 5.1Kb coding sequence, and result in truncation of the protein. A summary of the three Cry2Ab resistance alleles identified in H. armigera is presented in Fig 4a and 4b. Sequence analyses of the SP15, 6–364 and 6–798 resistant individuals confirmed that no other INDELs or nonsense mutations were present in coding regions of the ABCA2 gene. Similarly, additional sequence analyses from multiple susceptible H. armigera individuals showed the predicted fully functional (non-truncated) ABCA2 gene, while nucleotide variation (<1%) between Cry2Ab susceptible H. armigera ‘GR’ lines resulted in nine amino acid changes, six of which involved nonsynonymous substitutions between amino acids with hydrophobic side-chains (e.g., valine (V), isoleucine (I), tyrosine (Y), phenylalanine (F), methionine (M), and leucine (L)), and one amino acid substitutions of each between lysine (K) and threonine (T), alanine (A) and proline (P), and glycine (G) and glutamic acid (E) (S2 Fig).
Fig. 4. A summary of the three Cry2Ab resistance alleles (Ha2Ab-R01, R02, R03) in the ABCA2 gene in Helicoverpa armigera, and one H. punctigera Cry2Ab resistance allele (Hp2Ab-R04). 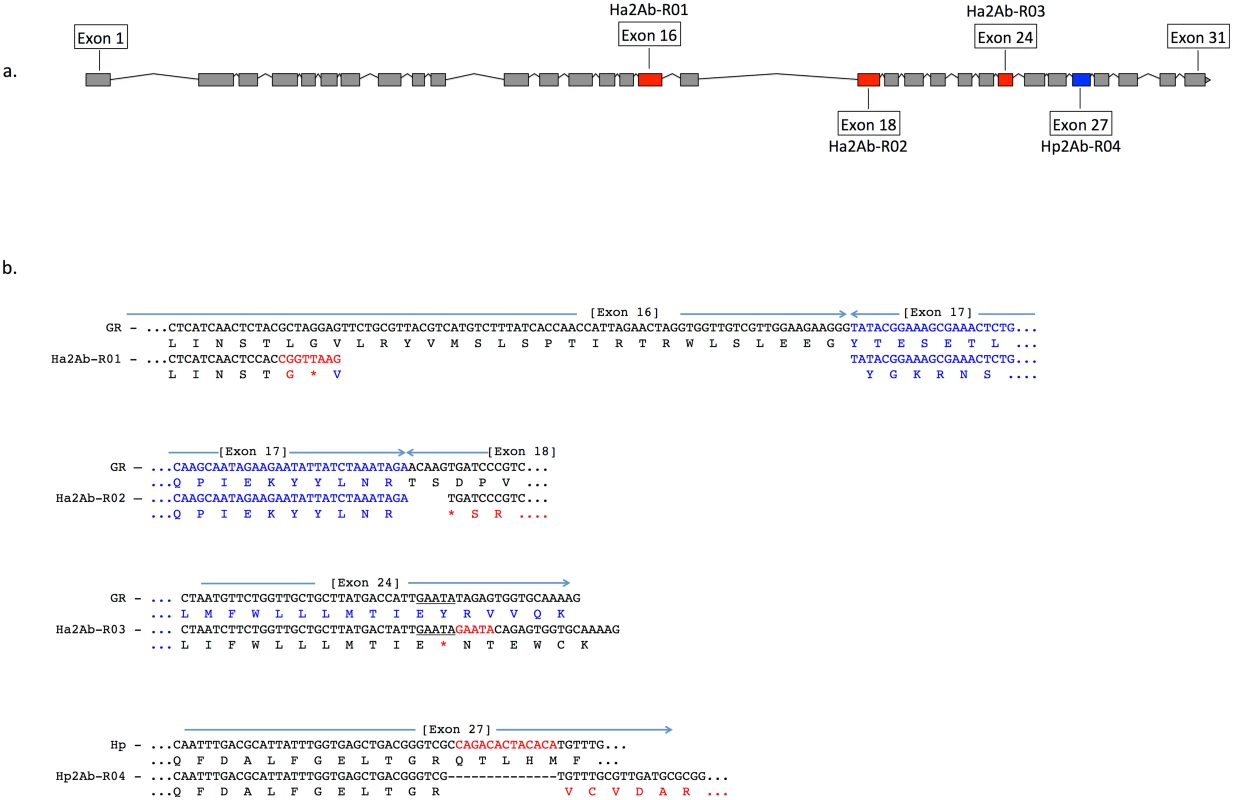
The ABCA2 gene consists of 31 exons (Fig 4a), with mutations at exons 16, 18, 24 and 27 indicated by red (for H. armigera) and blue (for H. punctigera) boxes. Fig 4b: The Ha2Ab-R01 allele was the result of a 73 base pair (bp) deletion at the c-terminus of exon 16, and the insertion of a 8bp ‘CGGTTAAG’ sequence, and resulted in a glycine (G) replacing the leucine (L) amino acid followed by a premature stop codon (*) (in red). The Ha2Ab-R02 allele was the result of a 5bp ‘ACAAG’ deletion at the start of exon 18 that resulted in a premature stop codon. The Ha2Ab-R03 allele was the result of a 5bp ‘GAATA’ target site duplication signature reminiscent of past insertions by transposable elements. This duplication mutation also resulted in a premature stop codon in exon 24. The H. punctigera Hp2Ab-R04 allele was the result of a 14bp deletion, and resulted in missense mutations. Absence of HaABCA2 mutations in susceptible field-collected insects
To confirm the significance of the HaABCA2 mutations in resistant lines, we asked whether susceptible individuals collected from the field carried the same INDEL mutations or other inactivating mutations in the H. armigera ABCA2 gene. Starting with single pairs of field-collected insects, F1 pools from each mating pair generated F2 progenies (n = 90) that were screened against a discriminating dose of Cry2Ab and identified those pairs whose F2 progeny all died as carrying only susceptible alleles. This test is enough to exclude any resistance-conferring alleles (occurring as heterozygotes) amongst the grandparents. Ten individual 3rd instar larvae representing 10 different field-collected susceptible populations were RNA extracted and RT-PCR used to generate cDNA. To screen for any evidence of mutations in the ABCA2 gene, PCR of the cDNA and sequencing using appropriate primer pairs (see S1 Table) were performed. No evidence for any inactivating mutations was found in any of these 10 individuals, i.e. all contained untruncated transcripts at exons 16, 18 and 24 where Ha2Ab-R01, R02 and R03 alleles were detected, respectively.
Protein domain prediction
A total of two transmembrane domains (TMDs; i.e., TMD 1, TMD 2) with each consisting of six transmembrane helices (TM I-VI in TMD 1; TM VII-XII in TMD 2, Fig 5) were predicted from the HaABC2 sequence that corresponded to those characteristic of the ABC transporter subfamily A. As for other members of this subfamily, N-glycosylation sites were also predicted for both of the extracellular domain (ECD) loops between TM I and TM II, and between TM VII and TM VIII (Figs 3 and 5). The intracellular loop between TMD 1 and TMD 2 (i.e., between TM VI and TM VII) and after TM XII contained the highly conserved regions for ATP Nucleotide Binding Fold 1 (NBF1) (including the Transporter signature Motif 1; TpM1), and NBF2 (and TpM2), respectively.
Fig. 5. Diagram of the ABCA2 protein structure and location of mutations in H. armigera and H. punctigera. 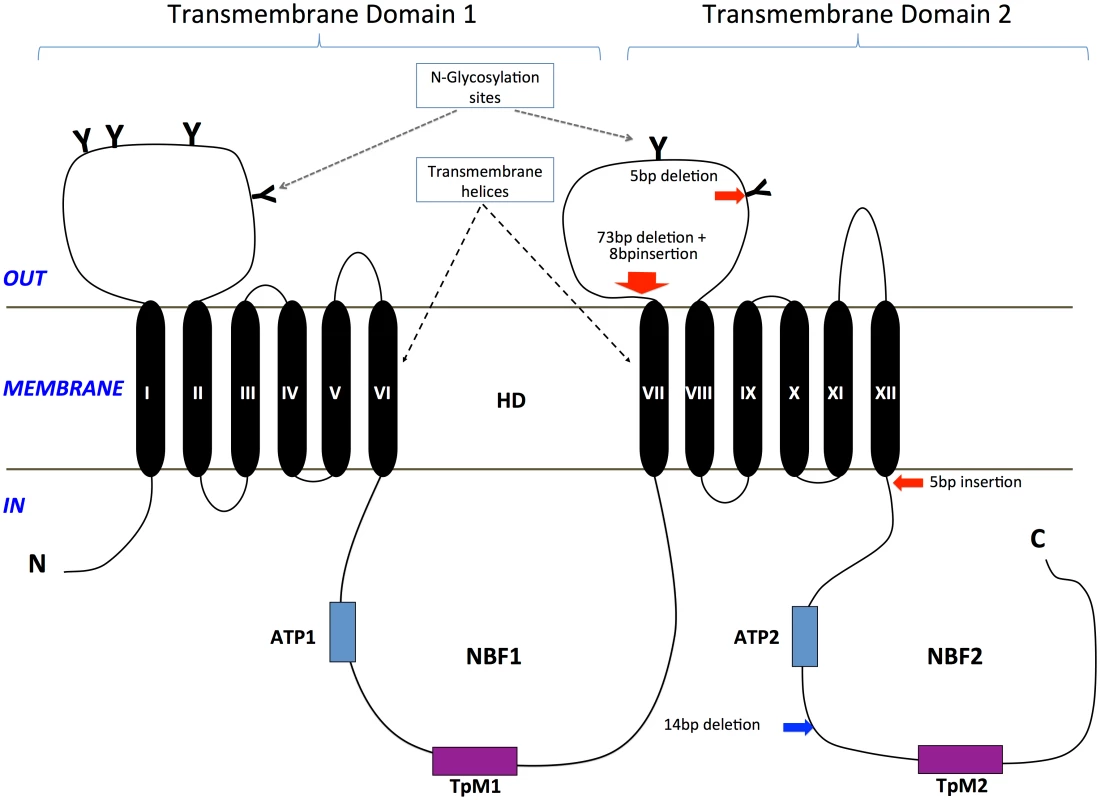
Glycosylation sites on the two large extracellular loops are represented by ‘Y’. Two highly conserved ATP Nucleotide Binding Folds (NBF1, NBF2) that included the Transporter signature motifs 1 and 2 (TpM1, TpM2) are present in the intracellular environment. The Helicoverpa ABCA2 protein structure consists of two transmembrane domains (TMD 1, TMD 2), each with six transmembrane helices (TM I-VI in TMD 1; TM VII-XII in TMD 2). The approximate positions of the mutations in H. armigera and H. punctigera are indicated by red and blue arrows, respectively. Each of the mutant alleles Ha2Ab-R01, Ha2Ab-R02 and Ha2Ab-R03 introduced stop codons into the reading frame, causing significant truncations of the HaABCA2 protein (Fig 4). The first two mutations introduced stop codons in the extracellular loop between TM VII and TM VIII of TMD 2, whereas the 5bp insertion that resulted in the Ha2Ab-R03 allele occurred just one amino acid after TM XII of TMD 2. All three mutations therefore truncated the protein before the second nucleotide-binding domain NBF2 (Fig 5), which would render the ABC transporter completely inactive, even if the protein were expressed and integrated into the cell membrane.
Analysis of a mutation in H. punctigera
In the Cry2Ab-resistant H. punctigera Hp4-13 strain, the allele (Hp2Ab-R04) encoding the homolog of HaABCA2 was found to contain a 14bp deletion (Fig 4). This deletion disrupts the coding region of the transcript by introducing frame shifts that lead to a missense mutation and to the loss of the TpM2 transporter motif at the NBF2 (Figs 4 and 5). Although linkage mapping was not performed to conclusively associate this mutation with the resistant phenotype in H. punctigera, the fact that the same gene is mutated in a resistant strain in this species strongly supports the role of this gene as the target of mutations conferring resistance to Cry2Ab.
Identification of ABCA1 and ABCA2 orthologs in other lepidopteran genomes
We next asked whether sequences for these ABCA proteins existed in genomes of other Lepidoptera, including some (H. virescens [65], Plutella. xylostella [66]; B. mori [67]) known to be susceptible to Cry2Ab. We examined the published genome sequences of Danaus plexippus [68], Heliconius melpomene [69], and P. xylostella [70,71]. Existing predictions of the genes were often inaccurate, e.g., as with the two partial predictions for BmABCA2, and ABCA1 was predicted as two separate partial proteins for the D. plexippus genome [68]. We used Scipio [72] and FGENESH [73] as well as additional transcriptomic data to generate complete predictions (GenBank Accession numbers KP219762-KP219770). In each species, the ABCA1 and ABCA2 genes were present, and situated adjacently in a tail-to-tail orientation just as in B. mori. Orthology was further confirmed by conserved flanking genes; in each species the ortholog of B. mori mitochondrial ribosomal protein S7 (GenBank Accession XP_004927481) occurred upstream of ABCA2, and the ortholog of B. mori ubiquitin carboxyl-terminal hydrolase (GenBank Accession XP_004927480) occurred upstream of ABCA1. An alignment of the predicted ABCA1 and ABCA2 proteins along with the respective Drosophila protein showing greatest similarity (S1 Fig) was used to construct a phylogenetic (ML) tree. This tree indicated that the gene duplication creating ABCA1 and ABCA2 likely occurred in the common ancestor of the Lepidoptera shown (Fig 6).
Fig. 6. Phylogenetic tree showing clustering of ABCA1 and ABCA2 proteins of various Lepidoptera, with Drosophila homolog. 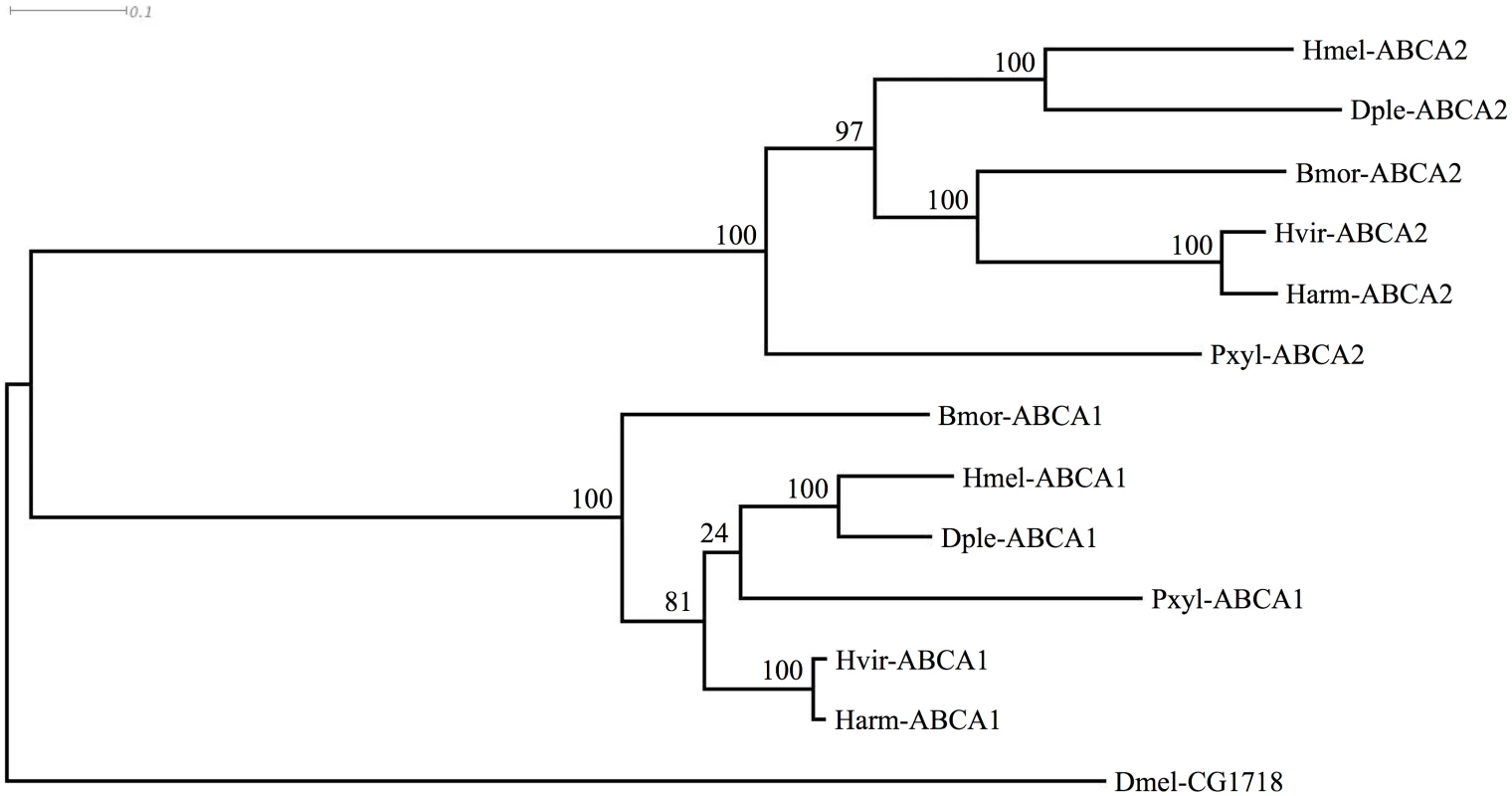
Species abbreviations are: Bmor, Bombyx mori; Harm, Helicoverpa armigera; Hvir, Heliothis virescens; Pxyl, Plutella xylostella; Dple, Danaus plexippus; Hmel, Heliconius melpomene; Dmel, Drosophila melanogaster. The tree is based on the alignment shown in S1 Fig Discussion
The importance of Bt toxins for insect pest and disease control has stimulated enormous interest in the study of their mode of action. For Cry1A toxins, there is specific and saturable binding to membrane targets, and a sequential mode of action has been proposed [25,39]. The toxin first binds to the 12-cadherin domain protein, resulting in processing and accelerated oligomerization before binding to membrane-bound glycosylated proteins such as aminopeptidases, alkaline phosphatase and other glycoproteins [25,26]; the integral membrane ABCC2 protein then facilitates pore insertion. Cry1A and Cry2A proteins have comparable three-domain structures [74,75], making them likely to act in similar ways as pore-forming toxins. Specific and saturable binding to membranes was also recently shown for Cry2Ab [22,75], and resistance is associated with a loss of binding [21]. Despite these similarities, toxicity of Cry2Ab in general is unaffected by mutations conferring Cry1Ac resistance. Specifically, mutations in APN or cadherin or ABCC2 do not render insects resistant to Cry2Ab, so that this toxin must be binding to one or more different targets.
The identification of ABCA2 suggests a mode of action of Cry2Ab differing slightly from that proposed for Cry1A toxins. ABCA2 carries two extracellular domains that are present as long loops between helices TM I and TM II, and between helices TM VII and TM VIII (Figs 3 and 5). Both of these loops are glycosylated in mammals [76], and six glycosylation sites are predicted for HaABCA2 (Fig 3). In contrast, for the lepidopteran ABCC2, the corresponding loops are very short and contain no glycosylation signals [34]. We hypothesize that Cry2Ab also has a sequential mode of action in which the ABCA2 protein itself is able to provide both binding and pore insertion functions. Specifically, Cry2A toxins would, upon activation [75] bind to the glycosylated ECD loops in TMD 1 and/or TMD 2. This binding could form the basis of oligomerization and bring the pre-pore structure close to the TMDs for pore insertion, as proposed for ABCC2 [34]. It is possible that other proteins may also be involved in Cry2Ab binding and pore formation, particularly since mammalian ABCAs have been suggested to occur in multi-protein complexes in the membrane [77]. Interestingly, the ABCA2 mutations confer resistance to very high concentrations of Cry2A [42], as would be expected if both receptor and pore insertion functions are simultaneously blocked. Similarly, in H. virescens the ABCC2 mutation results in higher levels of resistance to Cry1Ac than does the mutation in cadherin, but when both are homozygous in the same strain and both receptor and pore insertion functions are blocked, extremely high resistance levels result [34].
To what extent are these findings likely to apply to other Bt toxins of the Cry2A family? Phylogenetic analyses based on the shared common three-domain structure [78] showed that Cry2Aa and Cry2Ai are sister toxin groups and occupy a basal position to both Cry2Ab/Cry2Ag and Cry2Ae/Cry2Ah clades. Cross-resistance between Cry2Ab and Cry2Aa has been demonstrated in the SP15 strain of H. armigera [42], and between Cry2Ab and Cry2Ae in both H. armigera and H. punctigera [21]. Resistance to Cry2Aa has also been identified in H. virescens [53,79], in H. zea [15], in P. gossypiella [40], and in Ostrinia nubilalis [80]; ABCA2 remains to be investigated in these species. Cry2A toxins are also toxic to some Diptera [81,82] (but see [83]), with Cry2Ab recently shown to be effective against the malaria mosquito vector Anopheles gambiae [82]. Cry2Ab was ineffective against Aedes aegypti [82] but the similar toxin Cry2Ag was highly effective [84].
Seven ABC transporters of the A subfamily are present in the B. mori genome (three on BmChr17, and one each on chromosomes 5, 14, 16, and 19; see [61,62] for analysis of the initial gene models), and similar numbers have been found in other Lepidoptera with sequenced genomes. This subfamily has been well characterised in vertebrates; there are 12 members known in humans [85] and a similar number in the mouse; the nomenclature for these differs from that of insects. The HaABCA2 gene and the other insect genes shown in Fig 6 belong to an insect-specific clade within this subfamily, with no direct orthologs among the vertebrate genes [86]. The human ABCA genes have been extensively analysed and are expressed in a variety of tissues, with most being involved in lipid transport and trafficking. Mutations in human ABCA2 (not orthologous to lepidopteran ABCA2) are associated with early-onset of Alzheimer’s disease [87,88]. The mouse ABCA2 (an ortholog of the human ABCA2) has a possible role in regulating cholesterol homeostasis and low-density lipoprotein receptor metabolism in N2 neuroblastoma cells [89], with knock-out causing a ‘shaky’ (tremor) phenotype [90]. Mutations in the mouse ABCA3 gene, which is expressed in lung tissues, are associated with a foetal surfactant deficiency that is fatal. However, in H. armigera (but not H. punctigera), the Cry2Ab resistant line homozygous for ABCA2-inactivating mutation has no demonstrated substantial fitness costs compared to Cry2Ab susceptible insects [91]. Whether this is due to functional redundancy with another of the midgut-expressed ABCAs remains to be determined.
The frequency distribution of resistant ABCA2 alleles identified to-date is non-uniform. Seven resistant H. armigera lines, isolated independently from the field between 2002 and 2012, produced three resistant alleles. The Ha2Ab-R01 allele was present in five lines: SP15, 5–405, 9–4802, 10–485 and 12–2169; the Ha2Ab-R02 allele was homozygous in 6–364 and present in heterozygotes in 6–798; the Ha2Ab-R03 allele was the alternative allele in the heterozygous 6–798 line. A single mutant allele, Hp2Ab-R04, was found in the one H. punctigera resistant line HP4-13. Thus some alleles in H. armigera were common enough to be recovered several times from the field. Whether this is due to some selection by an unknown agent in the Australian environment as proposed [92] remains to be tested.
The still-rare resistance-conferring alleles identified in field populations occur at a limited number of locations in the gene. If confirmed by studies of further alleles, this raises the possibility that DNA-based screens will allow monitoring of the spread of Bt resistance in H. armigera and H. punctigera. Although PCR-based screens [29,93] for mutations in the 12-cadherin domain protein of H. armigera that confers resistance to Cry1Ab and Cry1Ac identified the same allele (r1) in field material from northern China as that originally identified as conferring resistance [12], this has not always been the case. For H. virescens [94], for example, different mutations in the same gene were identified by the F1 screen (i.e., mating field-caught individuals with an existing homozygous resistant strain and testing the F1 offspring). For P. gossypiella, screening in India [95] found different cadherin gene mutations to those originally identified in Arizona [96].
Every Cry2Ab-resistant line from an F2 screen that has been molecularly characterized has shown mutations in the ABCA2 gene. This confirms the value of using the less expensive F1 screen with ABCA2-mutant lines to extend the estimation of Cry2Ab resistance allele frequencies in Australia. Incorporating PCR-based screening will further improve detection efficacies of ABCA2-based resistance in the field, enabling more accurate and faster estimates of resistance allele frequencies, and is especially relevant for the analysis of historical field material generated through F1/F2 screening methods [43,92], and for tracking spatial and temporal movement patterns of resistance alleles across the landscape. Further characterisation of resistance-conferring ABCA2 alleles will also help to resolve the current discrepancy between the F2 screen and the F1 screen in estimating allele frequencies [91]. It will be important to determine whether Cry2Ab-resistance-conferring ABCA2 mutations occur in H. armigera elsewhere in its geographic range, including its recent incursions into the Americas [97,98]. Finally, examination of ABCA2 may provide insight in several species where the Cry2A resistance mechanism is still unknown, including H. virescens [53], P. gossypiella [40], H. zea [41], and Trichoplusia ni [99].
Materials and Methods
Linkage mapping—Strains and families
As a result of an F2 screen in 2002, the first H. armigera Cry2Ab resistant strain (Sp15) was established from a single pair of moths collected as eggs on corn near Griffith, New South Wales (NSW), Australia [42]. Detailed descriptions of the techniques employed have been provided [42,92]. F1 progeny from that pair were intercrossed and the resultant F2 larvae exposed to a screening concentration of Cry2Ab in ground leaf material of the cotton variety Sicala V-2 transformed with the B. thuringiensis variety kurstaki cry2Ab gene construct. Survivors among the F2 formed the basis of the resistant colony Sp15. Since 2003, F2 screens with H. armigera and H. punctigera performed as part of a resistance monitoring program have isolated additional lines [10]. The isolated H. punctigera Cry2Ab resistant line (Hp4-13) was established from eggs collected at St George, Queensland, Australia in 2004 [43]. Complementation tests for allelism established that one or more alleles at the same locus conferred resistance to Cry2Ab in five lines of H. armigera derived from the field in Australia from 2002 to 2006, including SP15 and 5–405 (previously named NA405) [100].
Identification of candidate Cry2Ab resistance AFLP linkage group
Assignment of the Cry2Ab resistance locus to an AFLP linkage group was carried out using the resistant line SP15 and the susceptible GR line. The initial cross was a SP15 Cry2Ab r/r ♂ x GR Cry2Ab s/s-♀, yielding family G. An F1 female from family G was crossed to an SP15 male to produce the female informative backcross family F2031. Backcross progeny were bioassayed using the discriminatory Bt Cry2Ab concentration [92] to select for homozygous resistant (Cry2Ab r/r) individuals. Additional backcross progeny were not exposed to Cry2Ab, to serve as controls. AFLPs [101] from genomic DNA of grandparents, parents and 59 progeny of family F2301 were analysed for linkage using the method of Heckel et al. [18]. Twenty-nine progeny were survivors of exposure to Cry2Ab and 30 were untreated controls. AFLPs were grouped using the program DBM3Lnk.p as in Heckel et al. [18]. As expected from achiasmatic oogenesis in female Lepidoptera, no recombinants were found within AFLP linkage groups. Linkage to resistance was tested for each linkage group using signed interaction chi-squared tests with one degree of freedom [102], with a Bonferroni correction for 31 linkage groups.
One AFLP band from the only linkage group with a significant association with resistance was cut out of the gel, reamplified, cloned and sequenced (GenBank Accession No. KJ419919). The insert was hybridized to a Southern blot made from an unrelated Bt-susceptible H. armigera family in which several ribosomal protein genes had previously been mapped, enabling comparison to the homologous ribosomal protein genes of Bombyx mori. This showed AFLP group 8 in family F2301 to correspond to B. mori chromosome 17 (BmChr17).
Additional linkage mapping in a male-informative backcross (G2016) and two F2 families (G2020, G2029) was performed to further localize the resistance locus. Offspring from these families that had survived the discriminating concentration and were presumably homozygous for the SP15-derived resistance allele were examined for recombinants at marker loci. H. armigera homologs of ribosomal protein genes RpL22 and RpS24 on B. mori BmChr17 were sequenced to identify polymorphisms to be used in mapping. The gene bre-5 on BmChr17 was considered a candidate for the resistance gene because of its role in Cry4B resistance in the nematode Caenorhabditis elegans [19], and was also mapped using sequence variation in the coding region and a PCR-RFLP using a polymorphic PstI restriction site.
Mapping of the Cry2Ab candidate resistance locus using male-informative cross
To establish an appropriate mapping family, a GR Cry2Ab susceptible homozygous male (Cry2Ab s/s ♂) was mated with a SP15 Cry2Ab resistant homozygous female (Cry2Ab r/r ♀). The F1 susceptible heterozygous male (Cry2Ab r/s ♂) was back-crossed to a SP15 female to obtain F2 offspring of either homozygous resistant (Cry2Ab r/r) or heterozygous susceptible (Cry2Ab r/s) genotypes in equal proportions. Approximately 300 F2 offspring were bioassayed using the discriminatory Bt Cry2Ab concentration [92] to select for homozygous resistant (Cry2Ab r/r) individuals. Control (n = 100) F2 offspring were not bioassayed and were included in subsequent genotyping experiments using EPIC-PCR markers as described below.
Chromosome walk by EPIC markers to ascertain recombination rates
EPIC PCR markers used in this study were designed using the primer designing criteria previously reported by [103] for H. armigera. Briefly EPIC-PCR primers were designed using the primer analysis software Oligo Version: 7.17 (Molecular Biology Insights, Inc., Cascade, CO 80809, USA) and avoiding false primer annealing sites for both forward and reverse primer, with no or minimal hairpin structures and primer dimmer formation. We also designed the EPIC-PCR primers with intron amplicon of typically less than 500bp such that polymorphisms in F2 cross can be easily scored. Intron sizes were estimated based on B. mori gene annotation. EPIC-PCR primers were optimised prior to having a fluorescent tag (FAM, HEX or TET) attached to the 5’ end of the forward primer. Amplicons of the mapping family from individual EPIC-PCR primer pairs were visualised on 1–1.5% agarose gels prior to being purified by acetic acid/ethanol precipitation and sent to Genetic Analysis Facility (GAF) at James Cook University (JCU) for genotyping. PCR conditions, and genotyping procedures were previously described [103,104]. A list of all EPIC-PCR primers used in this study can be found in S2 Table.
Genomic DNA was extracted using the Qiagen Blood and Tissue extraction kit (Qiagen Cat. #69506). For the founding grandparents (i.e., F0) and parents (i.e., F1) one leg each was used in gDNA extraction, with gDNA eluted in 200°L of the AE buffer. Bioassayed and control F2 samples were collected as 3rd instar larvae and gDNA was extracted as for the parents and grandparents. All genotyping with EPIC-PCR markers involved screening of grandparents, F1 parents, 72 bioassayed (Cry2Ab r/r) offspring and 20 control F2 offspring (i.e., either Cry2Ab r/r or Cry2Ab r/s). Under the linkage mapping pattern, genome/chromosome walking towards the resistance gene should generate reduced recombination rates in the resistant F2 as one approaches the genomic region of interest.
Identification of candidate genes
Messenger RNA sequencing was done in order to generate full-length transcripts in H. armigera for candidate genes, identify resistant alleles where cDNA amplification failed and to identify the homologous candidate genes in H. punctigera. Total RNA was extracted from the midgut of third-instar larvae or whole larvae using the TRIzo Plus RNA purification kit (Life Technologies, Cat # 12183555) and dried down for shipping with RNAstable Tube Kit (Biometrica Cat. # 93221–001). RNAseq library preparation, sequencing and bioinformatic analysis was done according to standard Illumina protocols by the Beijing Genomics Institute (BGI) in Shenzen, China.
Except for the resistant line 7–183 which used gDNA as a template for sequencing, candidate genes from the remaining resistant lines were completely sequenced using a cDNA template from 3rd instar larvae prepared using an RNA extraction kit (Qiagen RNeasy mini kit, Cat. # 74106), and trace genomic DNA contaminants removed using the Qiagen RNase-Free DNase set (Cat. # 79254). First strand cDNA was synthesised using the Invitrogen SuperScript III RT First Strand Synthesis System for RT PCR (Cat. # 18080–051), in the presence of RNase H. All sequencing was performed at the John Curtin School of Medical Research, Australian National University (ANU), and used the ABI BigDye v3 chemistry. Contig assembly used the Staden pregap4 and Gap4 software [105] and was visualised using Artemis (Release 12.0) [106]. Sequences generated and used in this study have been deposited in GenBank (Accession numbers KP259910, KP259911, KP259912).
Prediction of ABCA2 transmembrane domains
The amino acid sequence predicted from a complete mRNA sequence from a Cry2Ab susceptible individual belonging to the GR-line was used to predict the domain structure of the H. armigera ABCA2 protein. The protein prediction software Split V3.5 <http://split4.pmfst.hr/split/4/> [107] was used to search for transmembrane protein secondary structure (i.e., transmembrane helices). In the mouse RmP ABC transporter, several N-glycosylation sites were predicted on the protein’s extracellular domains [76,108]. We used the NetNGlyc 1.0 Server <http://www.cbs.dtu.dk/services/NetNGlyc/> developed to predict N-Glycosylation sites in human proteins for the purpose of predicting N-Glycosylation sites in the protein sequences. The software uses artificial neural networks to examine for Asn-Xaa-Ser/Thr sequence context.
Identification of sequences and phylogenetic analyses
Sequences in the transcriptome databases corresponding to candidate genes were identified by standalone BLAST. Homologs in GenBank were identified using BLAST and homologous gene clusters identified in NCBI and in Ensembl. Orthologous genes from other lepidopteran genomes were retrieved using their online databases from public domains as cited in the appropriate sections below. Protein sequences were aligned using Multiple Alignment using Fast Fourier Transform (MAFFT) [109] <http://www.ebi.ac.uk/Tools/msa/mafft/> and phylogenetic tree (maximum likelihood (ML) with rapid bootstrapping) inference using RAxML-HPC2 on XSEDE (8.0.24) (available at the CIPRES Science Gateway V3.3) <http://www.phylo.org/sub_sections/portal/> [110–112], and redrawn using Dendroscope version 2.4 [113].
Supporting Information
Zdroje
1. Sanahuja G, Banakar R, Twyman RM, Capell T, Christou P. Bacillus thuringiensis: a century of research, development and commercial applications. Plant Biotechnol J. 2011;9(3):283–300. doi: 10.1111/J.1467-7652.2011.00595.X ISI:000288630900001. 21375687
2. Farias JR, Andow DA, Horikoshi RJ, Sorgatto RJ, Fresia P, dos Santos AC, et al. Field-evolved resistance to Cry1F maize by Spodoptera frugiperda (Lepidoptera: Noctuidae) in Brazil. Crop Prot. 2014;64 : 150–8. WOS:000341470000021.
3. Gassmann AJ, Petzold-Maxwell JL, Clifton EH, Dunbar MW, Hoffmann AM, Ingber DA, et al. Field-evolved resistance by western corn rootworm to multiple Bacillus thuringiensis toxins in transgenic maize. P Natl Acad Sci USA. 2014;111(14):5141–6. doi: 10.1073/Pnas.1317179111 WOS:000333985200034.
4. Tabashnik BE, Brevault T, Carriere Y. Insect resistance to Bt crops: lessons from the first billion acres. Nat Biotechnol. 2013;31(6):510–21. doi: 10.1038/nbt.2597 23752438.
5. Mitter C, Poole RW, Matthews M. Biosystematics of the Heliothinae (Lepidoptera, Noctuidae). Annu Rev Entomol. 1993;38 : 207–25. WOS:A1993KF69700010.
6. Matthews M. Heliothine Moths of Australia. Melbourne: CSIRO Publishing; 1999. x+320 p.
7. Hardwick DF. The Corn Earworm complex. Mem Entomol Soc Can. 1965;40 : 1–248.
8. Behere GT, Tay WT, Russell DA, Heckel DG, Appleton BR, Kranthi KR, et al. Mitochondrial DNA analysis of field populations of Helicoverpa armigera (Lepidoptera: Noctuidae) and of its relationship to H. zea. BMC Evol Biol. 2007;7 : 117.
9. Downes S, Parker T, Mahon R. Incipient Resistance of Helicoverpa punctigera to the Cry2Ab Bt Toxin in Bollgard II® Cotton. PLoS One. 2010;5(9):e12567. doi: 10.1371/journal.pone.0012567 ISI:000281631300007. 20830203
10. Downes S, Parker TL, Mahon RJ. Characteristics of resistance to Bacillus thuringiensis toxin Cry2Ab in a strain of Helicoverpa punctigera (Lepidoptera: Noctuidae) isolated from a field population. J Econ Entomol. 2010;103(6):2147–54. ISI:000286845900027. 21309238
11. Xu XJ, Yu LY, Wu YD. Disruption of a cadherin gene associated with resistance to Cry1Ac δ-Endotoxin of Bacillus thuringiensis in Helicoverpa armigera. Appl Environ Microb. 2005;71(2):948–54. ISI:000227043400045.
12. Yang YJ, Chen HY, Wu SW, Yang YH, Xu XJ, Wu YD. Identification and molecular detection of a deletion mutation responsible for a truncated cadherin of Helicoverpa armigera. Insect Biochem Molec. 2006;36(9):735–40. doi: 10.1016/J.Ibmb.2006.06.003 ISI:000240783300006.
13. Zhao J, Jin L, Yang YH, Wu YD. Diverse cadherin mutations conferring resistance to Bacillus thuringiensis toxin Cry1Ac in Helicoverpa armigera. Insect Biochem Molec. 2010;40(2):113–8. doi: 10.1016/J.Ibmb.2010.01.001 ISI:000275972800003.
14. Dhurua S, Gujar GT. Field-evolved resistance to Bt toxin Cry1Ac in the pink bollworm, Pectinophora gossypiella (Saunders) (Lepidoptera: Gelechiidae), from India. Pest Manag Sci. 2011;67(8):898–903. doi: 10.1002/Ps.2127 WOS:000293327300004. 21438121
15. Burd AD, Gould F, Bradley JR, Van Duyn JW, Moar WJ. Estimated frequency of nonrecessive Bt resistance genes in bollworm, Helicoverpa zea (Boddie) (Lepidoptera: Noctuidae) in eastern North Carolina. J Econ Entomol. 2003;96(1):137–42. ISI:000180967000020. 12650356
16. Anilkumar KJ, Rodrigo-Simon A, Ferre J, Pusztai-Carey M, Sivasupramaniam S, Moar WJ. Production and characterization of Bacillus thuringiensis Cry1Ac-resistant cotton bollworm Helicoverpa zea (Boddie). Appl Environ Microb. 2008;74(2):462–9. ISI:000252453100016.
17. Tabashnik BE, Gassmann AJ, Crowder DW, Carriere Y. Insect resistance to Bt crops: evidence versus theory. Nature Biotechnol. 2008;26(2):199–202. doi: 10.1038/nbt1382 WOS:000253193000021.
18. Heckel DG, Gahan LJ, Liu YB, Tabashnik BE. Genetic mapping of resistance to Bacillus thuringiensis toxins in diamondback moth using biphasic linkage analysis. P Natl Acad Sci USA. 1999;96(15):8373–7. doi: 10.1073/Pnas.96.15.8373 ISI:000081589400019.
19. Griffitts JS, Whitacre JL, Stevens DE, Aroian RV. Bt toxin resistance from loss of a putative carbohydrate-modifying enzyme. Science. 2001;293(5531):860–4. doi: 10.1126/science.1062441 WOS:000170241000044. 11486087
20. Gonzalez-Cabrera J, Farinos GP, Caccia S, Diaz-Mendoza M, Castanera P, Leonardi MG, et al. Toxicity and mode of action of Bacillus thuringiensis Cry proteins in the Mediterranean corn borer, Sesamia nonagrioides (Lefebvre). Appl Environ Microb. 2006;72(4):2594–600. ISI:000236749400041.
21. Caccia S, Hernandez-Rodriguez CS, Mahon RJ, Downes S, James W, Bautsoens N, et al. Binding site alteration is responsible for field-isolated resistance to Bacillus thuringiensis Cry2A insecticidal proteins in two Helicoverpa species. PLoS One. 2010;5(3):e9975. doi: 10.1371/journal.pone.0009975 ISI:000276418200048.
22. Gouffon C, Van Vliet A, Van Rie J, Jansens S, Jurat-Fuentes JL. Binding sites for Bacillus thuringiensis Cry2Ae toxin on Heliothine brush border membrane vesicles are not shared with Cry1A, Cry1F, or Vip3A toxin. Appl Environ Microb. 2011;77(10):3182–8. doi: 10.1128/Aem.02791-10 ISI:000290473200003.
23. Jurat-Fuentes JL, Karumbaiah L, Jakka SRK, Ning CM, Liu CX, Wu KM, et al. Reduced levels of membrane-bound alkaline phosphatase are common to lepidopteran strains resistant to Cry toxins from Bacillus thuringiensis. PLoS One. 2011;6(3):e17606. doi: 10.1371/journal.pone.0017606 ISI:000287932100025. 21390253
24. Heckel DG, Gahan LJ, Baxter SW, Zhao JZ, Shelton AM, Gould F, et al. The diversity of Bt resistance genes in species of Lepidoptera. J Invertebr Pathol. 2007;95(3):192–7. doi: 10.1016/J.Jip.2007.03.008 ISI:000247850700008. 17482643
25. Bravo A, Likitvivatanavong S, Gill SS, Soberon M. Bacillus thuringiensis: A story of a successful bioinsecticide. Insect Biochem Molec. 2011;41(7):423–31. doi: 10.1016/J.Ibmb.2011.02.006 ISI:000293041200002.
26. Vachon V, Laprade R, Schwartz JL. Current models of the mode of action of Bacillus thuringiensis insecticidal crystal proteins: A critical review. J Invertebr Pathol. 2012;111(1):1–12. doi: 10.1016/J.Jip.2012.05.001 ISI:000307797300001. 22617276
27. Gahan LJ, Gould F, Heckel DG. Identification of a gene associated with Bt resistance in Heliothis virescens. Science. 2001;293 : 857–60. 11486086
28. Morin S, Biggs RW, Sisterson MS, Shriver L, Ellers-Kirk C, Higginson D, et al. Three cadherin alleles associated with resistance to Bacillus thuringiensis in pink bollworm. P Natl Acad Sci USA. 2003;100(9):5004–9. doi: 10.1073/Pnas.0831036100 ISI:000182612600010.
29. Yang YJ, Chen HY, Wu YD, Yang YH, Wu SW. Mutated cadherin alleles from a field population of Helicoverpa armigera confer resistance to Bacillus thuringiensis toxin Cry1Ac. Appl Environ Microb. 2007;73(21):6939–44. doi: 10.1128/Aem.01703-07 ISI:000250700600028.
30. Fabrick JA, Mathew LG, Tabashnik BE, Li X. Insertion of an intact CR1 retrotransposon in a cadherin gene linked with Bt resistance in the pink bollworm, Pectinophora gossypiella. Insect Mol Biol. 2011;20(5):651–65. doi: 10.1111/J.1365-2583.2011.01095.X ISI:000295089500009. 21815956
31. Liu CX, Xiao YT, Li XC, Oppert B, Tabashnik BE, Wu KM. Cis-mediated down-regulation of a trypsin gene associated with Bt resistance in cotton bollworm. Sci Rep. 2014;4 : 7219. doi: 10.1038/Srep07219 WOS:000346252800002. 25427690
32. Xiao Y, Zhang T, Liu C, Heckel DG, Li X, Tabashnik BE, et al. Mis-splicing of the ABCC2 gene linked with Bt toxin resistance in Helicoverpa armigera. Sci Rep. 2014;4 : 6184. doi: 10.1038/srep06184 25154974; PubMed Central PMCID: PMCPMC4143771.
33. Zhang SP, Cheng HM, Gao YL, Wang GR, Liang GM, Wu KM. Mutation of an aminopeptidase N gene is associated with Helicoverpa armigera resistance to Bacillus thuringiensis Cry1Ac toxin. Insect Biochem Molec. 2009;39(7):421–9. doi: 10.1016/J.Ibmb.2009.04.003 ISI:000267790700001.
34. Gahan LJ, Pauchet Y, Vogel H, Heckel DG. An ABC transporter mutation is correlated with insect resistance to Bacillus thuringiensis Cry1Ac toxin. PLoS Genet. 2010;6(12):e1001248. doi: 10.1371/journal.pgen.1001248 21187898; PubMed Central PMCID: PMCPMC3002984.
35. Baxter SW, Badenes-Perez FR, Morrison A, Vogel H, Crickmore N, Kain W, et al. Parallel evolution of Bacillus thuringiensis toxin resistance in Lepidoptera. Genetics. 2011;189(2):675–9. doi: 10.1534/Genetics.111.130971 ISI:000296158500022. 21840855
36. Atsumi S, Miyamoto K, Yamamoto K, Narukawa J, Kawai S, Sezutsu H, et al. Single amino acid mutation in an ATP-binding cassette transporter gene causes resistance to Bt toxin Cry1Ab in the silkworm, Bombyx mori. P Natl Acad Sci USA. 2012;109(25):E1591–E8. doi: 10.1073/pnas.1120698109 WOS:000306061400005.
37. Park Y, Gonzalez-Martinez RM, Navarro-Cerrillo G, Chakroun M, Kim Y, Ziarsolo P, et al. ABCC transporters mediate insect resistance to multiple Bt toxins revealed by bulk segregant analysis. BMC Biol. 2014;12 : 46. doi: 10.1186/1741-7007-12-46 24912445; PubMed Central PMCID: PMCPMC4071345.
38. Tanaka S, Miyamoto K, Noda H, Jurat-Fuentes JL, Yoshizawa Y, Endo H, et al. The ATP-binding cassette transporter subfamily C member 2 in Bombyx mori larvae is a functional receptor for Cry toxins from Bacillus thuringiensis. FEBS Journal. 2013;280(8):1782–94. doi: 10.1111/febs.12200 WOS:000317609000005. 23432933
39. Heckel DG. Learning the ABCs of Bt: ABC transporters and insect resistance to Bacillus thuringiensis provide clues to a crucial step in toxin mode of action. Pestic Biochem Phys. 2012;104(2):103–10. doi: 10.1016/J.Pestbp.2012.05.007 ISI:000311068900005.
40. Tabashnik BE, Unnithan GC, Masson L, Crowder DW, Li X, Carriere Y. Asymmetrical cross-resistance between Bacillus thuringiensis toxins Cry1Ac and Cry2Ab in pink bollworm. Proc Natl Acad Sci U S A. 2009;106(29):11889–94. doi: 10.1073/pnas.0901351106 19581574; PubMed Central PMCID: PMCPMC2706268.
41. Tabashnik BE, Carriere Y. Field-evolved resistance to Bt cotton: bollworm in the U.S. and pink bollworm in India. Southwest Entomol. 2010;35(3):417–24. doi: 10.3958/059.035.0326 ISI:000284301400025.
42. Mahon RJ, Olsen KM, Garsia KA, Young SR. Resistance to Bacillus thuringiensis toxin Cry2Ab in a strain of Helicoverpa armigera (Lepidoptera: Noctuidae) in Australia. J Econ Entomol. 2007;100(3):894–902. ISI:000246950300034. 17598553
43. Downes S, Parker TL, Mahon RJ. Frequency of alleles conferring resistance to the Bacillus thuringiensis toxins Cry1Ac and Cry2Ab in Australian populations of Helicoverpa punctigera (Lepidoptera: Noctuidae) from 2002 to 2006. J Econ Entomol. 2009;102(2):733–42. ISI:000264899500035. 19449655
44. Andow DA, Alstad DN. F2 screen for rare resistance alleles. J Econ Entomol. 1998;91(3):572–8. ISI:000074273400003.
45. Gould F, Anderson A, Jones A, Sumerford D, Heckel DG, Lopez J, et al. Initial frequency of alleles for resistance to Bacillus thuringiensis toxins in field populations of Heliothis virescens. P Natl Acad Sci USA. 1997;94(8):3519–23. 11038613; PubMed Central PMCID: PMC20471.
46. Liu F, Xu Z, Chang J, Chen J, Meng F, Zhu YC, et al. Resistance allele frequency to Bt cotton in field populations of Helicoverpa armigera (Lepidoptera: Noctuidae) in China. J Econ Entomol. 2008;101(3):933–43. 18613597.
47. Yue BS, Huang FN, Leonard BR, Moore S, Parker R, Andow DA, et al. Verifying an F1 screen for identification and quantification of rare Bacillus thuringiensis resistance alleles in field populations of the sugarcane borer, Diatraea saccharalis. Entomol Exp Appl. 2008;129(2):172–80. WOS:000260009400008.
48. Downes S, Mahon R. Successes and challenges of managing resistance in Helicoverpa armigera to Bt cotton in Australia. GM crops & food. 2012;3(3):228–34. doi: 10.4161/gmcr.20194 22572906.
49. Downes S, Mahon R. Evolution, ecology and management of resistance in Helicoverpa spp. to Bt cotton in Australia. J Invertebr Pathol. 2012;110(3):281–6. doi: 10.1016/j.jip.2012.04.005 22537836.
50. Downes S. 2014–15 End of season resistance monitoring report. Australian Government Cotton Research and Development Corporation, 2015.
51. Shimomura M, Minami H, Suetsugu Y, Ohyanagi H, Satoh C, Antonio B, et al. KAIKObase: an integrated silkworm genome database and data mining tool. BMC Genomics. 2009;10 : 486. doi: 10.1186/1471-2164-10-486 19843344; PubMed Central PMCID: PMCPMC2770533.
52. Sahara K, Yoshido A, Shibata F, Fujikawa-Kojima N, Okabe T, Tanaka-Okuyama M, et al. FISH identification of Helicoverpa armigera and Mamestra brassicae chromosomes by BAC and fosmid probes. Insect Biochem Molec. 2013;43(8):644–53. doi: 10.1016/J.Ibmb.2013.04.003 ISI:000322561700003.
53. Gahan LJ, Ma YT, Coble MLM, Gould F, Moar WJ, Heckel DG. Genetic basis of resistance to Cry1Ac and Cry2Aa in Heliothis virescens (Lepidoptera: Noctuidae). J Econ Entomol. 2005;98(4):1357–68. ISI:000231056100037. 16156591
54. Knight PJK, Knowles BH, Ellar DJ. Molecular-cloning of an insect aminopeptidase-N that serves as a receptor for Bacillus thuringiensis Cry1A(C) toxin. J Biol Chem. 1995;270(30):17765–70. WOS:A1995RM26600023. 7629076
55. Hossain DM, Shitomi Y, Moriyama K, Higuchi M, Hayakawa T, Mitsui T, et al. Characterization of a novel plasma membrane protein, expressed in the midgut epithelia of Bombyx mori, that binds to Cry1A toxins. Appl Environ Microb. 2004;70(8):4604–12. WOS:000223290100026.
56. Mauchamp B, Royer C, Garel A, Jalabert A, Da Rocha M, Grenier A-M, et al. Polycalin (chlorophyllid A binding protein): A novel, very large fluorescent lipocalin from the midgut of the domestic silkworm Bombyx mori L. Insect Biochem Molec. 2006;36(8):623–33. WOS:000240084100003.
57. Jin L, Wei Y, Zhang L, Yang Y, Tabashnik BE, Wu Y. Dominant resistance to Bt cotton and minor cross-resistance to Bt toxin Cry2Ab in cotton bollworm from China. Evolutionary applications. 2013;6(8):1222–35. doi: 10.1111/eva.12099 24478804; PubMed Central PMCID: PMC3901552.
58. Wei J, Guo Y, Liang G, Wu K, Zhang J, Tabashnik BE, et al. Cross-resistance and interactions between Bt toxins Cry1Ac and Cry2Ab against the cotton bollworm. Sci Rep. 2015;5 : 7714. doi: 10.1038/srep07714 25586723; PubMed Central PMCID: PMCPMC4293620.
59. Gould F, Martinez-Ramirez A, Anderson A, Ferre J, Silva FJ, Moar WJ. Broad-spectrum resistance to Bacillus thuringiensis toxins in Heliothis virescens. P Natl Acad Sci USA. 1992;89(17):7986–90. 11607319; PubMed Central PMCID: PMC49840.
60. Carriere Y, Crickmore N, Tabashnik BE. Optimizing pyramided transgenic Bt crops for sustainable pest management. Nat Biotechnol. 2015;33(2):161–8. doi: 10.1038/nbt.3099 25599179.
61. Liu SM, Zhou S, Tian L, Guo EN, Luan YX, Zhang JZ, et al. Genome-wide identification and characterization of ATP-binding cassette transporters in the silkworm, Bombyx mori. BMC Genomics. 2011;12 : 491 : 491. doi: 10.1186/1471-2164-12-491 ISI:000296333500003; PubMed Central PMCID: PMCPMC3224256. 21981826
62. Xie XD, Cheng TC, Wang GH, Duan J, Niu WH, Xia QY. Genome-wide analysis of the ATP-binding cassette (ABC) transporter gene family in the silkworm, Bombyx mori. Mol Biol Rep. 2012;39(7):7281–91. doi: 10.1007/S11033-012-1558-3 ISI:000304404600007. 22311044
63. Xia QY, Wang J, Zhou ZY, Li RQ, Fan W, Cheng DJ, et al. The genome of a lepidopteran model insect, the silkworm Bombyx mori. Insect Biochem Molec. 2008;38(12):1036–45. doi: 10.1016/J.Ibmb.2008.11.004 ISI:000264262200002.
64. Tay WT, Behere GT, Batterham P, Heckel DG. Generation of microsatellite repeat families by RTE retrotransposons in lepidopteran genomes. BMC Evol Biol. 2010;10 : 144. doi: 10.1186/1471-2148-10-144 20470440; PubMed Central PMCID: PMCPMC2887409.
65. Greenplate JT, Mullins JW, Penn SR, Dahm A, Reich BJ, Osborn JA, et al. Partial characterization of cotton plants expressing two toxin proteins from Bacillus thuringiensis: relative toxin contribution, toxin interaction, and resistance management. J Appl Entomol. 2003;127(6):340–7. doi: 10.1046/j.1439-0418.2003.00766.x WOS:000183511900006.
66. Li Y, Romeis J, Wang P, Peng Y, Shelton AM. A comprehensive assessment of the effects of Bt cotton on Coleomegilla maculata demonstrates no detrimental effects by Cry1Ac and Cry2Ab. PLoS One. 2011;6(7):e22185. doi: 10.1371/journal.pone.0022185 21765949; PubMed Central PMCID: PMCPMC3134477.
67. Ohsawa M, Tanaka M, Moriyama K, Shimazu M, Asano S, Miyamoto K, et al. A 50-Kilodalton Cry2A peptide is lethal to Bombyx mori and Lymantria dispar. Appl Environ Microb. 2012;78(13):4755–7. doi: 10.1128/Aem.07123-11 WOS:000305376600028.
68. Zhan S, Merlin C, Boore JL, Reppert SM. The monarch butterfly genome yields insights into long-distance migration. Cell. 2011;147(5):1171–85. doi: 10.1016/J.Cell.2011.09.052 ISI:000297376600026. 22118469
69. The Heliconius Genome Consortium. Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature. 2012;487(7405):94–8. Epub 2012/06/23. doi: 10.1038/nature11041 22722851; PubMed Central PMCID: PMC3398145.
70. You M, Yue Z, He W, Yang X, Yang G, Xie M, et al. A heterozygous moth genome provides insights into herbivory and detoxification. Nat Genet. 2013;45(2):220–5. Epub 2013/01/15. doi: 10.1038/ng.2524 23313953.
71. Jouraku A, Yamamoto K, Kuwazaki S, Urio M, Suetsugu Y, Narukawa J, et al. KONAGAbase: a genomic and transcriptomic database for the diamondback moth, Plutella xylostella. BMC Genomics. 2013;14 : 464. Epub 2013/07/11. doi: 10.1186/1471-2164-14-464 23837716; PubMed Central PMCID: PMC3711893.
72. Keller O, Odronitz F, Stanke M, Kollmar M, Waack S. Scipio: using protein sequences to determine the precise exon/intron structures of genes and their orthologs in closely related species. BMC Bioinformatics. 2008;9 : 278. doi: 10.1186/1471-2105-9-278 18554390; PubMed Central PMCID: PMCPMC2442105.
73. Salamov AA, Solovyev VV. Ab initio gene finding in Drosophila genomic DNA. Genome Res. 2000;10(4):516–22. doi: 10.1101/Gr.10.4.516 ISI:000086744300014. 10779491
74. Pigott CR, Ellar DJ. Role of receptors in Bacillus thuringiensis crystal toxin activity. Microbiol Mol Biol R. 2007;71(2):255–81. doi: 10.1128/Mmbr.00034-06 ISI:000247273300001.
75. Hernandez-Rodriguez CS, Van Vliet A, Bautsoens N, Van Rie J, Ferre J. Specific binding of Bacillus thuringiensis Cry2A insecticidal proteins to a common site in the midgut of Helicoverpa species. Appl Environ Microb. 2008;74(24):7654–9. doi: 10.1128/Aem.01373-08 ISI:000261513700025.
76. Bungert S, Molday LL, Molday RS. Membrane topology of the ATP binding cassette transporter ABCR and its relationship to ABC1 and related ABCA transporters—Identification of N-linked glycosylation sites. J Biol Chem. 2001;276(26):23539–46. ISI:000169531100044. 11320094
77. Kaminski WE, Piehler A, Wenzel JJ. ABC A-subfamily transporters: structure, function and disease. Bba-Mol Basis Dis. 2006;1762(5):510–24. doi: 10.1016/J.Bbadis.2006.01.011 ISI:000237878100003.
78. Crickmore N, Baum J, Bravo A, Lereclus D, Narva K, Sampson K, et al. Bacillus thuringiensis toxin nomenclature 2014 [cited 2014 15 November]. Available from: <http://www.btnomenclature.info/> (accessed 10-Aug-15).
79. Jurat-Fuentes JL, Gould FL, Adang MJ. Dual resistance to Bacillus thuringiensis Cry1Ac and Cry2Aa toxins in Heliothis virescens suggests multiple mechanisms of resistance. Appl Environ Microb. 2003;69(10):5898–906. ISI:000185881300022.
80. Li HR, Oppert B, Higgins RA, Huang FN, Buschman LL, Zhu KY. Susceptibility of Dipel-resistant and -susceptible Ostrinia nubilalis (Lepidoptera: Crambidae) to individual Bacillus thuringiensis protoxins. J Econ Entomol. 2005;98(4):1333–40. ISI:000231056100034. 16156588
81. Morse RJ, Yamamoto T, Stroud RM. Structure of Cry2Aa suggests an unexpected receptor binding epitope. Structure. 2001;9(5):409–17. ISI:000168684000007. 11377201
82. McNeil BC, Dean DH. Bacillus thuringiensis Cry2Ab is active on Anopheles mosquitoes: single D block exchanges reveal critical residues involved in activity. Fems Microbiol Lett. 2011;325(1):16–21. doi: 10.1111/J.1574-6968.2011.02403.X ISI:000297208500003. 22092857
83. Nicholls CN, Ahmad W, Ellar DJ. Evidence for 2 different types of insecticidal P2 toxins with dual specificity in Bacillus thuringiensis subspecies. J Bacteriol. 1989;171(9):5141–7. ISI:A1989AM27300083. 2570060
84. Liang HX, Liu Y, Zhu J, Guan P, Li SC, Wang SQ, et al. Characterization of Cry2-type genes of Bacillus thuringiensis strains from soil-isolated of Sichuan Basin, China. Braz J Microbiol. 2011;42(1):140–6. ISI:000286320600018. doi: 10.1590/S1517-83822011000100018 24031615
85. Albrecht C, Viturro E. The ABCA subfamily—gene and protein structures, functions and associated hereditary diseases. Pflug Arch Eur J Phy. 2007;453(5):581–9. ISI:000244303300005.
86. Dermauw W, Van Leeuwen T. The ABC gene family in arthropods: comparative genomics and role in insecticide transport and resistance. Insect Biochem Molec. 2014;45 : 89–110. doi: 10.1016/j.ibmb.2013.11.001 24291285.
87. Mace S, Cousin E, Ricard S, Genin E, Spanakis E, Lafargue-Soubigou C, et al. ABCA2 is a strong genetic risk factor for early-onset Alzheimer's disease. Neurobiol Dis. 2005;18(1):119–25. doi: 10.1016/J.Nbd.2004.09.011 ISI:000226452500011. 15649702
88. Mack JT, Townsend DM, Beljanski V, Tew KD. The ABCA2 transporter: Intracellular roles in trafficking and metabolism of LDL-derived cholesterol and sterol-related compounds. Curr Drug Metab. 2007;8(1):47–57. doi: 10.2174/138920007779315044 ISI:000243054800005. 17266523
89. Davis W. The ATP-binding cassette transporter-2 (ABCA2) regulates cholesterol homeostasis and low-density lipoprotein receptor metabolism in N2a neuroblastoma cells. Biochimica Et Biophysica Acta. 2011;1811(12):1152–64. doi: 10.1016/J.Bbalip.2011.07.010 ISI:000298521700018. 21810484
90. Mack JT, Beljanski V, Soulika AM, Townsend DM, Brown CB, Davis W, et al. "Skittish" Abca2 knockout mice display tremor, hyperactivity, and abnormal myelin ultrastructure in the central nervous system. Mol Cell Biol. 2007;27(1):44–53. ISI:000243136800003. 17060448
91. Mahon RJ, Young S. Selection experiments to assess fitness costs associated with Cry2Ab resistance in Helicoverpa armigera (Lepidoptera: Noctuidae). J Econ Entomol. 2010;103(3):835–42. 20568630.
92. Mahon RJ, Olsen KM, Downes S, Addison S. Frequency of alleles conferring resistance to the bt toxins Cry1Ac and Cry2Ab in Australian populations of Helicoverpa armigera (Lepidoptera: noctuidae). J Econ Entomol. 2007;100(6):1844–53. ISI:000251700400015. 18232402
93. Zhang H, Tian W, Zhao J, Jin L, Yang J, Liu C, et al. Diverse genetic basis of field-evolved resistance to Bt cotton in cotton bollworm from China. P Natl Acad Sci USA. 2012;109(26):10275–80. doi: 10.1073/pnas.1200156109 22689968; PubMed Central PMCID: PMC3387040.
94. Gahan LJ, Gould F, Lopez JD, Micinski S, Heckel DG. A polymerase chain reaction screen of field populations of Heliothis virescens for a retrotransposon insertion conferring resistance to Bacillus thuringiensis toxin. J Econ Entomol. 2007;100(1):187–94. doi: 10.1603/0022-0493(2007)100[187:Apcrso]2.0.Co;2 ISI:000243917200026. 17370827
95. Fabrick JA, Ponnuraj J, Singh A, Tanwar RK, Unnithan GC, Yelich AJ, et al. Alternative splicing and highly variable cadherin transcripts associated with field-evolved resistance of pink bollworm to bt cotton in India. PLoS One. 2014;9(5):e97900. doi: 10.1371/journal.pone.0097900 24840729; PubMed Central PMCID: PMC4026531.
96. Tabashnik BE, Fabrick JA, Henderson S, Biggs RW, Yafuso CM, Nyboer ME, et al. DNA screening reveals pink bollworm resistance to Bt cotton remains rare after a decade of exposure. J Econ Entomol. 2006;99(5):1525–30. ISI:000241240400003. 17066779
97. Tay WT, Soria MF, Walsh T, Thomazoni D, Silvie P, Behere GT, et al. A brave new world for an Old World pest: Helicoverpa armigera (Lepdioptera: Noctuidae) in Brazil. Plos One. 2013;8(11):e80134. doi: 10.1371/journal.pone.0080134 24260345; PubMed Central PMCID: PMCPMC3832445.
98. Kriticos DJ, Ota N, Hutchison WD, Beddow J, Walsh T, Tay WT, et al. The potential distribution of invading Helicoverpa armigera in North America: Is it just a matter of time? Plos One. 2015;10(3):e0119618. doi: 10.1371/Journal.Pone.0119618 WOS:000352138500121. 25786260
99. Song X, Kain W, Cassidy D, Wang P. Resistance to Bacillus thuringiensis toxin Cry2Ab in Trichoplusia ni is conferred by a novel genetic mechanism. Appl Environ Microb. 2015;81(15):5184–95. doi: 10.1128/Aem.00593-15 WOS:000357668600032.
100. Mahon RJ, Olsen KM, Downes S. Isolations of Cry2Ab resistance in Australian populations of Helicoverpa armigera (Lepidoptera: Noctuidae) are allelic. J Econ Entomol. 2008;101(3):909–14. ISI:000258337200034. 18613594
101. Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M, et al. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 1995;23(21):4407–14. doi: 10.1093/nar/23.21.4407 7501463; PubMed Central PMCID: PMCPMC307397.
102. Baxter SW, Zhao J-Z, Shelton AM, Vogel H, Heckel DG. Genetic mapping of Bt-toxin binding proteins in a Cry1A-toxin resistant strain of diamondback moth Plutella xylostella. Insect Biochem Molec. 2008;38(2):125–35. doi: 10.1016/j.ibmb.2007.09.014 WOS:000253564200001.
103. Tay WT, Behere GT, Heckel DG, Lee SF, Batterham P. Exon-primed intron-crossing (EPIC) PCR markers of Helicoverpa armigera (Lepidoptera: Noctuidae). Bull Entomol Res. 2008;98(5):509–18. doi: 10.1017/S000748530800583X 18826667.
104. Behere GT, Tay WT, Russell DA, Kranthi KR, Batterham P. Population Genetic Structure of the Cotton Bollworm Helicoverpa armigera (Hubner) (Lepidoptera: Noctuidae) in India as Inferred from EPIC-PCR DNA Markers. PLoS One. 2013;8(1):e53448. doi: 10.1371/journal.pone.0053448 WOS:000314705800045. 23326431
105. Staden R, Beal KF, Bonfield JK. The Staden package, 1998. Methods Mol Biol. 2000;132 : 115–30. 10547834.
106. Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, et al. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16(10):944–5. doi: 10.1093/Bioinformatics/16.10.944 11120685.
107. Juretic D, Zoranic L, Zucic D. Basic charge clusters and predictions of membrane protein topology. J Chem Inf Comp Sci. 2002;42(3):620–32. doi: 10.1021/Ci010263s ISI:000175920000024.
108. Azarian SM, Travis GH. The photoreceptor rim protein is an ABC transporter encoded by the gene for recessive Stargardt's disease (ABCR). Febs Lett. 1997;409(2):247–52. ISI:A1997XF37400027. 9202155
109. Katoh K, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 2008;9(4):286–98. Epub 2008/03/29. doi: 10.1093/bib/bbn013 18372315.
110. Stamatakis A, Ott M, Ludwig T. RAxML-OMP: An efficient program for phylogenetic inference on SMPs. Lect Notes Comput Sc. 2005;3606 : 288–302. ISI:000232251100025.
111. Stamatakis A, Hoover P, Rougemont J. A Rapid Bootstrap Algorithm for the RAxML Web Servers. Syst Biol. 2008;57(5):758–71. doi: 10.1080/10635150802429642 ISI:000259995600008. 18853362
112. Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312–3. doi: 10.1093/Bioinformatics/Btu033 ISI:000336095100024. 24451623
113. Huson DH, Richter DC, Rausch C, Dezulian T, Franz M, Rupp R. Dendroscope: An interactive viewer for large phylogenetic trees. BMC Bioinformatics. 2007;8 : 460. doi: 10.1186/1471-2105-8-460 18034891; PubMed Central PMCID: PMCPMC2216043.
114. Xia Q, Cheng D, Duan J, Wang G, Cheng T, Zha X, et al. Microarray-based gene expression profiles in multiple tissues of the domesticated silkworm, Bombyx mori. Genome Biol. 2007;8(8):R162. Epub 2007/08/09. doi: 10.1186/gb-2007-8-8-r162 17683582; PubMed Central PMCID: PMC2374993.
Štítky
Genetika Reprodukční medicína
Článek A Hereditary Enteropathy Caused by Mutations in the Gene, Encoding a Prostaglandin TransporterČlánek Exocyst-Dependent Membrane Addition Is Required for Anaphase Cell Elongation and Cytokinesis inČlánek Spindle-F Is the Central Mediator of Ik2 Kinase-Dependent Dendrite Pruning in Sensory Neurons
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 11
-
Všechny články tohoto čísla
- Agricultural Genomics: Commercial Applications Bring Increased Basic Research Power
- Ernst Rüdin’s Unpublished 1922-1925 Study “Inheritance of Manic-Depressive Insanity”: Genetic Research Findings Subordinated to Eugenic Ideology
- Convergent Evolution During Local Adaptation to Patchy Landscapes
- The Locus Controls Age at Maturity in Wild and Domesticated Atlantic Salmon ( L.) Males
- A Hereditary Enteropathy Caused by Mutations in the Gene, Encoding a Prostaglandin Transporter
- Absence of Maternal Methylation in Biparental Hydatidiform Moles from Women with Maternal-Effect Mutations Reveals Widespread Placenta-Specific Imprinting
- Calibrating the Human Mutation Rate via Ancestral Recombination Density in Diploid Genomes
- Anaplastic Lymphoma Kinase Acts in the Mushroom Body to Negatively Regulate Sleep
- Connecting Replication and Repair: YoaA, a Helicase-Related Protein, Promotes Azidothymidine Tolerance through Association with Chi, an Accessory Clamp Loader Protein
- UFBP1, a Key Component of the Ufm1 Conjugation System, Is Essential for Ufmylation-Mediated Regulation of Erythroid Development
- Mosaic and Intronic Mutations in Explain the Majority of TSC Patients with No Mutation Identified by Conventional Testing
- Members of the Epistasis Group Contribute to Mitochondrial Homologous Recombination and Double-Strand Break Repair in
- QTL Mapping of Sex Determination Loci Supports an Ancient Pathway in Ants and Honey Bees
- Genetic Interactions Implicating Postreplicative Repair in Okazaki Fragment Processing
- Genomics of Cancer and a New Era for Cancer Prevention
- Adaptation to High Ethanol Reveals Complex Evolutionary Pathways
- Dynamics of Transcription Factor Binding Site Evolution
- Exocyst-Dependent Membrane Addition Is Required for Anaphase Cell Elongation and Cytokinesis in
- Enhancer Runaway and the Evolution of Diploid Gene Expression
- Cattle Sex-Specific Recombination and Genetic Control from a Large Pedigree Analysis
- Drosophila Mutants Model Cornelia de Lange Syndrome in Growth and Behavior
- Pleiotropic Effects of Immune Responses Explain Variation in the Prevalence of Fibroproliferative Diseases
- Leaderless Transcripts and Small Proteins Are Common Features of the Mycobacterial Translational Landscape
- Tissue-Specific Effects of Reduced β-catenin Expression on Mutation-Instigated Tumorigenesis in Mouse Colon and Ovarian Epithelium
- Genus-Wide Comparative Genomics of Delineates Its Phylogeny, Physiology, and Niche Adaptation on Human Skin
- Mapping of Craniofacial Traits in Outbred Mice Identifies Major Developmental Genes Involved in Shape Determination
- Conserved Genetic Interactions between Ciliopathy Complexes Cooperatively Support Ciliogenesis and Ciliary Signaling
- Metabolomic Quantitative Trait Loci (mQTL) Mapping Implicates the Ubiquitin Proteasome System in Cardiovascular Disease Pathogenesis
- DNA Repair Cofactors ATMIN and NBS1 Are Required to Suppress T Cell Activation
- Spindle-F Is the Central Mediator of Ik2 Kinase-Dependent Dendrite Pruning in Sensory Neurons
- Ernst Rüdin and the State of Science
- ABCs of Insect Resistance to Bt
- Epigenetic Control of O-Antigen Chain Length: A Tradeoff between Virulence and Bacteriophage Resistance
- Encodes Dual Oxidase, Which Acts with Heme Peroxidase Curly Su to Shape the Adult Wing
- The Fanconi Anemia Pathway Protects Genome Integrity from R-loops
- Controls Quantitative Variation in Maize Kernel Row Number
- Genome-Wide Association Study of Golden Retrievers Identifies Germ-Line Risk Factors Predisposing to Mast Cell Tumours
- Insect Resistance to Toxin Cry2Ab Is Conferred by Mutations in an ABC Transporter Subfamily A Protein
- A Cytosine Methytransferase Modulates the Cell Envelope Stress Response in the Cholera Pathogen
- Conserved piRNA Expression from a Distinct Set of piRNA Cluster Loci in Eutherian Mammals
- The Multi-allelic Genetic Architecture of a Variance-Heterogeneity Locus for Molybdenum Concentration in Leaves Acts as a Source of Unexplained Additive Genetic Variance
- The lncRNA Controls Cryptococcal Morphological Transition
- Sae2 Function at DNA Double-Strand Breaks Is Bypassed by Dampening Tel1 or Rad53 Activity
- A Tandem Duplicate of Anti-Müllerian Hormone with a Missense SNP on the Y Chromosome Is Essential for Male Sex Determination in Nile Tilapia,
- Ectodysplasin/NF-κB Promotes Mammary Cell Fate via Wnt/β-catenin Pathway
- The QTL within the Complex Involved in the Control of Tuberculosis Infection in Mice Is the Classical Class II Gene
- Identifying Loci Contributing to Natural Variation in Xenobiotic Resistance in
- Variation in Rural African Gut Microbiota Is Strongly Correlated with Colonization by and Subsistence
- A Flexible, Efficient Binomial Mixed Model for Identifying Differential DNA Methylation in Bisulfite Sequencing Data
- Competition between Heterochromatic Loci Allows the Abundance of the Silencing Protein, Sir4, to Regulate Assembly of Heterochromatin
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- UFBP1, a Key Component of the Ufm1 Conjugation System, Is Essential for Ufmylation-Mediated Regulation of Erythroid Development
- Metabolomic Quantitative Trait Loci (mQTL) Mapping Implicates the Ubiquitin Proteasome System in Cardiovascular Disease Pathogenesis
- Genus-Wide Comparative Genomics of Delineates Its Phylogeny, Physiology, and Niche Adaptation on Human Skin
- Encodes Dual Oxidase, Which Acts with Heme Peroxidase Curly Su to Shape the Adult Wing
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání




