-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaA Transposable Element Insertion Confers Xenobiotic Resistance in Drosophila
Given the predictions of future environmental fluctuations, it is crucial to understand how organisms adapt to changing environments. The fruit fly Drosophila melanogaster is an ideal model organism to study environmental adaptation because of our deep understanding of developmental, physiological, and metabolic networks, as well as the ease of experimental manipulation. In this study, we showed that a previously identified putatively adaptive mutation, the insertion of the transposable element FBti0019627, mediates resistance to both natural and synthetic xenobiotics in Drosophila melanogaster. By combining experimental and computational approaches, we further elucidated the molecular and the biochemical mechanisms underlying this natural adaptive mutation. Our results should be relevant for other organisms as well since there are many similarities between species in the way cells respond to stress.
Published in the journal: . PLoS Genet 10(8): e32767. doi:10.1371/journal.pgen.1004560
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004560Summary
Given the predictions of future environmental fluctuations, it is crucial to understand how organisms adapt to changing environments. The fruit fly Drosophila melanogaster is an ideal model organism to study environmental adaptation because of our deep understanding of developmental, physiological, and metabolic networks, as well as the ease of experimental manipulation. In this study, we showed that a previously identified putatively adaptive mutation, the insertion of the transposable element FBti0019627, mediates resistance to both natural and synthetic xenobiotics in Drosophila melanogaster. By combining experimental and computational approaches, we further elucidated the molecular and the biochemical mechanisms underlying this natural adaptive mutation. Our results should be relevant for other organisms as well since there are many similarities between species in the way cells respond to stress.
Introduction
Understanding the functional consequences of naturally occurring mutations is one of the key challenges in modern biology. Recent years have seen an explosion in the availability of genomic data that have opened up the possibility of searching for adaptive mutations on an unprecedented scale [1]. Although there are some examples in which adaptive mutations have been connected to their phenotypic effects [2]–[5], our knowledge of the functional consequences of particular genetic variants is still very limited. Mapping genotype to phenotype is a difficult task due to the large number of genes that contribute to some phenotypes, to the pervasiveness of genetic interactions, and to the complex environmental influences on the phenotypic outcome [6], [7].
Current efforts in genotype-phenotype mapping include projects in several model organisms [8]. Among them, Drosophila melanogaster is one of the most promising cases due to the high quality gene annotation, deep understanding of developmental, physiological, and metabolic networks, and the availability of genetic resources. Because genes tend to work in evolutionarily conserved pathways, genotype-phenotype insights obtained in D. melanogaster provide valuable information that is relevant for other organisms as well [7]. Most ongoing projects in Drosophila focus on mapping SNP variants to a given set of phenotypic traits such as olfactory behavior or stress resistance [9]–[12]. While SNPs certainly contribute to ecologically relevant phenotypes, these efforts ignore other types of mutations, such as those caused by transposable element (TE) insertions.
TEs have the ability to generate mutations of great variety and magnitude, ranging from subtle regulatory mutations to large genomic rearrangements that can have complex phenotypic effects. Additionally, TEs have been shown to be susceptible and responsive to environmental changes; as such, they might have an important role in environmental adaptation [13]–[15]. We have recently used TEs as a tool to identify putatively adaptive mutations to the out-of-Africa environments in D. melanogaster on a genome-wide scale [16], [17]. We screened 763 TEs and identified 18 putatively adaptive TEs based on their population dynamics [16], [18]. For a subset of the candidate TEs, we also demonstrated that they show signatures of selective sweeps [16], [19], evidence of population differentiation [17], and two of them, FBti0019430 and FBti0018880, have already been linked to adaptive fitness effects [20]–[22]. Thus, putatively adaptive TEs in this set are good candidates to perform follow-up experiments that should allow us to map genotype to phenotype and to identify the underlying mechanisms of adaptive mutations.
In this study, we focused on mapping one of the previously identified putatively adaptive insertions, the 186 bp POGON1 element FBti0019627, to its ecologically relevant phenotype. FBti0019627 is inserted in the 3′ UTR region of kinetochore Mis12-Ndc80 network component 1 (Kmn1) gene, and it is closely located to CG11699, a gene of unknown function (Figure 1A) [23]. Kmn1 and CG11699 genes partially overlap and encode cis-natural antisense transcripts [24]. FBti0019627 has recently increased in frequency in out-of-African populations most likely due to positive selection, as suggested by the signatures of a selective sweep in the flanking regions of this TE, including CG11699 and Kmn1 coding sequences [16]. Here, we used an integrative approach, that combines gene structure and gene expression analyses, protein modeling and docking simulations, enzymatic activity and stress resistance assays, to map genotype to phenotype while disentangling the molecular and biochemical mechanisms underlying the adaptive effect of FBti0019627 insertion. We show that, besides being incorporated into Kmn1 transcript, FBti0019627 affects the choice of polyadenylation signal of CG11699 and as a result, only the short 3′ UTR transcript of this gene is produced. These structural changes are associated with increased CG11699 expression in flies with the insertion, leading to xenobiotic resistance through increased ALDH-III activity. Xenobiotic resistance is an ecologically relevant phenotypic trait that provides a plausible explanation for the recent increase in the frequency of FBti0019627 insertion due to positive selection [16], [17].
Fig. 1. FBti0019627 is inserted in the 3′ UTR of Kmn1 and affects the length of Kmn1 and CG11699. 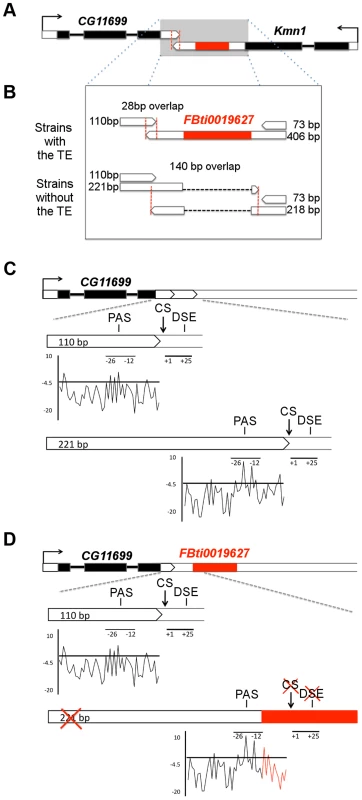
(A) Genomic location of FBti0019627 insertion. Exons are depicted as black boxes, UTRs as white boxes and the TE as a red box. The arrows indicate the direction of gene transcription. Red vertical dashed lines delimit the overlap region between the two genes. (B) 3′ UTRs of CG11699 and Kmn1 transcripts in flies with and without FBti0019627 insertion. Size of the different 3′UTRs and overlap region between the two genes is indicated. (C) Localization of the PolyAdenylation Signal (PAS), cleavage site (CS) and GU rich Downstream Sequence Element (DSE) for the two CG11699 transcripts in flies without the insertion. A graphical representation of the scores obtained for the PAS are also given. (D) Localization of the PAS, CS and DSE in flies with the insertion and graphical representation of the scores obtained for the PAS. FBti0019627 is inserted only 7 bp downstream from the distal PAS and 12 bp upstream the distal CS. Results
FBti0019627 affects the structure of two nearby genes
To confirm that the TE is inserted in the Kmn1 transcript (Figure 1A), we performed 3′ RACE experiments in flies from outbred-1 populations with and without FBti0019627 insertion (see Material and Methods). As expected, we found that the TE is incorporated into the Kmn1 transcript in flies with the insertion, while flies without the insertion have a 188 bp shorter transcript due to the absence of the TE (Figure 1B). Additionally, we discovered a previously unreported transcript with a much shorter 3′ UTR, only 73 bp long, that is present both in flies with and without the insertion (Figure 1B).
To check whether the TE also affects the structure of CG11699, we carried out 3′ RACE experiments. We found that while flies with FBti0019627 insertion have only one transcript with a 110 bp long 3′ UTR, flies without the insertion have two transcripts that differ in the length of their 3′ UTRs: 110 bp long and 221 bp long (Figure 1B). We further analyzed whether the difference in CG11699 transcripts present in flies with and without the insertion is due to FBti0019627 insertion. We identified the cleavage site of each transcript and performed a motif search analysis to identify the polyadenylation signals (PASs) and GU-rich downstream sequence element (DSEs) that are most likely being used to generate the short and the long 3′ UTR transcripts [25]. We found a weak proximal PAS and a strong distal PAS and their corresponding DSEs upstream and downstream respectively of the two cleavage sites (Figure 1C). In flies with the insertion, the TE is inserted between the PAS and the distal cleavage site disrupting the DSE (Figure 1D). In flies with the insertion, the distal cleavage site is not used; as a consequence, only the transcript with the short 3′ UTR is produced.
Because flies with and without the insertion differ in CG11699 transcript isoforms, the length of the overlapping region between this gene and Kmn1 is also different: 28 bp in flies with the insertion and 140 bp in flies without the insertion (Figure 1B). Overall, our results indicated that, besides being incorporated into Kmn1 transcript, FBti0019627 insertion affects the PAS choice of CG11699. As a result, flies with and without this insertion differ in their CG11699 transcript isoforms and in the length of the overlap between CG11699 and Kmn1. We decided to focus on CG11699 for further investigation.
FBti0019627 affects the relative abundance of transcript isoforms and increases the total level of expression of CG11699
To confirm that only the short 3′ UTR transcript is produced in flies with the insertion, and to determine the relative abundance of the short and long 3′ UTR transcripts in flies without the insertion, we performed transcript-specific qRT-PCR in flies from outbred-1 populations (see Material and Methods). We found that in flies with the insertion, the long 3′ UTR transcript is barely detectable. This confirmed that when the TE is present, only the short 3′ UTR transcript is produced (t-test p-value = 0.003 and 0.004 male and female respectively, Figure S1). On the other hand, in flies without the insertion ∼70% of the total CG11699 expression is due to the longer transcript (t-test p-value = 0.008 and 0.013 male and female respectively, Figure S1). These results are in accordance with the computational prediction of the distal PAS being stronger than the proximal PAS (Figure 1C).
Short 3′UTR transcript isoforms usually show increased relative expression levels compared to long 3′UTR transcript isoforms [26], [27]. Thus, we expected that flies with the insertion would have a higher level of expression of CG11699 compared to flies without the insertion, because 100% of the CG11699 isoforms are short in flies with the insertion, while only 30% of the isoforms are short in flies without the insertion. Indeed, our results showed that flies with the insertion have an increased level of expression of CG11699: ∼2.6 fold (t-test p-value = 0.011) in males and ∼2.3 in females (t-test p-value = 0.029) (Figure 2). Thus, FBti0019627 insertion affects the relative abundance of the short and long 3′ UTR transcripts and it is also associated with an overall increased expression of CG11699.
Fig. 2. Flies with FBti0019627 insertion show increased CG11699 expression. 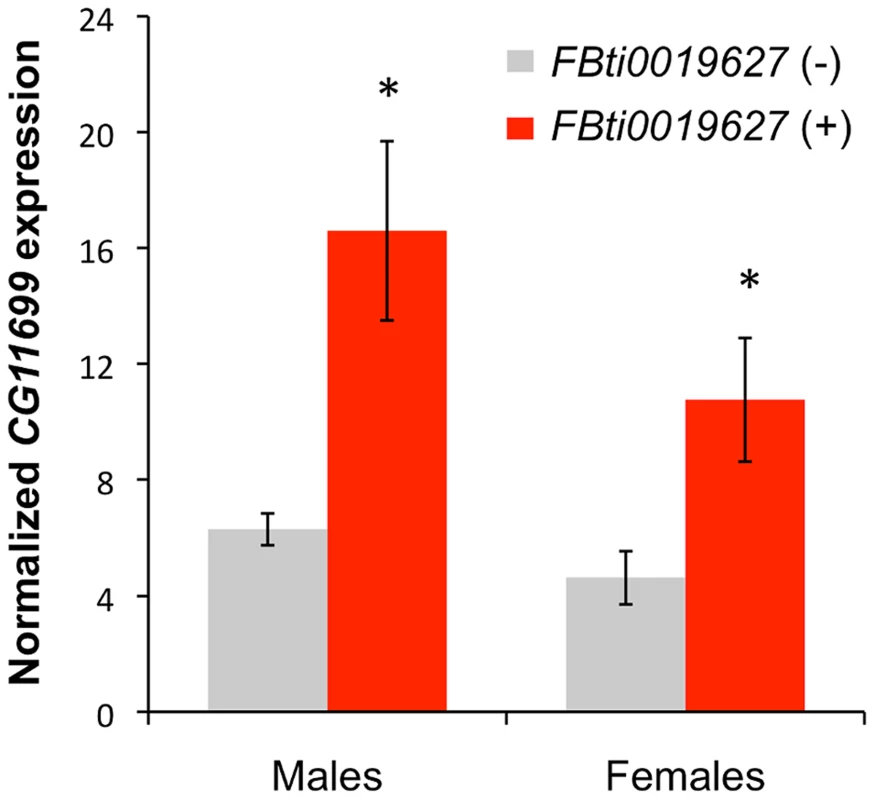
Real-Time PCR quantification of CG11699 transcript levels in flies without the FBti0019627 insertion (gray) and with the insertion (red) are shown for males and females. Average copy number of CG11699 relative to Act5C with error bars representing the S.E.M for three biological replicas are given. FBti0019627 is associated with increased ALDH-III activity
CG11699 encodes a transmembrane protein of unknown function that physically interacts with Aldehyde dehydrogenase III (ALDH-III) [28]. It has been shown that over-expression of CG11699 increases ALDH-III activity in phosphorylated membrane extracts [29]. We hypothesized that flies with FBti0019627 insertion, which have increased CG11699 expression, would have increased ALDH-III activity in the membrane. To test this hypothesis, we measured ALDH activity using different concentrations of benzaldehyde, which is a highly reactive substrate of this enzyme (see Material and Methods; [29], [30]). We compared ALDH-III substrate-activity curves in flies with and without the insertion from the outbred-1 populations. In agreement with our expectations, we found that flies with the insertion have significantly higher ALDH-III enzymatic activity than flies without the insertion (p-value = 0.0042) (Figure 3).
Fig. 3. Flies with FBti0019627 insertion show increased ALDH-III activity. 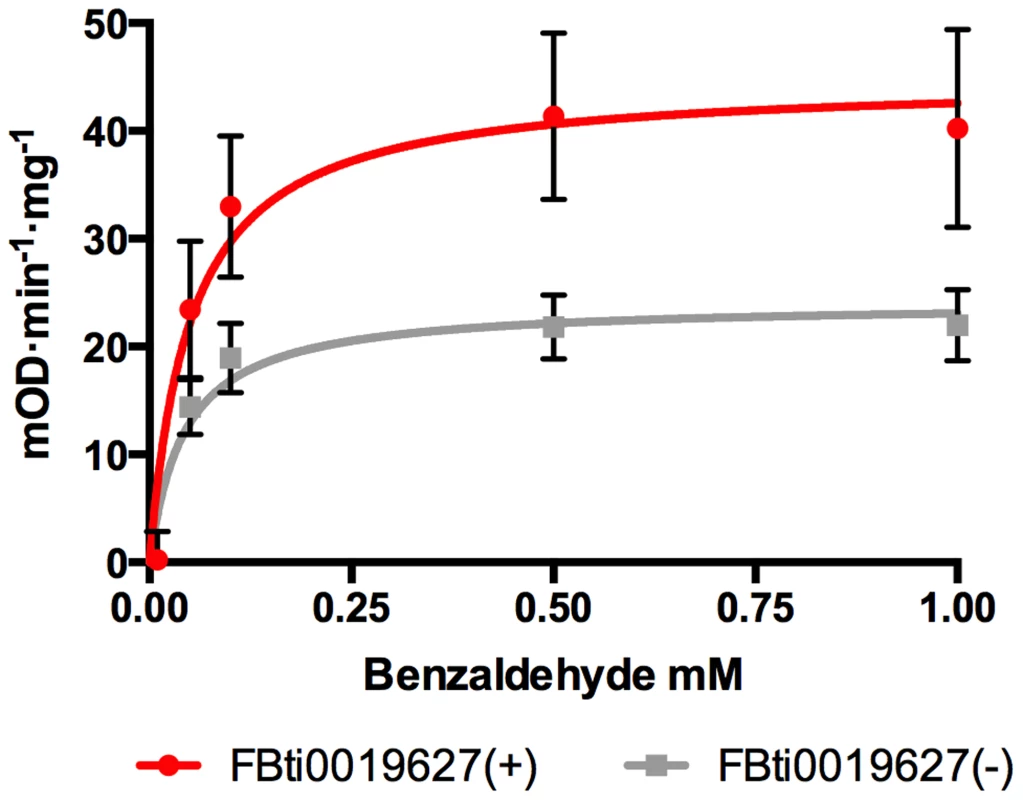
Substrate-velocity curves for ALDH-III activity in flies with (red line) and without (grey line) FBti0019627 insertion. Each data point is the average reaction rate of three biological replicates with corresponding standard error bars. Vmax (95% confidence interval) is 44.73 mOD·min−1·mg−1 (32.15 to 57.32 mOD·min−1·mg−1) for flies with FBti0019627 insertion and 24.06 mOD·min−1·mg−1 (18.65 to 29.48 mOD·min−1·mg−1) for flies without the insertion. FBti0019627 confers resistance to high doses of benzaldehyde
Benzaldehyde is highly toxic when present at concentrations that are too high to be rapidly eliminated because it readily form adducts with DNA, RNA, and proteins [31]. Additionally, benzaldehyde generates reactive oxygen species (ROS) that induce lipid peroxidation in the membrane [32]. ALDH-III not only metabolizes exogenous aldehydes, such as benzaldehyde, but also plays a protective role against endogenous aldehydes generated as a result of lipid peroxidation [30], [33]. Therefore, flies with FBti0019627 insertion that show increased ALDH-III activity (Figure 3) should be more resistant to high doses of benzaldehyde. To test this hypothesis, we compared the survival rate of outbred-1 populations with and without FBti0019627 insertion after an acute exposure to benzaldehyde (see Material and Methods). We analyzed 3 replicas of 50 flies each per sex and per strain for unstressed and stressed conditions (1,200 flies total). While there were no differences in survival rate between flies with and without the insertion in unstressed conditions, we found that flies with the insertion showed increased survival rate compared to flies without the insertion when exposed to high concentrations of benzaldehyde (females t-test p-value = 0.0035, odds-ratio (95% confidence intervals) = 3.11 (1.48–6.50), and males t-test p-value = 0.026, odds-ratio = 4.48 (2.24–8.96); Figure 4). We confirmed these results by replicating the experiment using a larger sample size (9 replicas of 50 flies each: 3,600 flies in total) (Figure 4). Again, both male and female flies with the insertion were more resistant to benzaldehyde than flies without the insertion (females t-test p-value≪0.001, odds-ratio = 7.53 (5.42–10.46) and males t-test p-value = 0.0017 odds-ratio = 6.94 (5.17–9.31); Figure 4). These results suggest that FBti0019627 mediates resistance to benzaldehyde, which is consistent with increased CG11699 expression (Figure 2) and increased ALDH-III activity (Figure 3) observed in flies with this insertion.
Fig. 4. FBti0019627 is associated with increased resistance to benzaldehyde. 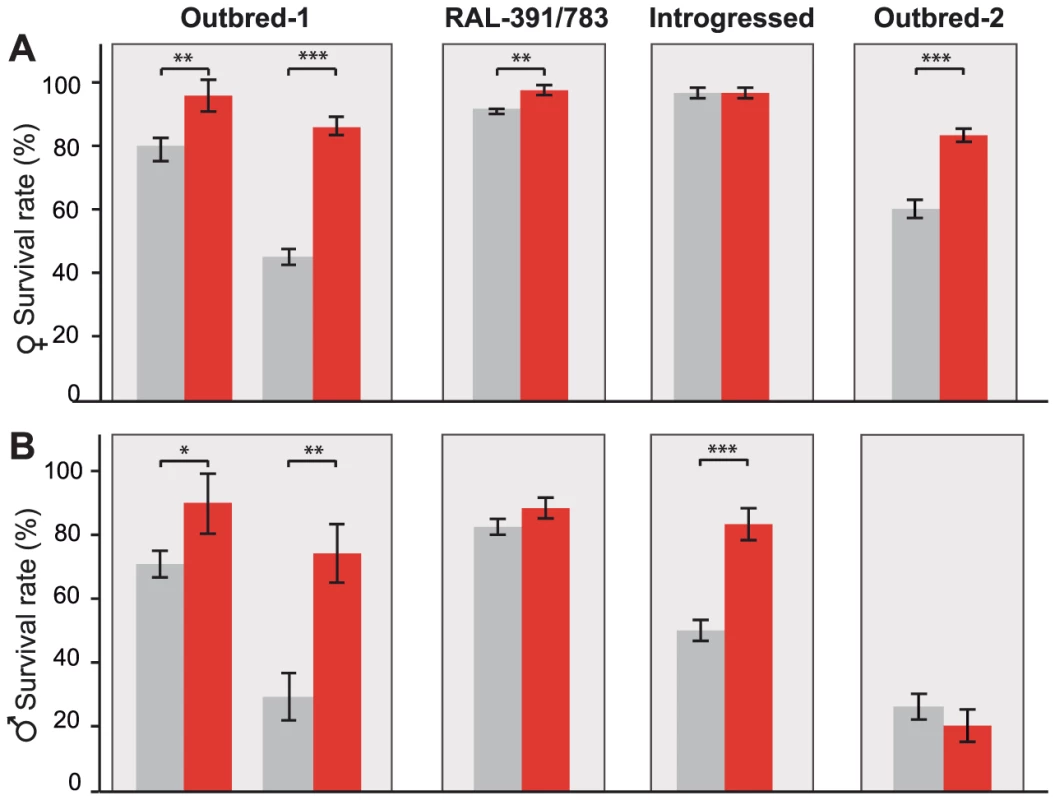
Average survival rate of females (A) and males (B) flies from outbred-1 population, DGRP strains (RAL-391 and RAL-783), introgressed strains, and outbred-2 population, after an acute exposure to benzaldehyde. Error bars indicate the S.E.M of the different replicas performed. To further confirm that resistance to benzaldehyde is due to the insertion and not to any other background mutation present in the outbred-1 populations, we performed the acute exposure to benzaldehyde experiment using flies with three different genetic backgrounds: two DGRP inbred strains, two introgressed strains, and outbred-2 populations (see Material and Methods). We found that females with the insertion showed a significant increase in survival rate compared to females without the insertion in two of the three new backgrounds analyzed: the DGRP strains (Mann-Whitney p-value = 0.001 odds ratio = 8.04 (4.42–14.63)) and the outbred-2 populations (t-test p-value≪0.001 odds ratio = 3.29 (2.14–5.05); Figure 4A). Both introgressed females with and without the TE were highly resistant to benzaldehyde and did not show statistically significant differences (Mann-Whitney, p-value>0.05) (Figure 4A). Similar results were obtained for males; DGRP males with the insertion showed a higher survival rate compared to males without the insertion, although the difference was not statistically significant (t-test, p-value = 0.12). Introgressed males with the insertion were more resistant to benzaldehyde than introgressed males without the insertion (t-test, p-value≪0.001 odds-ratio = 6.94 (3.08–7.06)). Finally, outbred-2 males with and without the insertion were both highly sensitive to benzaldehyde (t-test, p-value>0.05). While the differences in survival rate between flies with and without the insertion were not always significant, when they were, they were consistent with our expectations. These results strongly suggest FBti0019627 insertion mediates resistance to benzaldehyde and that mutations other than the FBti0019627 insertion also affect this phenotype.
FBti0019627 confers resistance to a carbamate insecticide
Aldehydes are present in decomposing fruits, a common food source for D. melanogaster in nature [34]. However, it is not clear whether flies in nature are exposed to such high concentrations of aldehydes as we used in our acute exposure experiments [35]. We searched for other ecologically relevant compounds for D. melanogaster natural populations that could also interact with ALDH-III.
Insecticides and herbicides such as carbamates and thiocarbamates are known to inhibit ALDH2 in humans and rats by covalent modification of the nucleophilic active site residue [36], [37]. ALDH enzymes share a wide range of common physiological functions and substrates and are predicted to have very similar catalytic site structures [37], [38]. It is thus possible that carbofuran, a carbamate insecticide, could also react with the active site of D. melanogaster ALDH-III inhibiting this enzyme. We built a homology-based model of this protein and we performed preliminary docking studies with aldi1, a known ALDH-III inhibitor, and with carbofuran. We found that the size and the shape of carbofuran molecule fits in the catalytic funnel of ALDH-III (Figure 5a). The aromatic rings and the oxo groups of both compounds are located in the same regions (Figure 5b) and the distance between the electrophilic group of carbofuran and the nucleophilic active site residue of ALDH-III is similar to the distance found for the known inhibitor (Figure 5B). Therefore, these preliminary docking results are compatible with carbofuran being a possible ALDH-III inhibitor.
Fig. 5. Carbofuran might be an Aldh-III inhibitor. 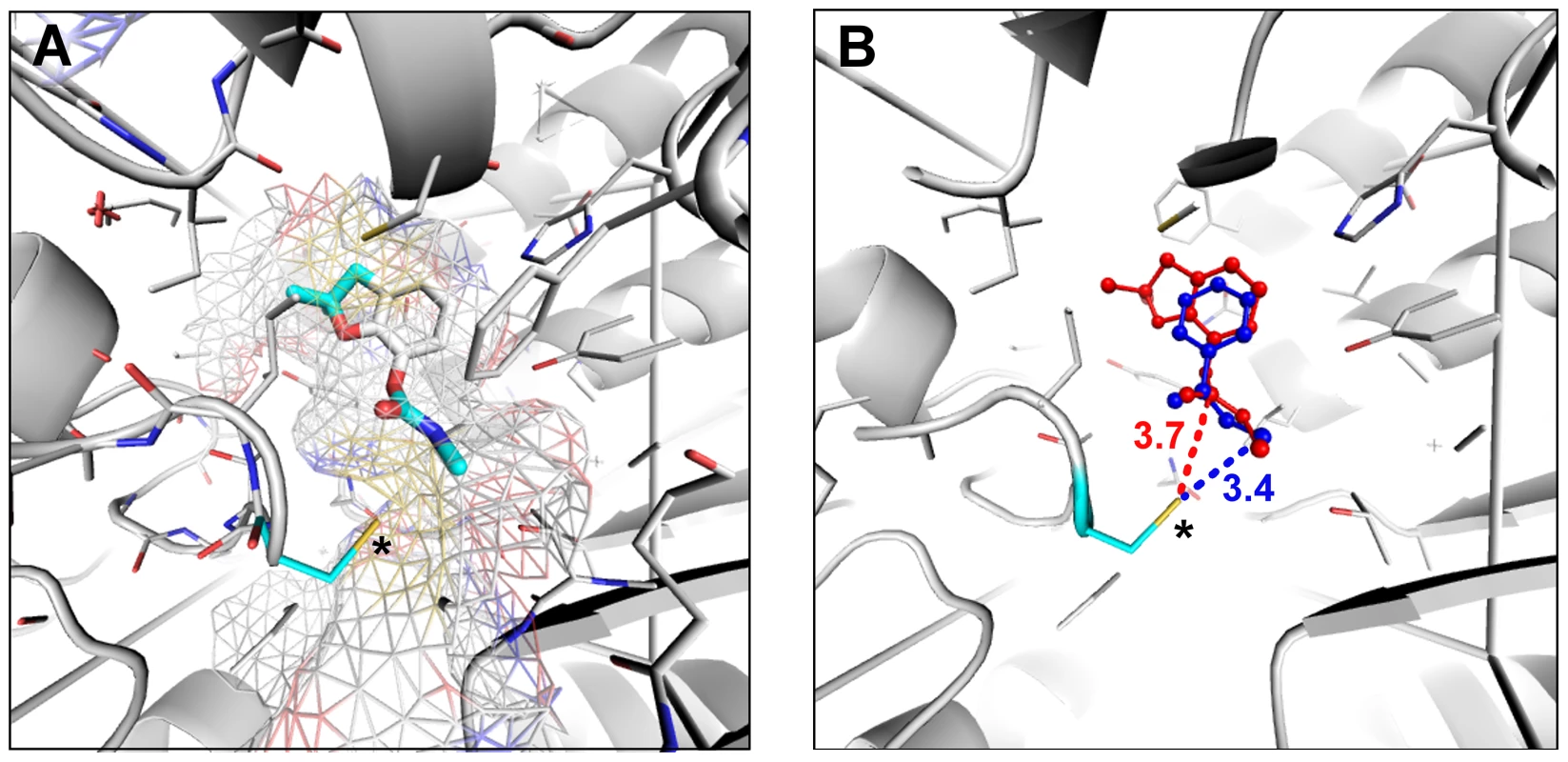
(A) The surface of ALDH-III catalytic funnel is represented as a mesh with carbon atoms colored in white, nitrogen atoms in blue, oxygen atoms in red, and sulfur atoms in yellow. The docked structure of carbofuran is represented with thicker sticks. The nucleophilic thiol group of the active site cysteine is indicated by an asterisk. (B) The best docked pose of carbofuran and aldi1 are shown in red and blue, respectively. Note that the aromatic rings and the oxo groups of both compounds are located in the same regions. The distance of the carbamate electrophilic carbon and the distance of the vinyl ketone of aldi1 to the nucleophilic thiol group of the active site cysteine are given. Although we cannot conclude that carbofuran is an ALDH-III inhibitor, increased ALDH-III activity could also lead to increased carbofuran resistance because carbofuran is an electrophilic molecule that causes lipid peroxidation through the generation of reactive oxygen species (ROS) [39]–[41]. As we have previously mentioned, ALDH3 is known to efficiently metabolize lipid peroxidation derived aldehydes [30] and could therefore play a protective role against carbofuran toxic effects. We hypothesized that flies with the insertion, which show increased ALDH-III activity, could have increased resistance to this carbamate insecticide. In order to evaluate this hypothesis, we compared the survival curves of flies with and without the insertion (3,200 flies in total) that were exposed to concentrations of carbofuran similar to those used in the field (http://www.epa.gov/oppsrrd1/REDs/carbofuran_red.pdf). We used flies with four different genetic backgrounds: outbred-1 populations, DGRP strains and introgressed strains previously used for the benzaldehyde experiments, and two new DGRP strains (see Material and Methods). We found that both males and females flies with the insertion were more resistant to carbofuran than flies without the insertion (Figure 6) (Table 1). Only introgressed males with and without the insertion did not show differences in survival rate (Figure 6B) (Table 1). The magnitude of the effect varied across backgrounds: the effect size was bigger for outbred-1 and RAL-391/783 compared to introgressed and RAL-810/857 (Table 1) strongly suggesting that mutations other than FBti0019627 influence this phenotype.
Fig. 6. FBti0019627 is associated with increased resistance to carbofuran. 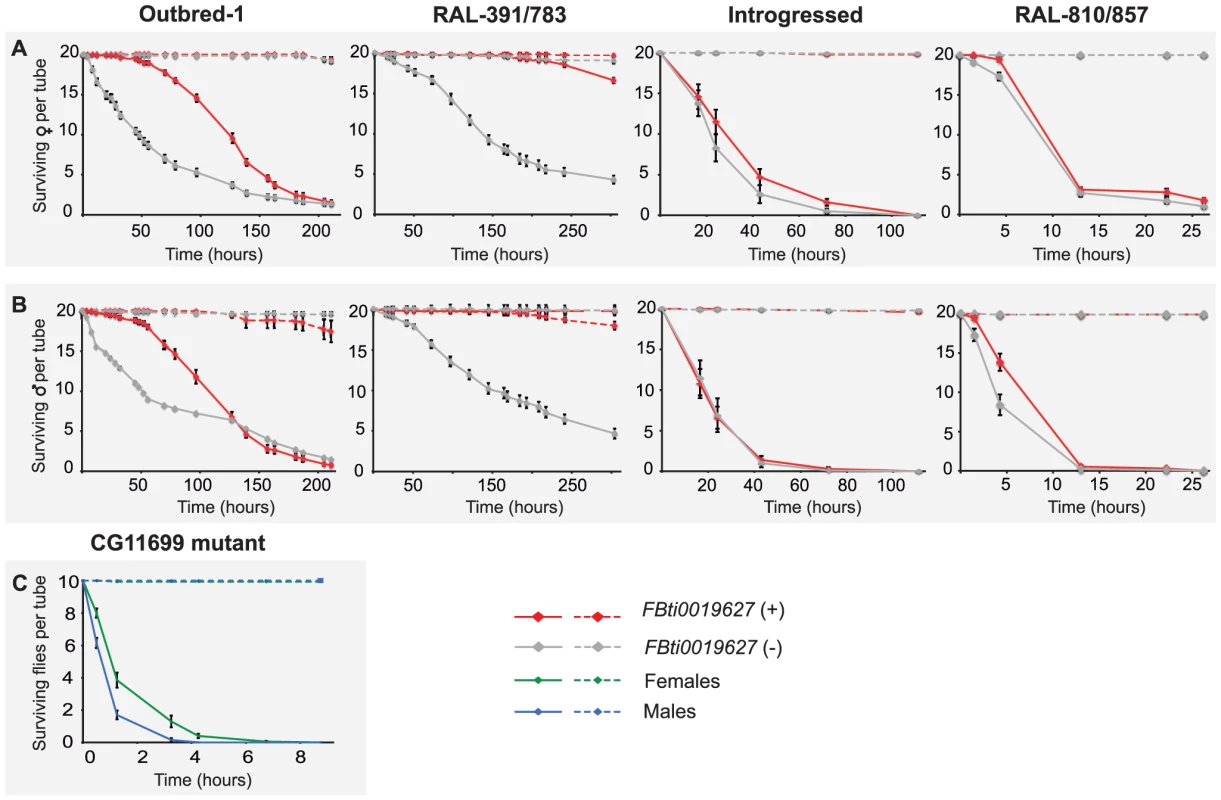
Survival curves of female (A) and male (B) flies from the four different genetic backgrounds analyzed. Solid lines represent survival curves of flies exposed to carbofuran and dashed lines correspond to survival curves of flies in control conditions. Each data point represents the average of surviving flies of 20 replicas of 20 individuals each with error bars indicating the S.E.M. (C) Survival curves for CG11699 mutant flies exposed to carbofuran (solid lines) and for flies kept in control conditions (dashed lines). Tab. 1. Log-rank test p-value and effect size of the mutation on carbofuran resistance for the four different backgrounds analyzed. 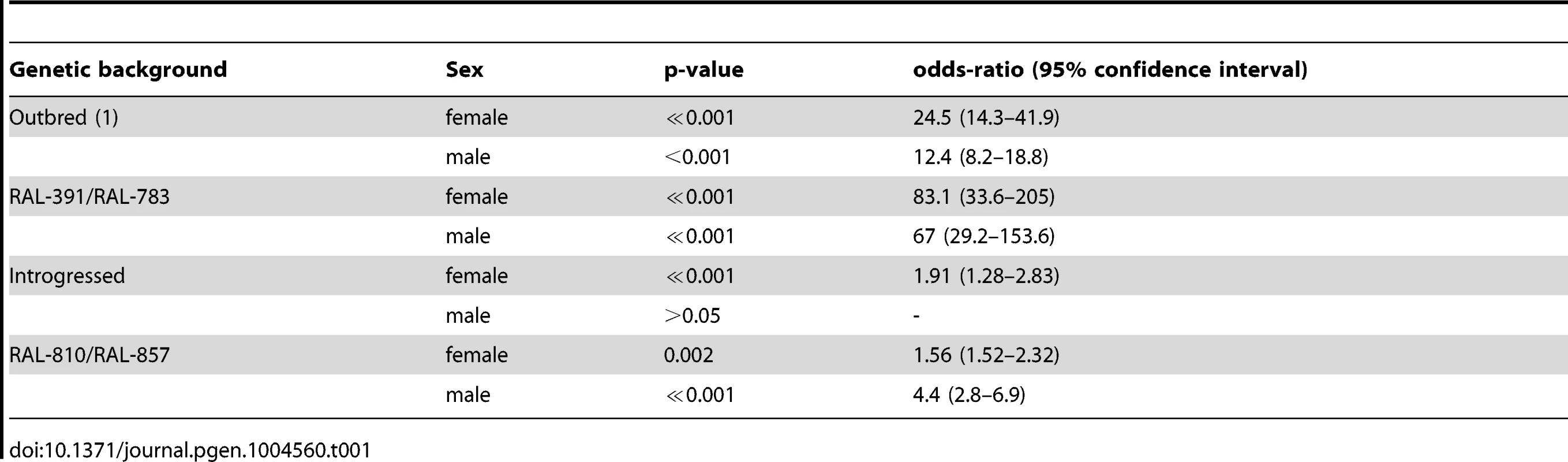
Finally, we also expect that CG11699 mutant flies, which have been previously shown to be highly sensitive to high doses of benzaldehyde [29], should also be highly sensitive to carbofuran. These mutant flies showed reduced or null CG11699 expression levels [42], and thus reduced ALDH-III activity [29]. As expected, our results showed that all CG11699 mutant flies died in the first 7 hours of treatment confirming the predicted high sensitivity of these flies to the insecticide (Figure 6).
Our results obtained from four different genetic backgrounds showed that FBti0019627 insertion mediates resistance to carbofuran insecticide, which is consistent with increased CG11699 expression (Figure 2) leading to increased ALDH-III activity (Figure 3). Similar to the results obtained with benzaldehyde, we also found differences in the magnitude of the effect between backgrounds; this is most likely explained by the contribution of other mutations to this phenotype. Additionally, results obtained with CG11699 lab mutants further confirmed the association between CG11699 expression levels, ALDH-III activity levels, and xenobiotic resistance.
FBti0019627 is not associated with resistance to oxidative stress induced by H2O2
In Drosophila, there is a common oxidative stress response and a specific oxidative stress response that varies depending on the oxidative stress-inducing agent [43]. Both benzaldehyde and carbofuran are lipophilic electrophiles that induce the generation of reactive oxygen species (ROS) leading to lipid peroxidation [31], [39]–[41]. To test whether FBti0019627 insertion confers resistance to other oxidative stress-inducing agents with different physicochemical properties than carbofuran and benzaldehyde, we used H2O2 to induce oxidative stress. While both carbofuran and benzaldehyde are lipophilic compounds, H2O2 is a small polar molecule that is not expected to directly interact with membranes [44]. We compared the survival curves of outbred-1 populations and DGRP strains with and without the insertion by analyzing 20 replicas of 20 flies each per sex and per strain, for unstressed and stressed conditions (3,200 flies in total). Female outbred-1 flies with the insertion were more sensitive than females without the insertion (log-rank p-value = 0.001, odds-ratio = 1.4 (1–1.8)) while males with the insertion were more resistant (log-rank p-value = 0.019, odds-ratio = 1.5 (1.1–2) (Figure 7). In both cases, the lower confidence interval of the odds-ratio was 1 or close to 1 indicating that these results barely reach statistical significance (see Material and Methods). On the other hand, DGRP strains with the insertion were more sensitive to H2O2 than strains without the insertion (log-rank p-value≪0.001, both for male and female flies) (Figure 7). However, this result is explained by the presence in RAL-783 of a TE insertion named Bari-Jheh that confers resistance to oxidative stress [22]. Flies with FBti0019627 insertion were equally or more sensitive to H2O2 compared to flies without the insertion, suggesting that FBti0019627 does not play a role in resistance to H2O2 (Figure 7).
Fig. 7. FBti0019627 is not associated with increased resistance to H2O2 induced oxidative stress. 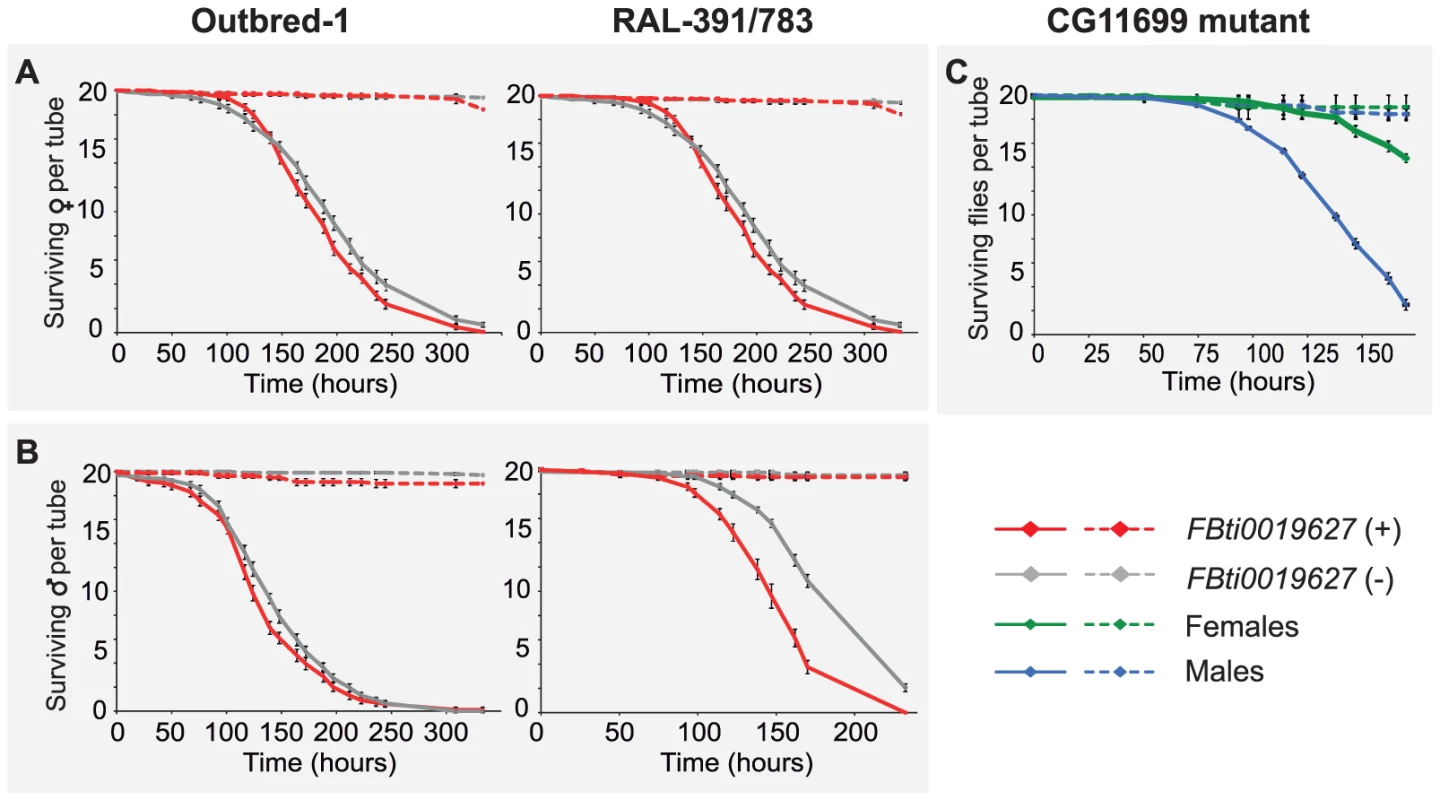
Survival curves of flies with FBti0019627 insertion (red) and without the insertion (gray) for female (A) and male (B) flies from outbred-1 population and DGRP strains (RAL-391 and RAL-783). Solid lines represent survival curves of flies exposed to H2O2 and dashed lines correspond to survival curves of flies in control conditions. Each data point represents the average of surviving flies of 20 replicas of 20 individuals each with error bars indicating the S.E.M. (C) Survival curves for CG11699 mutant flies exposed to H2O2 (solid lines) and for flies kept in control conditions (dashed lines). If the functional interplay of CG11699 and ALDH-III plays a role in general response to oxidative stress, we would expect CG11699 mutant flies to be highly sensitive to oxidative stress induced by H2O2. However, after 138 hours of treatment, 50% of the mutant males and 90% of the mutant females were alive (Figure 7C). These results contrast with the high sensitivity of CG11699 mutant flies to carbofuran: all flies were dead after only 7 hours of stress exposure (Figure 6C). Taken together, our results indicate that resistance to benzaldehyde and carbofuran in flies with the insertion is due to a specific oxidative stress response induced by lipophilic electrophiles and mediated by ALDH-III.
Discussion
FBti0019627 insertion mediates resistance to xenobiotics
In this study, we showed that FBti0019627 insertion mediates resistance to xenobiotics by increasing CG11699 expression leading to increased ALDH-III activity (Figure 2 and Figure 3). Flies with FBti0019627 insertion show increased survival in response to benzaldehyde (Figure 4) and to carbofuran (Figure 6) compared to flies without the insertion. Benzaldehyde is an aromatic aldehyde found in fruits in decomposition, and carbofuran is a carbamate insecticide that has been widely used in nature [41]. Thus, both fatty and aromatic aldehydes and carbamate insecticides found in D. melanogaster habitats are likely agents of selection driving the previously reported increase in FBti0019627 frequencies [16], [17]. Note that other ALDH-III substrates present in natural D. melanogaster habitats could also be acting as agents of selection of this mutation.
We confirmed that xenobiotic resistance is due to FBti0019627 insertion and not to any other background mutation by performing experiments using flies with five different genetic backgrounds: two pairs of outbred populations, two pairs of DGRP inbred strains, and one pair of introgressed strains. Although outbred populations, inbred strains, and introgressed strains differ in their patterns of linkage disequilibrium, in the composition and site frequency distribution of alleles, and in the presence/absence of heterozygous individuals, we consistently observed that flies with the insertion showed increased resistance to xenobiotics compared to flies without the insertion (Figure 4 and Figure 6). Differences in survival rate between flies with and without the insertion were not always significant. However, when they were, they were consistent with our expectations, suggesting that FBti0019627 mediates resistance to xenobiotics. The lack of consistent patterns among backgrounds when a different selective agent was used, i.e. oxidative stress induced by H2O2, further reinforces the role of FBti0019627 in xenobiotic resistance. Effect size of the mutation also varied across backgrounds indicating that genes other than the one affected by the TE insertion are also contributing to the xenobiotic resistance phenotype. These results contrast with previous findings in which the putatively causative mutations of several quantitative traits could not be replicated between strains [12]. While epistatic interactions do not appear to dominate the effect of FBti0019627, they probably play an important role.
Although there are a few examples of TE insertions mediating insecticide resistance in Drosophila [20], [22], [45]–[48], previous evidence linking TEs and resistance to natural xenobiotics was only indirect, i.e. based on the observation that TEs are enriched within or close to resistance genes [49], [50]. Therefore, our results provide the first experimental evidence for a role of TEs in both natural and synthetic xenobiotic resistance in eukaryotes.
Given the widespread distribution of TEs across the tree of life, the conservation of stress response pathways across organisms, and the ubiquitous presence of natural and/or synthetic xenobiotics in the environment, it is likely that TEs are involved in resistance to xenobiotic stress in organisms other than D. melanogaster.
Changes in alternative polyadenylation of CG11699 underlie the adaptive effect of FBti0019627 insertion
Our results indicate that the insertion of FBti0019627 interferes with the choice of CG11699 polyadenylation signal (PAS). As a result, in flies with the insertion, only the short 3′ UTR transcript is produced. As expected, the change in the length of the 3′ UTR leads to increased CG11699 expression levels. Shorter 3′ UTRs isoforms are less likely to possess microRNA binding site and/or other regulatory sequences such as AU-rich elements; consequently, they produce higher levels of transcripts and of protein [26], [27], [51].
Alternative polyadenylation, which leads to transcripts with 3′ UTRs of different lengths, is emerging as a major player in controlling gene regulation [27]. Deciphering the mechanisms behind the choice of alternative polyadenylation sites is considered to be one of the most interesting questions that remains to be answered. Our results provide evidence for TEs playing a role in this selection.
FBti0019627 insertion also affects the transcript length of Kmn1, which could lead to a change in the level of expression of this gene. Further experiments should help elucidate the effect of FBti0019627 on Kmn1, which would provide a more complete picture of the effect of this insertion.
Aldh-III plays a role in xenobiotic resistance in D. melanogaster
Besides elucidating the molecular mechanism underlying the adaptive effect of FBti0019627 insertion, in this analysis we also shed light on its biochemical underpinnings. We showed that increased CG11699 expression is associated with increased ALDH-III activity as was first proposed by Arthaud et al (2011) [29]. Flies with FBti0019627 insertion are more resistant to benzaldehyde (Figure 4) and carbofuran (Figure 6) but not to H2O2 (Figure 7) suggesting that resistance to benzaldehyde and carbofuran is due to a specific stress response induced by lipophilic electrophiles and mediated by ALDH-III. We also found that a lab mutant strain with null or low levels of CG11699 expression, which has been previously shown to be sensitive to benzaldehyde [29], is also highly sensitive to carbofuran but not to H2O2. This result reinforces the functional interplay between CG11699 expression, ALDH-III activity, and xenobiotic resistance. Insecticide resistance is an ongoing challenge for pest management and our results add ALDH-III to the list of previously reported enzymes that play a role in this resistance [49].
TEs as a tool to map genotype to phenotype in D. melanogaster
Mapping genotype to phenotype is currently one of the key challenges in biology [52]. Our approach to genotype-phenotype mapping in D. melanogaster combines a genome-wide screen for adaptive TE insertions, in which we gathered several lines of evidence suggesting their adaptive role, with hypothesis-driven mechanistic and functional analyses of the identified TEs [16], [17], [20], [22]. In this study, we further showed that this approach is able to identify true biological signals of selection and to provide a causal link between genotype and phenotype. By integrating results from gene structure and gene expression analyses, we were able to identify the molecular effect of the insertion (Figure 8A). We combined these results with the wealth of genetic and biochemical information available for Drosophila to construct mechanistic models and experimentally verify their predictions (Figure 8B). Our results provide a plausible explanation for the increase in frequency of FBti0019627 insertion in out-of-Africa populations (Figure 8C), and adds to the limited number of examples in which a natural TE insertion has been linked to its ecologically relevant phenotypic effect [21], [22], [53].
Fig. 8. Graphical summary of the results. 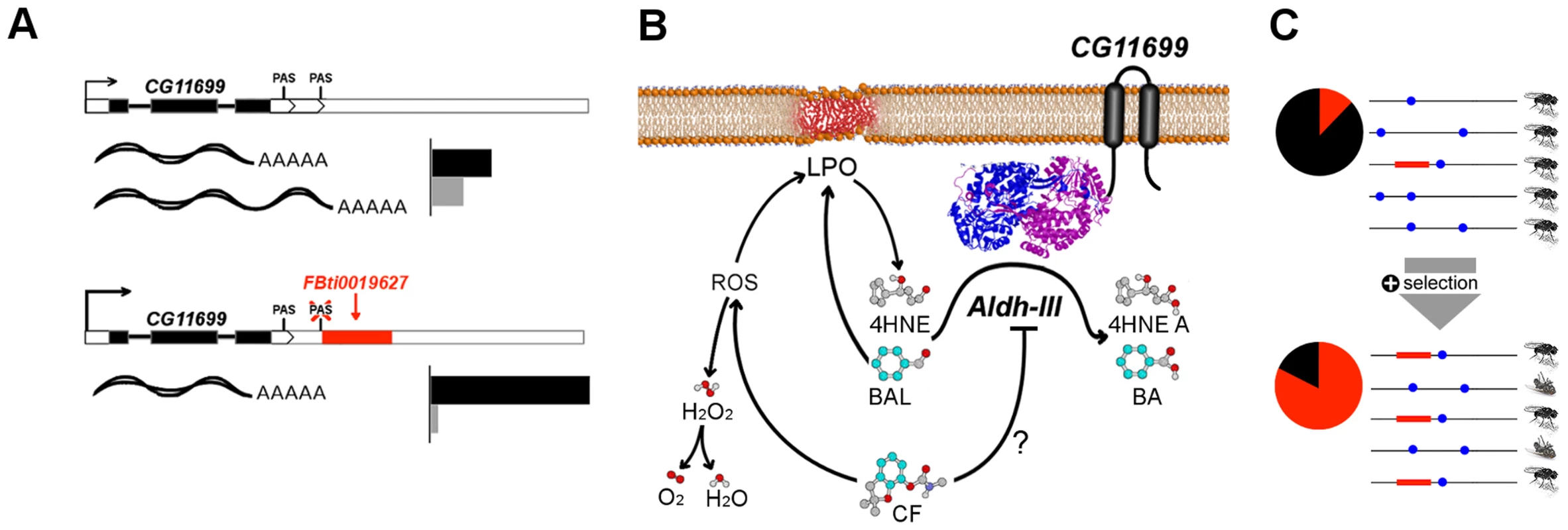
(A) FBti0019627 affects CG11699 PAS choice and, as a consequence, the relative abundance of CG11699 transcripts changes, and the overall expression of the gene increases (see Figure 1 legend). (B) CG11699 is a transmembrane protein that physically interacts with ALDH-III. Increased CG11699 expression is associated with increased ALDH-III activity that results in resistance to benzaldehyde and to carbofuran. Benzaldehyde (BAL) is oxidized by ALDH-III to benzoic acid (BA). Carbofuran (CF) could be inhibiting ALDH-III, as suggested by preliminary docking studies, and/or generating Reactive Oxygen Species (ROS) leading to lipid peroxidation (LPO) derived aldehydes, such as 4HNE that are substrates of ALDH-III (Figure 7b). Although ROS trigger the formation of H2O2, FBti0019627 does not confer resistance to oxidative stress induced by H2O2. This suggests that the effect of the insertion is mediated by ALDH-III. (C) Benzaldehyde and carbofuran are likely selective agents driving the differences in survival rate, and thus the increase in FBti0019627 frequencies in natural populations of D. melanogaster. Each horizontal line represents one haplotype. The red box represents FBti0019627 insertion and the blue dots represent other mutations. The pie charts show the frequency of flies with the TE (red) and without the TE (black) [16]. Besides the other candidate adaptive TE insertions already identified [16], [17], the increasing availability of next generation sequencing data and of computational pipelines to estimate the frequency of TEs in populations should lead to the identification of a larger set of candidate adaptive TEs in the near future [54]–[56]. The in-depth individual analysis of these TEs is very promising, and it should help us obtain a general picture of the adaptive process.
Materials and Methods
Fly strains
Outbred populations
Four outbred populations, two homozygous for the presence and two homozygous for the absence of FBti0019627 were created in the lab by mixing inbred lines from the Drosophila Genetic Reference Panel (DGRP; [11]. The present outbred-1 population was created by mixing RAL-40, RAL-177, RAL-405, RAL-461, and RAL-908; the absent outbred-1 was created by mixing RAL-21, RAL-75, RAL-383, RAL-441, and RAL-855. The present outbred-2 population contained flies from RAL - 776, RAL-801, RAL-802, RAL-822, and RAL-894; the absent oubred-2 population contained flies from RAL-88, RAL-195, RAL-716, RAL-820, and RAL-857. For each one of these four populations, we placed ten virgin females and ten males of each strain in a fly chamber; each outbred population was maintained by random mating during 10 (oubred-1) and 5 (outbred-2) generations before starting the experiments. The census size of each population was n≈800 per generation.
DGRP inbred strains
We also used four individual inbred strains from the Drosophila Genetic Reference Panel [11] to perform the phenotypic assays. Strains RAL-783 and RAL-810 are homozygous for the presence of FBti0019627 insertion. Strains RAL-391 and RAL-857 are homozygous for the absence of FBti0019627.
Introgressed strains
We created two introgressed stocks, one homozygous for the presence of FBti0019627 and the other homozygous for the absence. We individually crossed virgin females flies from stock RAL-783, homozygous for the presence of FBti0019627, with male flies from stock RAL-716, homozygous for the absence of the element. F1 virgin females were individually backcrossed to males form the RAL-716 parental stock. F2 virgin females were backcrossed to the parental males and after a few days were genotyped to check for the presence/absence of FBti0019627. Only the F3 progeny of crosses from females carrying the insertion were used to establish the next generation. Backcrosses were performed for 6 generations, and brother-sister mating was then established to obtain a strain homozygous for the presence and a strain homozygous for the absence of FBti0019627.
CG11699 mutant flies
We used the stock number 16374 from Bloomington Drosophila Stock Center that contains an insertion of a P-element in the 5′ UTR of CG11699 leading to a null or hypomorphic mutation in this gene [42].
3′ RACE experiments
Total RNA was extracted form 40 mg of embryos, 50 L3 larvae, 40 5-day-old males, 40 5-day-old females, and ovaries from 35 5-day-old females using Trizol and a PureLink RNA Mini kit (Ambion). RNA was treated on-column with DNase I (Invitrogen). Reverse transcription was carried out using 3 µg of total RNA for embryos, females, and larvae and 1.5 µg of total RNA for males and ovaries. cDNA was constructed using the SuperScript II RT First Strand Synthesis system for RT-PCR (Invitrogene). We amplified the cDNA 3′ends of CG11699 and Kmn1 genes using a Universal Amplification primer and two nested gene specific primers for CG11699 (5′-AGCCGCACCGATTTCGAGAGTCT-3′ and 5′-CTGGCAGCCTGGAACGAGGAATA-3′) and Kmn1 (5′-CATGATGGAGCTGCAGTGCAATA-3′ and 5′-CCAACGGTGACCCTAAGCTATGC-3′).
The 3′ RACE products were cloned using TOPO TA Cloning Kit for Sequencing (Invitrogene) following the manufacturer's instructions. When there were several 3′ RACE products, DNA from each individual band was extracted from the agarose gel before the cloning reaction. Several clones per 3′ RACE reaction were sequenced in both directions using M13 forward and reverse primers.
Polyadenylation Signal (PAS) and Downstream Sequence Element (DSE) motifs search
Position-specific scoring matrices were derived from the empirical analysis of D. melanogaster Polyadenylation Signal (PAS) and GU-Rich Downstream Element (DSE) motifs published in [25]. The log-likelihood matrix was computed assuming all nucleotides were equiprobable. Sliding windows of 6 bp were run along the 50 bp region upstream of the cleavage site to search for the occurrence of PAS motifs. Sliding windows of 7 bp were run along 50 bp region downstream of the cleavage site to search for occurrence of DSE motifs. We expected the PAS signal to be located between the nucleotide positions −26 to −12 upstream of the cleavage site and the DSE to be located between the positions +1 and +25 downstream of the cleavage site. The highest scoring motifs located in these regions were considered as the most probable PAS and DSE motifs being used.
Total and transcript specific qRT-PCR
Total RNA was extracted from three biological samples of 50 adult males and 50 adult females (4–6 days posteclosion) using Trizol reagent and PureLink RNA Mini kit (Ambion). RNA was then treated on-column with DNase I (Thermo) during purification, and then treated once more after purification. Reverse transcription was carried out using 500 ng and 300 ng of total RNA for females and males respectively using Anchored-oligo(dT) primer and Transcriptor First Strand cDNA Synthesis Kit (Roche). The resulting cDNA was used for qRT-PCR with SYBR green master-mix (BioRad) on an iQ5 Thermal cycler.
Total expression was measured using a pair of primers specific to a 118 bp cDNA amplicon spanning the exon2/exon3 junction of CG11699 present in both transcripts (5′-CTGGAAGCTATCCGGAGCCAA-3′ and 5′-CGTGAGACTCTCGAAATCGGTGCG-3′). Long 3′UTR isoform expression was measured using a pair of primers specific to a 91 bp cDNA amplicon located in the 3′ most region of CG11699 3′UTR and therefore, only present in the long transcript (5′-ACCAGAACATAAAACGAAACCTTTG-3′ and 5′-TGACCGAAACAAATGAAAACCG-3′). In both cases, expression was normalized using Act5C as an endogenous control gene (5′-GCGCCCTTACTCTTTCACCA-3′ and 5′-ATGTCACGGACGATTTCACG-3′). We used serial dilutions of plasmid DNA to derive standard curves for each amplicon. Each curve was then used to determine the quantity of the corresponding transcript relative to the reference gene taking into account the reaction efficiency of each primer pair in order to avoid spurious results caused by differences in the efficiency of the different primer pairs. Reaction efficiencies ranged between 91,4% and 99.7% (r2 larger than 0.99).
Protein extract preparation
Three replicates of 30 4-to-6 day old outbred females with and without the insertion were transferred to 1.5 ml microcentrifuge tubes under light CO2 anesthesia. Flies were homogenized with 1 ml of cold buffer (0.22 M sucrose, 0.12 M mannitol, 1 mM EDTA and 10 mM tricine, pH 7.2) [57] using a 2 ml glass tissue grinder on ice. The homogenate was briefly centrifuged at 4°C to pellet down whole cells and other debris. The supernatant was transferred to a clean microcentrifuge tube and centrifuged for 30 minutes at 13,000 rpm at 4°C in order to obtain a pellet enriched in membranes. The pellet was re-suspended in 1 ml of membrane-disrupting homogenization buffer containing 1% Triton X-100, incubated for 15 minutes on ice, and centrifuged again for 30 minutes at 13,000 rpm at 4°C (adapted from [58], [59]). The supernatant, containing the solubilized proteins that were bound to the membrane, was transferred to a clean microcentrifuge tube and was immediately used for protein quantification and enzymatic activity determination.
ALDH-III enzymatic activity determination
ALDH-III (EC 1.2.1.5) oxidizes benzaldehyde to benzoic acid using NAD(P)+ as an acceptor and producing NAD(P)H in the process. We measured ALDH-III activity by monitoring the increased in absorbance at 340 nm produced by the formation of NAD(P)H (Figure S2A). To control for differences in overall protein abundance between samples, we quantified the total protein content for each homogenate using Quick Start Bradford Protein Assay (BioRad) following the manufacturer's instructions. Each enzymatic activity determination was performed by mixing approximately 100 µg of membrane protein extract (100–200 µl) with 1 ml of reaction buffer (50 mM sodium phosphate, 1 mM NAD+, 1 mM NADP, pH 8 and benzaldehyde at 0.01, 0.05, 0.1, 0.5 or 1 mM) in 1.5 ml disposable cuvettes. Negative controls without substrate and negative controls without NAD(P)+ were run to ensure that the formation of NAD(P)H was specific for the assay conditions we wanted to test. We measured the formation of NAD(P)H every minute at 340 nm for 15 minutes using a UV-1700 PharmaSpec spectrophotometer (Schimadzu). The slope of the linear increase in absorbance over the measurement time for each condition (R2>0.98) was used as initial reaction rate. Once we confirmed that ALDH activity showed a linear relationship with the total amount of protein used in the assay (Figure S2B), we normalized the enzymatic activity measures by the total amount of protein in each sample (activity was expressed as mOD·min−1·mg−1).
In order to estimate Vmax in our samples, we fitted the Michaelis-Menten equation to our experimental data by least-squares method using GraphPad Prism version 6.0e for Mac OS X (GraphPad Software, La Jolla California USA). We performed a replicates test for lack of the fit and obtained a p-value greater than 0.05, indicating that there was no evidence to reject the Michaelis-Menten model. We estimated Km and Vmax and their corresponding 95% confidence intervals and performed a statistical analysis of the comparison between the three replicates of flies with and without the insertion.
Homology based modeling of D. melanogaster ALDH-III protein
The D. melanogaster protein sequence of ALDH-III isoform Q (NP_724562.2) was used to search for a structurally resolved closely related protein. The structure of human ALDH3A1 co-crystallized with aldi1, a covalent inhibitor of ALDH3A1, was selected as a template (PDB ID: 3SZB). The alignment of these two protein sequences was built using the pfam hidden markov model of ALDH family (Aldedh) and the package hmmer3/b 3.0. The final alignment had a 51.54% identity along 424 amino acids comprising the catalytic domain, the NAD binding domain, and part of the bridging domain of ALDH-III.
The ALDH-III model was built using the automodel class in Modeller 9.7 [60]. Energy minimization was carried out using VMD [61] and the extensions Automatic PSF builder and NAMDgui [62]. Default parameters were used except for the dielectric constant, which was set at 80 to simulate an implicit water environment. The stereochemical properties of the template and the model were evaluated using PROCHECK [63] and their pseudoenergetic profiles and z-score were calculated and compared using PROSA-II [64]. Superimposition of the model with human ALDH3A1 and retrieval of the corresponding structural alignment was performed using STAMP [65]. For the regions were the pseudo-energy in the model was higher than in the template, the PSI-PRED [66] predicted secondary structure of the model was compared with the description obtained using dssp [67].
Docking simulations
The docking simulations were performed for a known ALDH-III inhibitor, aldi1, and for carbofuran [68]. The protein structures and ligands were prepared for docking using the Autodock plugin for PyMol [69]. The energy-scoring grid was prepared as a 20 Å×20 Å×20 Å box centered around the catalytic cysteine of the ALDH-III model. The obtained ligands and receptor were used as the input for vina with default parameters [70]. The docking results were visualized and evaluated using PyMol. Redocking of aldi1 with human ALDH3A1 was used to verify that the docking parameters specified for this docking study were correct. The 10 highest scoring poses of 5 docking simulations were evaluated by comparing their localization in the catalytic pocket with the localization of aldi1 in a structural superimposition of ALDH-III model and ALDH3A1. The criteria that were used to select the best poses were: (i) co-planarity of the aromatic rings with Tyr-114 and aldi1; (ii) orientation of the carbamate group towards Cys-243; and (iii) distance between the carbamate electrophilic carbon and nucleophilic thiol group of the active site cysteine.
Phenotypic assays
Acute exposure to benzaldehyde
50 4-to-6 day-old females and males were placed in 50 ml fresh food vials the day before the acute exposure to benzaldehyde experiment was performed to allow flies to recover from the CO2. Three (first experiment) or nine (second experiment) vials of 50 individuals each per sex and per strain were exposed for five minutes to 10 µl of benzaldehyde (Sigma Aldrich) deposited on a cotton swab. Three consecutive exposures separated by 3-hour intervals were performed to each vial as reported in Arthaud et al (2011) [29]. The number of dead flies was counted 72 hours after the last exposure. As a negative control, flies were exposed to a cotton swab with 10 µl of water and the number of dead flies was also counted after 72 hours.
Carbofuran resistance assay
We first performed a dose-response assay using 0 µM 20 µM, 40 µM and 60 µM of carbofuran (PESTANAL, Sigma Aldrich) that was added to the food at 40°C to avoid loss of activity of the insecticide. For each carbofuran concentration, we performed 15 replicas with 10 flies per vial containing 4 ml of food each. Once we established the dose-response, we placed 20 8-to-10 day old males and females per vial, 20 replicas per sex and per strain, in food containing 60 µM of carbofuran. Flies were maintained at room temperature and dead flies were counted one to four times a day until the end of the experiment. Experiments were always done simultaneously for control flies to rule out other sources of variations in viability.
H2O2 resistance assay
We performed a dose-response assay using 0%, 0.25%, 0.50%, and 1% of H2O2 (35 wt. % in H2O, Sigma Aldrich). For each H2O2 concentration, we performed 15 replicas with 10 flies per vial containing 4 ml of food each. The experiment was then performed with a 1% final concentration of H2O2. 20 8-to-10 day old flies were placed per vial and 20 replicas per sex and per strain were performed. Flies were maintained at room temperature and dead flies were counted one to four times a day until the end of the experiment. Experiments were always done simultaneously for control flies to rule out other sources of variation in viability.
Supporting Information
Zdroje
1. StapleyJ, RegerJ, FeulnerPGD, SmadjaC, GalindoJ, et al. (2010) Adaptation genomics: the next generation. Trends Ecol Evol 25 : 705–712 doi:10.1016/j.tree.2010.09.002
2. KorvesTM, SchmidKJ, CaicedoAL, MaysC, StinchcombeJR, et al. (2007) Fitness effects associated with the major flowering time gene FRIGIDA in Arabidopsis thaliana in the field. Am Nat 169: E141–57 doi:10.1086/513111
3. TishkoffSA, ReedFA, RanciaroA, VoightBF, BabbittCC, et al. (2007) Convergent adaptation of human lactase persistence in Africa and Europe. Nat Genet 39 : 31–40 doi:10.1038/ng1946
4. SchluterD, MarchinkoKB, BarrettRDH, RogersSM (2010) Natural selection and the genetics of adaptation in threespine stickleback. Philos Trans R Soc Lond B Biol Sci 365 : 2479–2486 doi:10.1098/rstb.2010.0036
5. LinnenCR, PohY-P, PetersonBK, BarrettRDH, LarsonJG, et al. (2013) Adaptive evolution of multiple traits through multiple mutations at a single gene. Science 339 : 1312–1316 doi:10.1126/science.1233213
6. WuR (2012) Predicting the Genotype-Phenotype Map of Complex Traits. J Biom Biostat 03 : 3–5 doi:10.4172/2155-6180.1000e109
7. LehnerB (2013) Genotype to phenotype: lessons from model organisms for human genetics. Nat Rev Genet 14 : 168–178 doi:10.1038/nrg3404
8. HouleD, GovindarajuDR, OmholtS (2010) Phenomics: the next challenge. Nat Rev Genet 11 : 855–866 doi:10.1038/nrg2897
9. SwarupS, HuangW, MackayTFC, AnholtRRH (2013) Analysis of natural variation reveals neurogenetic networks for Drosophila olfactory behavior. Proc Natl Acad Sci U S A 110 : 1017–1022 doi:10.1073/pnas.1220168110
10. AyrolesJF, CarboneMA, StoneEA, JordanKW, LymanRF, et al. (2009) Systems genetics of complex traits in Drosophila melanogaster. Nat Genet 41 : 299–307 doi:10.1038/ng.332
11. MackayTFC, RichardsS, StoneEA, BarbadillaA, AyrolesJF, et al. (2012) The Drosophila melanogaster Genetic Reference Panel. Nature 482 : 173–178 doi:10.1038/nature10811
12. HuangW, RichardsS, CarboneMA, ZhuD, AnholtRRH, et al. (2012) Epistasis dominates the genetic architecture of Drosophila quantitative traits. Proc Natl Acad Sci U S A 109 : 15553–15559 doi:10.1073/pnas.1213423109
13. BiémontC, VieiraC (2006) Junk DNA as an evolutionary force. Nature 443 : 521–524 doi:10.1038/443521a
14. ChénaisB, CarusoA, HiardS, CasseN (2012) The impact of transposable elements on eukaryotic genomes: from genome size increase to genetic adaptation to stressful environments. Gene 509 : 7–15 doi: 10.1016/j.gene.2012.07.042
15. CasacubertaE, GonzálezJ (2013) The impact of transposable elements in environmental adaptation. Mol Ecol 22 : 1503–1517 doi:10.1111/mec.12170
16. GonzálezJ, LenkovK, LipatovM, MacphersonJM, PetrovDA (2008) High rate of recent transposable element-induced adaptation in Drosophila melanogaster. PLoS Biol 6: e251 doi:10.1371/journal.pbio.0060251
17. GonzálezJ, KarasovTL, MesserPW, PetrovDA (2010) Genome-wide patterns of adaptation to temperate environments associated with transposable elements in Drosophila. PLoS Genet 6: e1000905 doi:10.1371/journal.pgen.1000905
18. GonzálezJ, MacphersonJM, MesserPW, PetrovDA (2009) Inferring the strength of selection in Drosophila under complex demographic models. Mol Biol Evol 26 : 513–526 doi:10.1093/molbev/msn270
19. GonzálezJ, MacphersonJM, PetrovDA (2009) A recent adaptive transposable element insertion near highly conserved developmental loci in Drosophila melanogaster. Mol Biol Evol 26 : 1949–1961 doi:10.1093/molbev/msp107
20. AminetzachYT, MacphersonJM, PetrovDA (2005) Pesticide resistance via transposition-mediated adaptive gene truncation in Drosophila. Science 309 : 764–767 doi:10.1126/science.1112699
21. MagwireMM, BayerF, WebsterCL, CaoC, JigginsFM (2011) Successive increases in the resistance of Drosophila to viral infection through a transposon insertion followed by a Duplication. PLoS Genet 7: e1002337 doi: 10.1371/journal.pgen.1002337
22. GuioL, BarrónMG, GonzálezJ (2014) The transposable element Bari-Jheh mediates oxidative stress response in Drosophila. Mol Ecol 23 : 2020–2030 doi: 10.1111/mec.12711
23. MarygoldSJ, LeylandPC, SealRL, GoodmanJL, ThurmondJ, et al. (2013) FlyBase: improvements to the bibliography. Nucleic Acids Res 41: D751–7 doi:10.1093/nar/gks1024
24. OkamuraK, BallaS, MartinR, LiuN, LaiEC (2008) Two distinct mechanisms generate endogenous siRNAs from bidirectional transcription in Drosophila melanogaster. Nat Struct Mol Biol 15 : 581–590 doi:10.1038/nsmb.1438
25. RetelskaD, IseliC, BucherP, JongeneelCV, NaefF (2006) Similarities and differences of polyadenylation signals in human and fly. BMC Genomics 7 : 176 doi:10.1186/1471-2164-7-176
26. SandbergR, NeilsonJR, SarmaA, SharpPA, BurgeCB (2008) Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science 320 : 1643–1647 doi:10.1126/science.1155390
27. Di GiammartinoDC, NishidaK, ManleyJL (2011) Mechanisms and consequences of alternative polyadenylation. Mol Cell 43 : 853–866 doi:10.1016/j.molcel.2011.08.017
28. GiotL, BaderJS, BrouwerC, ChaudhuriA, KuangB, et al. (2003) A protein interaction map of Drosophila melanogaster. Science 302 : 1727–1736 doi:10.1126/science.1090289
29. ArthaudL, BenRokia-Mille S, RaadH, DombrovskyA, PrevostN, et al. (2011) Trade-off between toxicity and signal detection orchestrated by frequency - and density-dependent genes. PLoS One 6: e19805 doi:10.1371/journal.pone.0019805
30. MarchittiSA, BrockerC, OrlickyDJ, VasiliouV (2010) Molecular characterization, expression analysis, and role of ALDH3B1 in the cellular protection against oxidative stress. Free Radic Biol Med 49 : 1432–1443 doi:10.1016/j.freeradbiomed.2010.08.004
31. SinghAK, PandeyOP, SenguptaSK (2013) Synthesis, spectral and antimicrobial activity of Zn(II) complexes with Schiff bases derived from 2-hydrazino-5-[substituted phenyl]-1,3,4-thiadiazole and benzaldehyde/2-hydroxyacetophenone/indoline-2,3-dione. Spectrochim Acta A Mol Biomol Spectrosc 113 : 393–399 doi: 10.1016/j.saa.2013.04.045
32. MattiaCJ, AdamsJD, BondySC (1993) Free radical induction in the brain and liver by products of toluene catabolism. Biochem Pharmacol 46 : 103–110.
33. LindahlR, PetersenDR (1991) Lipid aldehyde oxidation as a physiological role for class 3 aldehyde dehydrogenases. Biochem Pharmacol 41 : 1583–1587.
34. R'KhaS, CapyP, DavidJR (1991) Host-plant specialization in the Drosophila melanogaster species complex: a physiological, behavioral, and genetical analysis. Proc Natl Acad Sci U S A 88 : 1835–1839.
35. MattheisJP, BuchananDA, FellmanJK (1992) Volatile compounds emitted by sweet cherries (Prunus avium Cv. Bing) during fruit development and ripening. J Agric Food Chem 40 : 471–474.
36. AllenEMG, AndersonDGR, FlorangVR, KhannaM, HurleyDT, et al. (2011) Relative inhibitory potency of molinate and metabolites with aldehyde dehydrogenase2: implications for the mechanism of enzyme inhibition. 23 : 1843–1850 doi:10.1021/tx100317q.Relative
37. KoppakaV, ThompsonDC, ChenY, EllermannM, NicolaouKC, et al. (2012) Aldehyde dehydrogenase inhibitors: a comprehensive review of the pharmacology, mechanism of action, substrate specificity, and clinical application. Pharmacol Rev 64 : 520–539 doi:10.1124/pr.111.005538
38. LiuZJ, SunYJ, RoseJ, ChungYJ, HsiaoCD, et al. (1997) The first structure of an aldehyde dehydrogenase reveals novel interactions between NAD and the Rossmann fold. Nat Struct Biol 4 : 317–326.
39. MilatovicD, GuptaRC, DekundyA, MontineTJ, DettbarnW-D (2005) Carbofuran-induced oxidative stress in slow and fast skeletal muscles: prevention by memantine and atropine. Toxicology 208 : 13–24 doi:10.1016/j.tox.2004.11.004
40. KambojA, KiranR, SandhirR (2006) N-acetylcysteine ameliorates carbofuran-induced alterations in lipid composition and activity of membrane bound enzymes. Mol Cell Biochem 286 : 107–114 doi:10.1007/s11010-005-9100-8
41. RaiDK, SharmaB (2007) Carbofuran-induced oxidative stress in mammalian brain. Mol Biotechnol 37 : 66–71 doi:10.1007/s12033-007-0046-9
42. LaFaveMC, SekelskyJ (2011) Transcription initiation from within P elements generates hypomorphic mutations in Drosophila melanogaster. Genetics 188 : 749–752 doi:10.1534/genetics.111.129825
43. GirardotF, MonnierV, TricoireH (2004) Genome wide analysis of common and specific stress responses in adult drosophila melanogaster. BMC Genomics 5 : 74 doi:10.1186/1471-2164-5-74
44. StępniakJ, LewińskiA, Karbownik-LewińskaM (2013) Membrane lipids and nuclear DNA are differently susceptive to Fenton reaction substrates in porcine thyroid. Toxicol In Vitro 27 : 71–78 doi:10.1016/j.tiv.2012.09.010
45. GahanLJ, GouldF, HeckelDG (2001) Identification of a gene associated with Bt resistance in Heliothis virescens. Science 293 : 857–860.
46. DabornPJ, YenJL, BogwitzMR, Le GoffG, FeilE, et al. (2002) A Single P450 Allele Associated with Insecticide Resistance in Drosophila. 297 : 2253–2256.
47. SchlenkeTA, BegunDJ (2004) Strong selective sweep associated with a transposon insertion in Drosophila simulans. Proc Natl Acad Sci U S A 101 : 1626–1631.
48. Rostant WG, Wedell N, Hosken DJ (2012) Transposable elements and insecticide resistance. 1st ed. Elsevier Inc. doi:10.1016/B978-0-12-394394-1.00002-X.
49. LiX, SchulerMA, BerenbaumMR (2007) Molecular mechanisms of metabolic resistance to synthetic and natural xenobiotics. Annu Rev Entomol 52 : 231–253 doi: 10.1146/annurev.ento.51.110104.151104
50. ChenS, LiX (2007) Transposable elements are enriched within or in close proximity to xenobiotic-metabolizing cytochrome P450 genes. BMC Evol Biol 7 : 46 doi: 10.1186/1471-2148-7-46
51. MayrC, BartelDP (2009) Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 138 : 673–684 doi:10.1016/j.cell.2009.06.016
52. BenfeyPN, Mitchell-OldsT (2008) From genotype to phenotype: systems biology meets natural variation. Science 320 : 495–497 doi:10.1126/science.1153716
53. BarrettRDH, HoekstraHE (2011) Molecular spandrels: tests of adaptation at the genetic level. Nat Rev Genet 12 : 767–780 doi:10.1038/nrg3015
54. Fiston-LavierA-S, CarriganM, PetrovDA, GonzálezJ (2011) T-lex: a program for fast and accurate assessment of transposable element presence using next-generation sequencing data. Nucleic Acids Res 39: e36 doi:10.1093/nar/gkq1291
55. KoflerR, BetancourtAJ, SchlöttererC (2012) Sequencing of pooled DNA samples (Pool-Seq) uncovers complex dynamics of transposable element insertions in Drosophila melanogaster. PLoS Genet 8: e1002487 doi: 10.1371/journal.pgen.1002487
56. Fiston-LavierA-S, BarronMG, PetrovDA, GonzalezJ (2014) T-lex2: genotyping, frequency estimation and re-annotation of transposable elements using single or pooled next-generation sequencing data. Cold Spring Harbor Labs Journals doi: 10.1101/002964
57. KhannaMR, StanleyBA, ThomasGH (2010) Towards a membrane proteome in Drosophila: a method for the isolation of plasma membrane. BMC Genomics 11 : 302 doi:10.1186/1471-2164-11-302
58. MontoothKL, SiebenthallKT, ClarkAG (2006) Membrane lipid physiology and toxin catabolism underlie ethanol and acetic acid tolerance in Drosophila melanogaster. J Exp Biol 209 : 3837–3850 Available: http://www.ncbi.nlm.nih.gov/pubmed/16985200. Accessed 28 April 2014.
59. FryJD, SaweikisM (2006) Aldehyde dehydrogenase is essential for both adult and larval ethanol resistance in Drosophila melanogaster. Genet Res 87 : 87–92.
60. Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, et al.. (2007) Comparative protein structure modeling using MODELLER. doi:10.1002/0471140864.ps0209s50.
61. HumphreyW, DalkeA, SchultenK (1996) VMD: visual molecular dynamics. J Mol Graph 14 : 33–38.
62. PhillipsJC, BraunR, WangW, GumbartJ, TajkhorshidE, et al. (2005) Scalable molecular dynamics with NAMD. J Comput Chem 26 : 1781–1802 doi: 10.1002/jcc.20289
63. LaskowskiRA, MacArthurMW, MossDB, ThorntonJM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst 26 : 283–291 doi:10.1107/S0021889892009944
64. WiedersteinM, SipplMJ (2007) ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res 35: W407–10 doi:10.1093/nar/gkm290
65. RussellRB, BartonGJ (1992) Multiple protein sequence alignment from tertiary structure comparison: assignment of global and residue confidence levels. Proteins 14 : 309–323 doi:10.1002/prot.340140216
66. JonesDT (1999) Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol 292 : 195–202.
67. KabschW, SanderC (1983) Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22 : 2577–2637.
68. KhannaM, ChenC-H, Kimble-HillA, ParajuliB, Perez-MillerS, et al. (2011) Discovery of a novel class of covalent inhibitor for aldehyde dehydrogenases. J Biol Chem 286 : 43486–43494 doi: 10.1074/jbc.M111.293597
69. SeeligerD, de GrootBL (2010) Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J Comput Aided Mol Des 24 : 417–422 doi: 10.1007/s10822-010-9352-6
70. TrottO, OlsonA (2011) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. 31 : 455–461 doi:10.1002/jcc.21334
Štítky
Genetika Reprodukční medicína
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 8
-
Všechny články tohoto čísla
- DNA Double Strand Break Repair in Mitosis Is Suppressed by Phosphorylation of XRCC4
- Inference of Transposable Element Ancestry
- The Population Genetics of Evolutionary Rescue
- Retinoic Acid Activates Two Pathways Required for Meiosis in Mice
- Pooled Segregant Sequencing Reveals Genetic Determinants of Yeast Pseudohyphal Growth
- Comprehensive Identification of Single Nucleotide Polymorphisms Associated with Beta-lactam Resistance within Pneumococcal Mosaic Genes
- SMA-Causing Missense Mutations in Display a Wide Range of Phenotypes When Modeled in
- Branch Migration Prevents DNA Loss during Double-Strand Break Repair
- Transcriptome Sequencing from Diverse Human Populations Reveals Differentiated Regulatory Architecture
- Genetic Deletion of SEPT7 Reveals a Cell Type-Specific Role of Septins in Microtubule Destabilization for the Completion of Cytokinesis
- Tethering Sister Centromeres to Each Other Suggests the Spindle Checkpoint Detects Stretch within the Kinetochore
- Global Genetic Variations Predict Brain Response to Faces
- Demography and the Age of Rare Variants
- The Response to High CO Levels Requires the Neuropeptide Secretion Component HID-1 to Promote Pumping Inhibition
- Sp6 and Sp8 Transcription Factors Control AER Formation and Dorsal-Ventral Patterning in Limb Development
- The Groucho Co-repressor Is Primarily Recruited to Local Target Sites in Active Chromatin to Attenuate Transcription
- A Transposable Element Insertion Confers Xenobiotic Resistance in Drosophila
- The Genomic Architecture of Population Divergence between Subspecies of the European Rabbit
- Human Social Genomics
- Gene Expansion Shapes Genome Architecture in the Human Pathogen : An Evolutionary Genomics Analysis in the Ancient Terrestrial Mucorales (Mucoromycotina)
- Canonical Non-Homologous End Joining in Mitosis Induces Genome Instability and Is Suppressed by M-phase-Specific Phosphorylation of XRCC4
- Roles of Type 1A Topoisomerases in Genome Maintenance in
- The TRIM-NHL Protein LIN-41 Controls the Onset of Developmental Plasticity in
- Wnt-Mediated Repression via Bipartite DNA Recognition by TCF in the Hematopoietic System
- KDM6 Demethylase Independent Loss of Histone H3 Lysine 27 Trimethylation during Early Embryonic Development
- Integration of UPR and Oxidative Stress Signaling in the Control of Intestinal Stem Cell Proliferation
- miR171-Targeted Scarecrow-Like Proteins Bind to GT -Elements and Mediate Gibberellin-Regulated Chlorophyll Biosynthesis under Light Conditions
- Syndecan-1 Is Required to Maintain Intradermal Fat and Prevent Cold Stress
- LIN-3/EGF Promotes the Programmed Cell Death of Specific Cells in by Transcriptional Activation of the Pro-apoptotic Gene
- A System for Genome-Wide Histone Variant Dynamics In ES Cells Reveals Dynamic MacroH2A2 Replacement at Promoters
- Multiple Regulation of Rad51-Mediated Homologous Recombination by Fission Yeast Fbh1
- A Genome-Wide Association Study of the Maize Hypersensitive Defense Response Identifies Genes That Cluster in Related Pathways
- The RNA Helicases AtMTR4 and HEN2 Target Specific Subsets of Nuclear Transcripts for Degradation by the Nuclear Exosome in
- Asymmetric Division and Differential Gene Expression during a Bacterial Developmental Program Requires DivIVA
- A Model-Based Approach for Identifying Signatures of Ancient Balancing Selection in Genetic Data
- Chromatin Insulator Factors Involved in Long-Range DNA Interactions and Their Role in the Folding of the Drosophila Genome
- Conditional Inactivation of Upstream Binding Factor Reveals Its Epigenetic Functions and the Existence of a Somatic Nucleolar Precursor Body
- Evidence for Divisome Localization Mechanisms Independent of the Min System and SlmA in
- Patterns of Admixture and Population Structure in Native Populations of Northwest North America
- Response Regulator Heterodimer Formation Controls a Key Stage in S Development
- A Genetic Strategy to Measure Circulating Insulin Reveals Genes Regulating Insulin Production and Secretion
- EVA-1 Functions as an UNC-40 Co-receptor to Enhance Attraction to the MADD-4 Guidance Cue in
- Dysfunction of the CNS-Heart Axis in Mouse Models of Huntington's Disease
- An Otx/Nodal Regulatory Signature for Posterior Neural Development in Ascidians
- Phosphorylation of a Central Clock Transcription Factor Is Required for Thermal but Not Photic Entrainment
- Genome-Wide Patterns of Genetic Variation within and among Alternative Selective Regimes
- EF-P Dependent Pauses Integrate Proximal and Distal Signals during Translation
- Ku-Mediated Coupling of DNA Cleavage and Repair during Programmed Genome Rearrangements in the Ciliate
- Functional Specialization Among Members Of Knickkopf Family Of Proteins In Insect Cuticle Organization
- Playing RNase P Evolution: Swapping the RNA Catalyst for a Protein Reveals Functional Uniformity of Highly Divergent Enzyme Forms
- The Translational Regulators GCN-1 and ABCF-3 Act Together to Promote Apoptosis in
- Meta-Analysis of Genome-Wide Association Studies in African Americans Provides Insights into the Genetic Architecture of Type 2 Diabetes
- A -Regulatory Mutation of Causes Silky-Feather in Chickens
- VIB1, a Link between Glucose Signaling and Carbon Catabolite Repression, Is Essential for Plant Cell Wall Degradation by
- A Population Genetic Signal of Polygenic Adaptation
- A Conserved Dopamine-Cholecystokinin Signaling Pathway Shapes Context–Dependent Behavior
- The MAP Kinase p38 Is Part of Circadian Clock
- The Cohesin Subunit Rad21 Is Required for Synaptonemal Complex Maintenance, but Not Sister Chromatid Cohesion, during Drosophila Female Meiosis
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Meta-Analysis of Genome-Wide Association Studies in African Americans Provides Insights into the Genetic Architecture of Type 2 Diabetes
- KDM6 Demethylase Independent Loss of Histone H3 Lysine 27 Trimethylation during Early Embryonic Development
- The RNA Helicases AtMTR4 and HEN2 Target Specific Subsets of Nuclear Transcripts for Degradation by the Nuclear Exosome in
- EF-P Dependent Pauses Integrate Proximal and Distal Signals during Translation
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



