-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaFemale Bias in and Regulation by the Histone Demethylase KDM6A
The Rhox cluster on the mouse X chromosome contains reproduction-related homeobox genes expressed in a sexually dimorphic manner. We report that two members of the Rhox cluster, Rhox6 and 9, are regulated by de-methylation of histone H3 at lysine 27 by KDM6A, a histone demethylase with female-biased expression. Consistent with other homeobox genes, Rhox6 and 9 are in bivalent domains prior to embryonic stem cell differentiation and thus poised for activation. In female mouse ES cells, KDM6A is specifically recruited to Rhox6 and 9 for gene activation, a process inhibited by Kdm6a knockdown in a dose-dependent manner. In contrast, KDM6A occupancy at Rhox6 and 9 is low in male ES cells and knockdown has no effect on expression. In mouse ovary where Rhox6 and 9 remain highly expressed, KDM6A occupancy strongly correlates with expression. Our study implicates Kdm6a, a gene that escapes X inactivation, in the regulation of genes important in reproduction, suggesting that KDM6A may play a role in the etiology of developmental and reproduction-related effects of X chromosome anomalies.
Published in the journal: . PLoS Genet 9(5): e32767. doi:10.1371/journal.pgen.1003489
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1003489Summary
The Rhox cluster on the mouse X chromosome contains reproduction-related homeobox genes expressed in a sexually dimorphic manner. We report that two members of the Rhox cluster, Rhox6 and 9, are regulated by de-methylation of histone H3 at lysine 27 by KDM6A, a histone demethylase with female-biased expression. Consistent with other homeobox genes, Rhox6 and 9 are in bivalent domains prior to embryonic stem cell differentiation and thus poised for activation. In female mouse ES cells, KDM6A is specifically recruited to Rhox6 and 9 for gene activation, a process inhibited by Kdm6a knockdown in a dose-dependent manner. In contrast, KDM6A occupancy at Rhox6 and 9 is low in male ES cells and knockdown has no effect on expression. In mouse ovary where Rhox6 and 9 remain highly expressed, KDM6A occupancy strongly correlates with expression. Our study implicates Kdm6a, a gene that escapes X inactivation, in the regulation of genes important in reproduction, suggesting that KDM6A may play a role in the etiology of developmental and reproduction-related effects of X chromosome anomalies.
Introduction
Homeobox (HOX) genes are known for their ability to regulate embryogenesis and guide tissue differentiation. These genes encode transcription factors that specify cell identity and regulate many embryonic programs including axis formation, limb development, and organogenesis [1]. Control of HOX gene expression via epigenetic modifications that include DNA methylation and histone modifications is critical to this process. Notably, tri-methylation of lysine residue 27 of histone H3 (H3K27me3) plays a major role in repression of HOX genes [2]. The histone demethylase KDM6A (also known as UTX) removes H3K27me3 from HOX genes to restore their activity [3]. KDM6A contains a tetratricopeptide motif predicted to mediate protein-protein interactions [4], and is a member of a stable multi-protein complex that not only de-methylates H3K27me3 but also methylates lysine 4 at histone H3 to facilitate gene expression [5], [6]. Different protein partners modulate KDM6A recruitment to specific chromatin regions since ectopic KDM6A expression does not result in significant reduction of genome-wide H3K27me3 levels but rather targets specific genes [3], [7], [8], [9]. For example, KDM6A regulates muscle-specific genes during myogenesis and is necessary for proper cardiac cell differentiation [10], [11]. KDM6A mutations have been discovered in patients with Kabuki syndrome, a rare syndrome associated with distinct facial features, intellectual disability, growth retardation, and skeletal anomalies [12], [13]. Recent studies have also implicated KDM6A as a candidate tumor suppressor gene whereby ectopic expression leads to enhanced expression of the RB (retinoblastoma) and RBL2 (retinoblastoma-like 2) genes [14]. KDM6A inactivating mutations have been discovered in acute promyelocytic leukemia and multiple other cancer types [15], [16], [17].
A large set of homeobox genes clustered on the X chromosome has been implicated in male and female reproduction. In mouse, this cluster called Rhox (reproductive homeobox X-linked) contains 33 adjacent genes organized into three sub-clusters: α, β, and γ [18]. The Rhox cluster evolved at a rapid pace in mammals: the rat cluster contains 11 genes, and the human cluster, only 3 genes. In mouse, members of each paralog family have nearly identical sequences and are thus considered to be functional, although few members have been studied in detail [18]. Rhox genes are selectively expressed in male and female reproductive tissues, including testis, ovary, and placenta [19]. Similar to other homeobox genes, Rhox genes are also expressed during early embryonic development [19], [20], [21], [22], [23]. Little is known about the biological significance of individual paralogs.
Epigenetic regulation of the Rhox gene cluster has been mainly focused on DNA methylation and histone H1 control in placenta and during embryonic development [24], [25]. It is unknown whether other histone modifications control Rhox expression and what histone modifiers might be responsible. An important contender is KDM6A, which is known to regulate the HOX cluster [3]. Interestingly, KDM6A is encoded by an X-linked gene that escapes X inactivation in somatic tissues of human and mouse [26], [27], [28]. Expression of most X-linked genes in somatic tissues is equalized between males (XY) and females (XX) by random silencing of one X chromosome in early development [29]. Genes that escape X inactivation represent exceptional genes with higher expression in females versus males, suggesting that they may be important for female-specific functions [29], [30], [31].
To explore the potential role of KDM6A in the sex-specific regulation of Rhox genes, chromatin analyses were done to follow KDM6A recruitment to the Rhox cluster in male and female ES cells. We focused on Rhox6 and 9, two members of the Rhox cluster we discovered to be most affected by Kdm6a knockdown. KDM6A was specifically recruited to Rhox6 and 9 in female but not male ES cells, resulting in removal of the repressive histone mark H3K27me3 and in increased expression. KDM6A was also bound to Rhox6 and 9 in ovary where these genes are highly expressed. We conclude that KDM6A is important for removal of a repressive histone mark at the bivalent promoters of Rhox6 and 9 to facilitate their expression in female ES cells and in ovary.
Results
Rhox6 and 9 and Kdm6a are expressed in a sexually dimorphic manner
Rhox6 and 9 expression levels were significantly higher in undifferentiated female versus male ES cells (Figure 1A). Expression was measured using quantitative RT-PCR (qRT-PCR) in two female (PGK12.1 and E8) and two male (WD44 and E14) ES cell lines before and after differentiation. ES cell differentiation and embryoid body formation were induced by removal of LIF (leukemia inhibitory factor). Rhox6 and 9 primers were verified to be gene-specific by cDNA sequencing (Figure S1). Sex-specific differences persisted to day 2 of ES cell differentiation (Figure 1B). Analyses of sexed 8-cell pre-implantation embryos confirmed higher female than male expression of Rhox6 and 9 in early development in vivo (Figure S2A). Furthermore, re-analyses of published microarray expression data [32] revealed a female bias in Rhox6 and 9 expression at later embryonic stages (11.5–13.5 dpc) in both germ cells (at all stages) and somatic cells (at 12.5–13.5 dpc) (Figure 1C). Note that expression was much higher in germ cells compared to somatic cells.
Fig. 1. Sexual dimorphism of Rhox6 and 9 and Kdm6a expression in ES cells and embryos. 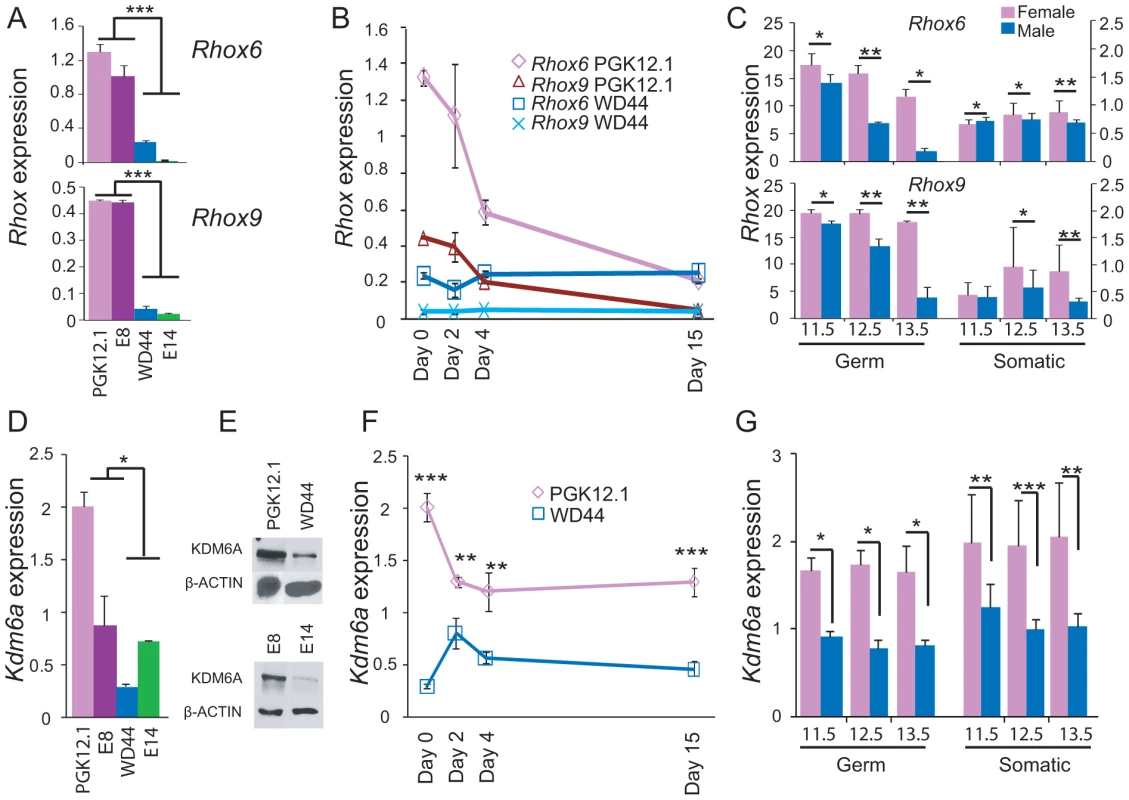
(A) Rhox6 and 9 expression measured by qRT-PCR is higher in female (PGK12.1 and E8) than male (WD44 and E14) undifferentiated ES cells (***p<0.0001). Gene expression was normalized to 18s levels. (B) Rhox6 and 9 expression measured by qRT-PCR in female PGK12.1 ES cells and in male WD44 ES cells during ES cell differentiation. Gene expression was normalized to 18s levels. (C) Re-analyses of published expression array data in germ cells and somatic cells in sexed embryos (11.5–13.5 dpc) shows higher Rhox6 and 9 expression in female than male embryos (*p<0.05, **p<0.001) (see also Figure S2A). Endothelial, mesenchymal, and follicle cells were analyzed together as somatic cells (12 samples total). Values were normalized to the array mean. (D) Kdm6a expression measured by qRT-PCR is higher in female (PGK12.1 and E8) than male (WD44 and E14) undifferentiated ES cells (*p<0.05). Gene expression was normalized to 18s levels. (E) Western blot analysis confirms higher protein levels in female (PGK12.1 and E8) versus male (WD44 and E14) ES cells. β-ACTIN is used as a control. (F) Kdm6a expression measured by qRT-PCR in female PGK12.1 ES cells and in male WD44 ES cells is higher in undifferentiated female than male ES cells throughout differentiation (**p<0.001, ***p<0.0001). Gene expression was normalized to 18s levels. (G) Re-analyses of published expression array data in germ cells and somatic cells in sexed embryos (11.5–13.5 dpc) shows higher Kdm6a expression in female than male embryos (*p<0.05, **p<0.001, ***p<0.0001). Endothelial, mesenchymal, and follicle cells were analyzed together as somatic cells (12 samples total). was higher. Values were normalized to the array mean. A female bias in Rhox6 and 9 expression in ES cells was unexpected because these genes are solely expressed from the maternal allele due to paternal imprinting [25]. Thus, the significantly higher expression we observed in undifferentiated female versus male ES cells (>6-fold for Rhox6 and >10-fold for Rhox9, respectively) must be due to another factor (Figure 1A). Interestingly, levels of the histone demethylase KDM6A known to play a role in HOX gene regulation were approximately two-fold higher in female compared to male ES cells as measured by qRT-PCR and western blot analyses (Figure 1D, 1E). This sex bias initially due to the presence of two active X chromosomes in undifferentiated female ES cells [33], [34] persisted throughout differentiation and at later embryonic stages, as expected for a gene that escapes X inactivation (Figure 1F, 1G). To determine whether KDM6A was involved in the sex-specific regulation of Rhox6 and 9 we measured occupancy using chromatin immunoprecipitation (ChIP) in male and female ES cells. KDM6A occupancy at the 5′ end of Rhox6 and 9 was greater in undifferentiated female than male ES cells as measured both by quantitative PCR (ChIP-qPCR) and by array analysis (ChIP-chip) (Figure 2A and Figure S2B). At day 2 of differentiation the female bias in KDM6A occupancy persisted, but following differentiation (day 15) KDM6A occupancy decreased (Figure 2C and S2B). These results are in agreement with the observed timing of changes in Rhox6 and 9 expression (Figure 1B). Furthermore, we observed corresponding changes in levels of H3K27me3, the histone modification removed by KDM6A. By ChIP-qPCR H3K27me3 enrichment mirrored changes in KDM6A occupancy at Rhox6 and 9 in female PGK12.1 ES cells during differentiation (Figure 2D). Quantitative analysis of H3K4me3 enrichment at Rhox6 and Rhox9 promoters revealed higher enrichment in female than male ES cells, as well as a decrease during differentiation correlating with expression changes (Figure 2B, Figure 1A and 1B).
Fig. 2. KDM6A is preferentially recruited to Rhox6 and 9 in female ES cells. 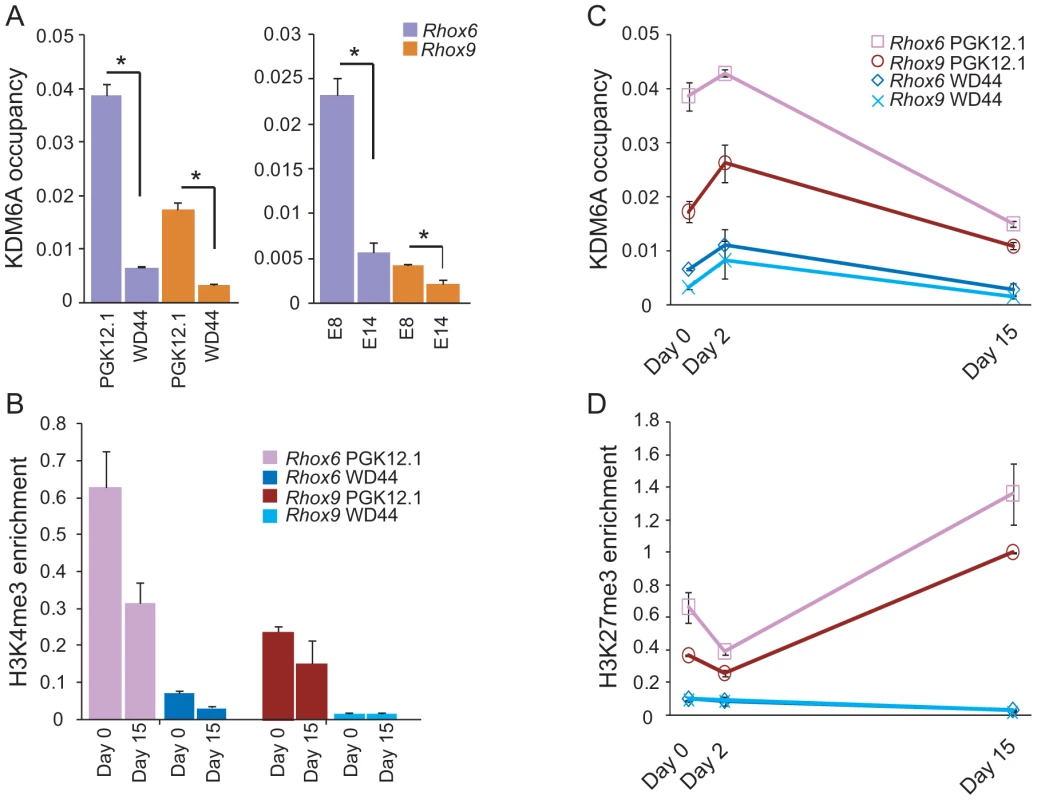
(A) ChIP-qPCR analysis of KDM6A occupancy at the 5′ end of Rhox6 and 9 is higher in female (PGK12.1 and E8) than male (WD44 and E14) undifferentiated ES cells (*p<0.05). (B) H3K4me3 enrichment during differentiation of female PGK12.1 and male WD44 ES cells shows lower levels in male ES cells and a decrease of between day 0 and 15 in agreement with gene silencing after differentiation of these ES cells (see also Figure 1). (C) KDM6A occupancy at the 5′ end of Rhox6 and 9 during differentiation of female PGK12.1 ES cells and male WD44 ES cells. (D) H3K27me3 levels at the 5′ end of Rhox6 and 9 mirror KDM6A occupancy changes. The increase at day 15 is due to X inactivation in female PGK12.1 ES cells (see also Figures S2B, S4, and S6). Average enrichment/occupancy for two separate ChIP experiments is shown as ChIP/input (A, B, C). During differentiation X inactivation initiates in female PGK12.1 ES cells, as confirmed by increased Xist expression and by the appearance of an Xist cloud detected by RNA-FISH in interphase nuclei of cells at day 15 (Figure S3) [35]. Concomitantly, H3K27me3 enrichment at Rhox6 and 9 increased almost 2–3-fold between day 0–2 and day 15 (Figure 2D). This increase was observed over the entire Rhox cluster, suggesting that the cluster is subject to silencing possibly by X inactivation (Figure S4) (see below) [29]. In male ES cells, H3K27me3 levels were very low at Rhox6 and 9 at all time points (Figure 2D). Taken together, our data indicate that KDM6A is specifically recruited to Rhox6 and 9 in undifferentiated female ES cells, which results in a 6–10-fold higher expression compared to male ES cells.
KDM6A regulates Rhox6 and 9 expression in female but not male ES cells
To directly assess the role of KDM6A in regulation of the Rhox cluster we performed knockdowns in two female and two male ES cell lines by RNAi. Using a pool of siRNAs to target multiple regions of Kdm6a RNA, we achieved a 60–80% knockdown in ES cells as shown by qRT-PCR and expression array analyses (Figure 3A). Immunoblots using two different antibodies confirmed a dramatic reduction (70–90%) in the amount of KDM6A protein after 48 h of knockdown (Figure 3A). Specificity of the siRNAs was confirmed using two individual siRNAs, each resulting in a ∼60% knockdown (Figure S5A).
Fig. 3. Kdm6a knockdown causes a female-specific decrease in Rhox6 and 9 expression in ES cells. 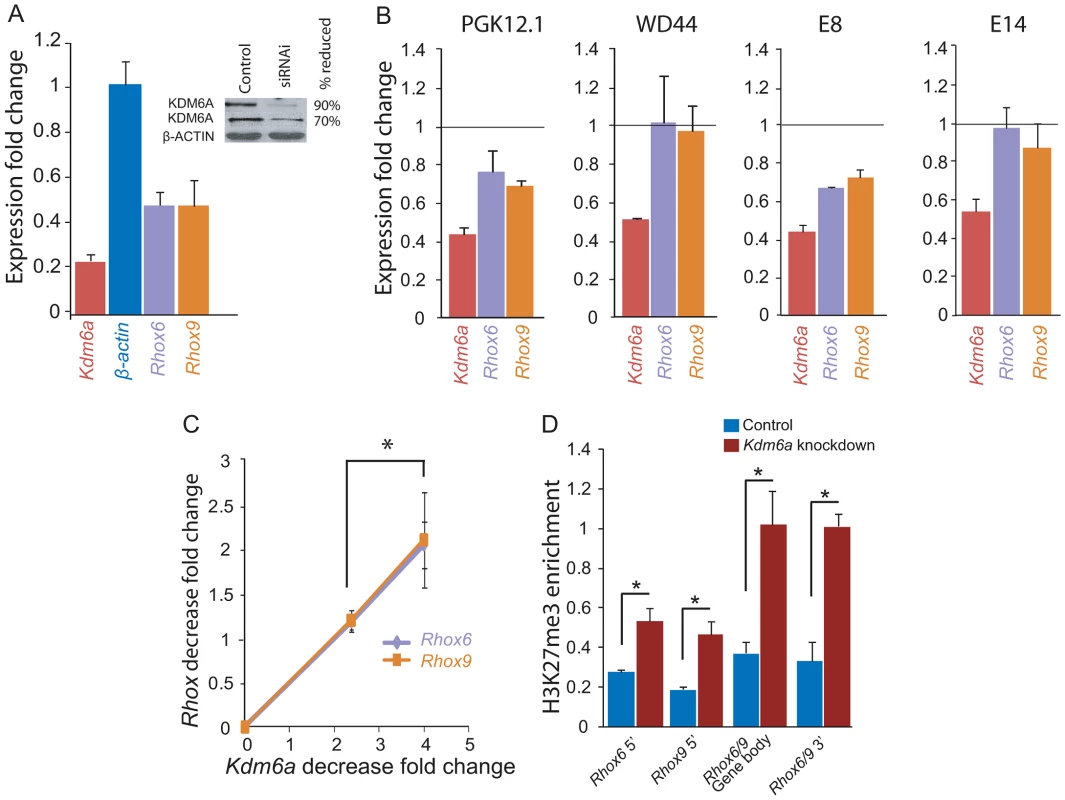
(A) Quantitative RT-PCR after Kdm6a knockdown in female PGK12.1 ES cells shows a 75% and a 52% decrease in Kdm6a and Rhox6 and 9 expression, respectively. Expression is shown relative to control levels obtained with scrambled siRNA. Control gene β-actin levels are set to 1. The inset shows a western blot using two different KDM6A antibodies, which confirms a ∼70–90% reduction in protein levels. β-ACTIN is used as a control. (B) Kdm6a knockdown results in a decrease in Rhox6 and 9 expression in female (PGK12.1 and E8) but not male (WD44 and E14) undifferentiated ES cells. Expression measured by array analysis (PGK12.1 and WD44) and qRT-PCR (E8 and E14) is shown as fold change compared to the control gene β-actin. Array results are from four independent experiments and qRT-PCR results are from three independent experiments. (C) The decrease in Rhox6 and 9 expression correlates with the level of Kdm6a knockdown in a dose dependent manner in PGK12.1 ES cells (*p<0.05). Fold changes in levels of Rhox6 and 9 measured by expression arrays are shown relative to fold changes in Kdm6a levels. (D) Kdm6a knockdown results in a significant (*p<0.05) increase in H3K27me3 enrichment at Rhox6 and 9 5′end, gene body, and 3′end. ChIP-qPCR of H3K27me3 enrichment in PGK12.1 ES cells treated with control scrambled siRNA and Kdm6a specific siRNA. Average enrichment for two separate ChIP experiments is shown as ChIP/input. Kdm6a knockdown caused a significant reduction in Rhox6 and 9 expression in the two female (PGK12.1 and E8) but not in the male (WD44 and E14) ES cell lines, indicating that the regulation of these genes by KDM6A is female-specific (Figure 3B). Rhox6 and 9 expression levels measured by qRT-PCR and by expression array analyses were diminished by 30–50% after Kdm6a knockdown whereas the control gene β-actin did not change (Figure 3A, 3B). By expression array analyses we found that among the Rhox genes, Rhox6 and 9 exhibited the highest expression decrease (>1.25 fold) (Table S1). The lesser decrease measured by expression arrays versus qRT-PCR can be attributed to the different methodologies; qRT-PCR was done using primers designed to be specific for either Rhox6 or Rhox9 (Figure S1), whereas expression changes measured by arrays may be dampened by cross-hybridization due to high sequence similarity between the genes. Importantly, Rhox6 and 9 expression depended on the amount of Kdm6a knockdown in a dose-sensitive manner, consistent with a sex-specific dosage effect (Figure 3C). Note that Rhox5 also showed a significant decrease after Kdm6a knockdown but its analysis was not pursued at this time. As expected, KDM6A occupancy was reduced at the 5′ end and gene body of Rhox6 and 9 after knockdown in female ES cells (Figure S5B). Conversely, H3K27me3 levels were significantly increased at the 5′ end, gene body, and 3′end of Rhox6 and 9, while there was no significant change in levels of H3K4me3 (Figure 3D and Figure S5C). Genes associated with pluripotency (e.g. Cd9, Nanog, Pou5f1, Stat3, Sox2) were not affected by Kdm6a knockdown in either female or male ES cells indicating no induction of differentiation (Figure S5D). We conclude that KDM6A plays a critical and dose-dependent role in regulating Rhox6 and 9 expression in female but not male ES cells.
Rhox6 and 9 are bivalently marked in undifferentiated ES cells
Chromatin domains that contain both activating and inactivating histone marks in undifferentiated ES cells have been termed bivalent and are thought to be poised for activation during development [36], [37]. Notably, bivalent genes include homeobox genes, such as HOX genes, suggesting that Rhox genes are also candidates for bivalency. ChIP-chip profiles in both female and male undifferentiated ES cells demonstrated that both Rhox6 and 9 in cluster β were enriched in H3K27me3 and H3K4me3, indicating that these genes are bivalent and thus poised for activity during development (Figure 4 and Figure S6). Quantitative measurements showed higher H3K4me3 and H3K27me3 levels in female versus male ES cells (Figure 2B, 2D). H3K4me3 levels decreased and H3K27 levels increased between day 0 and 15 of differentiation in female ES cells, consistent with a decrease in Rhox6 and 9 expression (Figure 2B, 2D). In contrast, levels remained low in male ES cells. KDM6A binding being clearly female-biased would explain the female bias in gene expression at day 0–2 of differentiation, as described above (Figure 1 and Figure 2).
Fig. 4. Rhox6 and 9 are bivalent and preferentially occupied by KDM6A in female ES cells. 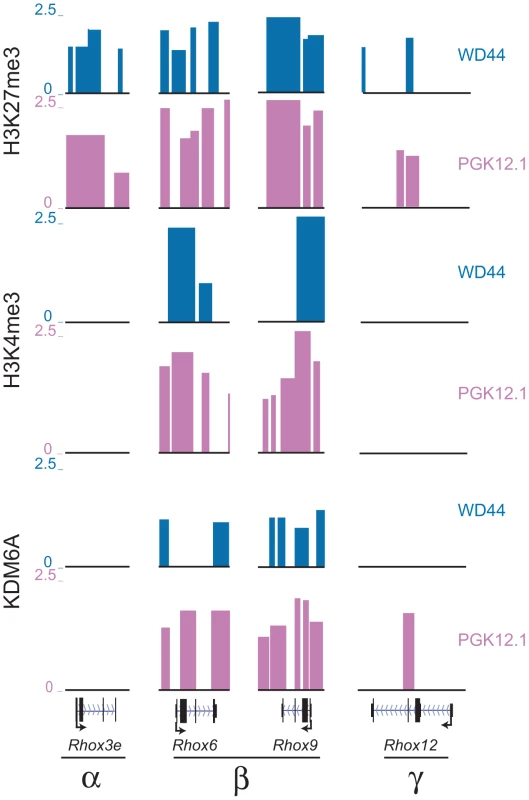
H3K27me3, H3K4me3 and KDM6A enrichment profiles in undifferentiated female PGK12.1 (pink) and male WD44 ES (blue) cells at representative genes from each Rhox subcluster (α, β, and γ) demonstrate that only Rhox6 and 9 are highly enriched with both histone modifications and are bound by KDM6A (see also Figure S6). Rhox3e (α cluster) is enriched in H3K27me3 but not H3K4me3 or KDM6A, and Rhox12 (γ cluster) shows little enrichment for the proteins analyzed. Significant enrichment peaks based on Nimblescan analysis (FDR score <.05) are shown. Data uploaded to UCSC genome browser (NCBI36/mm8). Other Rhox genes within the α - and γ-clusters did not appear to be bivalent but rather were enriched with the repressive mark H3K27me3, with no significant peaks of enrichment for the active mark H3K4me3, suggesting that these genes are mostly contained in a silenced chromatin domain in both female and male undifferentiated ES cells (Figure 4 and Figure S6). Inspection of representative genes from each cluster, Rhox6 and 9 in cluster β, Rhox3e in cluster α, and Rhox12 in cluster γ, confirmed these findings (Figure 4). Note that while Rhox1 and 7 also appeared bivalent at day 0, KDM6A was absent at their promoter, which may account for their low expression (Table S1).
Rhox6 and 9 are bound with KDM6A and highly expressed in ovary
Re-analyses of published expression array data confirmed that Rhox6 and 9 and Kdm6a are expressed at a higher level in ovary than in testis (Figure 5A) [18], [19]. This is consistent with measurements of expression in embryos, in which female germ cells have much higher expression than male germ cells (Figure 1C). As expected for a gene that escapes X inactivation, Kdm6a expression was higher in all female tissues examined in comparison to male tissues, including brain and sexual organs, as well as somatic and germ cells from embryos (Figure 5C and Figure 1G). To assess the in vivo binding of KDM6A to Rhox6 and 9 in reproductive tissues, chromatin extracted from adult mouse ovaries and testes was subjected to ChIP-qPCR. KDM6A occupancy was high at the promoters of Rhox6 and 9 in mouse ovary, consistent with high expression in this organ (Figure 5A, 5B) [38], [39]. In mouse testis where Rhox6 and 9 expression is lower, KDM6A was still bound but to a lesser extent (38% of occupancy in ovary), reflecting lower expression (Figure 5A, 5B). In mouse brain where the genes are not expressed [19], KDM6A occupancy was almost undetectable in females and completely undetectable in males (Figure 5B). Taken together, these data indicate that KDM6A occupancy is associated with Rhox6 and 9 expression in reproductive tissues, more significantly in females than in males.
Fig. 5. Rhox6 and 9 expression and KDM6A occupancy are high in ovary where the genes are imprinted. 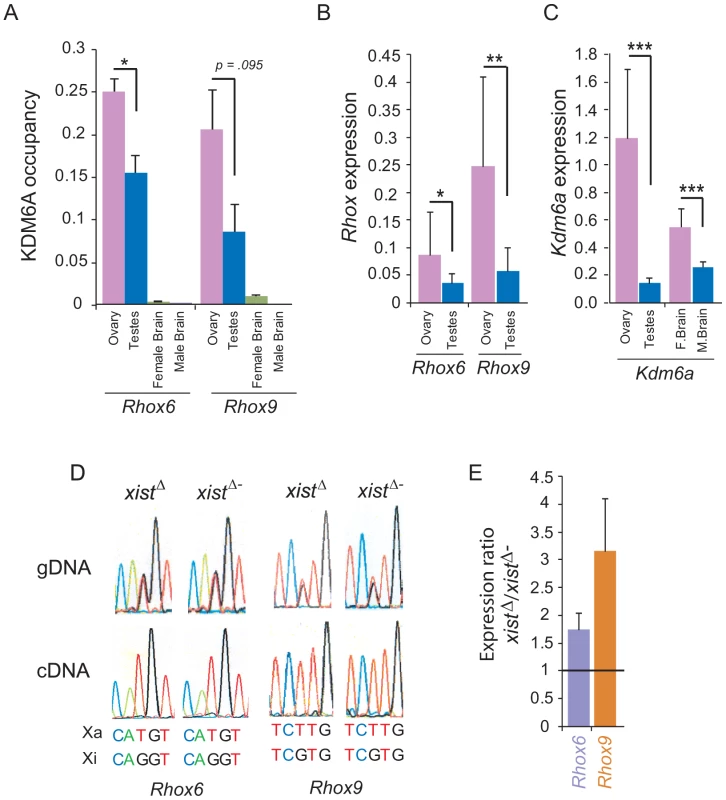
(A) Rhox6 and 9 have significantly higher expression in mouse ovary than in testis, based on re-analyses of published expression array data for 14 testis and 12 ovary specimens (*p<0.05, **p<0.001). Expression normalized to array mean (see also Figure 1C). (B) KDM6A occupancy measured by ChIP-qPCR at the 5′end of Rhox6 and 9 is higher in ovary than in testis, and is very low to undetectable in brain where these genes are not expressed [19]. Occupancy levels were normalized to input fractions. (C) Kdm6a has high expression in female tissues especially ovary based on analyses of published expression array data (***p<0.0001) (see also Figure 1F). (D) Rhox6 and 9 are expressed from the maternal allele only in ovary because of imprinting. DNA sequence chromatograms of gDNA and RT-PCR (cDNA) products derived from ovary from female F1 mice obtained by mating M. spretus males with C57BL/6J females with or without an Xist mutation (XistΔ and XistΔ−). SNPs to distinguish Rhox6 and 9 alleles on the active X (Xa) and on inactive X (Xi) are indicated below. In ovary from both XistΔ and XistΔ− mice the gDNA shows heterozygosity at the SNPs while the cDNA shows only the maternal allele, consistent with paternal imprinting. (E) By qRT-PCR Rhox6 and 9 are more highly expressed in ovary from XistΔ mice in which the maternal X chromosome is expressed in all cells, compared to XistΔ− mice in which there is random X inactivation (1.7-fold and 3-fold, respectively), suggesting that Rhox6 and 9 are silenced by X inactivation. Values represent the expression ratio between XistΔ and XistΔ− ovaries. Rhox6 and 9 are paternally imprinted in ovary
To determine the allele-specific expression of Rhox6 and 9 in ovary we employed F1 mice derived from crosses between C57BL/6J females with (XistΔ) or without (XistΔ−) an Xist mutation and Mus spretus males. In F1 animals that carry the mutant Xist (XistΔ), X inactivation is completely skewed towards the M. spretus X chromosome. SNPs between the mouse species were used to distinguish alleles after RT-PCR and Sanger sequencing. In F1 mice with (XistΔ) or without (XistΔ−) the Xist mutation expression of Rhox6 and 9 was exclusively from the maternal C57BL/6J allele, with no evidence of the M. spretus allele, consistent with imprinting of the paternal allele (Figure 5D). This is similar to what has been reported in mouse ES cells and placenta [25]. Control genomic DNA amplification confirmed the presence of the SNPs in the F1 mice (Figure 5D). Our results suggest that imprinting has taken place in the germ cells from adult ovary, as we did not observe any evidence of paternal allele expression. By qRT-PCR Rhox6 and 9 expression was higher (1.7-fold and 3-fold, respectively) in ovaries of F1 mice carrying the Xist mutation (XistΔ) in which the maternal allele is expressed in all cells (due to skewing of X inactivation), compared to ovaries from non-mutant F1 mice (XistΔ−) in which the maternal allele is expressed in half of the cells (due to random X inactivation) (Figure 5E). The X inactivation effect on Rhox6 and 9 expression would only be pertinent in somatic cells, but not in germ cells in which the inactive X chromosome is reactivated. This complicates interpretation of our data because expression was measured in whole ovary containing both germ cells with very high Rhox6 and 9 expression and supporting somatic cells with lower expression (Figure 1C). Additional studies in germ cells and somatic cells of the ovary are needed to fully understand the developmental regulation of Rhox6 and 9 in this organ. Nonetheless, we conclude that female biased expression of Rhox6 and 9 in ovary is not due to bi-allelic expression in this tissue but rather to recruitment of KDM6A to activate Rhox6 and 9 on the active maternal X chromosome.
Discussion
Rhox genes represent a set of X-linked homeobox genes specifically expressed in organs and cell types implicated in sexual development and reproduction [19], [20], [40], [41]. Here, we provide functional evidence identifying KDM6A, an enzyme that removes methylation at lysine 27 of histone H3, as an important regulator of a specific subset of Rhox genes, Rhox6 and 9, in female ES cells and in ovary. Interestingly, KDM6A is encoded by an X-linked gene that escapes X inactivation and has higher expression in females, which may indirectly facilitate its sex-specific role in enhancing Rhox6 and 9 expression [26], [27], [28]. Our knockdown experiments clearly support an important role for KDM6A in regulating Rhox6 and 9 in female but not male ES cells.
The 6–10 fold female bias in Rhox6 and 9 expression we measured in undifferentiated ES cells cannot be explained by the presence of two active X chromosomes in female ES cells prior to X inactivation since Rhox6 and 9 are paternally imprinted in these cells [25]. Rather, the female enhanced expression results from the specific recruitment of KDM6A at those genes to facilitate the transition from repressive to active histone modifications and to increase expression. A similar mechanism explains the female bias in Rhox6 and 9 expression in ovary where we demonstrate that the genes are imprinted as well. KDM6A is a member of a multi-protein complex that not only de-methylates H3K27me3 but also methylates lysine 4 at histone H3 to facilitate gene expression [5], [6]. KDM6A counterbalances polycomb activity by regulating H3K27me3 levels [9], which would help maintain Rhox6 and 9 expression in undifferentiated female ES cells and ovary. In differentiated female ES cells KDM6A occupancy decreases, which mirrors the accumulation of H3K27me3 at Rhox6 and 9, consistent with low expression in most somatic cell types as well as with the heavy DNA methylation reported for these genes during development [24]. Our analyses of 8-cell embryos suggest that a female bias in Rhox6 and 9 expression is already present at this early stage, prior to gonadal development. Somatic and germ cells from developing embryos still show a female bias in Rhox6 and 9 expression at later stages (11.5–13.5dpc) including those coincident with gonad differentiation, which confirms a previous study [20]. However, detailed analyses of sexed embryos at additional stages will be needed to fully follow developmental expression in multiple cell types.
Strikingly, within the Rhox cluster only Rhox6 and 9 show a marked increase in KDM6A in female ES cells. Tellingly, these genes share a high degree of sequence similarity but differ from the other Rhox genes in other sub-clusters [19], suggesting that they may contain a sequence motif to specifically recruit KDM6A. The question arises of which other histone demethylases would remove H3K27me3 at other Rhox genes to facilitate their expression in specific tissues. We determined that KDM6B, which also removes methylation at lysine 27 of histone H3, has low expression in undifferentiated male and female ES cells (data not shown). However, its expression increases after differentiation in both male and female ES cells, pointing towards a potential role for this enzyme in regulating expression of some of the other Rhox genes at later stages of development [42]. KDM6A binds to the promoter, gene body, and 3′end of Rhox6 and 9 in female ES cells, suggesting a mechanism of regulation at transcription initiation and elongation. Interestingly, recruitment of elongation factors to target genes has been demonstrated for KDM6B, in addition to its role in histone demethylation [43].
Epigenetic regulation of the Rhox cluster had been previously focused on DNA methylation [24], [44]. Rhox5 whose expression peaks at day 9 after ES cell differentiation is repressed by DNA methylation at later stages, while it remains unmethylated and highly expressed in extra-embryonic tissues. Similarly, Rhox6 and 9 are repressed following the establishment of CpG methylation by DNA methyltransferases DNMT3b and DNMT1 at their promoter regions in the embryo proper but not in extra-embryonic tissues [24]. Rhox5 is the only gene together with Xist known to be expressed from the paternal X chromosome (maternally imprinted) at early embryonic stages (until e6.5); surprisingly, it is expressed from the maternal X (paternally imprinted) in extra-embryonic tissues, like Rhox6 and 9 [18], [24], [25], [45]. Our results are consistent with paternal imprinting of Rhox6 and 9 in mouse ovary, in agreement with other studies in placenta and ES cells [25].
We found that Rhox6 and 9 are bivalently marked in undifferentiated ES cells as they are occupied by nucleosomes containing histone H3 methylated at both lysine 27 and lysine 4. Bivalent genes are usually silent while poised for expression [37]. However, Rhox6 and 9 are in fact expressed in undifferentiated ES cells, probably due to a specific recruitment of KDM6A in a portion of cells in which levels of H3K27me3 would be decreased. Bivalent modifications result from a dynamic equilibrium of negative and positive chromatin marks controlled by histone modifying enzymes such as KDM6A. Our findings of a H3K27me3 increase at Rhox6 and 9 after Kdm6a knockdown are in agreement with what has been reported for other bivalent genes and support a role for KDM6A in maintaining a balance between active and inactive marks at bivalent promoters [9]. Note that the extent of reduction in Rhox6 and 9 expression we measured in female ES cells is comparable to that reported for another HOX gene, HOXB1, after KDM6A knockdown in human cells [8]. Many homeobox genes important for specification of cell types and organs contain bivalent domains, suggesting that bivalency is an important part of stem cell differentiation and development [46], [47]. Except for Rhox6 and 9, most other Rhox genes are not occupied by bivalent marks in undifferentiated ES cells, thus Rhox6 and 9 may be specifically activated to influence lineage commitment. It will be interesting to determine which pathways and specific lineages are stimulated by RHOX6 and RHOX9 proteins. Rhox6 has been implicated in the determination of the germ cell lineage [48]. So far, only Rhox5 and 9 have been studied in vivo. Whereas Rhox5-null male mice exhibit increased germ cell apoptosis and sperm motility defects leading to sub-fertility, Rhox9-null male or female mice do not have any apparent phenotypes [19], [49]. It is possible that due to similarities in sequence, homeodomain, and expression patterns Rhox6 compensates for the loss of Rhox9 in these knockout mice [49], [50]. Additional evidence based on knockouts in mouse and rat epididymis, suggests that Rhox5 may act as a master regulator of many of its paralogs [51].
Both Rhox6 and 9 are highly expressed in ovary and to a lesser extent, in testis (this study) and [19]. An intriguing finding from our study is that KDM6A occupancy is high at Rhox6 and 9 in ovary and thus may serve to keep these two Rhox genes active. KDM6A is also bound to Rhox6 and 9 in testis, although at a lower level (1.7-fold and 2.5-fold lower in testis than in ovary, respectively), suggesting a threshold effect and/or another level of control in testis. Our knockdown experiments do indicate that KDM6A affects Rhox6 and 9 expression in a dose-dependent manner. In embryonic gonads the majority of Rhox genes are already expressed in a sexually dimorphic manner from an early stage. Specifically, Rhox6 and 9 are predominantly expressed in female versus male primordial germ cells at 12.5–15.5dpc (this study) and [19], [20], [38], [52]. In addition, Rhox6 and 9 are also expressed in somatic cumulus cells in ovary [53], [54]. Our findings indicate that Rhox6 and 9 are both imprinted on the paternal allele and subject to X inactivation. This implies the existence of a population of somatic cells without any Rhox6 and 9 expression, suggesting that cumulus cells tolerate such mosaicism. In contrast, all germ cells would express Rhox6 and 9 following X re-activation and subsequent imprinting of the paternal X chromosome. This is similar to what has been reported for some of the Xlr genes, a family of mouse genes also implicated in reproduction, some of which are also imprinted and subject to X inactivation [55].
In addition to removal of H3K27me3, KDM6A appears to have a demethylase-independent role in regulating chromatin structure [9], [56], [57]. Indeed, KDM6A and KDM6B regulate T-box family members through an interaction with SMARCA4-containing SWI/SNF complexes in T-cells [58]. Interestingly, Kdm6a knockout mice display a more severe phenotype at mid-gestation in female than male embryos [9], [59]. Thus, the Y-linked paralog Uty compensates for Kdm6a deficiency allowing survival of male embryos by a demethylase independent mechanism, since UTY does not have demethylase activity [9], [56], [59]. However, while some KDM6A-deficient male mice survive, most do not or are runted throughout adulthood, indicating that H3K27 demethylation remains an important function of KDM6A for survival and growth [56]. Additionally, histone demethylation appears to be the predominant mechanism required for activation of genes important in differentiation since mouse and human cells lacking KDM6A but retaining UTY fail to reprogram [60]. Furthermore, male primordial germ cells lacking KDM6A do not develop, as H3K27me3 levels are retained when compared to wild type [60]. Our knockdown experiments are consistent with a role for KDM6A in controlling levels of H3K27me3 and expression of Rhox6 and 9 in female ES cells. However, we cannot rule out the contribution of a demethylase independent mechanism since we did not test for one in the context of Rhox expression control.
It remains to be determined whether levels of KDM6A are critical for proper ovarian function. It is interesting that female mice with a single X chromosome, which would have a lower dose of KDM6A due to haploinsufficiency for Kdm6a, a gene that escapes X inactivation, have reduced fertility [26], [61], [62]. Furthermore, XO female mouse embryos are developmentally retarded when compared to XX littermates at early mid-gestation [63]. In human, the presence of a single X chromosome causes Turner syndrome associated with severe developmental defects and ovarian dysgenesis [62]. It will be important to determine whether any of the human RHOX genes are also regulated by KDM6A. Mutations of KDM6A in human cause Kabuki syndrome, associated with growth retardation, unique facial features, and severe intellectual disability. Both males with point mutations and females with complete heterozygous deletions have been reported [12], [13]. The Kabuki phenotype, present in females who have one deleted KDM6A copy but absent in Turner syndrome females, may be due to partial silencing of the normal copy by X inactivation in Kabuki females, while Turner females would have one expressed copy in all cells. It would be interesting to examine ovaries in these patients.
In summary, our study provides the first evidence that Rhox6 and 9 are regulated by the histone demethylase KDM6A in mouse ES cells and reproductive organs in a sex-specific manner. Our findings indicate that a gene that escapes X inactivation plays a sex-specific role in gene regulation in female ES cells and tissue. Higher female expression due to escape from X inactivation of Kdm6a may be favorable to Rhox6 and 9 expression in ovary.
Materials and Methods
ES cell culture and differentiation and mouse tissue collection
Male ES cells WD44 (from C. Ware, University of Washington, US) and E14 (BL6/Cast) (from J. Gribnau, Erasmus MC Rotterdam, NL), and female ES cells PGK12.1 [64] and E8 (BL6/Cast) (from J. Gribnau, Erasmus MC Rotterdam, NL) were grown in high glucose DMEM media supplemented with 15% fetal bovine serum (FBS), 1% non-essential amino acids, 10 mg/ml APS, 0.1 mM 2-mercaptoethanol and 25 mM L-glutamine. ES cells were maintained in the presence of 1000 U/ml leukemia inhibitory factor (LIF) (Millipore) on a mono-layer of chemically inactivated mouse embryonic fibroblasts (MEF) and grown in a humidified incubator at 37°C and 5% CO2. Plates were enriched for ES cells by incubation on 1% gelatin coated dishes for 30 min to allow MEFs to attach, followed by transfer to fresh gelatin coated plates for overnight culture. Differentiation was achieved by removing LIF and culturing on non-adherent dishes to facilitate the formation of embryoid bodies. After 7 days, embryoid bodies were transferred to cell culture dishes for 8 days. Cells were harvested on days 0, 2, 4, and 15 after LIF removal. To follow X inactivation in PGK12.1 cells before and after differentiation, Xist expression was determined by RT-PCR and by RNA-FISH using standard protocols with a probe for Xist (Vysis) (Figure S3). Ovary, testis, and whole brain were collected from adult female and male C57BL/6J mice. Additionally, ovaries were collected from female F1 obtained by mating M. spretus males (Jackson Labs) with females that carry an Xist mutation (XistΔ) (B6.Cg-Xist<tm5Sado>) (from T. Sado, Kyushu University, available from RIKEN) [65]. Female progeny were genotyped to verify inheritance of the mutant Xist allele. Female progeny with a XistΔ fail to silence the BL6 X chromosome and thus have complete skewing of inactivation of the spretus X chromosome. All procedures involving animals were reviewed and approved by the University Institutional Animal Care and Use Committee (IACUC), and were performed in accordance with the Guiding Principles for the Care and Use of Laboratory Animals.
Kdm6a siRNA knockdown
Stealth Select siRNAs (Invitrogen) were selected to target three different locations of the Kdm6a mRNA. The three siRNAs were transfected together or individually and oligonucleotides with no target were used as negative controls. For transfection, 5 µl of Lipofectamine RNAi Max Reagent (Invitrogen) mixed with 250 µl of Opti-MEM I Reduced Serum Medium (Invitrogen) containing 100 pmol of siRNAs were incubated for 30 min prior to addition to 6-well plates seeded with 1.5×105 ES cells in 2 ml of DMEM supplemented as stated above. Cells were harvested after 48 h of RNAi treatment. Knockdown was confirmed by qRT-PCR, expression arrays, and Western blotting using standard procedures. Immunoblot analysis was done using a KDM6A/UTX antibody either from K.Ge (NIDDK) or from Bethyl Labs. Three siRNAs were pooled and protein levels were measured after 48 h of treatment. Western blot band densities were measured using ImageJ software (http://rsbweb.nih.gov/ij/).
Chromatin immunoprecipitation (ChIP)
Tissues were homogenized using a glass homogenizer and ES cells were collected before, during, and after differentiation. Cells were incubated at room temperature for 15 min in 1% formaldehyde. Crosslinking was stopped by adding 50 µL glycine followed by a 5 min incubation at room temperature and cell lysis as described [66]. Chromatin was sonicated to yield fragments 300–1000 bp in length and was then pre-cleared with protein A agarose beads for 1 h at 4°C. An aliquot of 20 µL was kept to serve as the input fraction. Pre-cleared chromatin was incubated in immunoprecipitation buffer at 4°C overnight using the following antibodies: anti-KDM6A/UTX [7], anti-UTX (Bethyl Labs), anti-H3K27me3 (Millipore), and anti-H3K4me3 (Millipore). Samples were centrifuged at 1200 rpm for 1 min and a small portion of the suspension collected as the unbound fraction. Immunoprecipitated chromatin was collected and serially washed in increasingly stringent salt buffers. After elution, crosslinks were reversed in 5 M NaCl at 65°C overnight. DNA was purified using Qiaquick PCR purification kit (Qiagen) and subjected to PCR according to the following protocol: 95°C for 3 min followed by 35 cycles of 95°C for 30 sec, 56°C for 30 sec and 72°C 30 sec. Samples were incubated at 72°C for 10 min and analyzed by gel electrophoresis. Controls to assay the immunoprecipitation efficiency of KDM6A, H3K4me3, and H3K27me3 antibodies included an active gene (Kdm5c) and an inactive gene (Iqsec2) (Table S2).
Quantitative real-time and allele-specific RT–PCR and PCR
For qRT-PCR, total RNA was prepared using the Qiagen RNeasy kit with on-column DNaseI digestion. For cDNA synthesis, 500 ng-1 µg of mRNA was reverse transcribed using the SuperScript First Strand Synthesis system (Invitrogen) according to manufacturer's protocol. Table S2 lists the RT-PCR primers specific for Rhox6, Rhox9, and Kdm6a. Quantitative PCR was performed using a SYBR green master mix (Roche) and a standard curve for each primer pair. Data normalized to the 18s housekeeping gene were averaged for 2 to 3 separate reactions each assayed in duplicate. For chromatin analyses, ChIP DNA was subjected to real-time PCR using primers listed in Table S2. Rhox6R1 and Rhox9R1 amplify regions upstream of the transcription start site of their respective gene. Rhox6/9R2 amplify regions in the 5′ gene body, and Rhox6/9R3 regions towards the 3′ end of both Rhox6 and Rhox9. After normalization to the input fraction, relative enrichment was calculated based on two separate immunoprecipitation reactions each assayed in duplicate. Following PCR amplification, melting curves were used to ensure only a single product was amplified. Western blots were done to confirm sexually dimorphic KDM6A protein levels in female and male ES cells using standard procedures. Briefly, nuclear protein was captured using an anti-KDM6A antibody (Bethyl Labs) using 1∶5000 dilution. Anti-β-ACTIN was used at 1∶10,000 dilution (Sigma) as a loading control. KDM6A protein was detected using HRP conjugated donkey anti-rabbit IgG, and β-ACTIN was detected using HRP conjugated goat anti-mouse IgG.
Allele-specific expression was determined by Sanger sequencing of RT-PCR products and control PCR of genomic DNA using primers listed in Table S2. For Rhox6, the SNP (T>G) that distinguishes between the maternal C57BL/6J (Xa) and paternal M. spretus (Xi) alleles is at nucleotide position 35180550 (NCBI37/mm9 build). For Rhox9, the SNP (T>G) that distinguishes between the maternal C57BL/6J (Xa) and paternal M. spretus (Xi) alleles is at nucleotide position 35254278 (NCBI37/mm9 build).
Expression array analyses
cDNA was hybridized to Affymetrix 1.0 ST and Affymetrix 430 2.0 mouse arrays. Array hybridizations were done at the Microarray Center or at the Center on Human Development and Disability (University of Washington, Seattle WA). The raw data files from the 430 2.0 arrays were analyzed by the Affymetrix software (GCOS 1.1) to produce the data in .CHP format (Excel), while raw data files from the 1.0 ST arrays were analyzed with Affymetrix Expression Console Software (http://www.affymetrix.com). Data was normalized with the RMA method as implemented in the Bioconductor Affymetrix package. Microarray quality control metrics were included according to the manufacturer's recommended guidelines. For analyses of published array data [32], [38], [39], spots were normalized by dividing the signal intensity against the average fluorescent intensity of each array [67]. Expression array data have been deposited in NCBI's GEO database [68] and are accessible through series accession number GSE45034 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE45034).”
ChIP tiling array analyses
Following ChIP, DNA was amplified by whole genome amplification using the GenomePlex Complete Whole Genome Amplification Kit (Sigma) with modifications previously described [69]. ChIP DNA was lyophilized and re-suspended in 10 µl of water. Library preparation buffer and stabilization buffer were added (2 µl and 1 µl, respectively), and samples incubated at 95°C for 2 min. After addition of library preparation enzyme, samples were incubated in a thermal cycler according to the following protocol: 16°C for 20 min, 24°C for 20 min, 37°C for 20 min, 75°C for 5 min. For amplification of the library, a master mix containing amplification master mix, water, and WGA DNA polymerase was added and samples subjected to 15 cycles of: 95°C for 3 min, 94°C for 15 sec, and 65°C for 5 min. Samples were purified using the Qiaquick PCR purification kit. ChIP and input fractions were labeled according to the standard Nimblegen sample labeling protocol prior to hybridization to HD2 Nimblegen tiling arrays for the entire mouse X chromosome (Roche). Enrichment profiles were generated (Genomics Resource Center, Fred Hutchinson Cancer Research Center, Seattle WA). Peak maps generated by the Nimblescan software consist of significant peaks (FDR score <0.05). Tiling array data have been deposited in NCBI's GEO database [68] and are accessible through series accession number GSE45390 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE45390).”
Statistics
All p-values shown represent paired two-tailed Student's t-tests.
Supporting Information
Zdroje
1. WeatherbeeSD, HalderG, KimJ, HudsonA, CarrollS (1998) Ultrabithorax regulates genes at several levels of the wing-patterning hierarchy to shape the development of the Drosophila haltere. Genes Dev 12 : 1474–1482.
2. WangJ, MagerJ, SchnedierE, MagnusonT (2002) The mouse PcG gene eed is required for Hox gene repression and extraembryonic development. Mamm Genome 13 : 493–503.
3. LanF, BaylissPE, RinnJL, WhetstineJR, WangJK, et al. (2007) A histone H3 lysine 27 demethylase regulates animal posterior development. Nature 449 : 689–694.
4. SwigutT, WysockaJ (2007) H3K27 demethylases, at long last. Cell 131 : 29–32.
5. ChoYW, HongT, HongS, GuoH, YuH, et al. (2007) PTIP associates with MLL3 - and MLL4-containing histone H3 lysine 4 methyltransferase complex. J Biol Chem 282 : 20395–20406.
6. IssaevaI, ZonisY, RozovskaiaT, OrlovskyK, CroceCM, et al. (2007) Knockdown of ALR (MLL2) reveals ALR target genes and leads to alterations in cell adhesion and growth. Mol Cell Biol 27 : 1889–1903.
7. HongS, ChoYW, YuLR, YuH, VeenstraTD, et al. (2007) Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc Natl Acad Sci U S A 104 : 18439–18444.
8. AggerK, CloosPA, ChristensenJ, PasiniD, RoseS, et al. (2007) UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature 449 : 731–734.
9. WelsteadGG, CreyghtonMP, BilodeauS, ChengAW, MarkoulakiS, et al. (2012) X-linked H3K27me3 demethylase Utx is required for embryonic development in a sex-specific manner. Proc Natl Acad Sci U S A 109 : 13004–13009.
10. SeenundunS, RampalliS, LiuQC, AzizA, PaliiC, et al. (2010) UTX mediates demethylation of H3K27me3 at muscle-specific genes during myogenesis. EMBO J 29 : 1401–1411.
11. LeeS, LeeJW, LeeSK (2011) UTX, a Histone H3-Lysine 27 Demethylase, Acts as a Critical Switch to Activate the Cardiac Developmental Program. Dev Cell 22 ((1)): 25–37.
12. MiyakeN, MizunoS, OkamotoN, OhashiH, ShiinaM, et al. (2013) KDM6A Point Mutations Cause Kabuki Syndrome. Hum Mutat 34 : 108–110.
13. LedererD, GrisartB, DigilioMC, BenoitV, CrespinM, et al. (2012) Deletion of KDM6A, a histone demethylase interacting with MLL2, in three patients with Kabuki syndrome. Am J Hum Genet 90 : 119–124.
14. TerashimaM, IshimuraA, YoshidaM, SuzukiY, SuganoS, et al. (2010) The tumor suppressor Rb and its related Rbl2 genes are regulated by Utx histone demethylase. Biochem Biophys Res Commun 399 : 238–244.
15. van HaaftenG, DalglieshGL, DaviesH, ChenL, BignellG, et al. (2009) Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat Genet 41 : 521–523.
16. WartmanLD, LarsonDE, XiangZ, DingL, ChenK, et al. (2011) Sequencing a mouse acute promyelocytic leukemia genome reveals genetic events relevant for disease progression. J Clin Invest 121 : 1445–1455.
17. MarBG, BullingerL, BasuE, SchlisK, SilvermanLB, et al. (2012) Sequencing histone-modifying enzymes identifies UTX mutations in acute lymphoblastic leukemia. Leukemia 26 ((8)): 1881–3.
18. MacLeanJA2nd, WilkinsonMF (2010) The Rhox genes. Reproduction 140 : 195–213.
19. MacleanJA2nd, ChenMA, WayneCM, BruceSR, RaoM, et al. (2005) Rhox: a new homeobox gene cluster. Cell 120 : 369–382.
20. DaggagH, SvingenT, WesternPS, van den BergenJA, McClivePJ, et al. (2008) The rhox homeobox gene family shows sexually dimorphic and dynamic expression during mouse embryonic gonad development. Biol Reprod 79 : 468–474.
21. HogeveenKN, Sassone-CorsiP (2005) Homeobox galore: when reproduction goes RHOX and roll. Cell 120 : 287–288.
22. SpitzF, HerkenneC, MorrisMA, DubouleD (2005) Inversion-induced disruption of the Hoxd cluster leads to the partition of regulatory landscapes. Nat Genet 37 : 889–893.
23. ZhanM, MiuraT, XuX, RaoMS (2005) Conservation and variation of gene regulation in embryonic stem cells assessed by comparative genomics. Cell Biochem Biophys 43 : 379–405.
24. OdaM, YamagiwaA, YamamotoS, NakayamaT, TsumuraA, et al. (2006) DNA methylation regulates long-range gene silencing of an X-linked homeobox gene cluster in a lineage-specific manner. Genes Dev 20 : 3382–3394.
25. MacleanJ, BettegowdaA, KimBJ, LouCH, YangSM, et al. (2011) The Rhox Homeobox Gene Cluster is Imprinted and Selectively Targeted for Regulation by Histone H1 and DNA Methylation. Mol Cell Biol 31 ((6)): 1275–87.
26. GreenfieldA, CarrelL, PennisiD, PhilippeC, QuaderiN, et al. (1998) The UTX gene escapes X inactivation in mice and humans. Hum Mol Genet 7 : 737–742.
27. XuJ, BurgoynePS, ArnoldAP (2002) Sex differences in sex chromosome gene expression in mouse brain. Hum Mol Genet 11 : 1409–1419.
28. JohnstonCM, LovellFL, LeongamornlertDA, StrangerBE, DermitzakisET, et al. (2008) Large-scale population study of human cell lines indicates that dosage compensation is virtually complete. PLoS Genet 4: e9 doi:10.1371/journal.pgen.0040009.
29. HeardE, DistecheCM (2006) Dosage compensation in mammals: fine-tuning the expression of the X chromosome. Genes Dev 20 : 1848–1867.
30. DistecheCM (1995) Escape from X inactivation in human and mouse. Trends Genet 11 : 17–22.
31. BerletchJB, YangF, XuJ, CarrelL, DistecheCM (2011) Genes that escape from X inactivation. Hum Genet 130 : 237–245.
32. JamesonSA, NatarajanA, CoolJ, DeFalcoT, MaatoukDM, et al. (2012) Temporal transcriptional profiling of somatic and germ cells reveals biased lineage priming of sexual fate in the fetal mouse gonad. PLoS Genet 8: e1002575 doi:10.1371/journal.pgen.1002575.
33. LinH, GuptaV, VermilyeaMD, FalcianiF, LeeJT, et al. (2007) Dosage compensation in the mouse balances up-regulation and silencing of X-linked genes. PLoS Biol 5: e326 doi:10.1371/journal.pbio.0050326.
34. LinH, HalsallJA, AntczakP, O'NeillLP, FalcianiF, et al. (2011) Relative overexpression of X-linked genes in mouse embryonic stem cells is consistent with Ohno's hypothesis. Nat Genet 43 : 1169–1170; author reply 1171–1162.
35. XuN, TsaiCL, LeeJT (2006) Transient homologous chromosome pairing marks the onset of X inactivation. Science 311 : 1149–1152.
36. AzuaraV, PerryP, SauerS, SpivakovM, JorgensenHF, et al. (2006) Chromatin signatures of pluripotent cell lines. Nat Cell Biol 8 : 532–538.
37. BernsteinBE, MikkelsenTS, XieX, KamalM, HuebertDJ, et al. (2006) A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125 : 315–326.
38. GallardoTD, JohnGB, ShirleyL, ContrerasCM, AkbayEA, et al. (2007) Genomewide discovery and classification of candidate ovarian fertility genes in the mouse. Genetics 177 : 179–194.
39. RottscheidtR, HarrB (2007) Extensive additivity of gene expression differentiates subspecies of the house mouse. Genetics 177 : 1553–1567.
40. MacleanJA2nd, HayashiK, TurnerTT, WilkinsonMF (2012) The Rhox5 Homeobox Gene Regulates the Region-Specific Expression of Its Paralogs in the Rodent Epididymis. Biol Reprod 86 ((6)): 189.
41. SongHW, DannCT, McCarreyJ, MeistrichM, CornwallG, et al. (2012) Dynamic expression pattern and subcellular localization of the RHOX10 homeobox transcription factor during early germ cell development. Reproduction 143 ((5)) 611–24.
42. Hailesellasse SeneK, PorterCJ, PalidworG, Perez-IratxetaC, MuroEM, et al. (2007) Gene function in early mouse embryonic stem cell differentiation. BMC Genomics 8 : 85.
43. ChenS, MaJ, WuF, XiongLJ, MaH, et al. (2012) The histone H3 Lys 27 demethylase JMJD3 regulates gene expression by impacting transcriptional elongation. Genes Dev 26 : 1364–1375.
44. LiQ, BartlettDL, GorryMC, O'MalleyME, GuoZS (2009) Three epigenetic drugs up-regulate homeobox gene Rhox5 in cancer cells through overlapping and distinct molecular mechanisms. Mol Pharmacol 76 : 1072–1081.
45. SasakiH, HamadaT, UedaT, SekiR, HigashinakagawaT, et al. (1991) Inherited type of allelic methylation variations in a mouse chromosome region where an integrated transgene shows methylation imprinting. Development 111 : 573–581.
46. MiquelajaureguiA, Varela-EchavarriaA, CeciML, Garcia-MorenoF, RicanoI, et al. (2010) LIM-homeobox gene Lhx5 is required for normal development of Cajal-Retzius cells. J Neurosci 30 : 10551–10562.
47. YangJM, SimSM, KimHY, ParkGT (2010) Expression of the homeobox gene, HOPX, is modulated by cell differentiation in human keratinocytes and is involved in the expression of differentiation markers. Eur J Cell Biol 89 : 537–546.
48. LiuC, TsaiP, GarciaAM, LogemanB, TanakaTS (2011) A possible role of Reproductive homeobox 6 in primordial germ cell differentiation. Int J Dev Biol 55 : 909–916.
49. TakasakiN, RankinT, DeanJ (2001) Normal gonadal development in mice lacking GPBOX, a homeobox protein expressed in germ cells at the onset of sexual dimorphism. Mol Cell Biol 21 : 8197–8202.
50. ChunJY, HanYJ, AhnKY (1999) Psx homeobox gene is X-linked and specifically expressed in trophoblast cells of mouse placenta. Dev Dyn 216 : 257–266.
51. MacleanJA2nd, HayashiK, TurnerTT, WilkinsonMF (2012) The rhox5 homeobox gene regulates the region-specific expression of its paralogs in the rodent epididymis. Biol Reprod 86 : 189.
52. ThorrezL, Van DeunK, TrancheventLC, Van LommelL, EngelenK, et al. (2008) Using ribosomal protein genes as reference: a tale of caution. PLoS ONE 3: e1854 doi:10.1371/journal.pone.0001854.
53. SuYQ, SugiuraK, WigglesworthK, O'BrienMJ, AffourtitJP, et al. (2008) Oocyte regulation of metabolic cooperativity between mouse cumulus cells and oocytes: BMP15 and GDF9 control cholesterol biosynthesis in cumulus cells. Development 135 : 111–121.
54. SugiuraK, SuYQ, LiQ, WigglesworthK, MatzukMM, et al. (2010) Estrogen promotes the development of mouse cumulus cells in coordination with oocyte-derived GDF9 and BMP15. Mol Endocrinol 24 : 2303–2314.
55. RaefskiAS, O'NeillMJ (2005) Identification of a cluster of X-linked imprinted genes in mice. Nat Genet 37 : 620–624.
56. ShpargelKB, SengokuT, YokoyamaS, MagnusonT (2012) UTX and UTY Demonstrate Histone Demethylase-Independent Function in Mouse Embryonic Development. PLoS Genet 8: e1002964 doi:10.1371/journal.pgen.1002964.
57. WangC, LeeJE, ChoYW, XiaoY, JinQ, et al. (2012) UTX regulates mesoderm differentiation of embryonic stem cells independent of H3K27 demethylase activity. Proc Natl Acad Sci U S A 109 : 15324–15329.
58. MillerSA, MohnSE, WeinmannAS (2010) Jmjd3 and UTX play a demethylase-independent role in chromatin remodeling to regulate T-box family member-dependent gene expression. Mol Cell 40 : 594–605.
59. WangC, LeeJE, ChoYW, XiaoY, JinQ, et al. (2012) UTX regulates mesoderm differentiation of embryonic stem cells independent of H3K27 demethylase activity. Proc Natl Acad Sci U S A 109 ((38)): 15324–9.
60. MansourAA, GafniO, WeinbergerL, ZviranA, AyyashM, et al. (2012) The H3K27 demethylase Utx regulates somatic and germ cell epigenetic reprogramming. Nature 488 : 409–413.
61. BurgoynePS, BakerTG (1985) Perinatal oocyte loss in XO mice and its implications for the aetiology of gonadal dysgenesis in XO women. J Reprod Fertil 75 : 633–645.
62. BondyCA (2009) Turner syndrome 2008. Horm Res 71 Suppl 1 : 52–56.
63. IshikawaH, BanzaiM, YamauchiT (1999) Developmental retardation of XO mouse embryos at mid-gestation. J Reprod Fertil 115 : 263–267.
64. PennyGD, KayGF, SheardownSA, RastanS, BrockdorffN (1996) Requirement for Xist in X chromosome inactivation. Nature 379 : 131–137.
65. HokiY, KimuraN, KanbayashiM, AmakawaY, OhhataT, et al. (2009) A proximal conserved repeat in the Xist gene is essential as a genomic element for X-inactivation in mouse. Development 136 : 139–146.
66. NelsonJD, DenisenkoO, SovaP, BomsztykK (2006) Fast chromatin immunoprecipitation assay. Nucleic Acids Res 34: e2.
67. NguyenDK, DistecheCM (2006) Dosage compensation of the active X chromosome in mammals. Nat Genet 38 : 47–53.
68. EdgarR, DomrachevM, LashAE (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30 : 207–210.
69. O'GeenH, NicoletCM, BlahnikK, GreenR, FarnhamPJ (2006) Comparison of sample preparation methods for ChIP-chip assays. Biotechniques 41 : 577–580.
Štítky
Genetika Reprodukční medicína
Článek Attachment Site Selection and Identity in Bxb1 Serine Integrase-Mediated Site-Specific RecombinationČlánek Bck2 Acts through the MADS Box Protein Mcm1 to Activate Cell-Cycle-Regulated Genes in Budding YeastČlánek High-Resolution Transcriptome Maps Reveal Strain-Specific Regulatory Features of Multiple IsolatesČlánek Neuropeptides Function in a Homeostatic Manner to Modulate Excitation-Inhibition Imbalance inČlánek Implicates Tyrosine-Sulfated Peptide Signaling in Susceptibility and Resistance to Root Infection
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2013 Číslo 5
-
Všechny články tohoto čísla
- Functional Elements Are Embedded in Structurally Constrained Sequences
- RNA–Mediated Epigenetic Heredity Requires the Cytosine Methyltransferase Dnmt2
- Loss of Expression and Promoter Methylation of SLIT2 Are Associated with Sessile Serrated Adenoma Formation
- Attachment Site Selection and Identity in Bxb1 Serine Integrase-Mediated Site-Specific Recombination
- Human Genetics in Rheumatoid Arthritis Guides a High-Throughput Drug Screen of the CD40 Signaling Pathway
- Genome-Wide Analysis in German Shepherd Dogs Reveals Association of a Locus on CFA 27 with Atopic Dermatitis
- Liver X Receptors Protect from Development of Prostatic Intra-Epithelial Neoplasia in Mice
- Chromosomal Organization and Segregation in
- A Statistical Framework for Joint eQTL Analysis in Multiple Tissues
- Cell Polarity and Patterning by PIN Trafficking through Early Endosomal Compartments in
- Bck2 Acts through the MADS Box Protein Mcm1 to Activate Cell-Cycle-Regulated Genes in Budding Yeast
- High-Resolution Transcriptome Maps Reveal Strain-Specific Regulatory Features of Multiple Isolates
- Neuropeptides Function in a Homeostatic Manner to Modulate Excitation-Inhibition Imbalance in
- A Compendium of Nucleosome and Transcript Profiles Reveals Determinants of Chromatin Architecture and Transcription
- Wnt Signaling Regulates the Lineage Differentiation Potential of Mouse Embryonic Stem Cells through Tcf3 Down-Regulation
- Filamin and Phospholipase C-ε Are Required for Calcium Signaling in the Spermatheca
- The Specificity and Flexibility of L1 Reverse Transcription Priming at Imperfect T-Tracts
- Imputation-Based Meta-Analysis of Severe Malaria in Three African Populations
- Implicates Tyrosine-Sulfated Peptide Signaling in Susceptibility and Resistance to Root Infection
- Clathrin and AP2 Are Required for Phagocytic Receptor-Mediated Apoptotic Cell Clearance in
- Encodes CDF Transporters That Excrete Zinc from Intestinal Cells of and Act in a Parallel Negative Feedback Circuit That Promotes Homeostasis
- Global Properties and Functional Complexity of Human Gene Regulatory Variation
- DNA Binding of the Cell Cycle Transcriptional Regulator GcrA Depends on N6-Adenosine Methylation in and Other
- Side Effects: Substantial Non-Neutral Evolution Flanking Regulatory Sites
- From Paramutation to Paradigm
- From Mouse to Human: Evolutionary Genomics Analysis of Human Orthologs of Essential Genes
- Distinct Translational Control in CD4 T Cell Subsets
- Female Bias in and Regulation by the Histone Demethylase KDM6A
- ATM–Dependent MiR-335 Targets CtIP and Modulates the DNA Damage Response
- HDAC7 Is a Repressor of Myeloid Genes Whose Downregulation Is Required for Transdifferentiation of Pre-B Cells into Macrophages
- The Majority of Primate-Specific Regulatory Sequences Are Derived from Transposable Elements
- Identification of Meiotic Cyclins Reveals Functional Diversification among Plant Cyclin Genes
- EGL-13/SoxD Specifies Distinct O and CO Sensory Neuron Fates in
- Congruence of Additive and Non-Additive Effects on Gene Expression Estimated from Pedigree and SNP Data
- Using Extended Genealogy to Estimate Components of Heritability for 23 Quantitative and Dichotomous Traits
- Ikbkap/Elp1 Deficiency Causes Male Infertility by Disrupting Meiotic Progression
- Analysis of the Genetic Basis of Disease in the Context of Worldwide Human Relationships and Migration
- Duplication and Retention Biases of Essential and Non-Essential Genes Revealed by Systematic Knockdown Analyses
- Strong Purifying Selection at Synonymous Sites in
- , a Susceptibility Gene for Type 1 and Type 2 Diabetes, Modulates Pancreatic Beta Cell Apoptosis via Regulation of a Splice Variant of the BH3-Only Protein
- Chromosome Movements Promoted by the Mitochondrial Protein SPD-3 Are Required for Homology Search during Meiosis
- The Secretory Pathway Calcium ATPase PMR-1/SPCA1 Has Essential Roles in Cell Migration during Embryonic Development
- The Genomic Signature of Crop-Wild Introgression in Maize
- CDK4 T172 Phosphorylation Is Central in a CDK7-Dependent Bidirectional CDK4/CDK2 Interplay Mediated by p21 Phosphorylation at the Restriction Point
- Genome-Wide Identification of Regulatory RNAs in the Human Pathogen
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Using Extended Genealogy to Estimate Components of Heritability for 23 Quantitative and Dichotomous Traits
- HDAC7 Is a Repressor of Myeloid Genes Whose Downregulation Is Required for Transdifferentiation of Pre-B Cells into Macrophages
- Female Bias in and Regulation by the Histone Demethylase KDM6A
- ATM–Dependent MiR-335 Targets CtIP and Modulates the DNA Damage Response
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



