-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaKRAB–Zinc Finger Proteins and KAP1 Can Mediate Long-Range Transcriptional Repression through Heterochromatin Spreading
Krüppel-associated box domain-zinc finger proteins (KRAB–ZFPs) are tetrapod-specific transcriptional repressors encoded in the hundreds by the human genome. In order to explore their as yet ill-defined impact on gene expression, we developed an ectopic repressor assay, allowing the study of KRAB–mediated transcriptional regulation at hundreds of different transcriptional units. By targeting a drug-controllable KRAB–containing repressor to gene-trapping lentiviral vectors, we demonstrate that KRAB and its corepressor KAP1 can silence promoters located several tens of kilobases (kb) away from their DNA binding sites, with an efficiency which is generally higher for promoters located within 15 kb or less. Silenced promoters exhibit a loss of histone H3-acetylation, an increase in H3 lysine 9 trimethylation (H3K9me3), and a drop in RNA Pol II recruitment, consistent with a block of transcriptional initiation following the establishment of silencing marks. Furthermore, we reveal that KRAB–mediated repression is established by the long-range spreading of H3K9me3 and heterochromatin protein 1 β (HP1β) between the repressor binding site and the promoter. We confirm the biological relevance of this phenomenon by documenting KAP1–dependent transcriptional repression at an endogenous KRAB–ZFP gene cluster, where KAP1 binds to the 3′ end of genes and mediates propagation of H3K9me3 and HP1β towards their 5′ end. Together, our data support a model in which KRAB/KAP1 recruitment induces long-range repression through the spread of heterochromatin. This finding not only suggests auto-regulatory mechanisms in the control of KRAB–ZFP gene clusters, but also provides important cues for interpreting future genome-wide DNA binding data of KRAB–ZFPs and KAP1.
Published in the journal: . PLoS Genet 6(3): e32767. doi:10.1371/journal.pgen.1000869
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1000869Summary
Krüppel-associated box domain-zinc finger proteins (KRAB–ZFPs) are tetrapod-specific transcriptional repressors encoded in the hundreds by the human genome. In order to explore their as yet ill-defined impact on gene expression, we developed an ectopic repressor assay, allowing the study of KRAB–mediated transcriptional regulation at hundreds of different transcriptional units. By targeting a drug-controllable KRAB–containing repressor to gene-trapping lentiviral vectors, we demonstrate that KRAB and its corepressor KAP1 can silence promoters located several tens of kilobases (kb) away from their DNA binding sites, with an efficiency which is generally higher for promoters located within 15 kb or less. Silenced promoters exhibit a loss of histone H3-acetylation, an increase in H3 lysine 9 trimethylation (H3K9me3), and a drop in RNA Pol II recruitment, consistent with a block of transcriptional initiation following the establishment of silencing marks. Furthermore, we reveal that KRAB–mediated repression is established by the long-range spreading of H3K9me3 and heterochromatin protein 1 β (HP1β) between the repressor binding site and the promoter. We confirm the biological relevance of this phenomenon by documenting KAP1–dependent transcriptional repression at an endogenous KRAB–ZFP gene cluster, where KAP1 binds to the 3′ end of genes and mediates propagation of H3K9me3 and HP1β towards their 5′ end. Together, our data support a model in which KRAB/KAP1 recruitment induces long-range repression through the spread of heterochromatin. This finding not only suggests auto-regulatory mechanisms in the control of KRAB–ZFP gene clusters, but also provides important cues for interpreting future genome-wide DNA binding data of KRAB–ZFPs and KAP1.
Introduction
The proper control of gene expression is paramount to all cellular events, and is orchestrated through a sophisticated balance of activating and repressing influences. Krüppel-associated box domain zinc finger proteins (KRAB-ZFP) constitute the single largest group of transcriptional repressors encoded by the genomes of higher organisms. After appearing in early tetrapods, the KRAB-ZFP family has rapidly expanded and diverged through multiple rounds of gene and segmental duplications, to give rise to more than three hundred and fifty members annotated in both mice and humans [1]–[6]. In spite of their numerical abundance, wide range of tissue-specific expression and dynamic evolutionary history, the physiological functions of KRAB-ZFPs collectively remain ill-defined, and few of their targets have been identified [3],[7]. However, emerging evidence links KRAB/KAP1-mediated regulation to processes as essential and diverse as stem cell pluripotency, early embryonic development and differentiation, genomic imprinting, response to DNA damage and control of behavioral stress [8]–[14]. Furthermore, KAP1 controls endogenous retroviruses in embryonic stem cells, a process crucial for the maintenance of genomic stability [15].
KRAB-ZFPs all harbor a so-called KRAB domain situated upstream of an array of two to forty C2H2 zinc fingers, which provide sequence-specific DNA binding ability [2]. KRAB recruits KAP1 (KRAB-associated protein 1, also known as TRIM28, Tif1β or KRIP-1) [16]–[19], which acts as a scaffold for various heterochromatin-inducing factors, such as heterochromatin protein 1 (HP1), the histone methyltransferase SETDB1, the nucleosome-remodeling and histone deacetylation (NuRD) complex, the nuclear receptor corepressor complex 1 (N-CoR1) and, at least during early embryonic development, de novo DNA methyltransferases [20]–[27].
The phylogenetically conserved family of HP1 proteins is implicated in a variety of nuclear events, such as transcriptional repression and maintenance of chromosome structure [28]. HP1 harbors two major regions: the chromo domain, which binds to repressive di - and trimethylated histone 3 lysine 9 (H3K9me2 and H3K9me3, respectively) residues, and the chromo shadow domain, involved in HP1 homodimerization and recruitment of other partners [29]–[31]. Therefore, HP1 bridges histones with other chromatin-associated proteins, that promote heterochromatin spreading [32]. An advancing front of heterochromatization can thus be propagated by the creation of HP1 binding sites, through histone methyltransferase-mediated H3K9 methylation, followed by the HP1-mediated recruitment of more histone methyltransferase for another round of H3K9 methylation/HP1 binding [33]–[36]. Little is known about the kinetics, efficiency, self-perpetuating ability and action-range of this process. Tethering of a regulated KRAB repressor domain suggested that KRAB/KAP1 induced heterochromatin formation has a rather limited spreading potential in euchromatin, as silencing could be exerted no farther than 2–3 kilobases (kb) away from the repressor binding site [37]. Yet a more recent Chromatin IP (ChIP)-on-chip analysis performed in the human testicular carcinoma cell line Ntera2 revealed close to 7,000 KAP1 binding sites, a number of which were located at the 3′end of KRAB-ZFP genes [38]. While this suggests auto-regulatory negative feedback loops for these genes, such a process would imply that KRAB/KAP1 binding can affect promoters situated at very significant distances. In agreement with such a model, large heterochromatin domains associated with both HP1β and SUV39h1 were found on chromosome 19, where most of the KRAB-ZFP gene clusters reside [39].
To examine the genomic features of KRAB/KAP1-mediated transcriptional regulation, we developed an ectopic repressor assay. In our system, promoterless lentiviral vectors serve as gene traps to drive reporter expression from cellular promoters. Drug-controllable docking of an ectopic KRAB-based repressor then allows an assessment of the effects of KRAB/KAP1 recruitment at these loci. Using this system, we found that KRAB-induced silencing can repress promoters situated several tens of kilobases away from the repressor primary docking site through reduced RNA Pol II binding. Furthermore, we observed that this phenomenon is independent of promoter strength, facilitated by HP1 and associated with spreading of heterochromatin marks between repressor binding site and targeted promoters. Finally, we could document KAP1-mediated transcriptional repression at an endogenous KRAB-ZFP gene cluster by propagation of HP1β and H3K9me3 from the 3′ end of these genes to their transcriptional start site. Our results indicate that KRAB/KAP1 induce long-range repression through the spread of heterochromatin.
Results
KRAB–mediated silencing can act over several tens of kilobases
In order to study the features of KRAB/KAP1-induced silencing, we exposed lentivirally trapped cellular promoters to a drug-regulated KRAB-containing repressor. The tTRKRAB protein contains the KRAB domain of the human KOX1 ZFP fused to the E. coli tetracycline repressor (tTR), and binds to Tet operator sequences (TetO) in a doxycycline (dox)-controllable fashion [40],[41]. We engineered human immunodeficiency virus (HIV)-derived lentiviral (LV) gene trap vectors carrying tandem TetO repeats and a promoter-less puromycin resistance-GFP fusion reporter (puroR-GFP) downstream of a potent adenoviral splice acceptor. This design predicted that i) reporter expression would occur from the promoters of active genes targeted by the integrants, and ii) dox withdrawal would result in tTRKRAB binding to the TetO sites present in the proviruses, thus exposing the trapped promoters to potential KRAB/KAP1-mediated repression (Figure 1A).
Fig. 1. KRAB–mediated silencing can act over several tens of kilobases. 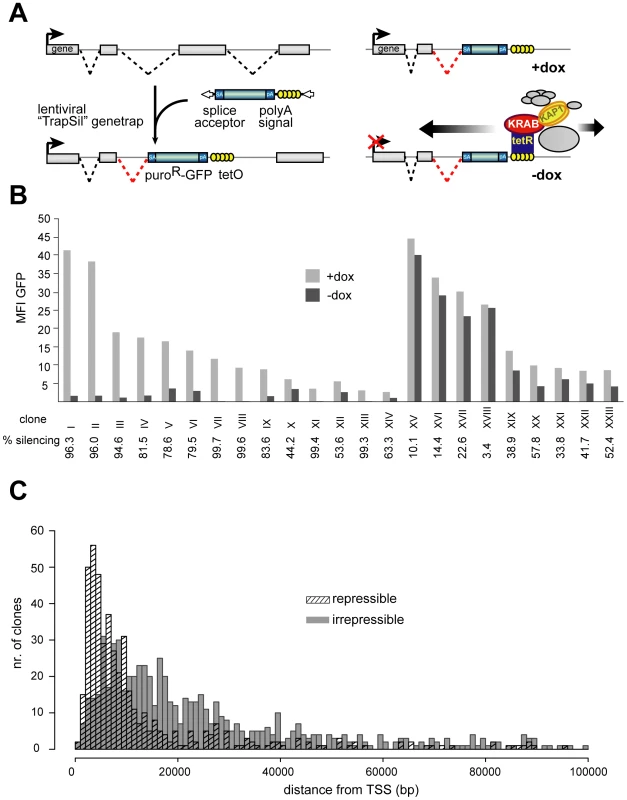
(A) Mechanism of how endogenous genes are targeted by tTRKRAB using the lentiviral vector-based “Trapping/Silencing” (TrapSil) system: TetO-containing gene traps carrying the promoterless puroR-GFP gene only express this reporter if after proviral integration they “trap” an actively transcribing gene. The TetO sites further allow binding of the ectopic repressor tTRKRAB to the gene traps after dox removal, while the trap reporter serves as a direct read-out for the effects of tTRKRAB–mediated “silencing”. (B) tTRKRAB–expressing HeLa cells were transduced with LV TrapSil vectors and 23 clones expressing the trap reporter were isolated. Mean fluorescence intensity (MFI) GFP values of these individual clones, cultured with and without dox, were determined and the ratios of these values were used to calculate the silencing efficiency (% silencing = 1- ((MFI GFP –dox)/(MFI GFP +dox))) depicted below the x-axis for each clone. (C) GFP-mediated cell sorting was used to isolate populations of HeLa cells exhibiting either a “repressible” (>90% silencing) or an “irrepressible” (<10% silencing) phenotype. 484 repressible and 699 irrepressible clonal integrations were mapped and the distance between them and their trapped promoter was plotted on a cumulative histogram. To study the effects of tTRKRAB recruitment to endogenous genes, we engineered HeLa cells stably expressing tTRKRAB and infected those with the TetO-containing LV trapping/silencing (“TrapSil”) vectors at a low multiplicity of infection to limit integrants to one per cell. Clones expressing the trap reporter puroR-GFP were then isolated and subjected to FACS analysis in the presence (-dox) or absence (+dox) of tTRKRAB binding. Comparing the mean fluorescence intensity values of GFP (MFI) in both conditions revealed heterogeneous effects of tTRKRAB recruitment, with clones exhibiting either high or low levels of reporter silencing (Figure 1B and Table S1). We saw no correlation between the strength of the trapped promoters and their susceptibility to tTRKRAB-mediated repression (Figure 1B). We could also verify proper recruitment of KRAB/KAP1 in different clones by ChIP targeting KAP1 (Figure S1 and data not shown). In addition, we analyzed the effects of tTRKRAB recruitment on populations of LV TrapSil clones and found that the level of GFP was decreased on both the protein and mRNA level upon repressor binding. We also found that the average silencing efficiency of the LV TrapSil population was at around 60%, which is consistent with repressible clones being more prevalent in this population than irrepressible clones (Figure S2).
HIV-derived vectors preferentially integrate in downstream regions of transcribed genes [42],[43]. Therefore, we evaluated the impact of distance between the tTRKRAB docking site and the trapped promoter on KRAB-induced repression in cell populations with distinct LV TrapSil integrants. For this, we used GFP-based cell sorting to purify subpopulations exhibiting trapped promoters, which were silenced by either more than 90% (“repressible”) or less than 10% (“irrepressible”). Proviral integration sites were mapped by ligation-mediated PCR on genomic DNA, followed by large scale pyrosequencing [44],[45]. To ensure an unequivocal analysis, we chose to consider only trapping events that mapped to characterized UCSC known genes with single transcriptional start sites. Using these criteria we were able to obtain 484 repressible integrants in 221 genes and 699 irrepressible integrants in 371 genes (Table S2). When we compared the distance between the proviral integration sites and their trapped promoters in both populations, we found that irrepressible promoters were on average located further away from the repressor-docking site than their repressible counterparts (Figure 1C). We observed a similar pattern when analyzing trapped genes harboring multiple integration events from both the repressible and the irrepressible groups. By analyzing six of the most targeted genes, we found that the provirus integrated closest to the promoter systematically induced a repressible phenotype, while integrants located further downstream often fell into the irrepressible group (Figure S3). The comparison between the effect of tTRKRAB-mediated silencing and the range of its action also revealed that repression could be exerted at several tens of kilobases away from the tTRKRAB binding site, albeit with a sharp decrease at around 15 kb (Figure 1C). Finally, the overlap between the two histograms indicates that distance between repressor-binding site and the targeted promoter was not the sole determinant of susceptibility to KRAB-mediated repression, but that sequence - or locus-dependent factors must also play a role.
Silenced promoters exhibit increased H3K9me3 and decreased H3-acetylation and RNA Pol II recruitment
To investigate the molecular bases of repressible and irrepressible phenotypes we characterized four clones (I, IX, XI and XVI) in more detail using 5′RACE to map the trapped fusion transcripts (Table S1) and by performing ChIP studies. All clones had the integrant within 10–20 kb from the trapped promoter, yet three (clones I, IX and XI) were repressible and one (clone XVI) was irrepressible (Figure 1B). The level of histone modifications known to be affected by KRAB/KAP1-mediated repression, namely histone H3-acetylation (H3Ac) and H3K9me3 was evaluated at each of the four trapped promoters. In all three repressible clones, H3Ac levels dropped significantly upon tTRKRAB-binding, consistent with a close to complete deacetylation of the trapped allele (Figure 2A). In contrast, the irrepressible promoter of the casein kinase 1d (CSNK1D) gene, trapped in clone XVI, exhibited constant levels of this chromatin mark (Figure 2A). An inverse image was obtained for H3K9me3, with increases of different magnitudes at the three silenced promoters upon dox removal. No such change in H3K9me3 levels was observed at the irrepressible promoter (Figure 2B). To exclude the possibility that increased H3K9me3 was due to chromatin condensation, we also monitored levels of total histone H3 at these promoters. Comparison of the dox+ and dox - conditions indicated that the observed H3K9me3 increases could not be solely attributed to changes in total H3 (Figure 2C). In order to probe the impact of KAP1-induced repression on transcriptional initiation, we measured the levels of hypophosphorylated RNA Pol II present at trapped promoters (Figure 2D). We found that tTRKRAB binding resulted in a significant decrease of the recruitment of this enzyme in repressible but not irrepressible clones. Noteworthy, the remaining levels of Pol II binding detected in the absence of dox most likely originated from the sister allele of the trapped promoter.
Fig. 2. Silenced promoters exhibit increased H3K9me3 and decreased H3-acetylation and RNA Pol II recruitment. 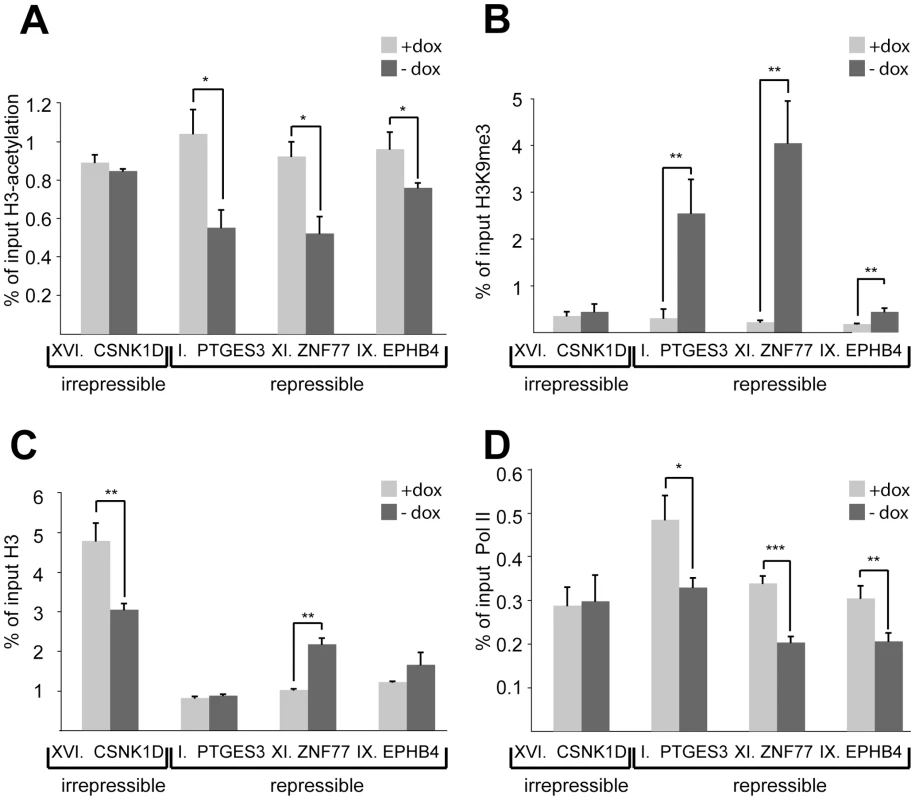
ChIP–based measurement of (A) H3-acetylation, (B) H3K9me3, (C) total histone H3, and (D) hypophosphorylated RNA Pol II at the irrepressible promoter of casein kinase 1d (CSNK1D) and at the repressible promoters of prostaglandin E synthase 3 (PTGES3), zinc finger protein 77 (ZNF77) and ephrin receptor B4 (EPHB4). The relative enrichment values (% of input) of H3-acetylation, total H3, and RNA Pol II were normalized to the relative enrichment at the GAPDH promoter. All values are expressed as means +SEM of triplicate experiments. The statistical test conducted was the student's t-test comparing +dox to –dox samples. If unequal variances were observed, the values were log10 transformed. The criterion for significance for all analyses was p<0.05. *: p<0.05, **, p<0.01, ***, p<0.005. HP1β and H3K9me3 spread from the KRAB–binding site to the promoter
To investigate further the mechanisms of this long-range repression, we measured levels of both H3K9me3 and HP1β enrichment between the promoters and the TrapSil integrants of the loci under study. It was previously demonstrated that all HP1 isoforms can interact with KAP1 [20],[22],[29],[46],[47]. However, recent FRET results revealed distinct physical interactions between KAP1 and HP1β or HP1γ during differentiation of embryonal carcinoma F9 cells to primitive endoderm-like cells [48]. Interestingly, only the KAP1-HP1β interaction induced a relocalization of genes from euchromatin to heterochromatin upon differentiation, suggesting a specific role for this isoform during KAP1-mediated gene silencing. In addition, both HP1β and KAP1 co-localize at the 3′end of KRAB-ZFP genes [38],[39], making HP1β a likely candidate to be involved in propagating KRAB/KAP1-mediated long-range repression.
Indeed, global levels of H3K9me3 and HP1β increased along the body of trapped genes upon tTRKRAB recruitment, and the magnitude and efficiency of propagation of this phenomenon correlated with its phenotypic outcome (Figure 3, Figure S4, Figure S5). Clone I, where the prostaglandin E synthase 3 (PTGES3) gene was trapped, exhibited high reporter GFP expression, which was completely silenced upon tTRKRAB binding (Figure 1B). Correspondingly, KRAB repressor binding led to a massive increase of both HP1β and H3K9me3 along this gene (Figure 3B). In comparison, the irrepressible CSNK1D gene body, showed smaller changes in HP1β and H3K9me3 loading, which gradually decreased towards the promoter (Figure 3A). The ChIP efficiencies between the +dox and the –dox samples were comparable for both factors, as shown by analysis of several control loci (Figure S6). We additionally monitored the levels of H3K9me3 and HP1β at the trapped and repressible KRAB-ZFP gene ZNF77. More specifically, tTRKRAB binding to the 3′end of ZNF77-1 led to an increase of both factors at the promoters and gene bodies of ZNF77-1, as well as the neighboring ZNF57 gene, which is located approximately 40 kb away (Figure S5). Therefore, we conclude that KRAB/KAP1 recruitment triggers the long-range spreading of silent chromatin marks. Efficient transcriptional silencing, however, is only established when this process extends over the promoter itself, interfering with proper RNA Pol II recruitment and transcriptional initiation (Figure 2D).
Fig. 3. HP1β and H3K9me3 spread from the KRAB–binding site to the promoter. 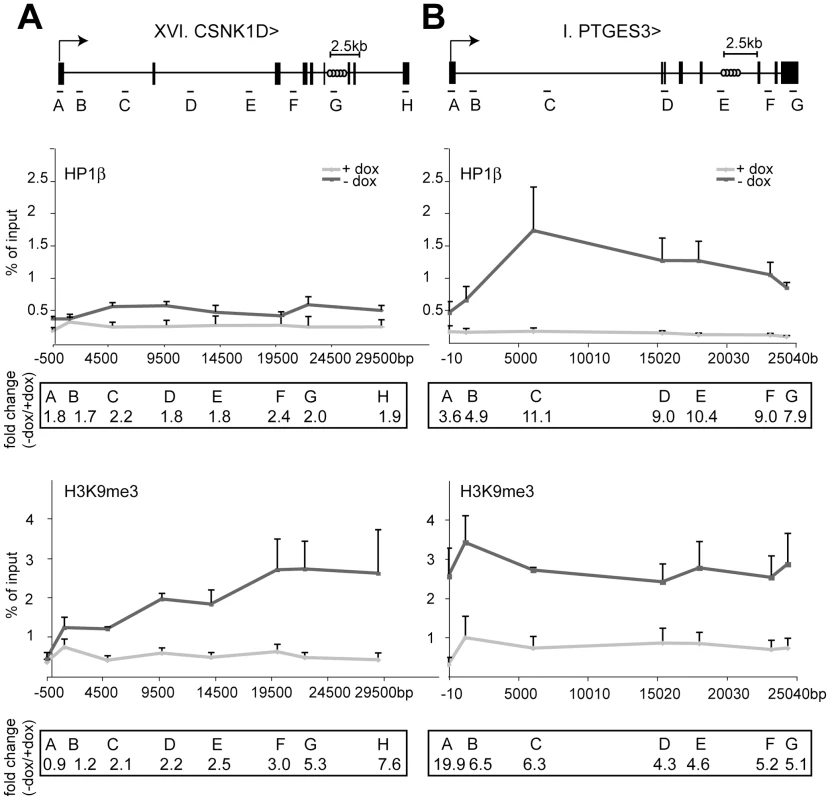
ChIP analyses quantifying the relative enrichment (% of input) of both H3K9me3 and HP1β were performed for the irrepressible clone XVI and the repressible clone I in the presence and absence of dox. The interrogated sequence spanned from the proviral tTRKRAB binding sites (light grey circles) to the indicated trapped promoters. (A) Enrichments of both modifications at the trapped casein kinase 1d (CSNK1D) locus. (B) Enrichments of both modifications at the prostaglandin E synthase 3 (PTGES3) locus. qPCR amplicons are depicted as letters and are not drawn to scale. All values are expressed as means +SEM of triplicate experiments. Fold changes were calculated as ratios of -dox/+dox enrichments, with the ratios of the respective positive controls set as 1. The controls consisted of p53BP2 for H3K9me3 and of ZNF556 for HP1β (Table S4). HP1 facilitates KRAB/KAP1–mediated long-range repression
We further explored the role of HP1 recruitment to KAP1 in KRAB/KAP1-mediated long-range repression by performing complementation studies in a KAP1 knockout mouse embryonic fibroblast (MEF) cell line stably expressing tTRKRAB. For this, we isolated two LV TrapSil clones, and used 5′RACE to map the fusion transcripts resulting from promoter trapping. Since both proviruses were intronic, we assigned integration to the middle of the targeted intron. This revealed that clone 1 had integrated in the gene Sestd1 (SEC14 and spectrin domains 1) at a distance of 15 kb from the promoter and that clone 2 had trapped the gene Prcp (prolylcarboxypeptidase) and was situated 25 kb away from its TSS (Table S1). Both clones were therefore suitable to serve in our study of KRAB/KAP1-mediated long-range repression. We found that GFP expression was independent of the presence of dox (Figure 4A), as predicted from the knockout of KAP1 (Figure 4B). However, silencing could be significantly restored in both clones by lentiviral vector-mediated complementation with wild type KAP1 (Figure 4A and 4B). By comparison, the transduction with a vector expressing a KAP1 mutant defective for HP1 binding (KAP1R487E, V488E) [22] only rescued tTRKRAB-mediated silencing to levels that were about six times lower than observed with wild type KAP1 in both clones (Figure 4A and 4B). This strongly suggests that HP1 recruitment by KAP1 facilitates the long-range impact of the KRAB/KAP1 silencing complex.
Fig. 4. HP1 facilitates KRAB/KAP1–mediated long-range repression. 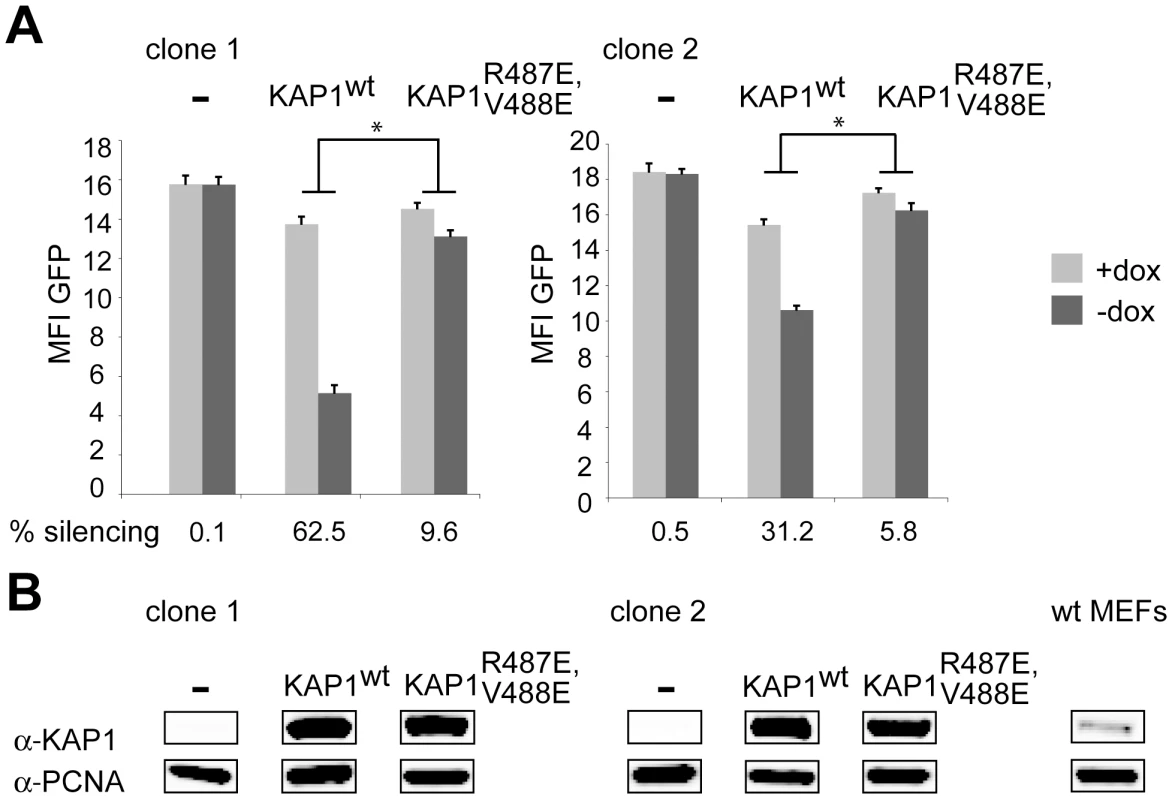
(A) Two LV TrapSil-selected, tTRKRAB–expressing KAP1−/− MEF clones were complemented by transduction with a lentiviral vector expressing either wild type KAP1 (KAP1wt) or HP1 binding-defective KAP1 (KAP1R487E, V488E), before FACS analysis in the presence or absence of dox. The mean fluorescence intensity values (MFI) of GFP and the silencing efficiency values are depicted as means of six independent measurements. The statistical analysis was performed using two-way ANOVA and, since the interactions were linked, the simple main effects were analyzed. The criterion for significance for all analyses was p<0.05. *: p<0.05. (B) Western blot analysis monitoring KAP1 protein levels, using PCNA as a loading control. The far right panel shows endogenous KAP1 expression in a MEF wild-type cell line. KAP1 represses KRAB-ZFP genes via 3′ to 5′ long-range heterochromatin spreading
Recent genome-wide studies have identified KAP1 and HP1β binding sites at the 3′end of KRAB-ZFP genes, suggesting long-range auto-regulatory feedback loops [38],[39]. To investigate this possibility directly, we generated a KAP1 knockdown HeLa cell line by transduction with an shRNA-expressing lentiviral vector. KAP1 mRNA and protein levels were about ten-fold lower in these cells than in their wild type control (Figure S7). We then analyzed a group of five KRAB-ZFPs located on human chromosome 19, where most members of the family reside in closely packed clusters (Figure 5, Figure 6, and Figure 7). In KAP1 knockdown cells, four out of five analyzed cluster-specific transcripts, namely ZNF554, ZNF555, ZNF556 and ZNF57, showed slight upregulation, which reached significance in the latter three genes, when compared to a cell line expressing an shRNA directed against GFP (Figure 5A). This is consistent with a de-repression of these transcripts following KAP1 depletion. The ZNF77 gene is predicted to produce two partly overlapping transcripts, ZNF77-1 and ZNF77-2. However, only ZNF77-1 was reliably detected in our cells and its levels were identical in KAP1 knockdown and control cells (Figure 5A and data not shown). We further verified that KAP1 depletion did not lead to a global decrease in heterochromatin protein expression, since four out of five analyzed genes displayed similar RNA levels in cells expressing either shKAP1 or shGFP (Figure 5B).
Fig. 5. KAP1 mediates transcriptional repression of KRAB–ZFPs. 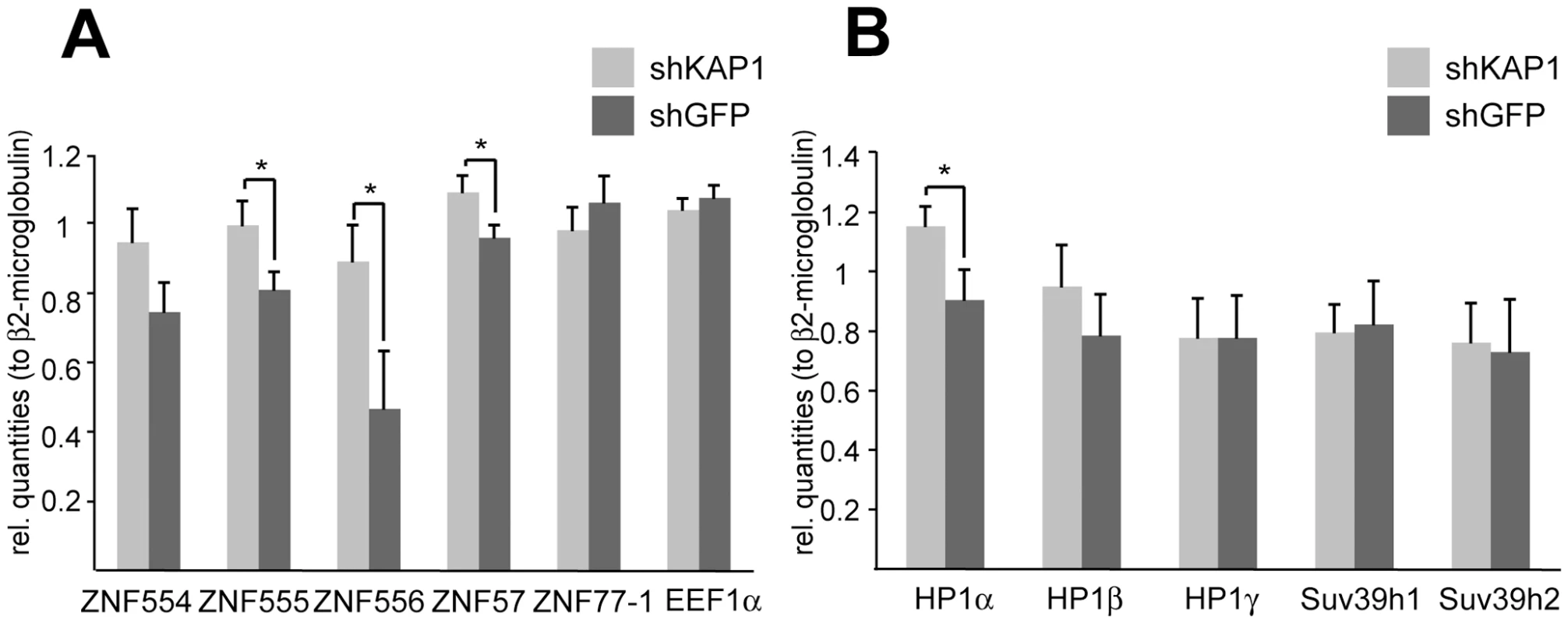
mRNA expression levels upon stable KAP1 (shKAP1) and GFP (shGFP) knockdown were assessed through qPCR in HeLa cells. The relative quantities were measured for (A) cluster-specific KRAB–ZFP transcripts and for (B) different heterochromatin protein expression levels. β2-microglobulin was used as a normalization gene, whereas the expression of the eukaryotic elongation factor 1α (EEF1α) served as a negative control. qPCR values are expressed as means +SEM of six experiments. The statistical test conducted was a t-test with Welch's correction, which assumes unequal variances: *: p<0.05. Fig. 6. KAP1 is enriched at the 3′end of KRAB–ZFPs. 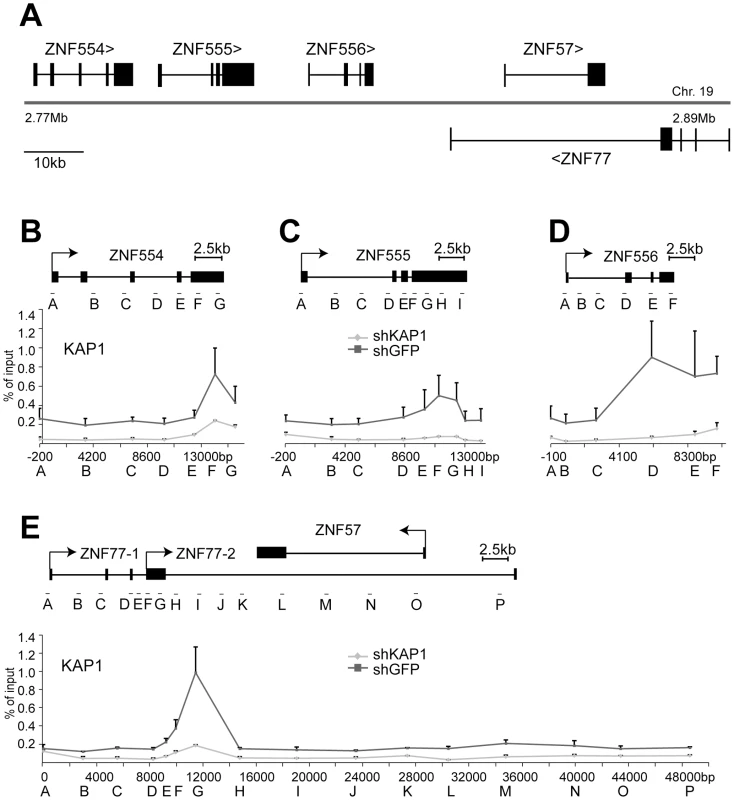
(A) Depiction of the KRAB–ZFP cluster under study, located on human chromosome 19. (B–E) Direct KAP1 binding to this cluster was assessed by ChIP for the (B) ZNF554 gene, (C) ZNF555 gene, (D) ZNF556 gene, and (E) the region containing ZNF77 and the overlapping ZNF57 gene. Values are the result of duplicate experiments and are depicted with +SEM error bars. Fig. 7. KAP1 mediates the spread of HP1β and H3K9me3 at KRAB–ZFP clusters. 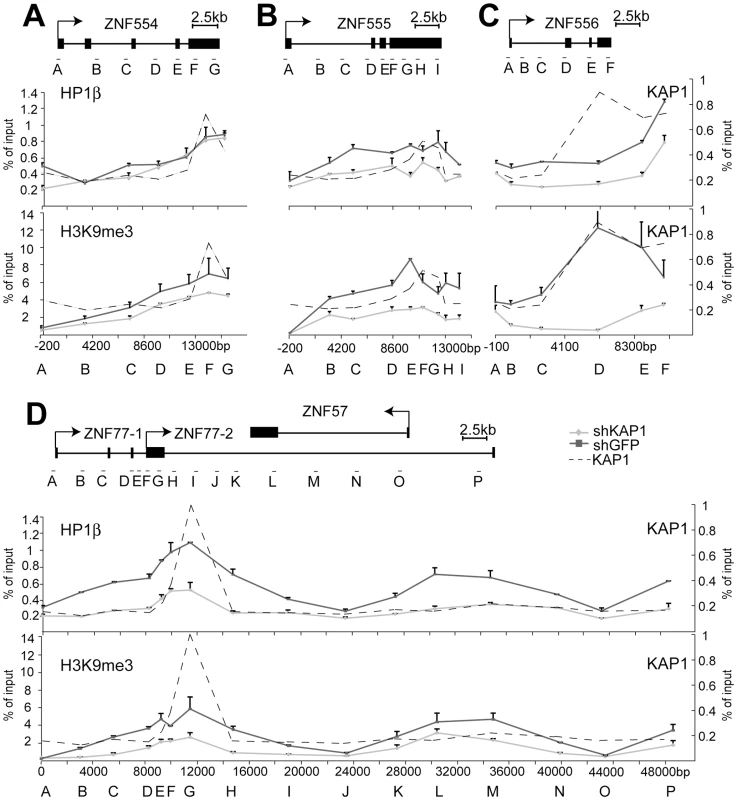
ChIP-mediated measurement of HP1β and H3K9me3 in control (shGFP) and KAP1 knockdown (shKAP1) HeLa cells at the (A) ZNF554 gene, (B) ZNF555 gene, (C) ZNF556 gene, and at the (D) ZNF77/ZNF57 genes. Dotted lines depict the location of KAP1 peaks determined (see Figure 6B–6E). All values are expressed as means +SEM of duplicate experiments. We ensured that comparable amounts of HP1β and H3K9me3 ChIP material were present in our shKAP1 and shGFP cell line by comparing relative enrichment levels at control loci (Figure S8). KAP1-specific ChIP identified KAP1 binding sites at the 3′end of ZNF554, ZNF555 and ZNF556, which are located more than 10 kb from the corresponding promoters, as well as within the region of ZNF77-1/2 overlap (Figure 6B–6E). We then examined the presence of HP1β and H3K9me3 throughout the five genes present in the cluster. In ZNF554, ZNF555, ZNF556 and ZNF77, we found peaks of HP1β and H3K9me3 enrichment at the KAP1 binding site, the intensity of which progressively decreased towards the promoter (Figure 7A–7D). We also observed elevated levels of the two silencing marks in the middle of ZNF57, for which no KAP1 association had been detected, possibly because it was not covered by our qPCR primer sets (Figure 7D). When KAP1 was targeted by shRNA, levels of HP1β and H3K9me3 decreased, consistent with KAP1 mediating the spread of silent chromatin at the affected gene bodies (Figure 7A–7D). Therefore, we conclude that KAP1 represses KRAB-ZFP gene clusters by mediating 3′ to 5′ spreading of HP1β and H3K9me3 from endogenous binding sites located at the distal end of these genes. This closely resembles results obtained with the ectopic TrapSil vectors, demonstrating the biological significance of KRAB/KAP1 in long-range repression through the propagation of heterochromatin-associated modifications.
Discussion
In the present work, we demonstrate that KRAB-ZFPs and their co-repressor KAP1 can induce long-range transcriptional repression through heterochromatin spreading. For this, we studied lentivirally-trapped promoters silenced by an ectopic KRAB-containing repressor, as well as the KAP1-mediated modulation of the chromatin landscape at an endogenous KRAB-ZFP gene cluster. We could document the propagation of HP1β and the silencing mark H3K9me3 from repressor binding sites to targeted promoters, and showed that gene silencing results from a block in transcriptional initiation. Furthermore, we could demonstrate that this silencing process is KAP1-dependent and facilitated by HP1.
Our results demonstrate that KRAB/KAP1 have the ability to repress promoters at far greater distances than suspected based on earlier publications. Indeed, it was previously found that recruitment of a regulatable KRAB-based repressor to an array of integrated reporter genes led to transcriptional repression of the promoter adjacent to the repressor binding site, but did not affect a transcriptional unit located 2–3 kb away [37]. Here, we frequently observed repression of promoters located several tens of kilobases away from the tTRKRAB binding site, although this process was most efficient for promoters located within 15 kb or less. These results are consistent with a recent KAP1 ChIP-chip analysis, which documented the association of this co-repressor with promoters and the 3′end of KRAB-ZFP genes, and corroborate data indicating that the KRAB-ZFP ZNF263 is often found within the body of genes [38],[49]. The novel approach used here, which allows targeting of chromatin-modifying proteins to hundreds of active genes at once, provides a functional correlation to these mapping studies.
Heterochromatin domains at KRAB–ZFP genes
The large family of KRAB-ZFP genes is the result of a recent evolutionary phenomenon, with iterative gene duplication events that have led to large sequence homologies between its different members. Therefore, it was proposed that the large heterochromatin domains containing HP1β and SUV39h1 at KRAB-ZFP genes act as a protection from deleterious recombination events, that such a homology would enhance [39]. We show here that KAP1, very likely in cooperation with specific KRAB-ZFPs, binds to the 3′end of KRAB-ZFP genes and mediates the spread of heterochromatin through their gene bodies, thus providing a molecular mechanism for the formation of these heterochromatic domains at KRAB-ZFP gene clusters. Furthermore, by examining a specific KRAB-ZFP gene cluster, we documented the KAP1-mediated control of its transcriptional units, by demonstrating increased expression of KRAB-ZFP genes upon KAP1 depletion. Importantly, the absence of silent chromatin marks at the promoters of these KRAB-ZFP genes, as well as the range of the transcriptional modulation observed, may be consistent with auto-regulatory feedback loops between different KRAB-ZFPs. We further propose that heterochromatic modifications concentrated in gene bodies of KRAB-ZFPs rather than in their promoter, ensure protection from recombination events, while preserving the transcriptional potential of KRAB-ZFP genes. Further analysis of specific KRAB-ZFPs and their binding profiles will provide more functional insight into this type of regulation.
Mechanism of KRAB/KAP1–induced heterochromatin spreading
Spreading is considered to be an intrinsic property of heterochromatin [50]. For mammals it has been proposed that initial heterochromatin assembly at the centromere is followed by a spreading process involving HP1 and H3K9me3 in a self-perpetuating loop. It remains unclear what brings about the initial H3K9me3 that nucleates heterochromatin formation. In addition, in mammalian cells spreading of heterochromatin relies on HP1 in cooperation with additional factors such as SUV39H, since neither H3K9me2 nor H3K9me3 alone is sufficient to recruit HP1 to chromatin [51]. By analogy, KRAB/KAP1-mediated silencing may depend on similar interactions between KAP1, HP1 and H3K9me3, since long-range repression is abrogated when KAP1-HP1 binding is lost, and effective silencing is only seen if levels of H3K9me3 increase at the affected promoters themselves. In agreement with this model, recent studies on the MEST promoter, a primary KAP1 target, showed that repression is rapidly lost upon disruption of the interaction between KAP1 and HP1, and is accompanied by a decrease in H3K9me3 [52].
Remarkably, we documented a decreasing gradient of HP1β and H3K9me3 from the KAP1 docking site to the promoter in both ectopically KRAB-repressed genes and at an endogenous KRAB-ZFP gene cluster. Although it is difficult to compare the binding curves of these different factors due to the non-linear binding kinetics of the antibodies used for their capture, this may suggest that KAP1 nucleates the spread of heterochromatin. Notably, KAP1 is enriched along the bodies of all KRAB-ZFP genes (Figure 6B–6E), consistent with a model of low levels of KAP1 spreading away from KAP1 primary binding sites. It has been proposed that the propagation of heterochromatin could occur by diffusion of an enzymatically-competent macromolecular complex from a source site to nearby lower affinity sites [53]. Our data suggest that KAP1, HP1 and specific histone methyltransferases may form such a complex to mediate the spread of silent chromatin.
Consequences of KRAB/KAP1–induced heterochromatin spreading
Our analyses of TrapSil-targeted genes and of an endogenous KRAB-ZFP gene cluster indicate not only that in both settings silent chromatin marks spread from sites of KAP1 recruitment, but also reveal that the transcriptional consequences of this event vary. Our finding that KAP1 depletion upregulated several transcripts within a KRAB-ZFP cluster, even in the absence of promoter silencing marks suggests that expression was modulated primarily by chromatin changes over the transcribed region of these genes. Therefore, our results imply broad functions for KRAB/KAP1-mediated long-range repression, encompassing not only transcriptional promoter silencing but also transcriptional dampening, as has previously been described for HP1 binding within the coding regions of genes [54],[55].
Our analysis of trapped cellular genes exposed to the tTRKRAB ectopic repressor revealed outcomes largely dictated by whether or not silencing marks reached their promoters. It is likely that the cumulative effect of increased H3K9me3 and decreased H3Ac interferes with binding of transcriptional activator proteins and the ultimate recruitment of RNA Pol II. This hypothesis is not only consistent with our finding of less hypophosphorylated RNA Pol II at trapped and silenced promoters, but was also recently put forward to explain how histone modifications regulate the expression of the L-type pyruvate kinase gene [56].
Analysis of trapped populations with different silencing phenotypes showed that the distance between sites of KRAB/KAP1 recruitment and trapped promoters had a strong influence on KRAB-mediated silencing, but was not the sole factor at play. Indeed, some promoters located at great distances from the tTRKRAB-binding site were efficiently shut off, while others situated close-by escaped this process. Several possibilities come to mind to explain this variation. First, distinct subgroups of promoters may exhibit intrinsically different susceptibility to KRAB/KAP1-mediated silencing. Second, counteracting influences may be exerted at some promoters, for instance downregulation of H3K9me3 levels by histone demethylases, as exemplified by the proposed action of JMJD1A and JMJD2C on pluripotency-associated genes in mouse ES cells [57],[58]. Third, an active nucleosome remodeling mechanism may protect a promoter from heterochromatization. This is consistent with our observation that the irrepressible trapped promoter of the casein kinase 1d locus exhibited lower levels of histone 3 upon tTRKRAB recruitment (Figure 2C). By analogy, the induction of the mammalian interferon-β promoter through external cues is also accompanied by nucleosome remodeling [59].
More generally, there may be genomic loci that exhibit chromatin environments permissive or restrictive to KRAB/KAP1-mediated long-range repression. Examples of both situations in this study include the long-range spreading of heterochromatin detected over a 50 kb long region harboring several KRAB-ZFP genes and the impaired spreading observed in the vicinity of the HNRNPA2B1-CBX3 ubiquitously-acting chromatin opening element (A2UCOE) [60]. In the first of these two cases the site of tTRKRAB recruitment at the 3′end of the ZNF77-1 gene coincided with an endogenous KAP1 binding site, so that the impact of tTRKRAB and KAP1 recruitment to this locus could be directly compared (Figure 6E and Figure S5). Their effect on promoters seemed to differ, as low basal levels of H3K9me3 at the ZNF77-1 and ZNF57 promoters dramatically increased upon tTRKRAB recruitment, which correlated with transcriptional repression. This result is consistent with our model that elevated H3K9me3 at active promoters leads to efficient transcriptional repression by KRAB/KAP1. If levels of this histone modification, however, are already low, then depletion of KAP1 will not affect the promoter as strongly. In contrast, we found that increases of both H3K9me3 and HP1β along the gene body were dependent on both KAP1 and tTRKRAB, consistent with the idea that both factors mediate the spread of heterochromatin from an initial binding site. We further speculate that specific regions, such as KRAB-ZFP gene clusters, are primed for KRAB/KAP1-mediated long-range repression, where this mechanism contributes to heterochromatin structure and long-range transcriptional repression.
In a second example we made use of trapped genes containing several TrapSil integrations with distinct silencing phenotypes (Figure S3). One of these genes is HNRNPA2B1, the promoter of which was shown to be part of a UCOE exhibiting a methylation-free CpG island (Figure S3E). This island encompasses the two divergently transcribing promoters of the housekeeping genes HNRNPA2B1 and CBX3 and allows for stable transgene expression in centromeric heterochromatin [60]. Although in our study the integrant closest to the promoter exhibited a repressible phenotype, the first irrepressible integrant was located at a distance of 3.5 kb, which is considerably lower than the observed average of 15 kb. Moreover, this gene contained a very high ratio of irrepressible/repressible integrants. It was recently shown that the DNA CpG-methylation free region spans 3 kb into the HNRNPA2B1 gene body, probably allowing for very efficient transcriptional initiation [61]. Therefore, it is conceivable that the high frequency of irrepressible clones found here is linked to the specific architecture of this promoter. We are currently trying to establish more detailed correlations between the effect of tTRKRAB-mediated silencing on genes and their individual chromatin environment.
In some cases, long-range acting elements such as enhancers might be the primary targets of KRAB/KAP1-induced chromatin modifications, with a secondary impact on promoters. More generally, specific cis-acting sequences or chromatin features located between KAP1 recruitment sites and transcriptional start sites may alter the spread of heterochromatin, either positively or negatively. Conversely, while the hereby observed long-range spreading of silencing heterochromatin suggests that some clusters of genes might be co-regulated through common KRAB-ZFP binding sites, it also implies the existence of limiting mechanisms to avoid that it systematically be the case.
Perspectives
In conclusion, our results reveal that KRAB-ZFPs, which constitute the largest family of transcription factors encoded by the human genome, mediate long-range repression through the spread of heterochromatin in a process influenced by the genomic context. Therefore, it is tempting to speculate that the long-range epigenetic silencing (LRES) of large chromosomal regions observed in cancer cells [62] may be due to aberrant spreading of heterochromatin stemming from the dysregulation of KRAB/KAP1-mediated epigenetic regulation. Indeed, the expression of KAP1 has been found to be altered in several types of malignancies, for instance lung cancer [63],[64]. Furthermore, since tTRKRAB recruitment results in long-range repression at permissive endogenous loci, its targeting to specific chromosomal regions, through the site-specific introduction of TetO sequences, could be a useful tool to elucidate functions of interesting regulatory landscapes. Finally, the controlled recruitment of KRAB-containing repressors should facilitate studies on the kinetics of heterochromatin spreading in different chromosomal environments, at distinct developmental stages, and in various physiopathogical conditions, thereby shedding light on the intricacies of transcriptional repression.
Materials and Methods
Cell lines and culture
HeLa, 293T and HCT116 cells (from ATCC, Manassas, VA) were cultured using standard methods. Mouse embryonic fibroblasts from homozygous KAP1 floxed mice were derived as described [27]. Doxycycline (Sigma Aldrich) was used at a concentration of 1 µg/mL. GFP expression was determined by flow cytometry using the Beckton Dickinson FACScan.
Vectors
LV - based gene trap vectors [65] were modified by introducing 7 TetO repeats. pLV-tTR-KRAB-Red was described [66]. LV-based plasmids expressing human wild type or mutant (R487E, V488E) KAP1 were a gift from Frank Rauscher (Wistar Institute). LV-based plasmids expressing shRNAs targeting KAP1 and GFP were purchased from Sigma and Addgene, respectively (for sequences see Table S3). LV - and MLV-based particles were produced as described [67].
Western blot
Cells were lysed using standard procedures and subjected to SDS-PAGE. Antibodies used were mouse monoclonal anti-PCNA (Oncogene Research Products, Boston, MA) and mouse monoclonal anti-KAP1 (MAB 3662, Chemicon), followed by secondary goat anti-mouse IRDye 800CW antibody (Li-cor). Imaging and quantification of the Western blot were performed on ODYSSEY infrared imager using the machine-specific software (Li-cor).
RNA procedures
Total RNA was isolated using an RNeasy plus kit (Qiagen).
For Northern blot 15 µg of RNA were run on a denaturing agarose gel and transferred onto a nylon membrane, which was pre-hybridized in ULTRAhyb buffer (Ambion) and after addition of a GFP-specific DIG-labelled probe (Roche, PCR DIG probe synthesis) hybridized overnight at 42°C. The membrane was then subjected to washing and detection procedures as recommended by the manufacturer (Roche, DIG Northern kit). For 5′RACE a previously described protocol was used [68]. Briefly, RNA was converted into cDNA with Super Script II Reverse Transcriptase (Invitrogen) and the trap-specific reporter primer RSP-6 or RSP-8 (for sequences see Table S3). Primary PCR reactions were performed using 2 µl of dA-tailed cDNA products and the reporter specific primers IRES(−) or RSP-6 and anchoring primers QA and QT. Nested PCR reactions were done using the reporter specific primers IRES1b or IRES(−) and anchoring primer QB. PCR products were gel purified and cloned into pCR4 by using a TOPO TA cloning kit (Invitrogen) and subsequently sequenced. Sequences were extracted from chromatograms using phred [69]. pCR4 and virus specific sequences were identified with BLAT [70] and removed prior to annotation. The location of each amplified cDNA fragment was determined with a blastn search for exact matches on the human genome (ensembl version, August 2007). Since most integrations were intronic and thus not present in the amplified cDNA, we arbitrarily assigned the proviral integration to the middle of the intron.
For quantitative Real-Time PCR (qPCR), RNA was converted into cDNA as in 5′RACE, however using random hexamer primers. For each sample a control reaction lacking RT was included. Diluted cDNA was used to assay the expression of each gene with 1x Power Sybr (Applied Biosystems), and a 400 nM concentration of each gene-specific primer. To verify specificity, each PCR was followed by a melting curve analysis. The increase in fluorescence was analysed with the SDS software, version 2.2.2 (Applied Biosystems). For all amplification plots the baseline data were set with the automatic cycle threshold function. A mean quantity was calculated from duplicate or triplicate PCRs for each sample, and a normalization factor was obtained by calculating the geometric mean of the values of the selected housekeeping genes, which was subsequently used to normalize the relative amounts of RNAs of interest [71].
Ligation-mediated PCR (LM–PCR)
Integration sites were mapped by LM-PCR using a previously described protocol [44],[45]. Integration site determination was processed by selecting integrations site amplicons satisfying all quality controls (4/5/4 primer sequence and co-amplified viral genome fragment) and using FetchGWI [72] for alignment on the March 2006 Human Genome assembly. The gene list used for integrant mapping was downloaded from the UCSC table browser by selecting “UCSC known genes”, further including SpID and genesymbol fields. TxStart and TxEnd were downloaded to determine the transcriptional start site and 3′ end for corresponding genes.
Chromatin immunoprecipitation (ChIP)
ChIP was performed according to the Upstate protocol with minor modifications (http://www.millipore.com/techpublications/tech1/mcproto407). About 107 cells were crosslinked with 1% formaldehyde for 15 min at RT, quenched by adding glycine and rinsed with PBS. Cells were then resuspended in lysis buffer and sheared by sonication with a Branson digital sonicator (model 250) on ice four times for 20 s. 100 µl of sonicated chromatin was directly de-crosslinked and used as the total input (TI) reference in quantitative PCR analysis at a dilution of 1∶350 or 1∶700. 100 µl of sonicated chromatin was used for each ChIP reaction and was diluted in 900 µl dilution buffer and precleared with 80 µl salmon-sperm DNA - proteinA agarose beads (Upstate). Precipitating antibodies H3Ac (Upstate, 06-599), H3K9me3 (Abcam ab 8898), total H3 (Abcam ab 1791), HP1β (Abcam, ab 49938), RNA Pol II 8WG16 (Abcam, ab 817) and rabbit polyclonal KAP1 (gift from D. Schultz) were added overnight (IP). Chromatin-antibody complexes were then captured, washed and eluted with 100 mM NaHCO3, 1% SDS. Cross-links between DNA and proteins were reversed by addition of NaCl (for H3Ac, H3K9me3, total H3, HP1β) and incubation at 65°C. DNA was precipitated after incubation with RNase A and Proteinase K and resuspended in 50 µl H2O and subjected to quantitative PCR analysis at a dilution of either 1∶4 or 1∶8. Quantitative PCR was done as stated in the RT-PCR section and primer sequences are available in Table S3. Negative control reactions without antibody were run for each sample and in all cases gave negligible results. To quantify the relative enrichment of proteins or specific histone modifications at a given sequence a ratio between the relative quantities of IP and TI was calculated with the help of a standard curve and expressed as the % of input value. In the case of H3Ac, total H3 and RNA Pol II these values were further normalized to the relative enrichment value of GAPDH. In all the other cases ChIP efficiency was comparable between samples as measured at specific control loci, including p53BP2 for H3K9me3 [73] and ZNF556 for HP1β (Figure S6 and Figure S8). Fold changes were calculated as ratios of -dox/+dox enrichments, with the ratios of the respective positive controls set as 1, the controls consisted of p53BP2 for H3K9me3 and of ZNF556 for HP1β (Table S4).
Supporting Information
Zdroje
1. DingG
LorenzP
KreutzerM
LiY
ThiesenHJ
2009 SysZNF: the C2H2 zinc finger gene database. Nucleic Acids Res 37 D267 273
2. EmersonRO
ThomasJH
2009 Adaptive evolution in zinc finger transcription factors. PLoS Genet 5 e1000325 doi:10.1371/journal.pgen.1000325
3. HuntleyS
BaggottDM
HamiltonAT
Tran-GyamfiM
YangS
2006 A comprehensive catalog of human KRAB-associated zinc finger genes: insights into the evolutionary history of a large family of transcriptional repressors. Genome Res 16 669 677
4. TadepallyHD
BurgerG
AubryM
2008 Evolution of C2H2-zinc finger genes and subfamilies in mammals: species-specific duplication and loss of clusters, genes and effector domains. BMC Evol Biol 8 176
5. UrrutiaR
2003 KRAB-containing zinc-finger repressor proteins. Genome Biol 4 231
6. VaquerizasJM
KummerfeldSK
TeichmannSA
LuscombeNM
2009 A census of human transcription factors: function, expression and evolution. Nat Rev Genet 10 252 263
7. HamiltonAT
HuntleyS
Tran-GyamfiM
BaggottDM
GordonL
2006 Evolutionary expansion and divergence in the ZNF91 subfamily of primate-specific zinc finger genes. Genome Res 16 584 594
8. CammasF
HerzogM
LerougeT
ChambonP
LossonR
2004 Association of the transcriptional corepressor TIF1beta with heterochromatin protein 1 (HP1): an essential role for progression through differentiation. Genes Dev 18 2147 2160
9. CammasF
MarkM
DolleP
DierichA
ChambonP
2000 Mice lacking the transcriptional corepressor TIF1beta are defective in early postimplantation development. Development 127 2955 2963
10. HuG
KimJ
XuQ
LengY
OrkinSH
2009 A genome-wide RNAi screen identifies a new transcriptional module required for self-renewal. Genes Dev 23 837 848
11. JakobssonJ
CorderoMI
BisazR
GronerAC
BusskampV
2008 KAP1-mediated epigenetic repression in the forebrain modulates behavioral vulnerability to stress. Neuron 60 818 831
12. LiX
ItoM
ZhouF
YoungsonN
ZuoX
2008 A maternal-zygotic effect gene, Zfp57, maintains both maternal and paternal imprints. Dev Cell 15 547 557
13. TianC
XingG
XieP
LuK
NieJ
2009 KRAB-type zinc-finger protein Apak specifically regulates p53-dependent apoptosis. Nat Cell Biol 11 580 591
14. ZhengL
PanH
LiS
Flesken-NikitinA
ChenPL
2000 Sequence-specific transcriptional corepressor function for BRCA1 through a novel zinc finger protein, ZBRK1. Mol Cell 6 757 768
15. RoweHM
JakobssonJ
MesnardD
RougemontJ
ReynardS
KAP1 controls endogenous retroviruses in embryonic stem cells. Nature 463 237 240
16. AbrinkM
OrtizJA
MarkC
SanchezC
LoomanC
2001 Conserved interaction between distinct Kruppel-associated box domains and the transcriptional intermediary factor 1 beta. Proc Natl Acad Sci U S A 98 1422 1426
17. FriedmanJR
FredericksWJ
JensenDE
SpeicherDW
HuangXP
1996 KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes Dev 10 2067 2078
18. Le DouarinB
NielsenAL
GarnierJM
IchinoseH
JeanmouginF
1996 A possible involvement of TIF1 alpha and TIF1 beta in the epigenetic control of transcription by nuclear receptors. Embo J 15 6701 6715
19. MoosmannP
GeorgievO
Le DouarinB
BourquinJP
SchaffnerW
1996 Transcriptional repression by RING finger protein TIF1 beta that interacts with the KRAB repressor domain of KOX1. Nucleic Acids Res 24 4859 4867
20. LechnerMS
BeggGE
SpeicherDW
RauscherFJ3rd
2000 Molecular determinants for targeting heterochromatin protein 1-mediated gene silencing: direct chromoshadow domain-KAP-1 corepressor interaction is essential. Mol Cell Biol 20 6449 6465
21. NielsenAL
OrtizJA
YouJ
Oulad-AbdelghaniM
KhechumianR
1999 Interaction with members of the heterochromatin protein 1 (HP1) family and histone deacetylation are differentially involved in transcriptional silencing by members of the TIF1 family. Embo J 18 6385 6395
22. RyanRF
SchultzDC
AyyanathanK
SinghPB
FriedmanJR
1999 KAP-1 corepressor protein interacts and colocalizes with heterochromatic and euchromatic HP1 proteins: a potential role for Kruppel-associated box-zinc finger proteins in heterochromatin-mediated gene silencing. Mol Cell Biol 19 4366 4378
23. SchultzDC
FriedmanJR
RauscherFJ3rd
2001 Targeting histone deacetylase complexes via KRAB-zinc finger proteins: the PHD and bromodomains of KAP-1 form a cooperative unit that recruits a novel isoform of the Mi-2alpha subunit of NuRD. Genes Dev 15 428 443
24. SchultzDC
AyyanathanK
NegorevD
MaulGG
RauscherFJ3rd
2002 SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev 16 919 932
25. SripathySP
StevensJ
SchultzDC
2006 The KAP1 corepressor functions to coordinate the assembly of de novo HP1-demarcated microenvironments of heterochromatin required for KRAB zinc finger protein-mediated transcriptional repression. Mol Cell Biol 26 8623 8638
26. UnderhillC
QutobMS
YeeSP
TorchiaJ
2000 A novel nuclear receptor corepressor complex, N-CoR, contains components of the mammalian SWI/SNF complex and the corepressor KAP-1. J Biol Chem 275 40463 40470
27. WiznerowiczM
JakobssonJ
SzulcJ
LiaoS
QuazzolaA
2007 The Kruppel-associated box repressor domain can trigger de novo promoter methylation during mouse early embryogenesis. J Biol Chem 282 34535 34541
28. DialynasGK
VitaliniMW
WallrathLL
2008 Linking Heterochromatin Protein 1 (HP1) to cancer progression. Mutat Res 647 13 20
29. NielsenAL
Oulad-AbdelghaniM
OrtizJA
RemboutsikaE
ChambonP
2001 Heterochromatin formation in mammalian cells: interaction between histones and HP1 proteins. Mol Cell 7 729 739
30. PlateroJS
HartnettT
EissenbergJC
1995 Functional analysis of the chromo domain of HP1. Embo J 14 3977 3986
31. ThiruA
NietlispachD
MottHR
OkuwakiM
LyonD
2004 Structural basis of HP1/PXVXL motif peptide interactions and HP1 localisation to heterochromatin. Embo J 23 489 499
32. KwonSH
WorkmanJL
2008 The heterochromatin protein 1 (HP1) family: put away a bias toward HP1. Mol Cells 26 217 227
33. AagaardL
LaibleG
SelenkoP
SchmidM
DornR
1999 Functional mammalian homologues of the Drosophila PEV-modifier Su(var)3-9 encode centromere-associated proteins which complex with the heterochromatin component M31. Embo J 18 1923 1938
34. BannisterAJ
ZegermanP
PartridgeJF
MiskaEA
ThomasJO
2001 Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410 120 124
35. LachnerM
O'CarrollD
ReaS
MechtlerK
JenuweinT
2001 Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410 116 120
36. NakayamaJ
RiceJC
StrahlBD
AllisCD
GrewalSI
2001 Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292 110 113
37. AyyanathanK
LechnerMS
BellP
MaulGG
SchultzDC
2003 Regulated recruitment of HP1 to a euchromatic gene induces mitotically heritable, epigenetic gene silencing: a mammalian cell culture model of gene variegation. Genes Dev 17 1855 1869
38. O'GeenH
SquazzoSL
IyengarS
BlahnikK
RinnJL
2007 Genome-wide analysis of KAP1 binding suggests autoregulation of KRAB-ZNFs. PLoS Genet 3 e89 doi:10.1371/journal.pgen.0030089
39. VogelMJ
GuelenL
de WitE
Peric-HupkesD
LodenM
2006 Human heterochromatin proteins form large domains containing KRAB-ZNF genes. Genome Res 16 1493 1504
40. DeuschleU
MeyerWK
ThiesenHJ
1995 Tetracycline-reversible silencing of eukaryotic promoters. Mol Cell Biol 15 1907 1914
41. MoosmannP
GeorgievO
ThiesenHJ
HagmannM
SchaffnerW
1997 Silencing of RNA polymerases II and III-dependent transcription by the KRAB protein domain of KOX1, a Kruppel-type zinc finger factor. Biol Chem 378 669 677
42. MitchellRS
BeitzelBF
SchroderAR
ShinnP
ChenH
2004 Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol 2 e234 doi:10.1371/journal.pbio.0020234
43. WuX
LiY
CriseB
BurgessSM
2003 Transcription start regions in the human genome are favored targets for MLV integration. Science 300 1749 1751
44. SchroderAR
ShinnP
ChenH
BerryC
EckerJR
2002 HIV-1 integration in the human genome favors active genes and local hotspots. Cell 110 521 529
45. WangGP
CiuffiA
LeipzigJ
BerryCC
BushmanFD
2007 HIV integration site selection: analysis by massively parallel pyrosequencing reveals association with epigenetic modifications. Genome Res 17 1186 1194
46. BrasherSV
SmithBO
FoghRH
NietlispachD
ThiruA
2000 The structure of mouse HP1 suggests a unique mode of single peptide recognition by the shadow chromo domain dimer. Embo J 19 1587 1597
47. MurzinaN
VerreaultA
LaueE
StillmanB
1999 Heterochromatin dynamics in mouse cells: interaction between chromatin assembly factor 1 and HP1 proteins. Mol Cell 4 529 540
48. CammasF
JanoshaziA
LerougeT
LossonR
2007 Dynamic and selective interactions of the transcriptional corepressor TIF1 beta with the heterochromatin protein HP1 isotypes during cell differentiation. Differentiation 75 627 637
49. FrietzeS
LanX
JinVX
FarnhamPJ
2009 Genomic targets of the KRAB and scan domain-containing zinc finger protein 263 (ZNF263). J Biol Chem
50. GrewalSI
JiaS
2007 Heterochromatin revisited. Nat Rev Genet 8 35 46
51. StewartMD
LiJ
WongJ
2005 Relationship between histone H3 lysine 9 methylation, transcription repression, and heterochromatin protein 1 recruitment. Mol Cell Biol 25 2525 2538
52. RicletR
ChendebM
VoneschJL
KoczanD
ThiesenHJ
2009 Disruption of the interaction between transcriptional intermediary factor 1{beta} and heterochromatin protein 1 leads to a switch from DNA hyper - to hypomethylation and H3K9 to H3K27 trimethylation on the MEST promoter correlating with gene reactivation. Mol Biol Cell 20 296 305
53. TalbertPB
HenikoffS
2006 Spreading of silent chromatin: inaction at a distance. Nat Rev Genet 7 793 803
54. de WitE
GreilF
van SteenselB
2007 High-resolution mapping reveals links of HP1 with active and inactive chromatin components. PLoS Genet 3 e38 doi:10.1371/journal.pgen.0030038
55. JohanssonAM
StenbergP
PetterssonF
LarssonJ
2007 POF and HP1 bind expressed exons, suggesting a balancing mechanism for gene regulation. PLoS Genet 3 e209 doi:10.1371/journal.pgen.0030209
56. BurkeSJ
CollierJJ
ScottDK
2009 cAMP prevents glucose-mediated modifications of histone H3 and recruitment of the RNA polymerase II holoenzyme to the L-PK gene promoter. J Mol Biol 392 578 588
57. CloosPA
ChristensenJ
AggerK
HelinK
2008 Erasing the methyl mark: histone demethylases at the center of cellular differentiation and disease. Genes Dev 22 1115 1140
58. LohYH
ZhangW
ChenX
GeorgeJ
NgHH
2007 Jmjd1a and Jmjd2c histone H3 Lys 9 demethylases regulate self-renewal in embryonic stem cells. Genes Dev 21 2545 2557
59. LomvardasS
ThanosD
2001 Nucleosome sliding via TBP DNA binding in vivo. Cell 106 685 696
60. AntoniouM
HarlandL
MustoeT
WilliamsS
HoldstockJ
2003 Transgenes encompassing dual-promoter CpG islands from the human TBP and HNRPA2B1 loci are resistant to heterochromatin-mediated silencing. Genomics 82 269 279
61. Lindahl AllenM
AntoniouM
2007 Correlation of DNA methylation with histone modifications across the HNRPA2B1-CBX3 ubiquitously-acting chromatin open element (UCOE). Epigenetics 2 227 236
62. ClarkSJ
2007 Action at a distance: epigenetic silencing of large chromosomal regions in carcinogenesis. Hum Mol Genet 16 Spec No 1 R88 95
63. BeerDG
KardiaSL
HuangCC
GiordanoTJ
LevinAM
2002 Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med 8 816 824
64. BhattacharjeeA
RichardsWG
StauntonJ
LiC
MontiS
2001 Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci U S A 98 13790 13795
65. De PalmaM
MontiniE
Santoni de SioFR
BenedicentiF
GentileA
2005 Promoter trapping reveals significant differences in integration site selection between MLV and HIV vectors in primary hematopoietic cells. Blood 105 2307 2315
66. WiznerowiczM
TronoD
2003 Conditional suppression of cellular genes: lentivirus vector-mediated drug-inducible RNA interference. J Virol 77 8957 8961
67. ZuffereyR
NagyD
MandelRJ
NaldiniL
TronoD
1997 Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat Biotechnol 15 871 875
68. ChenWV
SorianoP
2003 Gene trap mutagenesis in embryonic stem cells. Methods Enzymol 365 367 386
69. EwingB
HillierL
WendlMC
GreenP
1998 Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res 8 175 185
70. KentWJ
2002 BLAT–the BLAST-like alignment tool. Genome Res 12 656 664
71. VandesompeleJ
De PreterK
PattynF
PoppeB
Van RoyN
2002 Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3 RESEARCH0034
72. IseliC
AmbrosiniG
BucherP
JongeneelCV
2007 Indexing strategies for rapid searches of short words in genome sequences. PLoS One 2 e579 doi:10.1371/journal.pone.0000579
73. LiH
RauchT
ChenZX
SzaboPE
RiggsAD
2006 The histone methyltransferase SETDB1 and the DNA methyltransferase DNMT3A interact directly and localize to promoters silenced in cancer cells. J Biol Chem 281 19489 19500
Štítky
Genetika Reprodukční medicína
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2010 Číslo 3
-
Všechny články tohoto čísla
- Parental Genome Dosage Imbalance Deregulates Imprinting in
- Identification and Functional Analysis of the Vision-Specific BBS3 (ARL6) Long Isoform
- HAP2(GCS1)-Dependent Gamete Fusion Requires a Positively Charged Carboxy-Terminal Domain
- Initial Genomics of the Human Nucleolus
- Role of RecA and the SOS Response in Thymineless Death in
- PPS, a Large Multidomain Protein, Functions with Sex-Lethal to Regulate Alternative Splicing in
- Mislocalization of XPF-ERCC1 Nuclease Contributes to Reduced DNA Repair in XP-F Patients
- Transgenic Rat Model of Neurodegeneration Caused by Mutation in the Gene
- Human Population Differentiation Is Strongly Correlated with Local Recombination Rate
- Local-Scale Patterns of Genetic Variability, Outcrossing, and Spatial Structure in Natural Stands of
- Arginylation-Dependent Neural Crest Cell Migration Is Essential for Mouse Development
- HP1 Recruitment in the Absence of Argonaute Proteins in
- MiR-218 Inhibits Invasion and Metastasis of Gastric Cancer by Targeting the Robo1 Receptor
- Bias and Evolution of the Mutationally Accessible Phenotypic Space in a Developmental System
- Papillorenal Syndrome-Causing Missense Mutations in / Result in Hypomorphic Alleles in Mouse and Human
- Rapid Assessment of Genetic Ancestry in Populations of Unknown Origin by Genome-Wide Genotyping of Pooled Samples
- Regulation of Lifespan, Metabolism, and Stress Responses by the SH2B Protein, Lnk
- KRAB–Zinc Finger Proteins and KAP1 Can Mediate Long-Range Transcriptional Repression through Heterochromatin Spreading
- Identification of the Regulatory Logic Controlling Pathoadaptation by the SsrA-SsrB Two-Component System
- Drosophila Xpd Regulates Cdk7 Localization, Mitotic Kinase Activity, Spindle Dynamics, and Chromosome Segregation
- Multiple Signals Converge on a Differentiation MAPK Pathway
- In the Tradition of Science: An Interview with Victor Ambros
- Association of the Polymorphism His615Arg with Melanin Content in East Asian Populations: Further Evidence of Convergent Evolution of Skin Pigmentation
- Fatal Cardiac Arrhythmia and Long-QT Syndrome in a New Form of Congenital Generalized Lipodystrophy with Muscle Rippling (CGL4) Due to Mutations
- Deciphering Normal Blood Gene Expression Variation—The NOWAC Postgenome Study
- Derepression of the Plant Chromovirus Induces Germline Transposition in Regenerated Plants
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Deciphering Normal Blood Gene Expression Variation—The NOWAC Postgenome Study
- Transgenic Rat Model of Neurodegeneration Caused by Mutation in the Gene
- Papillorenal Syndrome-Causing Missense Mutations in / Result in Hypomorphic Alleles in Mouse and Human
- Fatal Cardiac Arrhythmia and Long-QT Syndrome in a New Form of Congenital Generalized Lipodystrophy with Muscle Rippling (CGL4) Due to Mutations
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



