-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaKidney Development in the Absence of and Requires
GDNF signaling through the Ret receptor tyrosine kinase (RTK) is required for ureteric bud (UB) branching morphogenesis during kidney development in mice and humans. Furthermore, many other mutant genes that cause renal agenesis exert their effects via the GDNF/RET pathway. Therefore, RET signaling is believed to play a central role in renal organogenesis. Here, we re-examine the extent to which the functions of Gdnf and Ret are unique, by seeking conditions in which a kidney can develop in their absence. We find that in the absence of the negative regulator Spry1, Gdnf, and Ret are no longer required for extensive kidney development. Gdnf−/−;Spry1−/− or Ret−/−;Spry1−/− double mutants develop large kidneys with normal ureters, highly branched collecting ducts, extensive nephrogenesis, and normal histoarchitecture. However, despite extensive branching, the UB displays alterations in branch spacing, angle, and frequency. UB branching in the absence of Gdnf and Spry1 requires Fgf10 (which normally plays a minor role), as removal of even one copy of Fgf10 in Gdnf−/−;Spry1−/− mutants causes a complete failure of ureter and kidney development. In contrast to Gdnf or Ret mutations, renal agenesis caused by concomitant lack of the transcription factors ETV4 and ETV5 is not rescued by removing Spry1, consistent with their role downstream of both RET and FGFRs. This shows that, for many aspects of renal development, the balance between positive signaling by RTKs and negative regulation of this signaling by SPRY1 is more critical than the specific role of GDNF. Other signals, including FGF10, can perform many of the functions of GDNF, when SPRY1 is absent. But GDNF/RET signaling has an apparently unique function in determining normal branching pattern. In contrast to GDNF or FGF10, Etv4 and Etv5 represent a critical node in the RTK signaling network that cannot by bypassed by reducing the negative regulation of upstream signals.
Published in the journal: . PLoS Genet 6(1): e32767. doi:10.1371/journal.pgen.1000809
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1000809Summary
GDNF signaling through the Ret receptor tyrosine kinase (RTK) is required for ureteric bud (UB) branching morphogenesis during kidney development in mice and humans. Furthermore, many other mutant genes that cause renal agenesis exert their effects via the GDNF/RET pathway. Therefore, RET signaling is believed to play a central role in renal organogenesis. Here, we re-examine the extent to which the functions of Gdnf and Ret are unique, by seeking conditions in which a kidney can develop in their absence. We find that in the absence of the negative regulator Spry1, Gdnf, and Ret are no longer required for extensive kidney development. Gdnf−/−;Spry1−/− or Ret−/−;Spry1−/− double mutants develop large kidneys with normal ureters, highly branched collecting ducts, extensive nephrogenesis, and normal histoarchitecture. However, despite extensive branching, the UB displays alterations in branch spacing, angle, and frequency. UB branching in the absence of Gdnf and Spry1 requires Fgf10 (which normally plays a minor role), as removal of even one copy of Fgf10 in Gdnf−/−;Spry1−/− mutants causes a complete failure of ureter and kidney development. In contrast to Gdnf or Ret mutations, renal agenesis caused by concomitant lack of the transcription factors ETV4 and ETV5 is not rescued by removing Spry1, consistent with their role downstream of both RET and FGFRs. This shows that, for many aspects of renal development, the balance between positive signaling by RTKs and negative regulation of this signaling by SPRY1 is more critical than the specific role of GDNF. Other signals, including FGF10, can perform many of the functions of GDNF, when SPRY1 is absent. But GDNF/RET signaling has an apparently unique function in determining normal branching pattern. In contrast to GDNF or FGF10, Etv4 and Etv5 represent a critical node in the RTK signaling network that cannot by bypassed by reducing the negative regulation of upstream signals.
Introduction
Signaling by the secreted protein GDNF through the RET receptor tyrosine kinase (RTK) and the GFRα1 co-receptor plays a central role in the initiating event of kidney development, the outgrowth of the ureteric bud (UB) from the Wolffian duct (WD) into the metanephric mesenchyme (MM). They are also important for the subsequent growth and branching of the UB to form the renal collecting duct system. This is apparent not only from the lack of UB development in Gdnf, Ret, and Gfra1 mutants in mice [1]–[5] and humans [6], but also from the observation that most of the other genes whose absence causes renal agenesis are upstream regulators of Gdnf or Ret expression [7]. We have recently reported that expression in the UB of the ETS transcription factors ETV4 and ETV5 is upregulated by GDNF/RET signaling, and that Etv4−/−;Etv5−/− double homozygous mice fail to develop kidneys. Thus, the effects of GDNF/RET signaling on UB branching morphogenesis are largely transduced via ETV4 and ETV5 [8].
The mechanism by which GDNF/RET signaling induces epithelial branching remains to be fully elucidated. In the WD, it initially promotes cell movements that precede and lead to the formation of the UB [9], and it then induces UB outgrowth from the duct [10],[11]. In the UB tips, it increases cell proliferation [12],[13], a likely prerequisite for branching. Furthermore, because GDNF is capable of acting as a chemoattractant for cultured kidney cells [14],[15], it has been suggested that GDNF may act as a chemoattractant for UB tips in vivo, thereby promoting and patterning their branching [10],[11],[16].
Here, we have further investigated the role of GDNF/RET signaling by identifying conditions under which the kidney can develop in the absence of either GDNF or RET. To achieve this, we employed a null allele of Sprouty1 (Spry1), a negative feedback inhibitor of RTK signaling, which modulates the response to GDNF during kidney development. Spry1−/− mutants show a pervasive defect in the development of the ureteric tree, including formation of supernumerary buds from the WD, which develop into multiplex ureters and kidneys, and an increase in the number and diameter of UB branches in the developing kidneys [17],[18]. The molecular mechanism of Sprouty protein function is incompletely understood. Engineered expression of Sprouty in cells leads to inhibition of signaling through the MAP kinase (MAPK) pathway, but effects have also been observed on the PI3K and PLCγ signaling pathways downstream of RTKs [19],[20].
It was previously found that removing one Spry1 allele corrected the renal hypoplasia in Gdnf+/ − heterozygous mice, and that removing one Gdnf allele corrected the abnormal UB branching in Spry1−/− mice. These findings demonstrated that the balance between GDNF and SPRY1 levels is critical for normal kidney development [17],[18]. We have now further tested this idea by examining the consequences of eliminating Gdnf and Spry1 (or all Ret and Spry1). Surprisingly, such doubly homozygous mutant mice developed two large and well-formed kidneys, each with a single, normally-positioned ureter. Thus, in the absence of GDNF/RET signaling, other factors must be able to support normal UB outgrowth and extensive UB branching, but only when SPRY1 is absent. We provide in vivo, genetic evidence that FGF10 is one such factor, consistent with the previous observation that exogenous FGFs are capable of inducing budding by the Wolffian duct in organ culture [21]. However, our data also reveal that the specific pattern of UB branching is abnormal in Ret−/−;Spry1−/− and Gdnf−/−;Spry1−/− double mutant kidneys. Therefore, although endogenous FGF10 and perhaps other factors can promote extensive UB branching, GDNF appears to serve a unique role in the patterning of UB branching morphogenesis. Finally, we show that, unlike the rescue of Gdnf or Ret mutations, the lack of both Etv4 and Etv5 cannot be overcome by removing Spry1. Thus, these two transcription factors represent a critical link in a signaling network downstream of Ret and other RTKs.
Results
Kidney development in the absence of Gdnf and Spry1, or Ret and Spry1
Gdnf−/− newborn (P0) mice display ∼80% renal agenesis and ∼20% severe renal hypodysplasia [1]–[3] (n = 28) (Figure 1A and 1B) (for statistical purposes, we count each of the two potential kidneys as a separate sample [22]). In contrast, we found that newborn Gdnf−/−;Spry1−/− (abbreviated GGSS) mice displayed only 11% renal agenesis (n = 18) and 89% of kidneys were normally shaped and only slightly smaller than controls (cross-sectional area 70±11% of wild-type) (Figure 1E). Unlike Spry1−/− mice (Figure 1D), GGSS newborns never showed hydroureter, although the bladder was often filled with urine (Figure 1E), indicating that the ureters were correctly connected to the bladder, and thus suggesting that the site of outgrowth of the UB from the WD had been normal [23],[24].
Fig. 1. Loss of Spry1 rescues kidney development in Gdnf−/− or Ret−/− mice. 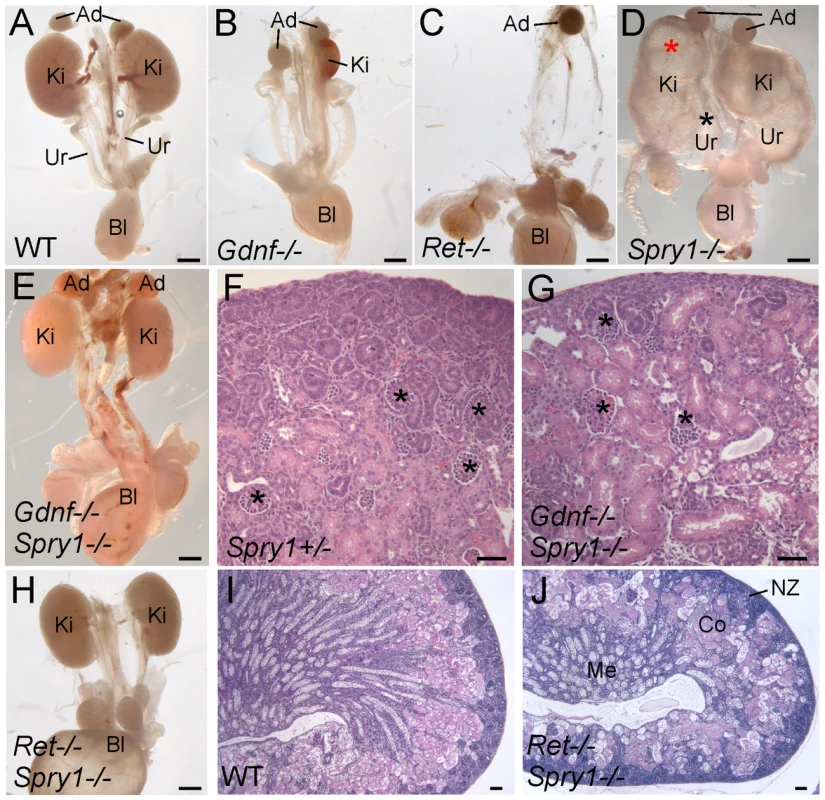
(A–E,H) excretory systems dissected from newborn mice of the indicated genotypes shown in whole mount. Note that the ureter in the Spry1−/− mutant is greatly expanded (black asterisk) and the kidneys are cystic (red asterisk). (F,G) H&E stained sections showing histology of cortex and nephrogenic zone in control (Spry1+/−) and Gdnf−/−;Spry1−/− kidneys, revealing a normal overall organization with well-differentiated glomeruli (*) (I,J) PAS-stained sections of wild-type and Ret−/−;Spry1−/− double mutant kidneys, showing normal overall organization with renal cortex, medulla, and outer nephrogenic zone. Abbreviations: Ad, adrenal gland; Bl, bladder; Co, cortex; Ki, kidney; Me, medulla, NZ, nephrogenic zone; Ur, ureter. Scale bars 1 mm in A-E and H, 100 µm in F,G,I,J. These observations raised the possibility that in the absence of Gdnf and Spry1, kidney development was supported by another GDNF-family ligand, such as Neurturin, which is expressed in the developing kidney [25]. To investigate this possibility we generated Ret−/−;Spry−/− (RRSS) newborn mice, as RET is the common signaling receptor for all GDNF family ligands[26]. Whereas Ret−/− newborn mice display renal agenesis (∼70%) or severe renal hypodysplasia (∼30%) (Figure 1C), 88% of the RRSS double mutants (n = 26) developed fairly large and well-formed kidneys (cross-sectional area 73±15% of wild-type) with apparently normal ureters (Figure 1H). Histological analysis of GGSS and RRSS kidneys indicated that the overall organization into renal papilla, medulla, cortex, and nephrogenic zone was essentially normal, with well differentiated collecting ducts, nephron epithelia and glomeruli in both double mutant genotypes (Figure 1F, 1G, 1I, and 1J and data not shown). Consistent with these findings, many podocalyxin-positive glomeruli were observed in the cortex of double mutant kidneys, although they were reduced in number (52±6% of wild-type in P0 RRSS mutants) (Figure 2A and 2B).
Fig. 2. Numerous nephron and normal nephron–UB connections are observed in double mutant kidneys. 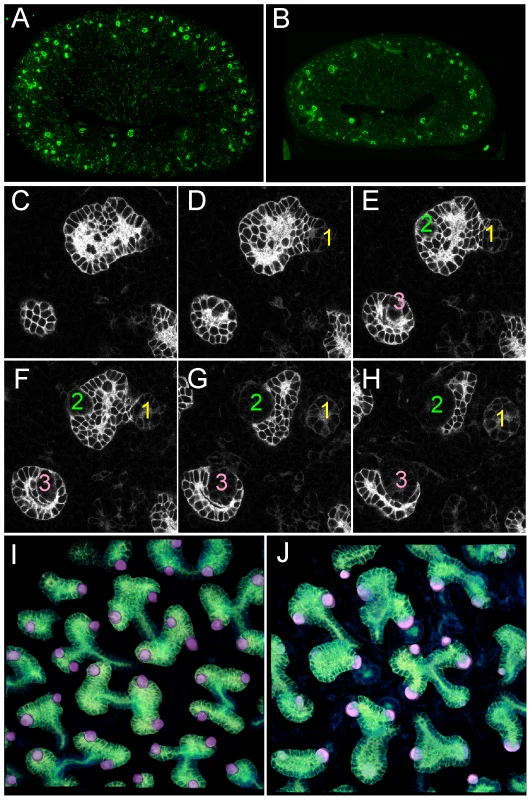
(A,B) Podocalyxin staining of nascent glomeruli in wild-type (A) and Ret−/−;Spry1−/− (B) kidneys at P0, showing numerous, cortically located nephrons in the double mutant, as in the wild-type. (C–H) Six optical sections at different Z-levels of a Gdnf−/−;Spry1−/− E15.5 kidney carrying Hoxb7/myrVenus. The sites where the UB connects to nephrons are visible as “holes” in the myrVenus-labeled UB, as the connecting tubule expresses little or no myrVenus. The connections of three nephrons (1, 2, 3) can be followed at different levels of the image stack. (I) Volume rendering of a wild-type kidney, with nephron connection sites indicated by the pink dots. (J) Volume rendering of double mutant kidneys shown in (C–H), showing normal number and positions of nephron connections per UB tip. Despite their apparently functional kidneys, GGSS and RRSS mice did not survive beyond 3–4 days after birth, presumably because removing Spry1 does not correct the multiple defects in the nervous system caused by lack of Gdnf or Ret [26].
UB branching is extensive, but abnormally patterned, in Gdnf−/−;Spry1−/− and Ret−/−;Spry1−/− kidneys
As GDNF/RET signaling is important for UB growth and branching, we crossed into the mutant backgrounds a Hoxb7/myrVenus transgene, which fluorescently labels the WD and UB lineage [27], to visualize UB branching in vivo or in cultured kidneys. In P0 wild-type kidneys, branching UB tips are numerous and regularly spaced over the kidney surface (Figure 3A). In Spry1−/− kidneys, the UB tips are likewise evenly spaced, but abnormally swollen (Figure 3D). In contrast, although there were numerous UB tips on the surface of GGSS and RRSS kidneys, indicating that the UB had branched very extensively even in the absence of Gdnf or Ret, the tips were irregularly and less densely arrayed, elongated, and abnormally shaped (Figure 3B and 3C). We also examined the kidneys at E15.5, when branching is less complex and the kidneys are small enough to image by confocal microscopy and perform 3D reconstruction (Figure 3E–3P). Volume rendering of the Hoxb7/myrVenus-positive UB tree showed that the double mutants had extensively branched, but the spacing and the branching geometry of UB tips was irregular (Figure 3F and 3G). In such samples, the points where UB tips connect to nephrons could be mapped in three dimensions, and were found to be essentially normal in GGSS mutants (Figure 2C–2J), indicating that the double mutant UB tips produce the factors necessary to connect to the nephrons. Higher magnification 3D reconstructions revealed a characteristic pattern of UB branching in wild-type kidneys (Figure 3I–3M), where successive branch generations occur at regular intervals, mostly at right angles to the parental branches (yellow dashed lines). In contrast, this regular pattern was rarely observed in GGSS or RRSS kidneys, where instead the UB tips were highly irregular in shape, orientation, and branching frequency (Figure 3J, 3K, 3N, and 3O). Spry1−/− UB tips resembled the wild type, except for an increased tip diameter (Figure 3L and 3P), indicating that the branching abnormalities in RRSS and GGSS are not due simply to lack of Spry1.
Fig. 3. Extensive but irregular UB branching in Gdnf−/−; Spry1−/− and Ret−/−; Spry1−/− double mutant kidneys. 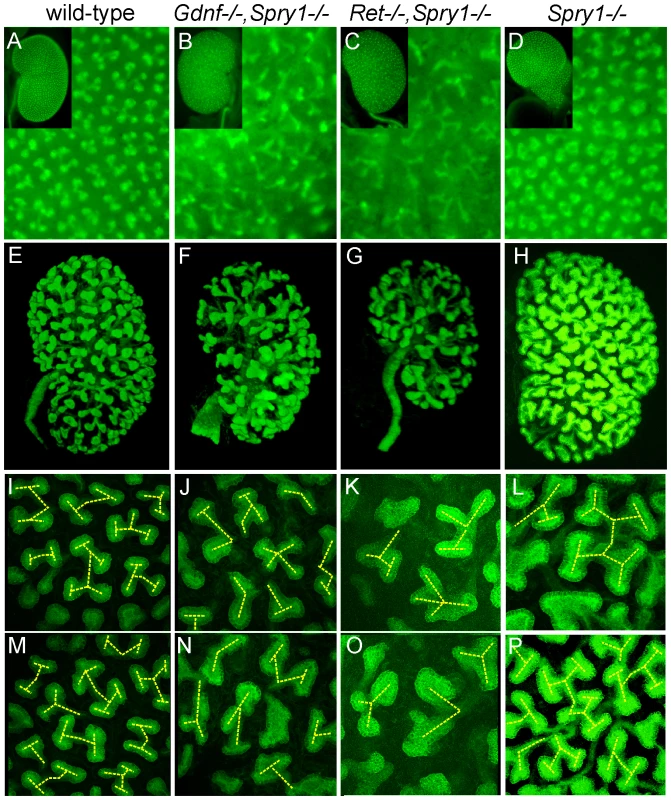
(A–D) Newborn stage kidneys, all carrying the Hoxb7/myrVenus transgene to label the UB branches. Each panel shows a high magnification view of the kidney surface, revealing the shape and organization of branching UB tips; insets show the entire kidney in whole mount. Wild-type kidneys (A) have evenly spaced UB tips with a regular branching pattern, whereas Gdnf−/−;Spry1−/− (B) and Ret−/−;Spry1−/− (C) double mutant kidneys have highly irregular branching. Spry1−/− kidneys (D) have regularly branched, but swollen UB tips. (E–P) 3D volume rendering of E15.5 kidneys. (E–H) Whole kidneys from embryos of the indicated genotypes, carrying Hoxb7/myrVenus. (I–P) Higher power views of two representative surface regions of each genotype. The 3D images were generated from confocal Z-stacks, using Volocity (E–H) or ImageJ (I–P). The yellow dashed lines indicate an interpretation of the branching patterns. While most UB branches in the wild-type (I,M) and Spry1−/− (L,P) kidneys show a reiterative pattern of terminal bifurcation, with branches forming at right angles to their predecessors, most UB branches in the double mutants (J,K,N,O) fail to conform to this pattern, and instead display a variety of abnormal shapes and branching patterns. To examine the initial branching events, we explanted the WD, ureter and kidney at E12.5. Consistent with what was observed in newborn GGSS and RRSS mutants, the ureter and kidney were nearly always present (88%, n = 26 and 100%, n = 16, respectively). In contrast, few Gdnf−/− or Ret−/− mutants had ureter and kidney at this stage (20%, n = 30 and 8%, n = 12, respectively). In none of the GGSS or RRSS mutants were duplicated ureters present, as they are in many Spry1−/− mutants. UB branching was somewhat delayed in the GGSS and RRSS kidneys compared to controls (Figure 4A versus Figure 4D, 0 hours; and data not shown). Several of the wild type, Spry1−/−, and GGSS E12.5 kidneys were cultured to examine the subsequent branching events. While the GGSS kidneys branched extensively in culture, some of the tips elongated abnormally without branching (Figure 4D, asterisks) and some tips grew too slowly (Figure 4D, arrowheads), resulting in an irregularly patterned tree. Thus, while GDNF/RET signaling is not required for the UB to undergo extensive growth and branching when Spry1 is also absent, it is necessary to impose a regular pattern on UB branching.
Fig. 4. Abnormal branching of double mutant kidneys in organ culture. 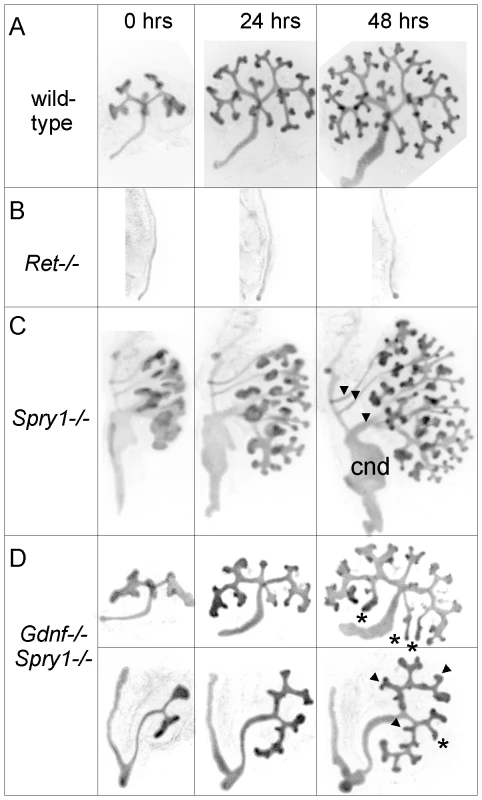
Kidneys of wild-type (A) and mutant genotypes (B–D), carrying Hoxb7/myrVenus, were excised at E12.5, cultured in vitro, and photographed at the indicated times. The Ret−/− Wolffian duct (B) failed to develop a ureter or kidney, while the Spry1−/− kidney (C) has multiple ureters (arrowheads), swollen UB tips and an enlarged common nephric duct (cnd). (D), in two examples of Gdnf−/−;Spry1−/− mutant kidneys, UB branching is retarded at E12.5, and subsequent branching in culture displays abnormal patterns (asterisks and arrowheads – see text) compared to wild-type. Differentiation of the UB into tip and trunk domains occurs in the absence of Gdnf/Ret and Spry1
The UB tips and trunks maintain different patterns of gene expression throughout kidney development, with many genes expressed specifically in one domain or the other [12],[28],[29]. Many tip-specific genes can be upregulated by exogenous GDNF, and their expression is reduced in a Ret hypomorphic mutant [8],[12],[30], suggesting that GDNF/RET signaling may be required to maintain the tip-specific pattern. However, we found that three tip-specific markers, Ret, Wnt11, and Etv4, all of which normally require wild-type levels of GDNF/RET signaling for expression in the UB, continued to be expressed in a tip-specific pattern in GGSS or RRSS double mutants (Figure 5A–5F). The trunk-specific marker Wnt7b [31] also retained its normal expression pattern in GGSS double mutants (Figure 5G and 5H), indicating that the lack of Wnt7b expression in the UB tip does not require GDNF/RET signaling. Therefore, there must be other, Ret-independent mechanisms that can establish and maintain tip/trunk differences in gene expression.
Fig. 5. Differential gene expression in tip and trunk domains is retained in Gdnf−/−;Spry1−/− and Ret−/−;Spry1−/− double mutant kidneys. 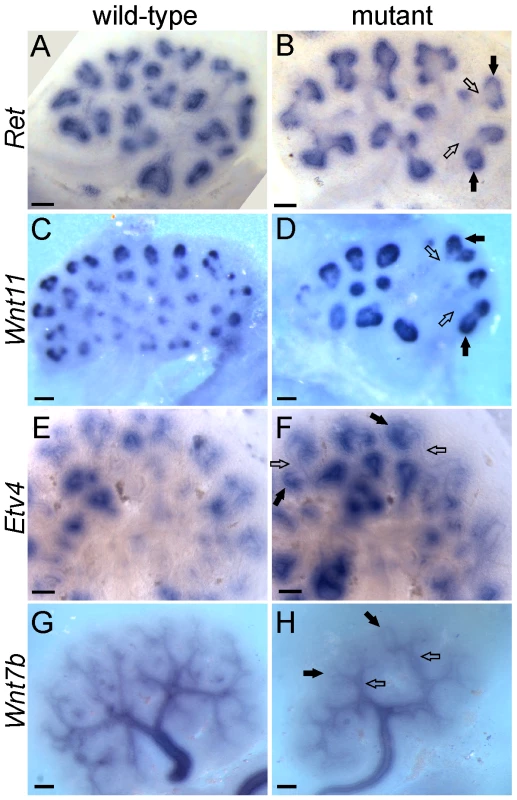
Whole mount in situ hybridization for the UB tip markers Ret, Wnt11 and Etv4 and the trunk marker Wnt7b, in wild-type (A,C,E,G) and double mutant E12.5 kidneys (B,F and H, Gdnf−/−;Spry1−/−, D, Ret−/−;Spry1−/−). Solid arrows indicate UB tips and open arrows indicate trunks. Scale bars 100 µm. Fgf10 cooperates with Gdnf to promote UB outgrowth and branching morphogenesis, and can largely compensate for the loss of Gdnf/Ret in the absence of Spry1
We next sought to determine what signaling molecule(s) support ureteric bud outgrowth from the WD, and subsequent growth and branching, in the absence of Gdnf/Ret and Spry1. The observation that kidney development is rescued in Gdnf−/− or Ret−/− embryos only when Spry1 is absent suggests that the signaling responsible for the rescue must itself be negatively regulated by Spry1. Since Sprouty genes are negative regulators of RTK signaling, the rescue most likely occurs through a RTK. According to this reasoning, FGF signaling is a strong candidate. Genetic studies in the mouse have identified FGF7 and FGF10, signaling through FGFR2, as important factors for normal UB branching [32],[33]; however, the effects of Fgf7 or Fgf10 knockouts (KOs) are far less severe than those caused by loss of Gdnf or Ret, indicating that these FGFs play a secondary role under normal conditions. Fgf7 mRNA was not detected in the kidney before E14.5 [34], whereas Fgf10, like Gdnf, is expressed in the MM at least as early as E10.5 (Figure 6A–6D), making Fgf10 a good candidate to participate in UB outgrowth and early branching morphogenesis. Fgf10−/− mice [35] have small kidneys at birth [33], and we found this to be reflected in reduced UB branching during kidney development (Figure 6E and 6F). The reduction in UB branching was comparable to that in kidneys lacking Fgfr2 (or both Fgfr1 and Fgfr2) in the UB lineage [36], suggesting that FGF10 is the major FGF signaling through FGFR2 in the UB. Furthermore, this defect could be corrected by deletion of one Spry1 allele (Figure 6G), indicating that Spry1 negatively regulates FGF10 (as well as GDNF) signaling.
Fig. 6. Fgf10 expression and function in early ureter and kidney development. 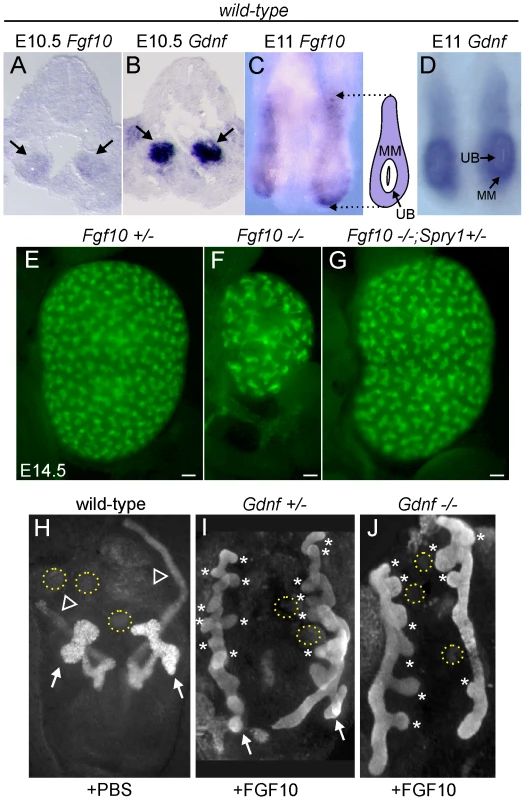
(A,B) In situ hybridization in transverse sections of E10.5 wild type embryos reveals that Fgf10 and Gdnf are expressed in metanephric mesenchyme (arrows). (C,D) Whole-mount in situ hybridization at E11.0 (dorsal view) shows that Fgf10 and Gdnf are expressed in metanephric mesenchyme (MM) surrounding the UB epithelium. The schematic diagram illustrates Fgf10 expression, with purple indicating where the hybridization signal was detected. (E–G) Visualization of Hoxb7/myrVenus shows (E) normal UB branching in an Fgf10+/− kidney, (F) reduced branching in an Fgf10−/− kidney, and (G) rescue of UB branching in an Fgf10−/− kidney when Spry1 dosage is reduced (Spry1+/−). Scale bars, 100 µm. (H–J) Induction of ectopic budding from the Wolffian duct by FGF10. Dissected E10.5 urogenital regions were cultured with control PBS-soaked beads (H) or beads soaked in FGF10 (I,J) placed between the two Wolffian ducts (dotted yellow circles). FGF10 induces multiple ectopic UB outgrowths (marked by asterisks) in both control Gdnf+/− (I) and Gdnf−/− (J) samples. Open arrowhead in H, Wolffian duct; arrows in H-I, normal ureteric buds. To examine the relationship between FGF10 and GDNF in kidney development, we performed gain - and loss-of-function studies. FGF10-soaked beads placed next to the WD of E10.5 embryos induced the formation of multiple ectopic buds (Figure 6H and 6I), as do GDNF beads [10]. To test whether FGF10 induced the ectopic buds indirectly, by up-regulating Gdnf, we performed the same experiment in Gdnf−/− embryos, but the result was similar (Figure 6J). Therefore, FGF10 is capable of inducing UB outgrowth, presumably by acting directly on the WD. The role of Fgf10 was also examined by performing genetic crosses between Fgf10 and Gdnf KO mice, and examining UB formation at early stages (E11.5–12.5) and kidney development in late fetal or newborn mice. Fgf10 heterozygotes always had normal ureters and kidneys (Figure 7A and 7B), whereas Gdnf heterozygotes had a low frequency (7–10%) of defective UB outgrowth or renal agenesis (Figure 7A). However, in Fgf10+/−;Gdnf+/− double heterozygotes, 81% of the UBs were missing or severely delayed at E11.5–E12.5 (e.g., Figure 7A and 7C–7E), and 58% of kidneys were absent at E17.5–P0 (e.g., Figure 7A, 7F, and 7G), roughly equivalent to what is observed in Gdnf null homozygotes with normal Fgf10 dosage (Figure 7A). Furthermore, although renal agenesis was rare in Fgf10 homozygotes (15%), removing one Gdnf allele (Fgf10−/−;Gdnf+/−) caused 100% agenesis (e.g., Figure 7A and 7H). Thus, while the consequences of deleting both Fgf10 alleles in a wild-type background are relatively mild, in a Gdnf+/− background the loss of even one Fgf10 allele causes more severe defects, and loss of both Fgf10 alleles is catastrophic, indicating that Fgf10 and Gdnf normally cooperate to promote UB outgrowth from the WD.
Fig. 7. Fgf10 and Gdnf cooperate to support UB outgrowth and kidney development. 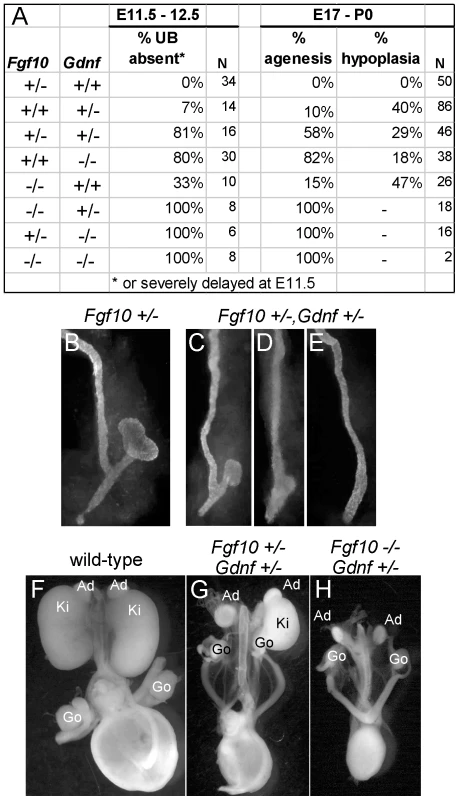
(A) Frequency of the failure of UB outgrowth at E11.5–12.5, and renal agenesis or hypoplasia at E17.5-P0. (B) Normal T-stage UB in an Fgf10+/− embryo at E11.5. (C–E) Three examples of UB formation or lack thereof in Fgf10+/−;Gdnf+/− E11.5 embryos. In (C), the UB is slightly retarded, in (D), the UB is severely delayed, and in (E) the UB is absent. (F–H), normal kidneys in wild-type and renal agenesis or hypoplasia in compound Fgf10/Gdnf mutants at P0. The wild-type in (F) has two normal kidneys, the double heterozygote in (G) has renal agenesis on one side and a hypoplastic kidney on the other, and the Fgf10−/−;Gdnf+/− example in (H) has bilateral agenesis. Ad, adrenal; Ki, kidney; Go, gonad. n = number of (potential) kidneys. To ask if it is FGF10 that rescues kidney development in Gdnf−/−;Spry1−/− mice, we next examined Gdnf−/−;Spry1−/− mice in which Fgf10 gene dosage was reduced. We found that removal of either one or both Fgf10 alleles resulted in 100% renal agenesis (Figure 8). These data conclusively demonstrate that FGF10 supports the extensive kidney development that occurs in Gdnf−/−;Spry1−/− mice.
Fig. 8. Fgf10 is required for ureter and kidney development in the absence of Gdnf and Spry1. 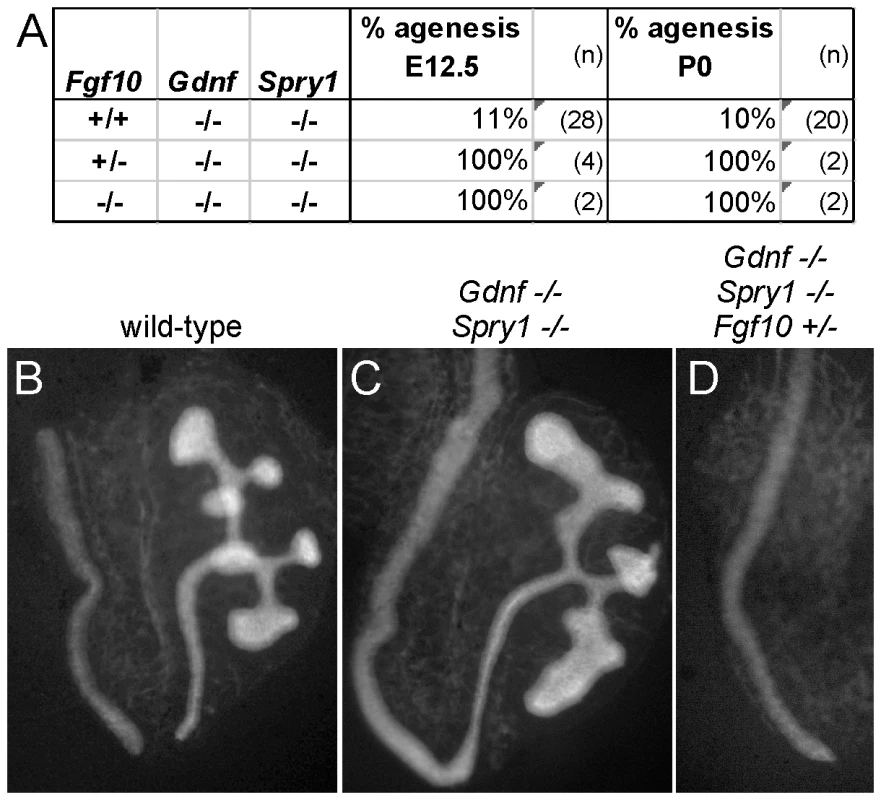
(A) Frequency of absence of the UB at E12.5 and renal agenesis at P0, in Gdnf−/−;Spry1−/− mice with 0, 1, or 2 Fgf10 null alleles. (B) Example of normally branched wild-type UB at E12.5. (C) Gdnf−/−;Spry1−/− UB with moderately reduced branching at E12.5. (D) Absence of the UB in a Gdnf−/−;Spry1−/−;Fgf10+/− embryo at E12.5. n = number of (potential) kidneys. Expression of the ETS transcription factors ETV4 and ETV5 in the UB in vivo requires normal levels of GDNF/RET signaling, and they can also be upregulated in kidney cultures by exogenous FGF10, suggesting that they function downstream of both RET and FGFR2 [8]. If ETV4 and ETV5 are needed to transduce both GDNF and FGF10 signals, removing Spry1 should be unable to rescue kidney development in Etv4−/−;Etv5−/− mice. In accordance with this prediction, the three triple mutant (Etv4−/−;Etv5−/−;Spry1−/−) mice obtained also lacked both kidneys, like Etv4−/−;Etv5−/− mice (Figure 9).
Fig. 9. Loss of Spry1 does not rescue kidney development in Etv4−/−;Etv5−/− mice. 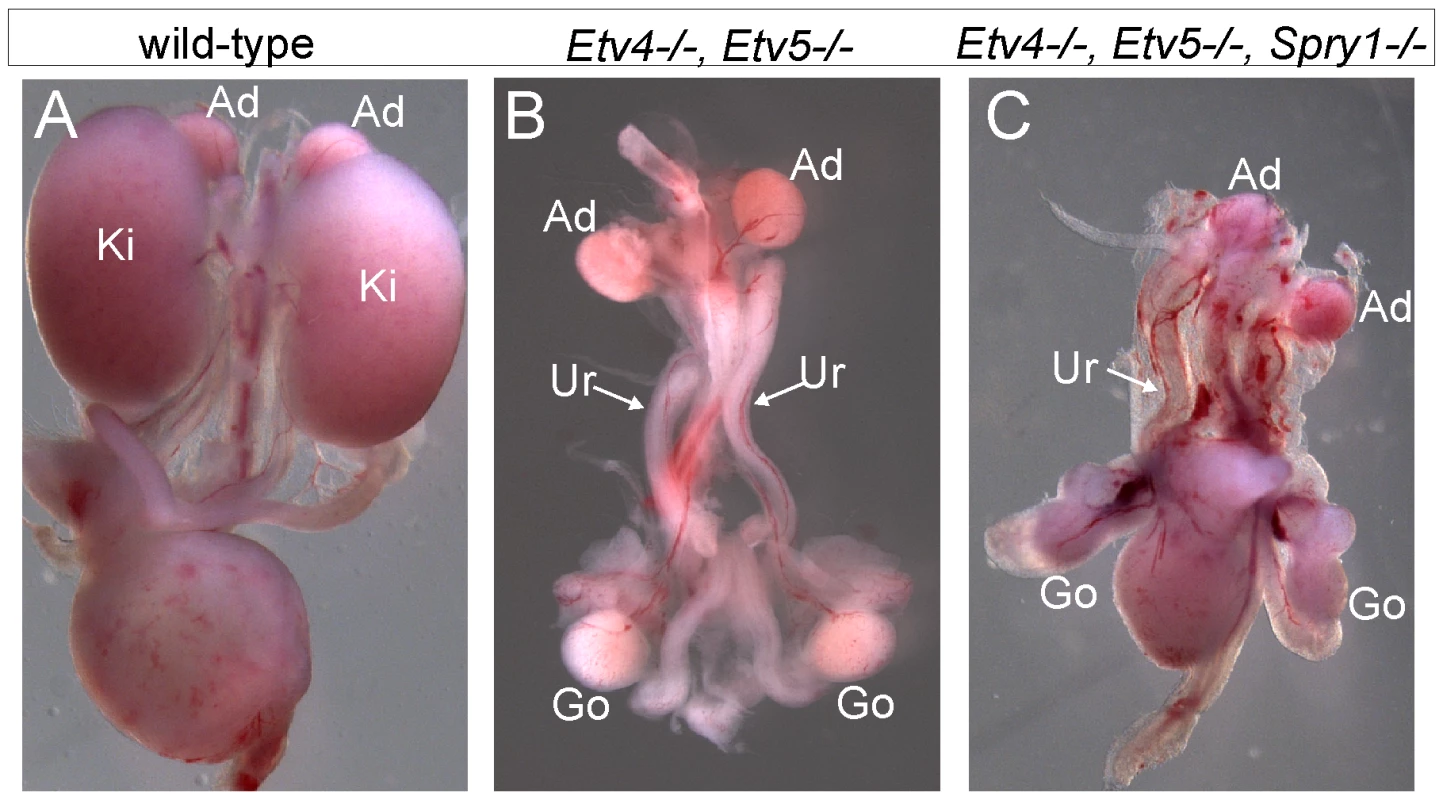
(A) Kidneys in a wild-type mouse at P0. (B) Etv4−/−;Etv5−/− mouse with two ureters but no kidneys. (C) Etv4−/−;Etv5−/−;Spry1−/− mouse with one ureter and no kidneys. Ki, kidney; Ad, adrenal gland; Ur, ureter; Go, gonad. Discussion
To investigate the roles of GDNF/RET signaling and negative regulation by Sprouty1 in branching morphogenesis of the Wolffian duct and ureteric bud during ureter and kidney development, we generated mice that lacked Spry1 and either Gdnf or Ret. We found, unexpectedly, that nearly all the double homozygous mutants developed two large, well organized kidneys, with normal ureters, a highly branched collecting duct system, and extensive nephrogenesis. Thus, it appears that for many aspects of ureter and kidney development, the balance between positive signaling via GDNF/RET and negative regulation via SPRY1 is more critical than the specific role of GDNF. These observations suggested that other signaling molecules, whose activity like that of GDNF is negatively regulated by SPRY1, must be able to perform many of the functions of GDNF, but only when SPRY1 is absent. We identified FGF10 as one such factor; although knockout of Fgf10 normally has relatively minor effects on kidney development, FGF10 plays a critical role when GDNF/RET signaling is reduced or absent. Close examination of Gdnf−/−;Spry1−/− and Ret−/−;Spry1−/− double mutant kidneys revealed that while the UB branches extensively, and proximal-distal UB patterning is retained, the characteristic branching pattern is significantly disrupted. Thus, although GDNF/RET signaling is not required for UB growth or branching per se (when SPRY1 is also absent), it has an apparently unique role in determining the normal branching pattern.
In a wild-type background (i.e., in the presence of SPRY1), GDNF/RET signaling is essential for the positioning and normal outgrowth of the UB from the WD. Not only does the UB usually fail to emerge in Ret−/− or Gdnf−/− mice, but when it does, its position is often abnormal, resulting in the lack of a normal connection to the bladder [37]. Furthermore, ectopic expression of Gdnf causes ectopic UBs to form along the WD [38]–[40]. This led to the model that the specific domain of Gdnf expression in the nephrogenic cord is critical for positioning the UB in the correct location [5],[16]. However, in the absence of SPRY1, mice lacking GDNF or RET make a normal UB that develops into a normal ureter connected to the bladder, as indicated by the absence of hydroureter. Therefore, signaling via another ligand/receptor that is also negatively regulated by SPRY1 must be able to properly position and induce outgrowth of the UB in the absence of GDNF or RET. We found that FGF10, presumably signaling via FGFR2, is an essential component of this alternative signaling, as removing either one or two Fgf10 alleles in a Gdnf−/−;Spry1−/− background caused failure of UB emergence, leading to renal agenesis. Moreover we showed that this FGF10/FGFR2 signaling is not dependent on GDNF signaling because FGF10 induces WD budding of the cultured mouse urogenital system, even in a Gdnf−/− embryo. The possibility remains that other factors (other FGFs, or other signaling molecules) are also involved in this process. It has been shown that several FGFs can induce UB outgrowth from cultured rat WD, and in this assay FGF10 had relatively weak activity whereas FGF7 and other FGFs were more active [21]. The ability of FGF7 to cooperate with or replace GDNF in this process remains to be tested genetically. Other mechanisms, such as the local inhibition of BMP4 by Gremlin1 [41],[42], also contribute to the normal positioning of the UB.
Unlike Gdnf−/− or Ret−/− ureteric buds on a wild-type (Spry1+/+) background, which grow and branch minimally if at all, the double mutant UBs (GGSS or RRSS) grew and branched extensively, leading to a kidney that was often close to normal in size, with an extensive collecting duct system, normal overall histoarchitecture and large numbers of nephrons connected to the collecting ducts. Therefore, GDNF/RET signaling does not have a unique ability to induce UB branching, including the predominant terminal bifurcations, nor is it required for the UB tips to induce nephrogenesis. As in the case of UB outgrowth from the WD, it appears that other factors are potentially redundant with GDNF in their ability to promote UB branching. Since loss of Fgf10 in a Gdnf−/−;Spry1−/− double mutant background eliminated initial UB outgrowth, it could not be determined to what extent FGF10 contributes to later UB branching in the absence of GDNF. However, the reduced UB branching in Fgf10−/− kidneys shows that FGF10 normally contributes significantly to UB branching, and is likely to be at least one of the factors that can promote this process in GGSS or RRSS double mutant mice. Other factors that might also be involved include HGF and EGF [43].
It was recently reported that the effects of a Ret-Y1062F point mutation, which causes renal agenesis or hypodysplasia similar to that observed in Ret knockout mice, can be rescued by removal of Spry1 [44]. The double mutant mice had kidneys of normal size, with normal glomerular number. The Y1062F mutation abolishes signaling through the PI3K-AKT and RAS-MAPK pathways, but does not affect signaling through PLC-γ or other pathways that potentially act downstream of RET (e.g., SRC) [45]. The authors speculated that in the double mutants, the ERK MAPK pathway might be activated by RET via an alternative pathway involving PLC-γ, allowing kidney development to proceed normally, and they did not suggest that other signaling molecules might substitute for GDNF under these conditions. However, in our Ret−/−;Spry1−/− double null mutant mice, the ability of RET to signal through alternative pathways was eliminated, which revealed the ability of other signaling molecules, including FGF10, to support kidney development in the absence of RET or GDNF.
Based on our findings, we propose a model (Figure 10) in which GDNF, FGF10 and probably other signaling molecules expressed in the MM signal through their cognate receptor tyrosine kinases in the UB epithelium to collectively promote budding from the Wolffian duct and subsequent growth and branching during kidney development. RET and FGFR2 (and probably other RTKs) activate a series of shared downstream signaling pathways, including RAS-MAPK, PI3K-AKT and PLC-γ-Ca++ [46], which together support UB branching morphogenesis. Spry1 expression is upregulated by these signals, and SPRY1 then provides negative feedback by regulating one or more of the shared signaling pathways downstream of RET and FGFR. In early kidney development, GDNF is the predominant signal, while FGF10 is much weaker (presumably due to lower expression) (Figure 10A). Expression of Etv4 and Etv5 is upregulated by these signals, thus controlling transcription of downstream genes required for UB growth and branching. Loss of Gdnf (Figure 10B) causes renal agenesis because in the presence of SPRY1 the level of FGF10 signaling via FGFR2 is not sufficient to produce the necessary responses, such as an appropriate level of Etv4 and Etv5 expression. Normally, loss of Fgf10 has relatively mild consequences because of the high level of GDNF signaling. When Spry1 is absent there is no brake on signaling via FGFR2 (Figure 10C), and GDNF can be removed without causing renal agenesis, due (at least in part) to the effects of FGF10, and to the restoration of Etv4/Etv5 expression; however, UB branching pattern is abnormal. If Fgf10 is also removed (Figure 10D) any remaining factors are insufficient to rescue kidney development, resulting in renal agenesis. In the absence of Etv4 and Etv5, removal of Spry1 is unable to rescue kidney development (Figure 10E). This suggests that Etv4 and Etv5 normally mediate the combined effects of several RTKs (RET, FGFR2 and probably others), and therefore elevated RTK signaling due to lack of SPRY1 cannot bypass the requirement for these transcription factors.
Fig. 10. Model: GDNF and FGF10 cooperate to promote ureteric bud branching morphogenesis, via Etv4 and Etv5, while Sprouty1 regulates signaling downstream of both RET and FGFR2. 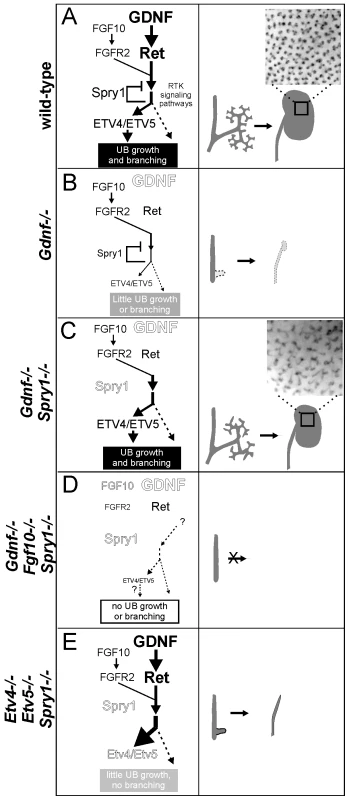
(A) In wild-type, GDNF/RET signaling plays a major role and FGF10/FGFR2 a minor role in promoting UB outgrowth and branching morphogenesis. The response to these signals is modulated by SPRY1, leading to a normal kidney at birth (right panel). The transcription factors ETV4 and ETV5 are downstream effectors of GDNF and FGF10 signaling. (B) In the absence of GDNF, there is presumably less SPRY1 produced [17] (indicated by smaller text), but FGF10 is insufficient to overcome negative regulation by SPRY1, causing reduced downstream signaling to induce UB budding and branching (indicated by thinner arrows), one manifestation of which is a severe reduction in Etv4/Etv5 expression [8]. Consequently, renal agenesis or severe hypodysplasia is observed. (C) When GDNF and SPRY1 are both absent, the lack of negative regulation of signaling by FGFR2 allows for Etv4/Etv5 expression, UB branching, and kidney development; however, the pattern of UB branching is altered, suggesting a unique role of GDNF in this process. (D) When FGF10 and GDNF are both absent, there is too little RTK signaling, even in the absence of negative regulation by SPRY1, to allow UB outgrowth from the Wolffian duct, resulting in renal agenesis (whether Etv4/Etv5 would be expressed is not known, as there is no ureter or kidney to analyze). (E) Renal agenesis in Etv4−/−;Etv5−/− mice is not rescued by loss of Spry1, showing that increased RTK signaling is insufficient for kidney development in the absence of Etv4 and Etv5 (dashed arrow). The observation that ureters develop in Etv4;Etv5;Spry1 triple mutants suggests that UB outgrowth, but not later branching, can occur independently of Etv4/Etv5. Insets in a and c show the pattern of branching UB tips in stage P0 wild-type and double mutant kidneys. The main abnormality observed in the GGSS and RRSS double mutant kidneys was in the specific pattern of branching. Instead of the regular terminal bifurcations in wild-type kidneys, which typically occur at right angles to the previous branching event, the double mutant branching UB tips were heterogeneous in shape, spacing, orientation, branch angle and frequency of branching. The defects were distinct from those caused by loss of Spry1 alone, which causes the UB tips to swell but does not alter branch orientation or tip spacing. Therefore, it appears that these specific defects in branching pattern are a consequence of the loss of GDNF/RET signaling, and reflect a function that cannot be replaced by FGF10 or other factors present in the double mutant kidneys.
How may GDNF/RET signaling influence the specific pattern of UB branching? One possibility is that GDNF in the metanephric mesenchyme acts as a chemoattractant to direct the growth of the UB tips toward local foci of GDNF expression [10],[14],[16], similar to the way in which FGF10 is thought to direct the branching of the developing lung epithelium [47],[48]. We have previously argued against such a model for several reasons [40]. First, the distribution of Gdnf mRNA in the MM is extremely diffuse; however, it remains possible that the protein is more limited in its spatial distribution than the mRNA. Second, we found that kidneys developed rather normally in Gdnf null mice in which Gdnf was misexpressed in the UB epithelium, suggesting that it is the presence, but not the location, of GDNF that is important [40]. However, the specific pattern of UB branching was not closely examined in those mutant/transgenic mice, and it remains possible that they had subtle branching defects similar to the GGSS and RRSS double mutant kidneys. Methods to locally and precisely manipulate the pattern of Gdnf expression will be needed to better test this model. If not through chemoattraction, then GDNF/RET signaling must in some other manner influence the specific pattern of growth and branching of the UB tips.
Methods
Ethics statement
All work on animals was conducted under PHS guidelines and approved by the relevant Institutional Animal Care and Use Committees.
Mouse strains
Ret [4], Gdnf [3], Spry1 [17], Fgf10 [35], Etv4 [49], Etv5 [50] and HoxB7/myrVenus [27] mutant mice have been described. These mice were maintained on a mixed background (129S1/SvmJ:C57BL/6). Embryo stage was estimated by considering noon of the day of the vaginal plug as embryonic day (E) 0.5, and more accurate staging was determined by counting somites. PCR genotyping of mice and embryos was done as described previously [3],[4],[8],[17],[35].
Whole-mount in situ hybridization
Whole-mount and section RNA in situ hybridization and detection of β-galactosidase activity were performed as described previously [38],[51] using digoxigenin-UTP-labeled anti-sense riboprobes.
Histological analysis and nephron counting
Newborn mice were sacrificed according to Institutional and NIH guidelines. Whole kidneys and urogenital tracts were dissected in PBS. Kidney cross-sectional area was determined from whole-mount photographs of 28 wt, 16 GGSS and 12 RRSS P0 kidneys using ImageJ. For histological analysis, 7–10 µm sections were prepared from paraffin-embedded samples fixed in 4% paraformaldehyde (PFA). De-waxed sections were stained with either haematoxylin and eosin (H&E) or Periodic Acid Schiff (PAS). To count glomeruli, five evenly-spaced sections across each kidney (two wild-type and four RRSS mutants) were stained with podocalyxin, and the number of glomeruli per section was averaged for each kidney.
Metanephric kidney explant cultures and immunohistochemistry
Intermediate mesoderm or metanephric kidneys were dissected from E10.5 to E14.5 embryos in PBS +Ca +Mg (Invitrogen). Explants were cultured at 37°C in DMEM/F12 (Invitrogen) supplemented with Glutamax, 100 U/ml penicillin, 100 µg/ml streptomycin, and 10% fetal bovine serum in a 5% CO2, humidified atmosphere at the medium-air interface on Costar Transwell filters (0.4 µm). After culture, explants were fixed in 4% PFA. For immunostaining, explants were incubated with goat anti-podocalyxin antibodies (R&D Systems), followed by Cy2 or Cy3 anti-goat Ig (Jackson ImmunoResearch). Images were captured on a Zeiss Axio Observer Z1.
For FGF bead experiments, posterior intermediate mesoderm was dissected from embryos at the 29 to 35 somite stage in Hanks Balanced Salt Solution, with 1% FBS. Two heparin acrylic beads (Sigma) soaked in FGF10 (R&D Systems), reconstituted in PBS at 1mg/ml) or in PBS were inserted between the WDs, and rudiments were cultured on Whatman Nuclepore Track-Etch Membrane filters (8 micron pore size) in 44% F12, 44% DMEM, 10% FBS, 1% glutamine, 1% Penstrep at the air-liquid interphase for 48–55 hours. Samples were fixed in cold 100% methanol and stained with anti-pan cytokeratin antibody (Sigma C9687).
Confocal imaging and 3D analysis
E15.5 metanephric kidneys were dissected in PBS and fixed overnight in 4% PFA. After clearing using FocusClear (CelExplorer), kidneys were mounted in MountClear (CelExplorer) and scanned using a Leica LS5 confocal microscope, and 3D rendering was performed using Volocity software.
Zdroje
1. MooreMW
KleinRD
FarinasI
SauerH
ArmaniniM
1996 Renal and neuronal abnormalities in mice lacking GDNF. Nature 382 76 79
2. PichelJG
ShenL
ShengHZ
GranholmAC
DragoJ
1996 Defects in enteric innervation and kidney development in mice lacking GDNF. Nature 382 73 76
3. SanchezMP
Silos-SantiagoI
FrisenJ
HeB
LiraSA
1996 Renal agenesis and the absence of enteric neurons in mice lacking GDNF. Nature 382 70 73
4. SchuchardtA
D'AgatiV
Larsson-BlombergL
CostantiniF
PachnisV
1994 Defects in the kidney and enteric nervous system of mice lacking the tyrosine kinase receptor Ret. Nature 367 380 383
5. CostantiniF
ShakyaR
2006 GDNF/Ret signaling and the development of the kidney. Bioessays 28 117 127
6. SkinnerMA
SaffordSD
ReevesJG
JacksonME
FreemermanAJ
2008 Renal aplasia in humans is associated with RET mutations. Am J Hum Genet 82 344 351
7. SchedlA
2007 Renal abnormalities and their developmental origin. Nat Rev Genet 8 791 802
8. LuB
2009 Etv4 and Etv5 are required downstream of GDNF and Ret for kidney branching morphogenesis. Nat Genet 41 1295 302
9. ChiX
MichosO
ShakyaR
RiccioP
EnomotoH
2009 Ret-dependent cell rearrangements in the Wolffian duct epithelium initiate ureteric bud morphogenesis. Dev Cell 17 199 209
10. SainioK
SuvantoP
DaviesJ
WartiovaaraJ
WartiovaaraK
1997 Glial-cell-line-derived neurotrophic factor is required for bud initiation from ureteric epithelium. Development 124 4077 4087
11. SariolaH
SainioK
1997 The tip-top branching ureter. Curr Opin Cell Biol 9 877 884
12. PepicelliCV
KispertA
RowitchDH
McMahonAP
1997 GDNF induces branching and increased cell proliferation in the ureter of the mouse. Dev Biol 192 193 198
13. MichaelL
DaviesJA
2004 Pattern and regulation of cell proliferation during murine ureteric bud development. J Anat 204 241 255
14. TangMJ
WorleyD
SanicolaM
DresslerGR
1998 The RET-glial cell-derived neurotrophic factor (GDNF) pathway stimulates migration and chemoattraction of epithelial cells. J Cell Biol 142 1337 1345
15. TangMJ
CaiY
TsaiSJ
WangYK
DresslerGR
2002 Ureteric bud outgrowth in response to RET activation is mediated by phosphatidylinositol 3-kinase. Dev Biol 243 128 136
16. SariolaH
SaarmaM
2003 Novel functions and signalling pathways for GDNF. J Cell Sci 116 3855 3862
17. BassonMA
AkbulutS
Watson-JohnsonJ
SimonR
CarrollTJ
2005 Sprouty1 is a critical regulator of GDNF/RET-mediated kidney induction. Dev Cell 8 229 239
18. BassonMA
Watson-JohnsonJ
ShakyaR
AkbulutS
HyinkD
2006 Branching morphogenesis of the ureteric epithelium during kidney development is coordinated by the opposing functions of GDNF and Sprouty1. Dev Biol 299 466 477
19. MasonJM
MorrisonDJ
BassonMA
LichtJD
2006 Sprouty proteins: multifaceted negative-feedback regulators of receptor tyrosine kinase signaling. Trends Cell Biol 16 45 54
20. AyadaT
TaniguchiK
OkamotoF
KatoR
KomuneS
2009 Sprouty4 negatively regulates protein kinase C activation by inhibiting phosphatidylinositol 4,5-biphosphate hydrolysis. Oncogene 28 1076 1088
21. MaeshimaA
SakuraiH
ChoiY
KitamuraS
VaughnDA
2007 Glial cell-derived neurotrophic factor independent ureteric bud outgrowth from the Wolffian duct. J Am Soc Nephrol 18 3147 3155
22. SchuchardtA
D'AgatiV
PachnisV
CostantiniF
1996 Renal agenesis and hypodysplasia in ret-k - mutant mice result from defects in ureteric bud development. Development 122 1919 1929
23. MackieGG
StephensFD
1975 Duplex kidneys: a correlation of renal dysplasia with position of the ureteral orifice. J Urol 114 274 280
24. IchikawaI
KuwayamaF
PopeJCt
StephensFD
MiyazakiY
2002 Paradigm shift from classic anatomic theories to contemporary cell biological views of CAKUT. Kidney Int 61 889 898
25. DaviesJA
MillarCB
JohnsonEMJr.
MilbrandtJ
1999 Neurturin: an autocrine regulator of renal collecting duct development. Dev Genet 24 284 292
26. AiraksinenMS
TitievskyA
SaarmaM
1999 GDNF family neurotrophic factor signaling: four masters, one servant? Mol Cell Neurosci 13 313 325
27. ChiX
HadjantonakisAK
WuZ
HyinkD
CostantiniF
2009 A transgenic mouse that reveals cell shape and arrangement during ureteric bud branching. Genesis 47 61 66
28. Schmidt-OttKM
YangJ
ChenX
WangH
ParagasN
2005 Novel regulators of kidney development from the tips of the ureteric bud. J Am Soc Nephrol 16 1993 2002
29. CaruanaG
Cullen-McEwenL
NelsonAL
KostouliasX
WoodsK
2006 Spatial gene expression in the T-stage mouse metanephros. Gene Expr Patterns 6 807 825
30. de GraaffE
SrinivasS
KilkennyC
D'AgatiV
MankooBS
2001 Differential activities of the RET tyrosine kinase receptor isoforms during mammalian embryogenesis. Genes Dev 15 2433 2444
31. YuJ
CarrollTJ
RajagopalJ
KobayashiA
RenQ
2009 A Wnt7b-dependent pathway regulates the orientation of epithelial cell division and establishes the cortico-medullary axis of the mammalian kidney. Development 136 161 171
32. QiaoJ
BushKT
SteerDL
StuartRO
SakuraiH
2001 Multiple fibroblast growth factors support growth of the ureteric bud but have different effects on branching morphogenesis. Mech Dev 109 123 135
33. OhuchiH
HoriY
YamasakiM
HaradaH
SekineK
2000 FGF10 acts as a major ligand for FGF receptor 2 IIIb in mouse multi-organ development. Biochem Biophys Res Commun 277 643 649
34. QiaoJ
UzzoR
Obara-IshiharaT
DegensteinL
FuchsE
1999 FGF-7 modulates ureteric bud growth and nephron number in the developing kidney. Development 126 547 554
35. MinH
DanilenkoDM
ScullySA
BolonB
RingBD
1998 Fgf-10 is required for both limb and lung development and exhibits striking functional similarity to Drosophila branchless. Genes Dev 12 3156 3161
36. ZhaoH
KeggH
GradyS
TruongHT
RobinsonML
2004 Role of fibroblast growth factor receptors 1 and 2 in the ureteric bud. Dev Biol 276 403 415
37. BatourinaE
ChoiC
ParagasN
BelloN
HensleT
2002 Distal ureter morphogenesis depends on epithelial cell remodeling mediated by vitamin A and Ret. Nat Genet 32 109 115
38. GrieshammerU
LeM
PlumpAS
WangF
Tessier-LavigneM
2004 SLIT2-mediated ROBO2 signaling restricts kidney induction to a single site. Dev Cell 6 709 717
39. KumeT
DengK
HoganBL
2000 Murine forkhead/winged helix genes Foxc1 (Mf1) and Foxc2 (Mfh1) are required for the early organogenesis of the kidney and urinary tract. Development 127 1387 1395
40. ShakyaR
JhoEH
KotkaP
WuZ
KholodilovN
2005 The role of GDNF in patterning the excretory system. Dev Biol 283 70 84
41. MichosO
PanmanL
VinterstenK
BeierK
ZellerR
2004 Gremlin-mediated BMP antagonism induces the epithelial-mesenchymal feedback signaling controlling metanephric kidney and limb organogenesis. Development 131 3401 3410
42. MichosO
GoncalvesA
Lopez-RiosJ
TieckeE
NaillatF
2007 Reduction of BMP4 activity by gremlin 1 enables ureteric bud outgrowth and GDNF/WNT11 feedback signalling during kidney branching morphogenesis. Development 134 2397 2405
43. IshibeS
KarihalooA
MaH
ZhangJ
MarlierA
2009 Met and the epidermal growth factor receptor act cooperatively to regulate final nephron number and maintain collecting duct morphology. Development 136 337 345
44. RozenEJ
SchmidtH
DolcetX
BassonMA
JainS
2009 Loss of Sprouty1 rescues renal agenesis caused by Ret mutation. J Am Soc Nephrol 20 255 259
45. JainS
EncinasM
JohnsonEMJr.
MilbrandtJ
2006 Critical and distinct roles for key RET tyrosine docking sites in renal development. Genes Dev 20 321 333
46. TakahashiM
2001 The GDNF/RET signaling pathway and human diseases. Cytokine Growth Factor Rev 12 361 373
47. BellusciS
GrindleyJ
EmotoH
ItohN
HoganBL
1997 Fibroblast growth factor 10 (FGF10) and branching morphogenesis in the embryonic mouse lung. Development 124 4867 4878
48. WeaverM
DunnNR
HoganBL
2000 Bmp4 and Fgf10 play opposing roles during lung bud morphogenesis. Development 127 2695 2704
49. LivetJ
SigristM
StroebelS
De PaolaV
PriceSR
2002 ETS gene Pea3 controls the central position and terminal arborization of specific motor neuron pools. Neuron 35 877 892
50. ChenC
OuyangW
GriguraV
ZhouQ
CarnesK
2005 ERM is required for transcriptional control of the spermatogonial stem cell niche. Nature 436 1030 1034
51. ZunigaA
MichosO
SpitzF
HaramisAP
PanmanL
2004 Mouse limb deformity mutations disrupt a global control region within the large regulatory landscape required for Gremlin expression. Genes Dev 18 1553 1564
Štítky
Genetika Reprodukční medicína
Článek Activation of Mutant Enzyme Function by Proteasome Inhibitors and Treatments that Induce Hsp70Článek Maternal Ethanol Consumption Alters the Epigenotype and the Phenotype of Offspring in a Mouse ModelČlánek Genetic Dissection of Differential Signaling Threshold Requirements for the Wnt/β-Catenin PathwayČlánek Distinct Type of Transmission Barrier Revealed by Study of Multiple Prion Determinants of Rnq1
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2010 Číslo 1
-
Všechny články tohoto čísla
- Irradiation-Induced Genome Fragmentation Triggers Transposition of a Single Resident Insertion Sequence
- A Major Role of the RecFOR Pathway in DNA Double-Strand-Break Repair through ESDSA in
- Kidney Development in the Absence of and Requires
- Modeling of Environmental Effects in Genome-Wide Association Studies Identifies and as Novel Loci Influencing Serum Cholesterol Levels
- Inverse Correlation between Promoter Strength and Excision Activity in Class 1 Integrons
- Activation of Mutant Enzyme Function by Proteasome Inhibitors and Treatments that Induce Hsp70
- Postnatal Survival of Mice with Maternal Duplication of Distal Chromosome 7 Induced by a / Imprinting Control Region Lacking Insulator Function
- The Werner Syndrome Protein Functions Upstream of ATR and ATM in Response to DNA Replication Inhibition and Double-Strand DNA Breaks
- Maternal Ethanol Consumption Alters the Epigenotype and the Phenotype of Offspring in a Mouse Model
- Understanding Gene Sequence Variation in the Context of Transcription Regulation in Yeast
- miR-30 Regulates Mitochondrial Fission through Targeting p53 and the Dynamin-Related Protein-1 Pathway
- Elevated Levels of the Polo Kinase Cdc5 Override the Mec1/ATR Checkpoint in Budding Yeast by Acting at Different Steps of the Signaling Pathway
- Alternative Epigenetic Chromatin States of Polycomb Target Genes
- Co-Orientation of Replication and Transcription Preserves Genome Integrity
- A Comprehensive Map of Insulator Elements for the Genome
- Environmental and Genetic Determinants of Colony Morphology in Yeast
- U87MG Decoded: The Genomic Sequence of a Cytogenetically Aberrant Human Cancer Cell Line
- The MCM-Binding Protein ETG1 Aids Sister Chromatid Cohesion Required for Postreplicative Homologous Recombination Repair
- Genetic Dissection of Differential Signaling Threshold Requirements for the Wnt/β-Catenin Pathway
- Differential Localization and Independent Acquisition of the H3K9me2 and H3K9me3 Chromatin Modifications in the Adult Germ Line
- Genetic Crossovers Are Predicted Accurately by the Computed Human Recombination Map
- Collaborative Action of Brca1 and CtIP in Elimination of Covalent Modifications from Double-Strand Breaks to Facilitate Subsequent Break Repair
- Distinct Type of Transmission Barrier Revealed by Study of Multiple Prion Determinants of Rnq1
- Genome-Wide Association Study Identifies as a Novel Susceptibility Gene for Osteoporosis
- and Regulate Reproductive Habit in Rice
- Nonsense-Mediated Decay Enables Intron Gain in
- Altered Gene Expression and DNA Damage in Peripheral Blood Cells from Friedreich's Ataxia Patients: Cellular Model of Pathology
- The Systemic Imprint of Growth and Its Uses in Ecological (Meta)Genomics
- The Gift of Observation: An Interview with Mary Lyon
- Genotype and Gene Expression Associations with Immune Function in
- The Elongator Complex Regulates Neuronal α-tubulin Acetylation
- Rising from the Ashes: DNA Repair in
- Mis-Spliced Transcripts of Nicotinic Acetylcholine Receptor α6 Are Associated with Field Evolved Spinosad Resistance in (L.)
- BRIT1/MCPH1 Is Essential for Mitotic and Meiotic Recombination DNA Repair and Maintaining Genomic Stability in Mice
- Non-Coding Changes Cause Sex-Specific Wing Size Differences between Closely Related Species of
- Evidence for Pervasive Adaptive Protein Evolution in Wild Mice
- Evolutionary Mirages: Selection on Binding Site Composition Creates the Illusion of Conserved Grammars in Enhancers
- VEZF1 Elements Mediate Protection from DNA Methylation
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- A Major Role of the RecFOR Pathway in DNA Double-Strand-Break Repair through ESDSA in
- Kidney Development in the Absence of and Requires
- The Werner Syndrome Protein Functions Upstream of ATR and ATM in Response to DNA Replication Inhibition and Double-Strand DNA Breaks
- Alternative Epigenetic Chromatin States of Polycomb Target Genes
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



