-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaLeaving the Past Behind
article has not abstract
Published in the journal: . PLoS Genet 4(10): e32767. doi:10.1371/journal.pgen.1000248
Category: Perspective
doi: https://doi.org/10.1371/journal.pgen.1000248Summary
article has not abstract
There is considerable interest from the wider scientific community in the heritability of epigenetic states across generations, and this has arisen as a result of a series of studies in mice [1],[2], flies [3], plants [4],[5], and yeast [6] over the past decade. These studies have identified genetic elements at which epigenetic states appear to be inherited through meiosis. The Lamarckian implications of these findings are hard to avoid. Transgenes, transposons, and other “foreign DNA” appear to be particularly prone to transgenerational epigenetic inheritance (reviewed in [7]). In this issue of PLoS Genetics, Singh et al. [8] describe the identification of a locus in the genome of maize at which a transposon, silenced by an RNAi-based mechanism, becomes reactivated over subsequent generations. This article reports an activating “position effect,” i.e., an integration site that is associated with the reversal of a previously established silent state in plants.
The authors have devised a clever system for studying position effects that involves a single transposon, MuDR, and a variant of MuDR, called Mu killer (Muk) [8]. The integration site of the MuDR can be altered by transposition. When MuDR and Muk are both present in one plant, the MuDR elements become epigenetically silenced as a result of a long hairpin RNA molecule produced from Muk that acts in trans to initiate DNA methylation of MuDR elements (Figure 1). Once the MuDR has been silenced, it generally remains so even after Muk segregates away in subsequent generations (Figure 1A). This is consistent with observations made by others studying the activity of endogenous genes or transgenes that have been silenced by RNA-directed mechanisms in plants [5],[9],[10] and with transgenes in mice [11]. However, at one particular integration site, they found that the opposite was true. Following the loss of Muk, the MuDR element reactivated, an event associated with loss of DNA methylation (Figure 1B). The integration site in this case turns out to be the 5′ untranslated region (UTR) of a gene of unknown function, designated Hemera [8].
Fig. 1. Locus-Specific Reactivation of the MuDR Transposon. 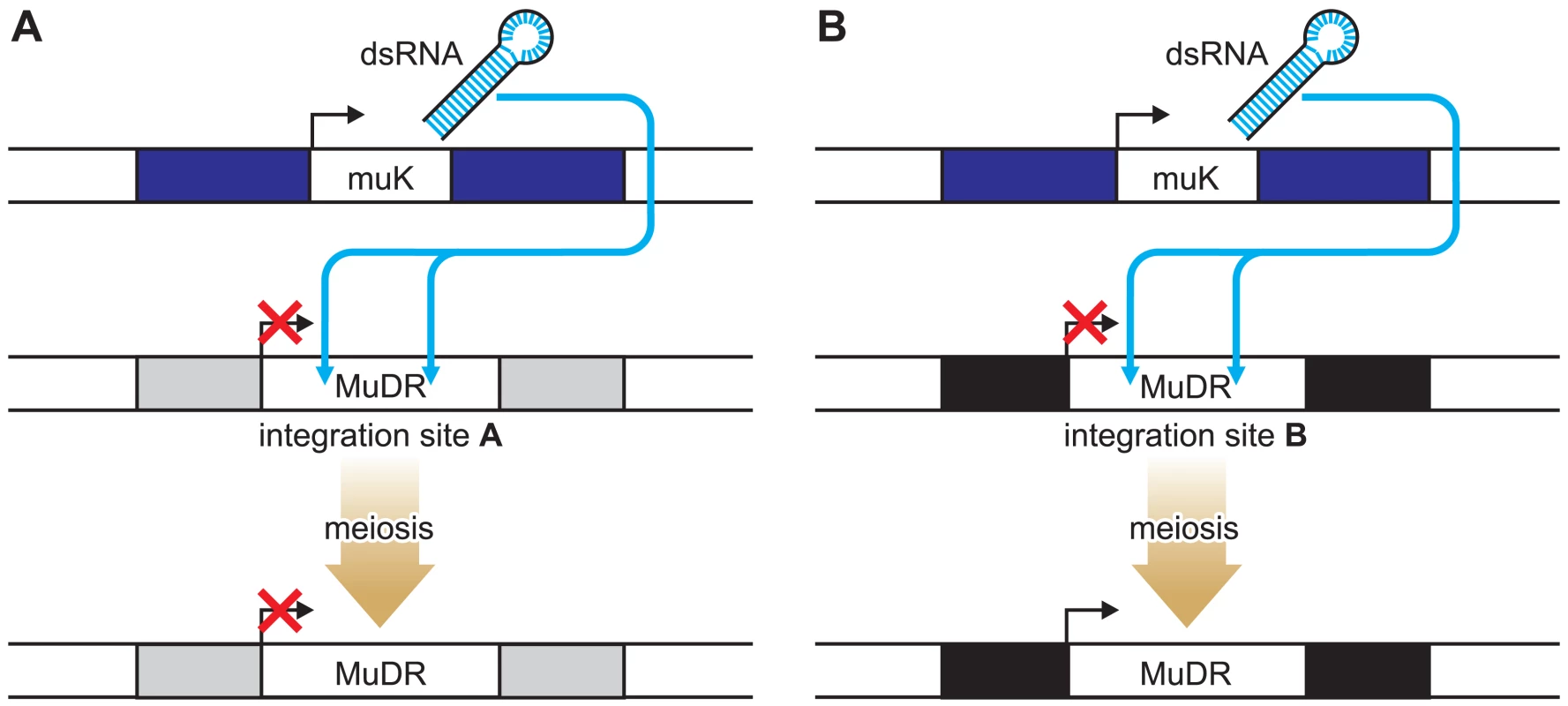
When MuDR and Muk are both present in one plant, the MuDR elements become epigenetically silenced as a result of a long hairpin RNA molecule produced from Muk that acts in trans to initiate DNA methylation of MuDR elements. At most loci, once the MuDR has been silenced it remains so even after Muk segregates away (A). In contrast (B), when inserted within the Hemera (black bar) locus, MuDR was reactivated in progeny plants that did not inherit MuK. Plant transposons frequently insert near or within transcribed genes, so what is special about this case? It is not known whether insertion of the transposon blocks Hemera activity, but if it does, then a trivial explanation for the reactivation of MuDR is that Hemera plays a role in maintaining silencing of targets of the RNA-directed DNA methylation pathway. A more likely and more interesting scenario is that reprogramming of Hemera during gamete formation or during the early stages of development of the subsequent embryo is associated with reactivation of the MuDR inserted within the 5′ UTR of this gene. The authors note that MuDR has inserted adjacent to a GA-rich sequence and suggest that this may be important for the reprogramming of both MuDR and Hemera during their passage to the next generation. This hypothesis could be readily tested using transgenic approaches to alter the sequences that flank MuDR in Hemera.
To plant epigeneticists, who focus mainly on transposons and transgenes, the reactivation of a silenced MuDR is a surprise. But to mammalian epigeneticists, it is not. In mice, for example, it is widely accepted that cis-acting sequences, e.g., promoters, are reprogrammed each generation so that the cells of the preimplantation embryo can acquire pluripotency. Indeed, for the mammalian epigeneticist, transgenerational epigenetic inheritance is the exception rather than the rule. Even the described cases of transgenerational epigenetic inheritance in mammals actually display considerable reprogramming of epigenetic state from generation to generation. The agouti viable yellow allele and the axin-fused allele are two well-characterised examples [1],[12]. It seems likely that there is epigenetic reprogramming of endogenous plant genes to ensure that the normal program of plant development is reiterated each generation (Figure 2), no matter what conditions the parental plant experienced. Indeed, it has recently been shown that the vernalization-induced epigenetic repression of the Arabidopsis FLC gene is reversed during pollen development or, when inherited through the maternal gamete, in the globular embryo [13].
Fig. 2. Sites of Potential Epigenetic Reprogramming during Maize Reproduction. 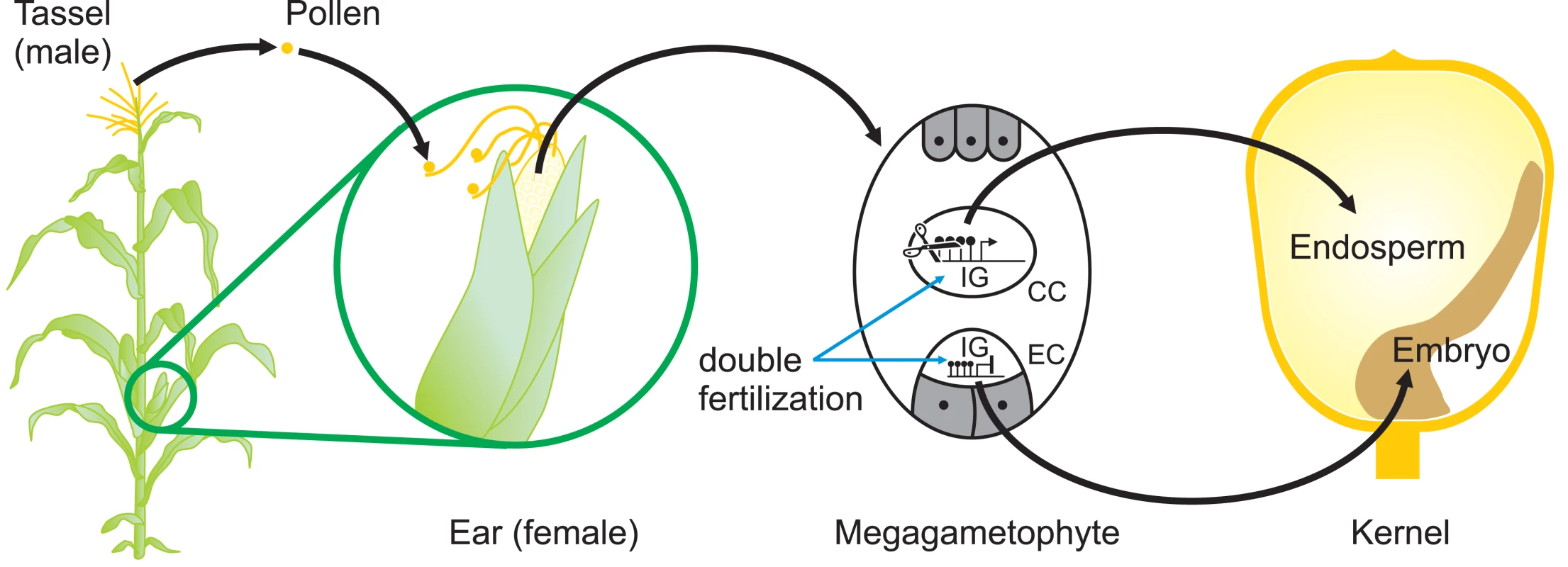
The reproductive organs, the ear, and the tassel of a maize plant arise when vegetative meristems differentiate to become inflorescence meristems. Pollen, formed in the tassel, falls onto the silks where it germinates. A pollen tube, containing two identical haploid sperm nuclei, grows down the silk until it reaches the megagametophyte containing the haploid egg cell (EC) and the diploid central cell (CC). One sperm nucleus fuses with the EC and the other fuses with the CC (double fertilization), giving rise to the zygote (diploid) and endosperm (triploid), which provides nutrients to the developing embryo. Epigenetic reprogramming that removes methylcytosine from the control regions of imprinted genes occurs in the CC but not in the EC, leading to differential expression of these genes in endosperm [16]. It is likely that other, as-yet uncharacterised, epigenetic reprogramming events occur during pollen or egg cell formation as well as during early stages of embryo or endosperm development. So what does this new finding tell us? It reaffirms the idea that the molecular mechanisms involved in the permanent silencing of foreign DNA have evolved from the mechanisms required for the successful development of an embryo. Consistent with this idea, random mutagenesis screens for modifiers of position effect variegation carried out in both Drosophila [14] and mouse [15] have found that most genes identified play critical roles in development. It has been difficult for plant biologists to study the developing embryo, because it is surrounded by developing endosperm and is embedded in the somatic tissue of the parent plant. In contrast preimplantation mouse embryos develop as unattached entities that can be flushed out of the uterus. As plant biologists acquire better methods of studying the zygote as it develops, they are likely to find more genetic elements of this type. For development to work at all, the genomes of multicellular organisms must leave the past behind.
Zdroje
1. MorganHD
SutherlandHG
MartinDI
WhitelawE
1999 Epigenetic inheritance at the agouti locus in the mouse. Nat Genet 23 314 318
2. RoemerI
ReikW
DeanW
KloseJ
1997 Epigenetic inheritance in the mouse. Curr Biol 7 277 280
3. CavalliG
ParoR
1998 The Drosophila Fab-7 chromosomal element conveys epigenetic inheritance during mitosis and meiosis. Cell 93 505 518
4. CubasP
VincentC
CoenE
1999 An epigenetic mutation responsible for natural variation in floral symmetry. Nature 401 157 161
5. AllemanM
SidorenkoL
McGinnisK
SeshadriV
DorweilerJE
2006 An RNA-dependent RNA polymerase is required for paramutation in maize. Nature 442 295 298
6. GrewalSI
KlarAJ
1996 Chromosomal inheritance of epigenetic states in fission yeast during mitosis and meiosis. Cell 86 95 101
7. ChongS
WhitelawE
2004 Epigenetic germline inheritance. Curr Opin Genet Dev 14 692 696
8. SinghJ
FreelingM
LischD
2008 A position effect on the heritability of epigenetic silencing. PLoS Genet 4(10) e1000216 doi:10.1371/journal.pgen.1000216
9. ChawlaR
NicholsonSJ
FoltaKM
SrivastavaV
2007 Transgene-induced silencing of Arabidopsis phytochrome A gene via exonic methylation. Plant J 52 et al.1105 1118
10. HuettelB
KannoT
DaxingerL
BucherE
van der WindenJ
2007 RNA-directed DNA methylation mediated by DRD1 and Pol IVb: a versatile pathway for transcriptional gene silencing in plants. Biochim Biophys Acta 1769 358 374
11. HadchouelM
FarzaH
SimonD
TiollaisP
PourcelC
1987 Maternal inhibition of hepatitis B surface antigen gene expression in transgenic mice correlates with de novo methylation. Nature 329 454 456
12. RakyanVK
ChongS
ChampME
CuthbertPC
MorganHD
2003 Transgenerational inheritance of epigenetic states at the murine Axin(Fu) allele occurs after maternal and paternal transmission. Proc Natl Acad Sci U S A 100 2538 2543
13. SheldonCC
HillsMJ
ListerC
DeanC
DennisES
2008 Resetting of FLOWERING LOCUS C expression after epigenetic repression by vernalization. Proc Natl Acad Sci U S A 105 2214 2219
14. SchottaG
EbertA
DornR
ReuterG
2003 Position-effect variegation and the genetic dissection of chromatin regulation in Drosophila. Semin Cell Dev Biol 14 67 75
15. BlewittME
VickaryousNK
HemleySJ
AsheA
BruxnerTJ
2005 An N-ethyl-N-nitrosourea screen for genes involved in variegation in the mouse. Proc Natl Acad Sci U S A 102 7629 7634
16. HermonP
SrilunchangK
ZouJ
DresselhausT
DanilevskayaON
2007 Activation of the imprinted polycomb group gene Fie1 gene in maize endosperm requires demethylation of the maternal allele. Plant Mol Biol 64 387 395
Štítky
Genetika Reprodukční medicína
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2008 Číslo 10
Nejčtenější v tomto čísle- Mutation and Evolutionary Rates in Adélie Penguins from the Antarctic
- Leaving the Past Behind
- Genetical Genomics: Spotlight on QTL Hotspots
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



