-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaA Six-Gene Signature Predicts Survival of Patients with Localized Pancreatic Ductal Adenocarcinoma
Background:
Pancreatic ductal adenocarcinoma (PDAC) remains a lethal disease. For patients with localized PDAC, surgery is the best option, but with a median survival of less than 2 years and a difficult and prolonged postoperative course for most, there is an urgent need to better identify patients who have the most aggressive disease.Methods and Findings:
We analyzed the gene expression profiles of primary tumors from patients with localized compared to metastatic disease and identified a six-gene signature associated with metastatic disease. We evaluated the prognostic potential of this signature in a training set of 34 patients with localized and resected PDAC and selected a cut-point associated with outcome using X-tile. We then applied this cut-point to an independent test set of 67 patients with localized and resected PDAC and found that our signature was independently predictive of survival and superior to established clinical prognostic factors such as grade, tumor size, and nodal status, with a hazard ratio of 4.1 (95% confidence interval [CI] 1.7–10.0). Patients defined to be high-risk patients by the six-gene signature had a 1-year survival rate of 55% compared to 91% in the low-risk group.Conclusions:
Our six-gene signature may be used to better stage PDAC patients and assist in the difficult treatment decisions of surgery and to select patients whose tumor biology may benefit most from neoadjuvant therapy. The use of this six-gene signature should be investigated in prospective patient cohorts, and if confirmed, in future PDAC clinical trials, its potential as a biomarker should be investigated. Genes in this signature, or the pathways that they fall into, may represent new therapeutic targets.
: Please see later in the article for the Editors' Summary
Published in the journal: . PLoS Med 7(7): e32767. doi:10.1371/journal.pmed.1000307
Category: Research Article
doi: https://doi.org/10.1371/journal.pmed.1000307Summary
Background:
Pancreatic ductal adenocarcinoma (PDAC) remains a lethal disease. For patients with localized PDAC, surgery is the best option, but with a median survival of less than 2 years and a difficult and prolonged postoperative course for most, there is an urgent need to better identify patients who have the most aggressive disease.Methods and Findings:
We analyzed the gene expression profiles of primary tumors from patients with localized compared to metastatic disease and identified a six-gene signature associated with metastatic disease. We evaluated the prognostic potential of this signature in a training set of 34 patients with localized and resected PDAC and selected a cut-point associated with outcome using X-tile. We then applied this cut-point to an independent test set of 67 patients with localized and resected PDAC and found that our signature was independently predictive of survival and superior to established clinical prognostic factors such as grade, tumor size, and nodal status, with a hazard ratio of 4.1 (95% confidence interval [CI] 1.7–10.0). Patients defined to be high-risk patients by the six-gene signature had a 1-year survival rate of 55% compared to 91% in the low-risk group.Conclusions:
Our six-gene signature may be used to better stage PDAC patients and assist in the difficult treatment decisions of surgery and to select patients whose tumor biology may benefit most from neoadjuvant therapy. The use of this six-gene signature should be investigated in prospective patient cohorts, and if confirmed, in future PDAC clinical trials, its potential as a biomarker should be investigated. Genes in this signature, or the pathways that they fall into, may represent new therapeutic targets.
: Please see later in the article for the Editors' SummaryIntroduction
Pancreatic ductal adenocarcinoma (PDAC), comprising over 90% of all pancreatic cancers, remains a lethal disease with an estimated 232,000 new cases, 227,000 deaths per year worldwide, and a less than 5% 5-y survival rate [1],[2]. Currently the standard of care for the 20% of patients with localized disease is surgery followed by chemotherapy, and in some cases radiation. Unfortunately, despite the use of adjuvant therapy, median survival remains at best 23 mo [3]. It is important to note, however, that up to 27% of patients with resected PDAC can survive for 5 y [4]–[10]. However, in studies examining actual long-term survivors [4]–[10], only two have found that adjuvant therapy was associated with improved survival [9],[10]. In addition, randomized controlled trials of gemcitabine-based chemotherapy demonstrate an improvement in median survival of at best 3 mo [3],[11]. One possible conclusion from these studies is that tumor biology dictates outcome and that our current adjuvant therapy has only a modest impact on altering a patient's course.
Hypothesizing that the dismal outcome of patients with localized disease is due to the presence of micrometastatic disease, current clinical investigation has focused on preoperative or neoadjuvant therapy [12],[13]. This approach, in which patients who cannot tolerate the stress of therapy or who develop metastatic disease during treatment are spared surgery, has demonstrated an overall survival of 34 mo in this highly selected patient population [12],[13]. Therefore the ability to select patients who would most benefit from a neoadjuvant approach may be important. One way to select these individuals is to define a prognostic gene signature that can identify patients with more aggressive tumor biology upfront.
Expression profiling of PDAC has lead to further studies of additional molecular diagnostic and prognostic markers [14]–[19]. However, the search for genes of biological significance in these large datasets continues to be challenging. One approach to identify genes or pathways that are biologically relevant is to study those that are of prognostic significance [20]. Lowe and colleagues found differential gene expression changes associated with nodal status in primary PDAC [21], suggesting that molecular differences in primary PDAC do exist. We hypothesized that by comparing primary PDAC tumors at the extremes of disease, we would identify molecular changes reflective of differences in biology within primary PDAC tumors.
Methods
Patients
PDAC samples from 15 patients with resected primary PDAC from the University of North Carolina at Chapel Hill (UNC) and 15 patients with metastatic PDAC from the University of Nebraska Medical Center Rapid Autopsy Pancreatic Program (NEB) were used to derive differentially expressed genes associated with metastatic disease. For the NEB samples, human pancreatic tumors from decedents who had previously been diagnosed with PDAC, and who generously consented to post mortem examinations, were obtained from the institutional review board (IRB)-approved NEB Tissue Bank. To ensure minimal degradation of tissue, organs were harvested within 3 h post mortem and the specimens flash frozen in liquid nitrogen.
The training cohort included 34 patients with resected PDAC from Johns Hopkins Medical Institutions (JHMI). The testing or validation cohort included patients from two institutions: 48 from Northwestern Memorial Hospital (NW) and 19 from NorthShore University HealthSystem (NSU). All samples were collected between 1999 and 2007 at the time of operation and flash frozen in liquid nitrogen after approval by each individual IRB. The UNC IRB approved use of all de-identified samples for this study. All available samples were reviewed by a single pathologist (KAV). De-identified data including tumor, node, and metastasis (TNM), grade or differentiation, margin status, and survival were available for the majority of patients.
RNA Isolation and Microarray Hybridization
All RNA isolation and hybridization was performed on Agilent (Agilent Technologies) human whole genome 4×44 K DNA microarrays and at UNC. RNA was extracted from macrodissected snap-frozen tumor samples using Allprep Kits (Qiagen) and quantified using nanodrop spectrophotometry (ThermoScientific). RNA quality was assessed with the use of the Bioanalyzer 2100 (Agilent Technologies). RNA was selected for hybridization using RNA integrity number and by inspection of the 18S and 28S ribosomal RNA. Similar RNA quality was selected across samples. One microgram of RNA was used as a template for DNA preparations and hybridized to Agilent 4×44 K whole human genome arrays (Agilent Technologies). cDNA was labeled with Cy5-dUTP and a reference control (Stratagene) was labeled with Cy3-dUTP using the Agilent (Agilent Technologies) low RNA input linear amplification kit and hybridized overnight at 65°C to Agilent 4×44 K whole human genome arrays (Agilent Technologies). Arrays were washed and scanned using an Agilent scanner (Agilent Technologies). The data are publicly available in Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE21501).
Microarray and Statistical Analysis
All array data were normalized using Lowess normalization. Data were excluded for genes with poor spot quality or genes that did not have mean intensity greater than 10 for one of the two channels (green and red) in at least 70% of the experiments. The log2 ratio of the mean red intensity over mean green intensity was calculated for each gene and went through LOWESS normalization [22]. Missing data were imputed using the k-nearest neighbors imputation (KNN) with k = 10 [23]. A distance weighted discrimination (DWD) was used to detect the systematic biases between the different datasets and then global adjustments made to remove these biases [24]. Genes that were significantly up - or down-regulated were identified using significance analysis of microarrays (SAM) [25]. Two centroids were created using the mean gene expression profile of this significant gene list from the derivation set and used to develop a single sample predictor (SSP, nearest centroid algorithm) [26] for an objective classifier. After DWD, the SSP was applied to a 34-patient training set where any new sample was compared to the resected centroid and assigned by the SSP distance function to the resected centroid using (1 − Pearson correlation coefficient). The X-Tile software program, which assigns a two-population log-rank value to each sample and then determines the best cut-point, was used to determine the best threshold for classifying samples into high - and low-risk categories [27]. X-Tile predicted that the (1 − Pearson correlation coefficient) distance of 1 would be the appropriate cut-point to stratify patients into a high - and low-risk group (p = 0.006). A second independent validation cohort was then used as a test set using this predetermined cut-point to evaluate outcome.
Survival analysis was performed using the statistical software programs R, the R-package “survival,” and SPSS (SPSS, Inc.). Overall survival (OS) was analyzed using the Kaplan-Meier product-limit method and the significance of our variables was measured by the log-rank test. The Fisher exact test was used to analyze associations between two variables, the Pearson Chi-square test was used to analyze association between more than two variables. Multivariable analysis and analysis of continuous and ordinal variables was performed using the Cox proportional hazards regression method.
Tissue Microarrays
Tissue microarrays (TMAs; UNC2) were prepared from formalin-fixed paraffin-embedded tissue sections using a 2-mm punch. The arrays contained triplicate cores of matched normal and tumor tissue as well as chronic pancreatitis when available, from each patient. We prepared 5-µm sections from each TMA block. Hematoxylin and eosin stained slides from each TMA block were reviewed by a pathologist (KAV) to ensure that tissues were cored accurately.
Immunohistochemistry
Slides with 5-µM sections from the paraffin-embedded specimens were deparaffinized and rehydrated. The slides were then subjected to alkaline heat antigen-retrieval using 1% Tris EDTA for 20 min in a steamer. All slides were incubated with 3% H2O2 for 5 min and washed with TBS. The slides were further treated with protein block solution (bovine serum albumin) for 20 min. The sections were incubated with primary KLF6 1∶150 antibody (sc-7158, Santa Cruz Biotechnology) for 60 min at room temperature. Following a TBS wash, the slides were incubated with secondary labeled Polymer-HRP anti-rabbit (Dako K4002) for 30 min. This was followed by a 5-min incubation with the substrate-chromogen, 3,3′-diaminobenzidine (Vector SK-4100). The sections were counterstained with Harris hematoxylin. Positive KLF6 staining was defined as when more than 5% of cells expressed the marker and graded from 0 (no staining) to 4 (strong staining). The results of each protein marker were then expressed as intensity (I) and proportion (P) of positive epithelial cells and the score as the product of I and P [28],[29]. All stained slides were reviewed in a blinded fashion (JMA).
Results
Patient and Tumor Characteristics
In order to study the extremes of PDAC tumor biology, we collected a diverse set of resected PDAC specimens from patients with and without metastases. As the tumor microenvironment is increasingly recognized to play a critical role in tumorigenesis [30]–[33], tissues were macrodissected in order to preserve the normal adjacent tissue and stroma of the tumors. The characteristics of the dataset used to derive the signature (derivation set) comprised 15 primary resected PDAC tumors (UNC1) and 15 primary tumors from patients with metastatic PDAC (NEB). The training set comprised 34 patients with primary PDAC and the independent validation test set comprised 67 patients with primary PDAC (Tables 1 and 2). There were no differences in RNA quality between the decedent and resected PDAC samples. Available treatment data of the patients in the training and test sets are also shown. One of 15 (7%) UNC1 patients received preoperative or neoadjuvant chemotherapy and 11/15 (73%) NEB patients received chemotherapy less than 6 mo prior to death. No patient in the 34-patient training set received neoadjuvant chemotherapy. Only 3% (2/67) of patients in the test set received neoadjuvant chemotherapy and 45% (30/67) of patients received postoperative or adjuvant chemotherapy.
Tab. 1. Patient, tumor, and treatment characteristics in the derivation set. 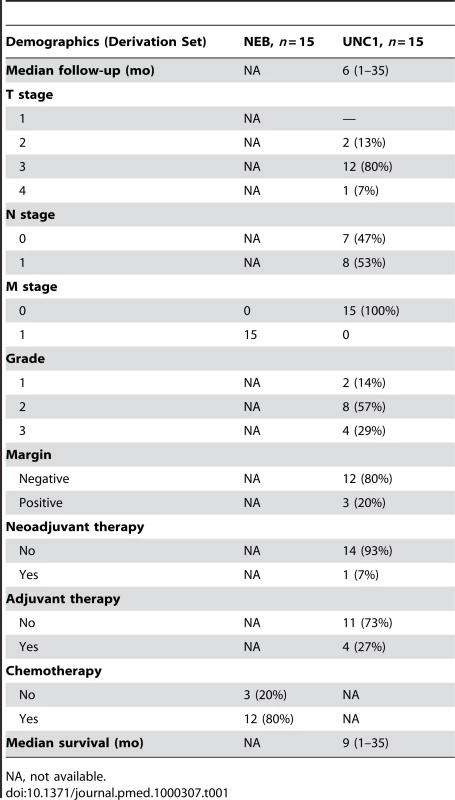
NA, not available. Tab. 2. Patient, tumor, and treatment characteristics in the training and testing sets. 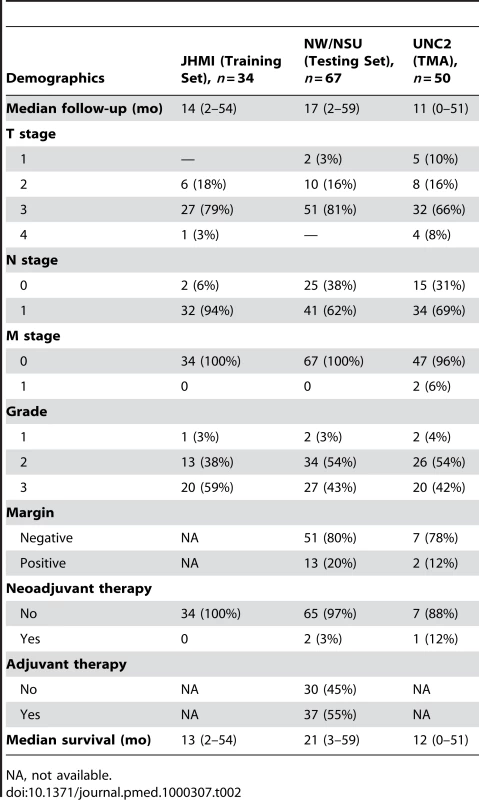
NA, not available. Gene Expression Differences in Nonmetastatic And Metastatic Primary Tumors
We hypothesized that we could enrich for molecular differences in primary PDAC, which may be clinically and biologically relevant, through examining primary tumors representing opposite spectrums of PDAC: early (localized) and late (metastatic) stage. To accomplish this, we compared nonmetastatic (UNC1) with metastatic (NEB) primary PDAC tumors. As the methods of procurement for these tumors differed, we used DWD to identify systematic biases between the two datasets [24]. This method has been used previously to successfully combine three breast cancer datasets across three microarray platforms [26], across species [34], and across multiple datasets [35],[36]. We therefore used DWD to adjust for the systematic biases between the UNC1 and NEB datasets by taking advantage of the fact that each dataset also had 15 normal pancreas samples assayed. In short, we used DWD to adjust these 15 tumor-normal pairs from both datasets to have similar distributions in principal component (PC) 1×PC 2 space. After the DWD adjustment, we used SAM to identify differentially expressed genes [22],[25]. Using a false discovery rate of 5%, we identified six genes that were differentially overexpressed between nonmetastatic and metastatic primary tumors: FBJ murine osteosarcoma viral oncogene homolog B (Fos B), Kruppel-like factor 6 (KLF6), nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta (NFKBIZ, IKBZ, MAIL), ATPase H+/K+ exchanging, alpha polypeptide (ATP4A), germ cell associated 1 (GSG1), and sialic acid binding Ig-like lectin 11 (SIGLEC11) (Figure 1A; Table S1).
Fig. 1. Identification, development, and application of a six-gene signature for PDAC. 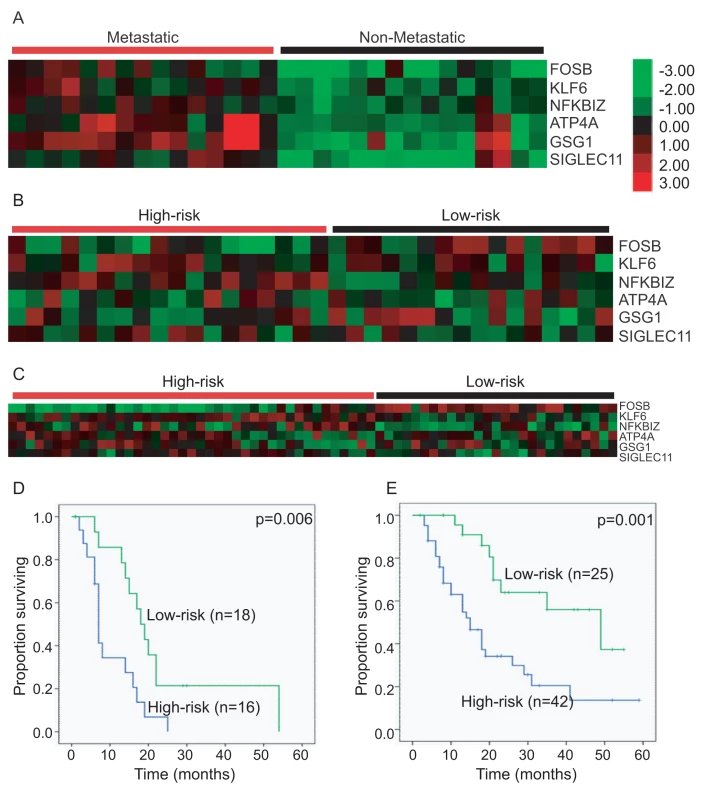
Clustering of (A) the six genes defined by SAM evaluation of the metastatic compared to nonmetastatic primary PDAC using a false discovery rate of 5%; (B) patient samples into high- and low-risk groups in a training set of 34 patients with localized and resected PDAC using the X-tile determined cut-point of a Pearson correlation coefficient of zero; (C) patient samples into high- and low-risk groups in an independent test set of 67 patients with localized and resected PDAC using the predetermined cut-point of zero. Kaplan-Meier overall survival of (D) the training set classified into high- and low-risk groups according to the X-tile determined cut-point of a Pearson correlation coefficient of zero; (E) and the independent test set classified into high- and low-risk groups according to the same predetermined cut-point. Development of a Classifier Using the Six-Gene Signature
We examined the relationship of our six-gene signature to outcome using a training set of 34 patients with localized and resected PDAC. After identifying and adjusting for systematic bias using DWD [24], a resected centroid-based predictor [26] was created using the 30 samples in the derivation dataset. The centroid was then applied to the DWD-adjusted training set of primary PDAC patients to determine the performance of the six-gene signature. X-tile [27] was used to determine the optimal distance function to the centroid cut-point for classifying this training set of patients into high-risk and low-risk groups on the basis of survival (Figure 1B and 1D). The optimal cut-point occurred at a Pearson correlation coefficient of zero (p = 0.006) with patients with Pearson correlation coefficients greater than zero in the low-risk and less than zero in the high-risk groups.
Application of the Six-Gene Signature to an Independent Validation Cohort of 67 Patients
In order to evaluate the performance of the cut-point determined by X-tile [27], we applied the cut-point to an independent validation test set of 67 patients with primary PDAC. Our predetermined Pearson correlation coefficient cut-point of zero distance to the centroid successfully stratified patients into high - (n = 42) and low-risk groups (n = 25) with a median overall survival (OS) of 15 versus 49 mo (p = 0.001) (Figure 1C and 1E). Patients in the high-risk group had 1-, 2-, and 3-y estimated survival rates of 55%, 34%, and 21%, compared to 91%, 64%, and 56% in the low-risk group.
Previous studies in PDAC have found that nodal status is the most predictive of outcome for patients with localized PDAC [37]. We compared our prognostic signature to current clinical prognostic benchmarks. We found that tumors that were node positive (p = 0.091) and grade 2 or 3 trended towards a shorter survival (p = 0.080). Neither T stage (p = 0.977) nor margin status (p = 0.223) were prognostic in this cohort. Treatment with adjuvant chemotherapy (p = 0.699) or with neoadjuvant chemotherapy (p = 0.409) was also not prognostic, although only two patients received neoadjuvant chemotherapy. We found no gene expression changes between the tumors of the two patients who received neoadjuvant chemotherapy and the tumors of patients who received no treatment prior to surgery.
An important feature of any prognostic signature is that it should be independent or additive to currently used clinicopathologic prognostic criteria. We therefore compared the prognostic importance of our molecular signature in the setting of grade (p = 0.417), nodal status (p = 0.381), T stage (p = 0.675), and margin status (p = 0.295). We found that our six-gene signature was the only independent predictor of survival in the 57 patients with complete data, with a hazard ratio of 4.1 (95% confidence interval 1.7–10.0) (Table 3).
Tab. 3. Cox proportional hazards regression analysis of the six-gene signature. 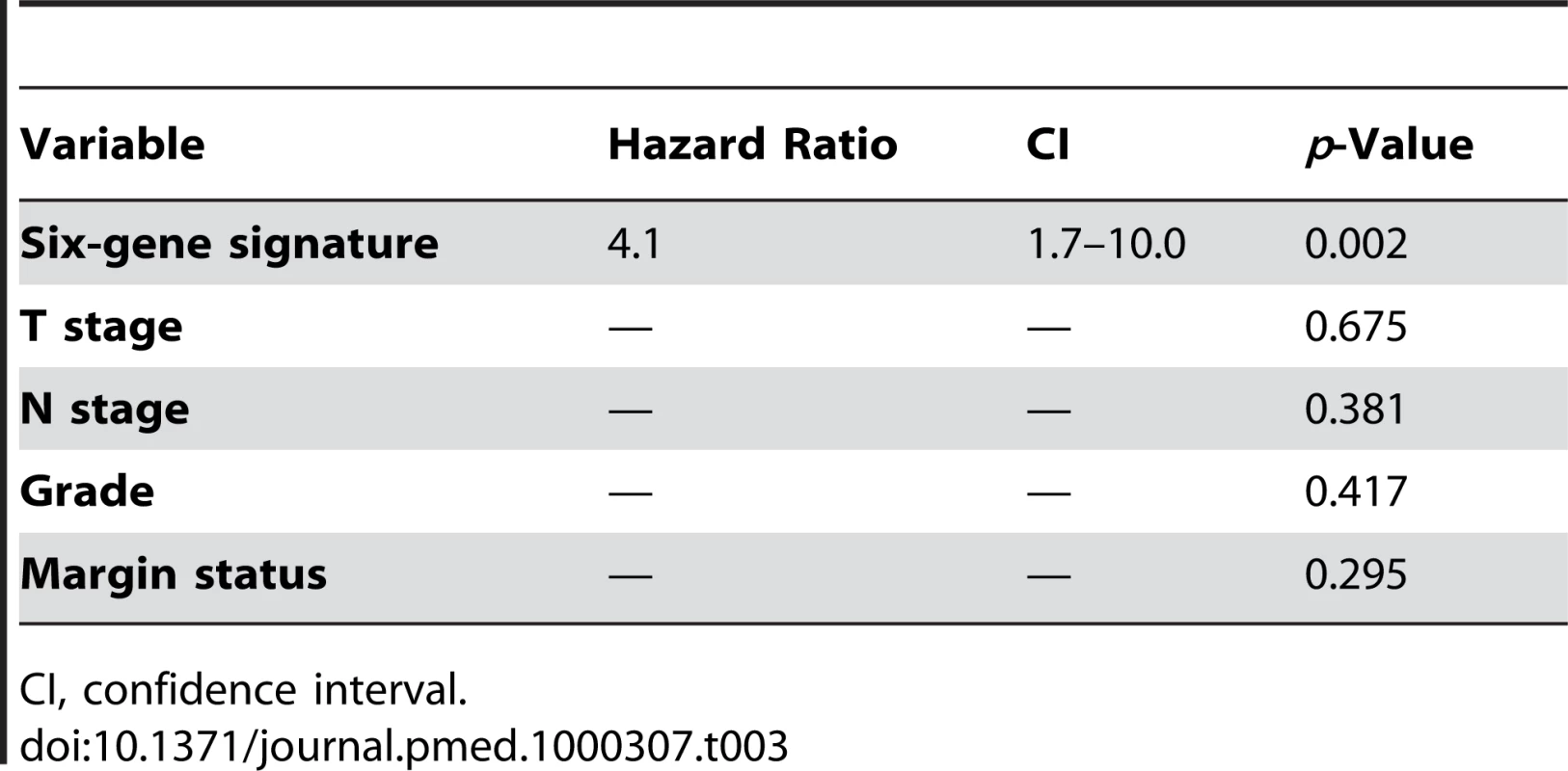
CI, confidence interval. We also looked at whether our six-gene signature was confounded by available clinicopathological variables. We found no association between our molecular signature, and tumor size, grade, margin status, nodal status, and neoadjuvant or adjuvant chemotherapy in our independent test set (Table 4).
Tab. 4. Relationship between the six-gene signature and clinicopathological variables. 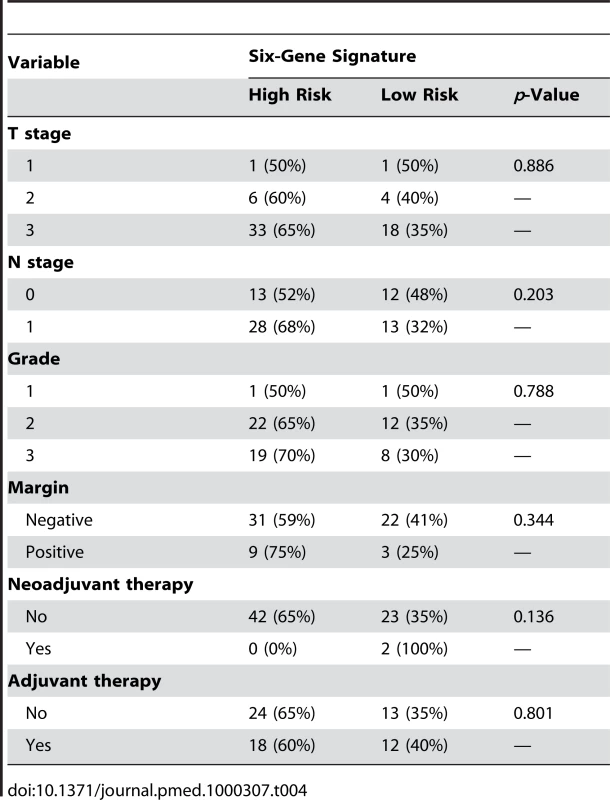
KLF6 Expression in Primary PDAC
In order to further validate the six-gene signature, we performed immunohistochemical analyses for KLF6, which showed a wide range of expression values between nonmetastatic versus metastatic samples (Figure 1A). To evaluate KLF6 protein expression, we obtained another independent dataset of 50 patients represented on a TMA with matched normal, chronic pancreatitis, and PDAC (UNC2, Table 2). First, using the median score of 1.5 as the cutoff, we found that KLF6 expression was much higher in tumors compared to normal pancreas (p<0.001) (Figure 2A and 2C). KLF6 expression was strong in normal islet cells in agreement with a previous study (Figure 2Ci, white arrowhead) [38]. Second, we found that KLF6 expression with a score greater than 1.5 (high) was associated with a shorter median survival of 11 mo compared to 24 mo for patients with KLF6 expression scores less than 1.5 (low) (p = 0.04) (Figure 2B).
Fig. 2. Significance of KLF6 and Fos B expression in primary PDAC. 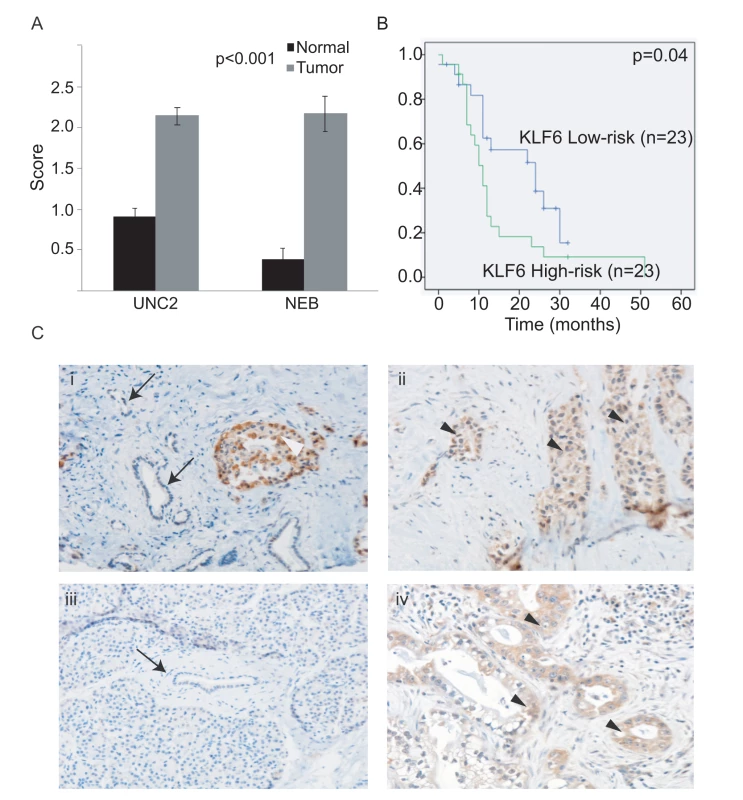
(A) KLF6 staining is significantly higher in PDAC compared to normal adjacent pancreas in an independent dataset of a 50-patient TMA (UNC2) as well as NEB samples used for the original analysis. (B) Kaplan-Meier overall survival of 50 patients classified by high and low KLF6 scores, using the median cutoff score of 1.5. (C) KLF6 immunostaining in the primary tumor of a patient who died of metastatic disease (ii) and in a resected primary tumor (iv). Minimal staining is seen in the matched normal adjacent tissue of both patients (i, iii). KLF6 immunostaining in islet cells (i, white arrowhead). Arrows illustrate normal ductal epithelium. Black arrowheads illustrate tumor. Discussion
We profiled and compared nonmetastatic and metastatic primary PDAC tumors and identified a six-gene signature. Although this signature was not derived on the basis of outcomes, we show that it was prognostic in a true test set of resectable PDAC patients. Importantly, our six-gene signature was independently predictive of survival, stratifying patients with median survivals of 15 compared to 49 mo, outperforming current pathological staging criteria, suggesting that our signature will be a powerful prognostic tool for patients with localized PDAC.
PDAC continues to be a devastating disease with few long-term survivors. Surgery remains the standard therapy for patients diagnosed with resectable PDAC [39]. Yet with a median survival only of less than 2 y after surgery, the attendant postoperative mortality rate of 2%–6% [40],[41], and postoperative complication and hospital readmission rates of 59% [41],[42], the decision for surgery should be made cautiously. Therefore, improved patient selection for therapy is necessary. For the majority of patients who cannot undergo surgery, gemcitabine chemotherapy remains the best option, yet only 5%–10% of patients respond to the treatment [43],[44]. Given the current therapeutic limitations, additional prognostic tools are needed to help a patient decide whether to have surgery, and/or neoadjuvant chemotherapy, or when to consider participation in a clinical trial.
Our analysis identified a surprisingly small number of genes with differential expression between early compared to late stage primary PDAC (Table S1). This finding suggests that primary PDAC may be largely homogenous from a global gene expression standpoint. Nonetheless, the differences that we identified appear to be clinically and therefore biologically important. Our findings of molecular differences in resected primary PDAC tumors suggest that there is subtle biological variation in these tumors that influences outcome. A review of previous published studies did not identify differential expression of our six genes [15],[21],[45]–[56]. This finding is not surprising, as previous studies examined differential gene expression changes between either normal pancreas or chronic pancreatitis and PDAC [15],[45]–[56]. Only one study has looked at gene expression changes between PDAC of different stages [21]. Ours was the first, to our knowledge, to study molecular differences between nonmetastatic versus metastatic primary tumors and identify and validate a prognostic signature for PDAC.
Of the six genes identified in this study, most do not have an obvious role in carcinogenesis. Three of the six genes demonstrated significantly higher expression in the poor prognostic groups (SIGLEC11, KLF6, NFKBIZ; Table S2). ATP4A, GSG1, and SIGLEC-11 have not been studied in cancer. SIGLEC-11 is thought to be expressed by tissue macrophages and also the brain microglia [57]. Interestingly, a missense mutation of SIGLEC-11 (S465A) was identified in the mutation discovery screen of the recent genome-wide sequencing of PDAC [58]. NFKBIZ, also called IkappaB zeta, binds to the p50 subunit of nuclear factor (NF)-kappaB and is important for interleukin-6 (IL-6) induction and may be induced by IL-1 receptor and Toll-like receptors [57]. Given the prevalence of chronic pancreatitis and high degree of stromal fibrosis, it is possible that NFKBIZ may play a role in PDAC and inflammation.
KLF6 is a transcription factor and its full length transcript is thought to be a tumor suppressor gene involved in prostate, lung, and ovarian carcinogenesis [59]. However a splice variant KLF6-SV1 has been shown to have oncogenic properties. The oligonucleotide probes used in the Agilent whole human genome array and the antibody against KLF6 did not differentiate between the full-length and splice variant. In agreement with a previous study [38], we found that KLF6 protein expression was higher in tumors than normal pancreas. In addition we found that higher KLF6 expression was associated with worse survival. Hartel et al. further investigated KLF6-SV1 expression in their study using real-time PCR and demonstrated that the higher KLF6 expression seen in tissues was associated with a higher ratio of KLF6-SV1 compared to full-length KLF6. Therefore our findings that KLF6 expression is higher in tumors and is prognostic is likely in agreement with this study.
Only one patient in the UNC1 cohort was treated with neoadjuvant chemotherapy compared to 80% of NEB patients who were treated with palliative chemotherapy. Although there is a possibility that our signature may be reflective of gemcitabine treatment or perhaps resistance, as NEB patients died of metastatic disease despite gemcitabine treatment, the successful application of our six-gene signature on an independent test set of patients where only 3% of patients with localized PDAC were treated with neoadjuvant therapy suggests that it is a rigorous predictor of prognosis in previously untreated patients. We found no association between our six-gene signature and whether a patient received adjuvant chemotherapy. In addition, chemotherapy treatment in this cohort, either pre - or postoperative, did not demonstrate a survival advantage.
Another concern is the validity of our hypothesis that gene expression changes at different stages of primary PDAC development may occur and be important for prognosis. Our study is in agreement with Lowe and colleagues' findings that differential gene expression changes can be identified within primary PDAC [21]. However, they did not address the prognostic value of their findings. Several studies have also suggested that gene expression changes in metastasis may be found in primary tumors. In a study of molecular differences between primary tumors and metastases, Golub and colleagues identified a gene expression signature of metastasis present that could be identified in primary tumors [60]. In addition, studies in melanoma have suggested that metastatic cells may be found in the parent primary tumor [61]. Finally studies in breast cancer have demonstrated that gene expression changes found in breast cancer cells with metastatic potential may be prognostic and predictive of patients who will develop metastasis [62]–[64]. Our study is the first to demonstrate that molecular differences in metastatic PDAC can be identified at earlier stages, and that these differences are predictive of future behavior. Whether these molecular changes are biologically associated with metastatic potential will require further investigation.
We have applied our six-gene signature to an independent dataset of 67 patients, and have validated its prognostic value. In addition, we have validated the protein expression of KLF6 in a 50-patient TMA. Although not nearly as powerful a predictor of prognosis as our six-gene signature, we found that KLF6 expression was prognostic in our 50-patient TMA. Further validation studies will be needed to see if KLF6 alone may be a useful prognostic marker as others have shown [38]. Our findings suggest that the prognostic value of KLF6 is strengthened in evaluating the six genes in their entirety.
Studies of patients with resectable PDAC demonstrate median survivals of up to 22 mo, equivalent to the median survival of patients in our training and testing cohorts [3],[11],[65]. Our finding that our six-gene signature is able to stratify patients, with startling differences in survival, suggests that it may be used to select patients for therapies. For example, for patients who are at high operative risk, knowledge of a median survival of 49 compared to 15 mo, may be helpful in the operative decision-making process. Similarly, patients who have a poor prognosis based on the six-gene signature may be considered for neoadjuvant therapy. Currently, the minority of centers use neoadjuvant therapy as a standard of care, most instead reserve this for patients with locally advanced unresectable or borderline resectable tumors. Therefore the current decision-making process is based on anatomical considerations. Our prognostic signature may refine this paradigm such that neoadjuvant therapy is offered to patients on the basis of biological considerations, regardless of resectability, and may allow us to further study and maximize the benefits of neoadjuvant treatment. In addition, as new therapies are developed, it may help to determine whether patients may require more or less aggressive treatment. Finally, our findings that there are molecular differences associated with late-stage primary tumors, which translate into differences in prognosis, suggest that the six genes in this signature should be further studied for their potential as biomarkers, and some of these genes, or the pathways that they fall into, may represent new therapeutic targets.
Supporting Information
Zdroje
1. BoyleP
LevinB
2008 World Cancer Report 2008. Lyon International Agency for Research on Cancer
2. ParkinDM
BrayF
FerlayJ
PisaniP
2005 Global cancer statistics, 2002. CA Cancer J Clin 55 74 108
3. NeuhausP
RiessH
PostS
GellertK
RidwelskiK
2008 CONKO-001: final results of the randomized, prospective, multicenter phase III trial of adjuvant chemotherapy with gemcitabine versus observation in patients with resected pancreatic cancer (PC). Journal of Clinical Oncology, 2008 ASCO Annual Meeting Proceedings 26 LBA4504
4. FerroneCR
BrennanMF
GonenM
CoitDG
FongY
2008 Pancreatic adenocarcinoma: the actual 5-year survivors. J Gastrointest Surg 12 701 706
5. ConlonKC
KlimstraDS
BrennanMF
1996 Long-term survival after curative resection for pancreatic ductal adenocarcinoma. Clinicopathologic analysis of 5-year survivors. Ann Surg 223 273 279
6. SchnelldorferT
WareAL
SarrMG
SmyrkTC
ZhangL
2008 Long-term survival after pancreatoduodenectomy for pancreatic adenocarcinoma: is cure possible? Ann Surg 247 456 462
7. ClearySP
GryfeR
GuindiM
GreigP
SmithL
2004 Prognostic factors in resected pancreatic adenocarcinoma: analysis of actual 5-year survivors. J Am Coll Surg 198 722 731
8. HanSS
JangJY
KimSW
KimWH
LeeKU
2006 Analysis of long-term survivors after surgical resection for pancreatic cancer. Pancreas 32 271 275
9. WinterJM
CameronJL
CampbellKA
ArnoldMA
ChangDC
2006 1423 pancreaticoduodenectomies for pancreatic cancer: a single-institution experience. J Gastrointest Surg 10 1199 1210; discussion 1210-1191
10. AhmadNA
LewisJD
GinsbergGG
HallerDG
MorrisJB
2001 Long term survival after pancreatic resection for pancreatic adenocarcinoma. Am J Gastroenterol 96 2609 2615
11. UenoH
KosugeT
MatsuyamaY
YamamotoJ
NakaoA
2009 A randomised phase III trial comparing gemcitabine with surgery-only in patients with resected pancreatic cancer: Japanese Study Group of Adjuvant Therapy for Pancreatic Cancer. Br J Cancer 101 908 915
12. EvansDB
VaradhacharyGR
CraneCH
SunCC
LeeJE
2008 Preoperative gemcitabine-based chemoradiation for patients with resectable adenocarcinoma of the pancreatic head. J Clin Oncol 26 3496 3502
13. VaradhacharyGR
WolffRA
CraneCH
SunCC
LeeJE
2008 Preoperative gemcitabine and cisplatin followed by gemcitabine-based chemoradiation for resectable adenocarcinoma of the pancreatic head. J Clin Oncol 26 3487 3495
14. KolbertCP
ChariS
SreekumarR
2008 Microarray technologies for gene transcript analysis in pancreatic cancer. Technol Cancer Res Treat 7 55 59
15. GrutzmannR
BorissH
AmmerpohlO
LuttgesJ
KalthoffH
2005 Meta-analysis of microarray data on pancreatic cancer defines a set of commonly dysregulated genes. Oncogene 24 5079 5088
16. GrutzmannR
SaegerHD
LuttgesJ
SchackertHK
KalthoffH
2004 Microarray-based gene expression profiling in pancreatic ductal carcinoma: status quo and perspectives. Int J Colorectal Dis 19 401 413
17. GogginsM
2007 Identifying molecular markers for the early detection of pancreatic neoplasia. Semin Oncol 34 303 310
18. GroteT
LogsdonCD
2007 Progress on molecular markers of pancreatic cancer. Curr Opin Gastroenterol 23 508 514
19. ToniniG
PantanoF
VincenziB
GabbrielliA
CoppolaR
2007 Molecular prognostic factors in patients with pancreatic cancer. Expert Opin Ther Targets 11 1553 1569
20. YehJJ
2009 Prognostic signature for pancreatic cancer: are we close? Future Oncol 5 313 321
21. KimHN
ChoiDW
LeeKT
LeeJK
HeoJS
2007 Gene expression profiling in lymph node-positive and lymph node-negative pancreatic cancer. Pancreas 34 325 334
22. YangYH
DudoitS
LuuP
LinDM
PengV
2002 Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res 30 e15
23. TroyanskayaO
CantorM
SherlockG
BrownP
HastieT
2001 Missing value estimation methods for DNA microarrays. Bioinformatics 17 520 525
24. BenitoM
ParkerJ
DuQ
WuJ
XiangD
2004 Adjustment of systematic microarray data biases. Bioinformatics 20 105 114
25. TusherVG
TibshiraniR
ChuG
2001 Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A 98 5116 5121
26. HuZ
FanC
OhDS
MarronJS
HeX
2006 The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics 7 96
27. CampRL
Dolled-FilhartM
RimmDL
2004 X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res 10 7252 7259
28. YehJJ
RouthED
RubinasT
PeacockJ
MartinTD
2009 KRAS/BRAF mutation status and ERK1/2 activation as biomarkers for MEK1/2 inhibitor therapy in colorectal cancer. Mol Cancer Ther 8 834 843
29. HoosA
Cordon-CardoC
2001 Tissue microarray profiling of cancer specimens and cell lines: opportunities and limitations. Lab Invest 81 1331 1338
30. TroesterMA
LeeMH
CarterM
FanC
CowanDW
2009 Activation of host wound responses in breast cancer microenvironment. Clin Cancer Res 15 7020 7028
31. AllinenM
BeroukhimR
CaiL
BrennanC
Lahti-DomeniciJ
2004 Molecular characterization of the tumor microenvironment in breast cancer. Cancer Cell 6 17 32
32. MuellerMM
FusenigNE
2004 Friends or foes - bipolar effects of the tumour stroma in cancer. Nat Rev Cancer 4 839 849
33. ComoglioPM
TrusolinoL
2005 Cancer: the matrix is now in control. Nat Med 11 1156 1159
34. HerschkowitzJI
SiminK
WeigmanVJ
MikaelianI
UsaryJ
2007 Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol 8 R76
35. OhDS
TroesterMA
UsaryJ
HuZ
HeX
2006 Estrogen-regulated genes predict survival in hormone receptor-positive breast cancers. J Clin Oncol 24 1656 1664
36. LuY
LemonW
LiuPY
YiY
MorrisonC
2006 A gene expression signature predicts survival of patients with stage I non-small cell lung cancer. PLoS Med 3 e467 doi:10.1371/journal.pmed.0030467
37. SohnTA
YeoCJ
CameronJL
KoniarisL
KaushalS
2000 Resected adenocarcinoma of the pancreas-616 patients: results, outcomes, and prognostic indicators. J Gastrointest Surg 4 567 579
38. HartelM
NarlaG
WenteMN
GieseNA
MartignoniME
2008 Increased alternative splicing of the KLF6 tumour suppressor gene correlates with prognosis and tumour grade in patients with pancreatic cancer. Eur J Cancer 44 1895 1903
39. YeoCJ
CameronJL
SohnTA
LillemoeKD
PittHA
1997 Six hundred fifty consecutive pancreaticoduodenectomies in the 1990s: pathology, complications, and outcomes. Ann Surg 226 248 257; discussion 257-260
40. EppsteinerRW
CsikeszNG
McPheeJT
TsengJF
ShahSA
2009 Surgeon volume impacts hospital mortality for pancreatic resection. Ann Surg 249 635 640
41. YermilovI
BentremD
SekerisE
JainS
MaggardMA
2009 Readmissions following pancreaticoduodenectomy for pancreas cancer: a population-based appraisal. Ann Surg Oncol 16 554 561
42. DeOliveiraML
WinterJM
SchaferM
CunninghamSC
CameronJL
2006 Assessment of complications after pancreatic surgery: a novel grading system applied to 633 patients undergoing pancreaticoduodenectomy. Ann Surg 244 931 937; discussion 937-939
43. Abou-AlfaGK
LetourneauR
HarkerG
ModianoM
HurwitzH
2006 Randomized phase III study of exatecan and gemcitabine compared with gemcitabine alone in untreated advanced pancreatic cancer. J Clin Oncol 24 4441 4447
44. Van CutsemE
VervenneWL
BennounaJ
HumbletY
GillS
2009 Phase III trial of bevacizumab in combination with gemcitabine and erlotinib in patients with metastatic pancreatic cancer. J Clin Oncol 27 2231 2237
45. Crnogorac-JurcevicT
EfthimiouE
CapelliP
BlaveriE
BaronA
2001 Gene expression profiles of pancreatic cancer and stromal desmoplasia. Oncogene 20 7437 7446
46. Crnogorac-JurcevicT
EfthimiouE
NielsenT
LoaderJ
TerrisB
2002 Expression profiling of microdissected pancreatic adenocarcinomas. Oncogene 21 4587 4594
47. KaranjawalaZE
IlleiPB
AshfaqR
InfanteJR
MurphyK
2008 New markers of pancreatic cancer identified through differential gene expression analyses: claudin 18 and annexin A8. Am J Surg Pathol 32 188 196
48. LoweAW
OlsenM
HaoY
LeeSP
Taek LeeK
2007 Gene expression patterns in pancreatic tumors, cells and tissues. PLoS ONE 2 e323 doi:10.1371/journal.pone.0000323
49. IshikawaM
YoshidaK
YamashitaY
OtaJ
TakadaS
2005 Experimental trial for diagnosis of pancreatic ductal carcinoma based on gene expression profiles of pancreatic ductal cells. Cancer Sci 96 387 393
50. SegaraD
BiankinAV
KenchJG
LanguschCC
DawsonAC
2005 Expression of HOXB2, a retinoic acid signaling target in pancreatic cancer and pancreatic intraepithelial neoplasia. Clin Cancer Res 11 3587 3596
51. LogsdonCD
SimeoneDM
BinkleyC
ArumugamT
GreensonJK
2003 Molecular profiling of pancreatic adenocarcinoma and chronic pancreatitis identifies multiple genes differentially regulated in pancreatic cancer. Cancer Res 63 2649 2657
52. Iacobuzio-DonahueCA
AshfaqR
MaitraA
AdsayNV
Shen-OngGL
2003 Highly expressed genes in pancreatic ductal adenocarcinomas: a comprehensive characterization and comparison of the transcription profiles obtained from three major technologies. Cancer Res 63 8614 8622
53. Iacobuzio-DonahueCA
MaitraA
OlsenM
LoweAW
van HeekNT
2003 Exploration of global gene expression patterns in pancreatic adenocarcinoma using cDNA microarrays. Am J Pathol 162 1151 1162
54. Iacobuzio-DonahueCA
MaitraA
Shen-OngGL
van HeekT
AshfaqR
2002 Discovery of novel tumor markers of pancreatic cancer using global gene expression technology. Am J Pathol 160 1239 1249
55. Crnogorac-JurcevicT
MissiagliaE
BlaveriE
GangeswaranR
JonesM
2003 Molecular alterations in pancreatic carcinoma: expression profiling shows that dysregulated expression of S100 genes is highly prevalent. J Pathol 201 63 74
56. GrutzmannR
PilarskyC
AmmerpohlO
LuttgesJ
BohmeA
2004 Gene expression profiling of microdissected pancreatic ductal carcinomas using high-density DNA microarrays. Neoplasia 6 611 622
57. AngataT
KerrSC
GreavesDR
VarkiNM
CrockerPR
2002 Cloning and characterization of human Siglec-11. A recently evolved signaling that can interact with SHP-1 and SHP-2 and is expressed by tissue macrophages, including brain microglia. J Biol Chem 277 24466 24474
58. JonesS
ZhangX
ParsonsDW
LinJC
LearyRJ
2008 Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 321 1801 1806
59. DiFeoA
MartignettiJA
NarlaG
2009 The role of KLF6 and its splice variants in cancer therapy. Drug Resist Updat 12 1 7
60. RamaswamyS
RossKN
LanderES
GolubTR
2003 A molecular signature of metastasis in primary solid tumors. Nat Genet 33 49 54
61. FidlerIJ
KripkeML
1977 Metastasis results from preexisting variant cells within a malignant tumor. Science 197 893 895
62. MinnAJ
GuptaGP
PaduaD
BosP
NguyenDX
2007 Lung metastasis genes couple breast tumor size and metastatic spread. Proc Natl Acad Sci U S A 104 6740 6745
63. MinnAJ
GuptaGP
SiegelPM
BosPD
ShuW
2005 Genes that mediate breast cancer metastasis to lung. Nature 436 518 524
64. MinnAJ
KangY
SerganovaI
GuptaGP
GiriDD
2005 Distinct organ-specific metastatic potential of individual breast cancer cells and primary tumors. J Clin Invest 115 44 55
65. NeoptolemosJP
DunnJA
StockenDD
AlmondJ
LinkK
2001 Adjuvant chemoradiotherapy and chemotherapy in resectable pancreatic cancer: a randomised controlled trial. Lancet 358 1576 1585
Štítky
Interní lékařství
Článek vyšel v časopisePLOS Medicine
Nejčtenější tento týden
2010 Číslo 7- Berberin: přírodní hypolipidemikum se slibnými výsledky
- Léčba bolesti u seniorů
- Příznivý vliv Armolipidu Plus na hladinu cholesterolu a zánětlivé parametry u pacientů s chronickým subklinickým zánětem
- Červená fermentovaná rýže účinně snižuje hladinu LDL cholesterolu jako vhodná alternativa ke statinové terapii
- Jak postupovat při výběru betablokátoru − doporučení z kardiologické praxe
-
Všechny články tohoto čísla
- Estimating the Number of Paediatric Fevers Associated with Malaria Infection Presenting to Africa's Public Health Sector in 2007
- HIV in Maternal and Child Heath: Concurrent Crises Demand Cooperation
- HIV Testing for Children in Resource-Limited Settings: What Are We Waiting For?
- Improving Implementation: Building Research Capacity in Maternal, Neonatal, and Child Health in Africa
- Evidence-Based Priority Setting for Health Care and Research: Tools to Support Policy in Maternal, Neonatal, and Child Health in Africa
- Three Adult Cases of Meningitis in Vietnam
- Integrative Genomic Analyses Identify as a Novel Lineage-Specific Oncogene in Lung Squamous Cell Carcinoma
- Association of Early Repolarization Pattern on ECG with Risk of Cardiac and All-Cause Mortality: A Population-Based Prospective Cohort Study (MONICA/KORA)
- Stable and Unstable Malaria Hotspots in Longitudinal Cohort Studies in Kenya
- A Model for the Roll-Out of Comprehensive Adult Male Circumcision Services in African Low-Income Settings of High HIV Incidence: The ANRS 12126 Bophelo Pele Project
- Left to Their Own Devices: Breakdowns in United States Medical Device Premarket Review
- A Six-Gene Signature Predicts Survival of Patients with Localized Pancreatic Ductal Adenocarcinoma
- Financing Maternal and Child Health—What Are the Limitations in Estimating Donor Flows and Resource Needs?
- Improving Prevention of Mother-to-Child Transmission of HIV Care and Related Services in Eastern Rwanda
- Social Relationships and Mortality Risk: A Meta-analytic Review
- PLOS Medicine
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Three Adult Cases of Meningitis in Vietnam
- Left to Their Own Devices: Breakdowns in United States Medical Device Premarket Review
- Social Relationships and Mortality Risk: A Meta-analytic Review
- Evidence-Based Priority Setting for Health Care and Research: Tools to Support Policy in Maternal, Neonatal, and Child Health in Africa
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



