-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaMaternal or zygotic: Unveiling the secrets of the Pancrustacea transcription factor
article has not abstract
Published in the journal: . PLoS Genet 14(3): e32767. doi:10.1371/journal.pgen.1007201
Category: Perspective
doi: https://doi.org/10.1371/journal.pgen.1007201Summary
article has not abstract
During metazoan early embryonic development, maternal mRNAs are gradually eliminated and replaced by zygotic ones during the maternal-to-zygotic transition (MZT), a stage where the zygotic genome takes control of developmental processes [1]. In 2006, the molecular basis of the MZT started to be uncovered by molecular and computational biology studies in the fruit fly Drosophila melanogaster. ten Bosch et al. showed that the heptamer consensus sequence CAGGTAG (i.e., the TAGteam) is overrepresented in regulatory regions of the earliest expressed zygotic genes [2]. Two years later, the identification of the zinc-finger (ZnF) transcription factor (TF) zelda (Zld; ZnF early Drosophila activator) as the TAGteam binding factor was a breakthrough in MZT research [3]. In 2011, two papers in PLOS Genetics showed that Zld binds to over 1,000 cis-regulatory regions during D. melanogaster early embryogenesis [4, 5]. Zld increases chromatin accessibility of early essential fly TFs (e.g., Dorsal and bicoid), resembling the role of pioneer vertebrate TFs (e.g., FoxA1 and Gata4) [6–9]. Recently, zld was shown to be important for the establishment of topologically associating domains (TADs) during zygotic genome activation [10].
Although many important roles of Zld in transcriptional regulation have been elucidated over the past 10 years, the biochemical and structural features that make Zld a master TF remain largely unknown. Previous biochemical and cell culture analysis revealed a low complexity region, corresponding to the transactivation domain, and four C-terminal ZnFs that are required for DNA binding and transcriptional activation in Drosophila [11]. Recently, a study in PLOS Genetics showed that Zld is conserved throughout Pancrustacea and comprises other widely conserved regions, including an acidic patch—which could be involved in the recruitment of chromatin remodeling proteins—and two N-terminal ZnFs, ZnF-Novel and ZnF1 [12]. Interestingly, despite the ZnF-Novel conservation in several insect groups, this domain has been eroded in dipterans [12]. In addition, zld is found as a single-copy gene in all insect genomes sequenced to date, indicating that it is sensitive to increased gene dosage [12].
In December 2017’s issue of PLOS Genetics, Hamm and collaborators [13] report the functional analysis of the D. melanogaster Zld conserved domains in vivo and in vitro. An elegant Cas9 engineering system was used to generate flies with point mutations in ZnF1, in the acidic patch, or in the ZnF2 JAZ-finger. This approach has the advantage of generating mutations in the native locus, avoiding issues with position effects of transgenic insertions. While mutations in the ZnF1 or in the acidic patch did not affect viability, mutations in the ZnF2 JAZ-finger domain led to a maternal-effect lethal phenotype. Hamm et al. also conducted a transcriptome analysis of the mutated zld ZnF2 JAZ-finger (zld ZnF2). They showed that the ZnF2 JAZ-finger acts as a maternal repressive domain during fly embryogenesis in vivo, being particularly involved in the transcriptional regulation of zygotic zld targets and maternal mRNA clearance. Collectively, Hamm et al. [13] show for the first time that Zld is a modular TF, with different ZnF domains playing distinct roles in transcriptional regulation during early embryogenesis (Fig 1).
Fig. 1. Protein domain architectures of Zld proteins from the red flour beetle Tribolium castaneum (representing the insect ancestral state) and the fruit fly D. melanogaster. 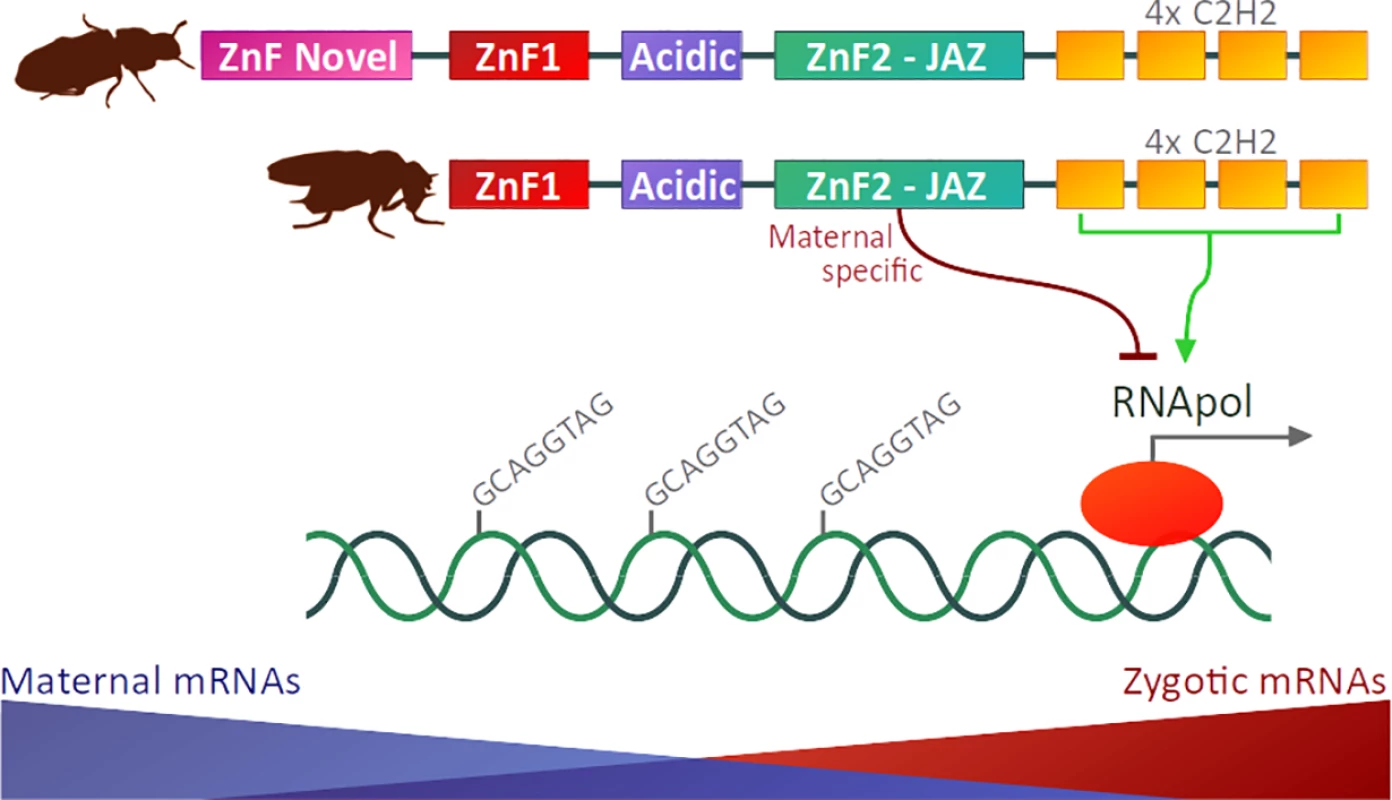
The ZnF-Novel domain is present in most insect orders but has been eroded in Diptera [12]. Zld is a multi-domain ZnF TF, and at least one of its zinc-fingers (i.e., the ZnF2 JAZ-finger) is required for viability via regulation of maternal mRNA clearance and zygotic genome activation [13]. TF, transcription factor; Zld, zelda; ZnF, zinc-finger. The work by Hamm et al. also poses intriguing questions. The observation that point mutations in the acidic patch and ZnF1 did not alter fly viability suggests at least three scenarios: 1) the amino acid changes did not impair the activity of the domains; 2) these domains are dispensable for Zld-mediated transcriptional activation in D. melanogaster (as hypothesized by the authors) or; 3) these domains are not essential in flies kept under standard laboratory conditions.
We believe that the former two possibilities are unlikely because Zld is a master TF, and the most important (and conserved) residues of the acidic patch and ZnF1 were precisely mutated. The latter hypothesis seems more appealing, as these domains have been conserved for over 300 million years in various insect lineages [12]. Detailed functional analysis of these mutant flies in different environments (e.g., under natural stress conditions) or genetic backgrounds may unveil other modular roles (like those of the ZnF2 JAZ-finger domain) and possible redundancies of these domains. Since the amount of zld expression has been reported to be critical for correct activation of target genes in time and space [4, 13], it will also be important to investigate if any of the mutant zld lines display altered gradients of key TFs (e.g., Dorsal and bicoid) by live imaging. Further, analysis of TAD boundaries by Hi-C in zldZnF2 mutants could provide new hints of Zld's function in the establishment of chromatin architectures [10].
Another interesting finding from Hamm et al. was that a subset of homozygous zldZnF2 male and female adults displayed small eyes [13], strongly supporting the idea that zld also performs post-embryonic roles, as previously reported in beetles [12]. Importantly, such roles are also supported by zld expression in eye and wing imaginal discs [13, 14]. Profound changes in cell proliferation, differentiation, and morphology take place during imaginal disc development, and the potential roles of zld in such processes are yet to be elucidated. Recently, zld was also described as an important factor for neuronal progenitor formation in flies, and its inhibition by the tumor suppressor TRIM-NHL protein Brain tumor (Brat) appears to be essential to avoid neuronal tumor progression [15]. Hence, unveiling whether zld gene regulatory networks are similar among these developmental processes will be of great value.
Finally, Hamm et al. open new perspectives in the elucidation of Zld functions. Comparative functional studies in other insect species may provide a unique opportunity to address the roles of zld in early transcriptional reprogramming and unlock further secrets about evolution of this master TF throughout the Pancrustacea.
Zdroje
1. Tadros W, Lipshitz HD. The maternal-to-zygotic transition: a play in two acts. Development. 2009;136(18):3033–42. doi: 10.1242/dev.033183 19700615
2. ten Bosch JR, Benavides JA, Cline TW. The TAGteam DNA motif controls the timing of Drosophila pre-blastoderm transcription. Development. 2006;133(10):1967–77. doi: 10.1242/dev.02373 16624855
3. Liang H-L, Nien C-Y, Liu H-Y, Metzstein MM, Kirov N, Rushlow C. The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature. 2008;456(7220):400–3. doi: 10.1038/nature07388 18931655
4. Nien C-Y, Liang H-L, Butcher S, Sun Y, Fu S, Gocha T, et al. Temporal coordination of gene networks by Zelda in the early Drosophila embryo. PLoS Genet. 2011;7(10):e1002339. doi: 10.1371/journal.pgen.1002339 22028675
5. Harrison MM, Li X-Y, Kaplan T, Botchan MR, Eisen MB. Zelda binding in the early Drosophila melanogaster embryo marks regions subsequently activated at the maternal-to-zygotic transition. PLoS Genet. 2011;7(10):e1002266. doi: 10.1371/journal.pgen.1002266 22028662
6. Xu Z, Chen H, Ling J, Yu D, Struffi P, Small S. Impacts of the ubiquitous factor Zelda on Bicoid-dependent DNA binding and transcription in Drosophila. Genes & Development. 2014;28(6):608–21. doi: 10.1101/gad.234534.113 24637116
7. Li X-Y, Harrison MM, Villalta JE, Kaplan T, Eisen MB. Establishment of regions of genomic activity during the Drosophila maternal to zygotic transition. eLife. 2014;3. doi: 10.7554/eLife.03737 25313869
8. Schulz KN, Bondra ER, Moshe A, Villalta JE, Lieb JD, Kaplan T, et al. Zelda is differentially required for chromatin accessibility, transcription factor binding, and gene expression in the early Drosophila embryo. Genome Res. 2015;25(11):1715–26. doi: 10.1101/gr.192682.115 26335634; PubMed Central PMCID: PMCPMC4617967.
9. Foo SM, Sun Y, Lim B, Ziukaite R, O'Brien K, Nien C-Y, et al. Zelda potentiates morphogen activity by increasing chromatin accessibility. Current Biology. 2014;24(12):1341–6. doi: 10.1016/j.cub.2014.04.032 24909324
10. Hug CB, Grimaldi AG, Kruse K, Vaquerizas JM. Chromatin Architecture Emerges during Zygotic Genome Activation Independent of Transcription. Cell. 2017;169(2):216–28 e19. doi: 10.1016/j.cell.2017.03.024 28388407.
11. Hamm DC, Bondra ER, Harrison MM. Transcriptional activation is a conserved feature of the early embryonic factor Zelda that requires a cluster of four zinc fingers for DNA binding and a low-complexity activation domain. The Journal of Biological Chemistry. 2015;290(6):3508–18. doi: 10.1074/jbc.M114.602292 25538246
12. Ribeiro L, Tobias-Santos V, Santos D, Antunes F, Feltran G, de Souza Menezes J, et al. Evolution and multiple roles of the Pancrustacea specific transcription factor zelda in insects. PLoS Genet. 2017;13(7):e1006868. doi: 10.1371/journal.pgen.1006868 28671979; PubMed Central PMCID: PMCPMC5515446.
13. Hamm DC, Larson ED, Nevil M, Marshall KE, Bondra ER, Harrison MM. A conserved maternal-specific repressive domain in Zelda revealed by Cas9-mediated mutagenesis in Drosophila melanogaster. PloS Genet. 2017.
14. Staudt N, Fellert S, Chung H-R, Jäckle H, Vorbrüggen G. Mutations of the Drosophila zinc finger-encoding gene vielfältig impair mitotic cell divisions and cause improper chromosome segregation. Molecular Biology of the Cell. 2006;17(5):2356–65. doi: 10.1091/mbc.E05-11-1056 16525017
15. Reichardt I, Bonnay F, Steinmann V, Loedige I, Burkard TR, Meister G, et al. The tumor suppressor Brat controls neuronal stem cell lineages by inhibiting Deadpan and Zelda. EMBO Rep. 2017. doi: 10.15252/embr.201744188 29191977.
Štítky
Genetika Reprodukční medicína
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2018 Číslo 3
-
Všechny články tohoto čísla
- Asgard archaea do not close the debate about the universal tree of life topology
- splicing defect in dogs with lethal acrodermatitis
- Asgard archaea are the closest prokaryotic relatives of eukaryotes
- 2017 Reviewer and Editorial Board Thank You
- Maternal or zygotic: Unveiling the secrets of the Pancrustacea transcription factor
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Asgard archaea do not close the debate about the universal tree of life topology
- Asgard archaea are the closest prokaryotic relatives of eukaryotes
- splicing defect in dogs with lethal acrodermatitis
- 2017 Reviewer and Editorial Board Thank You
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



