-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
Reklama"T-bet"-ing on autoimmunity variants
article has not abstract
Published in the journal: . PLoS Genet 13(9): e32767. doi:10.1371/journal.pgen.1006924
Category: Perspective
doi: https://doi.org/10.1371/journal.pgen.1006924Summary
article has not abstract
During the past decade, genome-wide association studies (GWAS) have linked autoimmune disorders to hundreds of candidate risk variants across the human genome. Yet, our understanding of how these variants confer disease risk has severely lagged behind our ability to identify them. Almost 90% of GWAS hits map to noncoding regions of the genome, which creates a challenge to extract mechanistic insight from the human genetics. Although mounting evidence suggests that the majority of disease-associated SNPs lie in enhancers and may alter transcription factor (TF) occupancy and target gene regulation [1], only a handful of noncoding disease variants have been experimentally validated [2–4]. In a recent issue of PLOS Genetics, Soderquest et al. examined how genetic variants might alter the binding landscape of T-bet (encoded by TBX21), a critical transcriptional regulator in immune cells [5].
CD4+ T cells, which are central in autoimmune pathology, can take on distinct effector functions in the context of inflammation, including Th1, Th2, Th17, and regulatory T cell (Treg) phenotypes. Each subset is defined by characteristic cytokines and cell surface receptors—programs that are driven by master TFs [6]. Disruption of these programs could lead to disease. In patients with Crohn’s disease, T cells from the lamina propria express high levels of T-bet, the master regulator of Th1 cells [7], potentially implicating this TF in inflammatory bowel disease (IBD) pathology.
Soderquest et al. asked whether common noncoding disease variants might contribute to autoimmune disease risk by altering T-bet occupancy and regulation of genomic targets [5]. The authors previously used chromatin immunoprecipitation sequencing (ChIP-seq) to map T-bet genomic binding sites in primary human Th1 cells [8–10]. Here, they found that GWAS risk variants linked with Crohn’s disease, and to a lesser extent ulcerative colitis and celiac disease, preferentially mapped to T-bet-bound Th1 regulatory elements. Interestingly, GWAS variants associated with rheumatoid arthritis and psoriasis were not enriched at T-bet binding sites, raising the possibility that genetic effects on the T-bet program may be disease-specific.
This enrichment of IBD and celiac risk variants at T-bet binding sites motivated further efforts to dissect the effects of noncoding variants. The authors developed OligoFlow to rapidly screen for variants that alter T-bet binding affinity. This new method relies on mixing cell lysate containing T-bet with bead-conjugated oligonucleotides corresponding to T-bet binding sites with or without a candidate disease-variant. A fluorochrome-labeled antibody against the TF of interest—in this case, T-bet—is then added to quantify the in vitro TF-oligo affinity by flow cytometry (Fig 1A). The authors used OligoFlow to test T-bet binding at a number of variants and demonstrated significant reduction of T-bet binding for 3 of them: rs1465321, rs1006353, and rs11135484. Interestingly, despite exhibiting altered T-bet binding, only 1 of these variants falls in a recognizable T-bet binding motif. The mechanisms by which the other variants alter T-bet binding remain unclear—they could affect noncanonical sequences that directly affect T-bet affinity, or they could influence the binding of neighboring TFs that contribute to T-bet recruitment. OligoFlow is likely to prove useful as a general tool to screen for the effects of sequence variation on DNA–TF interactions.
Fig. 1. Identifying noncoding variants that impair transcription factor (TF) binding. 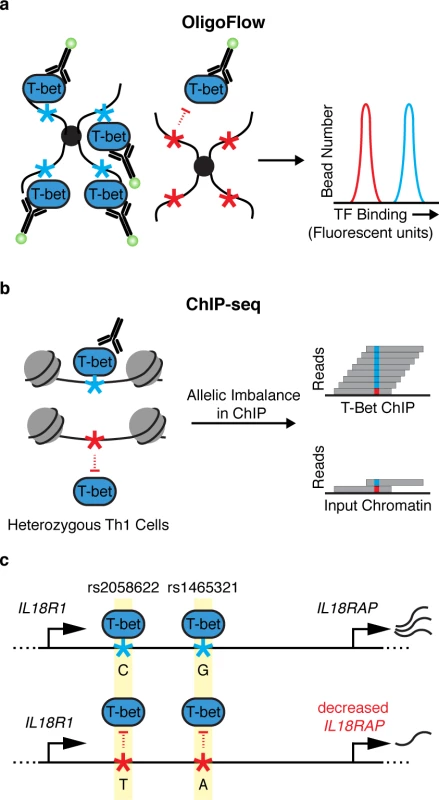
(a) OligoFlow is a flow-cytometry–based method for rapid screening of DNA variants that affect TF binding. In brief, cell lysate is mixed with bead-coupled oligonucleotides containing a T-bet binding site (light blue asterisk, left panel) or without (red asterisk, right panel) a candidate disease variant. Samples are incubated with a fluorochrome-labeled antibody against T-bet, and binding affinity is measured by flow cytometry. (b) chromatin immunoprecipitation sequencing (ChIP-seq) allows for the assessment of TF binding in its native chromatin context. Soderquest et al., confirmed allelic imbalance of T-bet binding to 2 linked SNPs (rs1465321 and rs2058622) at the immune risk locus IL18RAP (c). These SNPs are associated with decreased expression of the gene [11], suggesting that they affect T-bet–dependent regulation of IL18RAP. IL18R1/IL18RAP locus not drawn to scale. The authors went on to validate the effects of rs1465321 on T-bet occupancy and target gene regulation in its endogenous chromatin context. They performed ChIP-seq on in vitro polarized primary human Th1 cells from individuals who are heterozygous for rs1465321 and confirmed allelic imbalance of T-bet binding to the SNP as well as a nearby linked SNP (rs2058622) (Fig 1B and 1C). rs1465321 falls within a risk locus for celiac disease, which contains multiple SNPs that are associated with decreased expression of IL18RAP [11]. The authors also confirmed that Il18rap expression was significantly reduced in T-bet–deficient murine Th1 cells compared with wild-type cells. Taken together, the results highlight a T-bet–dependent gene regulatory circuit disrupted by a common human variant.
The field of human immunogenetics is coming to maturity. High-density genotyping arrays and new statistical algorithms have made it possible to confidently identify causal risk variants. The focus now more than ever is on how to determine the biological significance of disease variants. Soderquest et al. suggest that a subset of risk variants associated with IBD and celiac disease alter binding of the Th1 master regulatory TF T-bet. However, there are still important questions that remain unanswered. Further studies will be needed to examine if the effects of variants are specific to T-bet genome occupancy. Alternatively, there may be sets of key TFs with altered binding profiles contributing to the risk for distinct diseases. In addition, T-bet also contributes to gene regulatory programs in multiple immune cell types, including CD8+ T cells, innate lymphoid cells, natural killer (NK) cells, and even subsets of Tregs and Th17 cells [12]. Disease variants may have pleotropic effects on gene regulation in multiple cell types. Along with accelerating the acquisition of human genetic data, mechanistic studies are now possible with advanced chromatin mapping approaches and genome engineering technologies. OligoFlow offers a complementary technology to rapidly test for the effects of noncoding genetic variation. Studies integrating these emerging techniques have the potential to reveal underlying pathological mechanisms and, ultimately, could lead to targeted treatments for common human diseases.
Zdroje
1. Farh KK-H, Marson A, Zhu J, Kleinewietfeld M, Housley WJ, Beik S, et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature. 2015;518 : 337–343. doi: 10.1038/nature13835 25363779
2. Claussnitzer M, Dankel SN, Kim K-H, Quon G, Meuleman W, Haugen C, et al. FTO Obesity Variant Circuitry and Adipocyte Browning in Humans. New England Journal of Medicine. Massachusetts Medical Society; 2015;373 : 895–907. doi: 10.1056/NEJMoa1502214 26287746
3. Sur IK, Hallikas O, Vaharautio A, Yan J, Turunen M, Enge M, et al. Mice Lacking a Myc Enhancer That Includes Human SNP rs6983267 Are Resistant to Intestinal Tumors. Science. 2012;338 : 1360–1363. doi: 10.1126/science.1228606 23118011
4. Bauer DE, Kamran SC, Lessard S, Xu J, Fujiwara Y, Lin C, et al. An Erythroid Enhancer of BCL11A Subject to Genetic Variation Determines Fetal Hemoglobin Level. Science. American Association for the Advancement of Science; 2013;342 : 253–257. doi: 10.1126/science.1242088 24115442
5. Soderquest K, Hertweck A, Giambartolomei C, Henderson S, Mohamed R, Goldberg R, et al. Genetic variants alter T-bet binding and gene expression in mucosal inflammatory disease. Wells CA, editor. PLoS Genet. Public Library of Science; 2017;13: e1006587. doi: 10.1371/journal.pgen.1006587 28187197
6. Zhu J, Yamane H, Paul WE. Differentiation of Effector CD4 T Cell Populations. Annual Review of Immunology. Annual Reviews; 2010;28 : 445–489. doi: 10.1146/annurev-immunol-030409-101212 20192806
7. Neurath MF, Weigmann B, Finotto S, Glickman J, Nieuwenhuis E, Iijima H, et al. The Transcription Factor T-bet Regulates Mucosal T Cell Activation in Experimental Colitis and Crohn's Disease. The Journal of Experimental Medicine. 2002;195 : 1129–1143. doi: 10.1084/jem.20011956 11994418
8. Jenner RG, Townsend MJ, Jackson I, Sun K, Bouwman RD, Young RA, et al. The transcription factors T-bet and GATA-3 control alternative pathways of T-cell differentiation through a shared set of target genes. Proceedings of the National Academy of Sciences. 2009;106 : 17876–17881. doi: 10.1073/pnas.0909357106 19805038
9. Kanhere A, Hertweck A, Bhatia U, G kmen MR, Perucha E, Jackson I, et al. T-bet and GATA3 orchestrate Th1 and Th2 differentiation through lineage-specific targeting of distal regulatory elements. Nature Communications. Nature Publishing Group; 2012;3 : 1268. doi: 10.1038/ncomms2260 23232398
10. Hertweck A, Evans CM, Eskandarpour M, Lau JCH, Oleinika K, Jackson I, et al. T-bet Activates Th1 Genes through Mediator and the Super Elongation Complex. Cell Reports. 2016;15 : 2756–2770. doi: 10.1016/j.celrep.2016.05.054 27292648
11. Hunt KA, Zhernakova A, Turner G, Heap GAR, Franke L, Bruinenberg M, et al. Newly identified genetic risk variants for celiac disease related to the immune response. Nature Genetics. Nature Publishing Group; 2008;40 : 395–402. doi: 10.1038/ng.102 18311140
12. Lazarevic V, Glimcher LH, Lord GM. T-bet: a bridge between innate and adaptive immunity. Nature Reviews Immunology. 2013;13 : 777–789. doi: 10.1038/nri3536 24113868
Štítky
Genetika Reprodukční medicína
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2017 Číslo 9
Nejčtenější v tomto čísle- "T-bet"-ing on autoimmunity variants
- Analysis of nuclear and organellar genomes of in humans reveals ancient population structure and recent recombination among host-specific subpopulations
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



