A systematic genomic screen implicates nucleocytoplasmic transport and membrane growth in nuclear size control
Membrane-bound organelles are maintained at a size proportional to cell size during cell growth and division. How this is achieved is a little-understood area of cell biology. The nucleus is generally present in single copy within a cell and provides a useful model to study overall membrane-bound organelle growth and organelle size homeostasis. Previous mechanistic studies of nuclear size control have been limited to cell-free nuclear assembly systems. Here, we screened a near genome-wide fission yeast gene deletion collection for mutants exhibiting aberrant nuclear size, to identify, more systematically, components involved in nuclear size control. Roles for protein complexes previously implicated in nuclear mRNA export and membrane synthesis were identified. Molecular and genetic analysis of mRNA nuclear export gene mutant cells with enlarged nuclear size revealed that general accumulation of nuclear content, including bulk mRNA and proteins, accompanies the nuclear size increase which is dependent on new membrane synthesis. We propose that properly regulated nucleocytoplasmic transport and nuclear envelope expansion are critical for appropriate nuclear size control in growing cells.
Published in the journal:
. PLoS Genet 13(5): e32767. doi:10.1371/journal.pgen.1006767
Category:
Research Article
doi:
https://doi.org/10.1371/journal.pgen.1006767
Summary
Membrane-bound organelles are maintained at a size proportional to cell size during cell growth and division. How this is achieved is a little-understood area of cell biology. The nucleus is generally present in single copy within a cell and provides a useful model to study overall membrane-bound organelle growth and organelle size homeostasis. Previous mechanistic studies of nuclear size control have been limited to cell-free nuclear assembly systems. Here, we screened a near genome-wide fission yeast gene deletion collection for mutants exhibiting aberrant nuclear size, to identify, more systematically, components involved in nuclear size control. Roles for protein complexes previously implicated in nuclear mRNA export and membrane synthesis were identified. Molecular and genetic analysis of mRNA nuclear export gene mutant cells with enlarged nuclear size revealed that general accumulation of nuclear content, including bulk mRNA and proteins, accompanies the nuclear size increase which is dependent on new membrane synthesis. We propose that properly regulated nucleocytoplasmic transport and nuclear envelope expansion are critical for appropriate nuclear size control in growing cells.
Introduction
Much is known about the molecular mechanisms that underpin membrane trafficking and local membrane growth in eukaryotic cells [1], but how membrane-bound organelles determine their overall growth rate and maintain an appropriate size is not well understood. The simple shape of the nucleus, and the fact that it is generally present in single copy within a cell, makes it a useful model to study overall membrane-bounded organelle growth and organelle size homeostasis. Work in algae and sea urchin embryos led Hertwig in 1903 to propose that there is a constant karyoplasmic ratio characteristic of cells [2]; since then nuclear size has been reported to correlate with cell size across a range of cell types and species [2,3]. Budding and fission yeasts exhibit a nuclear size proportional to cell size, resulting in a constant ratio of nuclear and cellular volumes (N/C ratio) [4,5]. In fission yeast the N/C ratio remains constant throughout the cell cycle, and no increase in the ratio is observed during or after S phase; even a 16-fold increase in nuclear DNA content does not affect N/C ratio [5]. These results indicate that, contrary to the generally accepted view, nuclear size is not directly determined by nuclear DNA content. Increases in ploidy do result in enlarged nuclei but this occurs indirectly, via an increase in cell volume which results in an increase in nuclear size [5]. Study of multi-nucleated cells with nuclei that are unevenly distributed throughout the cell revealed that the volume of each nucleus is proportional to that of its surrounding cytoplasm [5]. Results of an in vitro study of Xenopus egg extracts demonstrated that the available space surrounding a nucleus determines nuclear expansion rate [6], consistent with the fission yeast results. Cytoplasmic effects on nuclear size were also observed when erythrocyte nuclei injected into the cytoplasm of larger HeLa cells were found to grow in size [7]. Similarly, HeLa nuclei increased in volume when injected into the cytoplasm of X. laevis oocytes [8].
These experiments indicate that nuclear size is determined by the overall size of the cell, and that the cytoplasmic content immediately surrounding a particular nucleus is important for determining its size. However, these studies have given no insight into the molecular mechanisms that control nuclear growth and nuclear size homeostasis. An important contribution to molecular mechanism was provided by Levy and Heald [9]. These authors studied nuclear assembly around exogenous DNA added to egg extracts from two species of Xenopus: X. laevis, with large nuclei, and X. tropicalis, with small nuclei. A GFP-NLS (nuclear localisation signal) fusion protein was found to be accumulated at a faster rate into nuclei assembled in extracts from X. laevis. The authors concluded that nuclear transport was key to establishing the differing nuclear sizes assembled in vitro in the egg extracts. The transport factor Impα2 (an importin) was found to be at a higher level in X. laevis extracts than in X. tropicalis extracts whereas the transport factor Ntf2 was found to have an inverse relationship. Increasing the level of Impα2 increased the size of the in vitro assembled nuclei and overexpression of Impα2 increased nuclear size in embryonic cells. Addition of Lamin B3, a cargo of Impα2, to extracts also increased the size of the nuclei assembled in vitro. This study led the authors to propose that Lamin B3, transported by Impα2 into the nucleus, plays a key role in determining nuclear size. A further study reported that total lamin concentration, rather than the level of a specific lamin, affects the size of nuclei assembled in X. laevis egg extracts as well as in Xenopus embryos and mammalian tissue culture cells, though different lamin concentrations have different effects in different cell types and developmental stages, sometimes increasing the lamin level increased nuclear size and sometimes decreased it [10].
Fission yeast cells lack lamins yet display nuclear size control [5], suggesting that there are other key players in nuclear size control that have not, as yet, been identified. To identify, more systematically, other proteins involved in controlling nuclear growth and nuclear size homeostasis, we have carried out a genetic screen to identify genes that when deleted alter the N/C ratio of growing fission yeast cells, and so are candidates for playing roles in nuclear size control. Our screen has identified new factors as having roles in this control, and our studies have led us to conclude that nucleocytoplasmic transport of both RNA and protein and nuclear membrane growth contribute to the overall control of nuclear growth and nuclear size homeostasis in growing cells.
Results
Screen for N/C ratio mutants
To identify genes involved in nuclear size control, we carried out a screen for fission yeast mutants exhibiting an abnormal N/C ratio. We screened a gene deletion collection consisting of 2,969 viable deletion mutants representing approximately 80% of S. pombe viable haploid gene deletion strains [11]. The screen was carried out in three stages (Fig 1A). A visual screen was carried out on solid agar, examining growing cells on the edges of colonies, using the lipophilic fluorescent dye DiOC6 [12] to visualise nuclear and cellular membranes; 366 strains were identified as potentially having N/C ratios greater or smaller than wild type. In a second stage, these potential N/C ratio mutants were visually screened with DiOC6 during steady state growth in liquid media; 97 potential N/C ratio mutants were retained. In a third stage a nuclear membrane marker Cut11-GFP was introduced into cells, and imaging was carried out during steady state growth in liquid media. Cut11 is a transmembrane nuclear pore complex protein orthologous to H. sapiens and S. cerevisiae NDC1. N/C ratios were determined as described previously [5]. We identified 14 gene deletion strains that displayed a N/C ratio at least 15% higher than that of wild type cells (Fig 1B, S1 Table); four of these, mlo3, caf1, dss1 and trm112 (Fig 1C), exhibited a N/C ratio greater than 0.100, which was more than 25% higher than the wild type value of 0.081. No strains with a N/C ratio significantly smaller than wild type were identified.
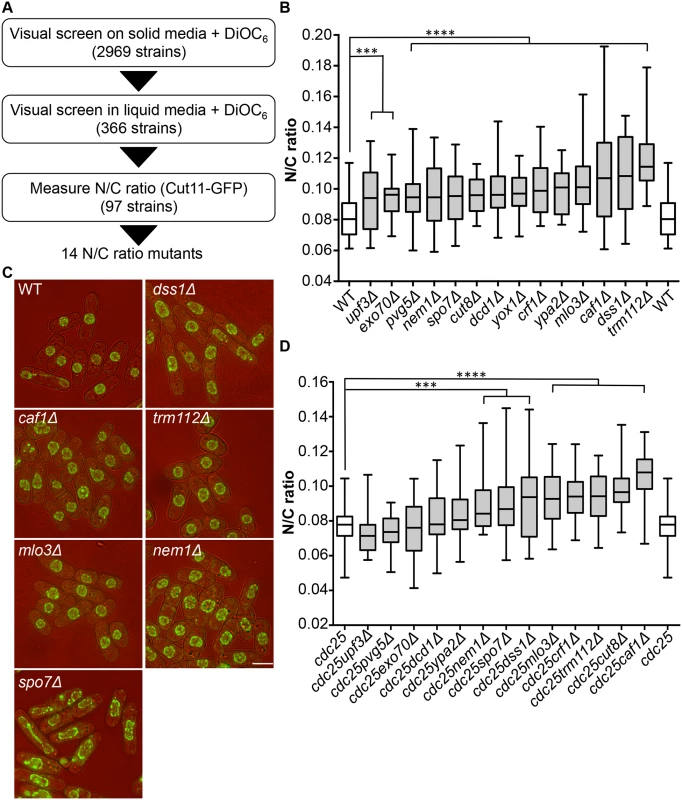
Mutants could generate aberrant N/C ratios as a consequence of asymmetric nuclear division instead of an interphase nuclear size control defect. If this were the case the N/C ratio would be expected to correct if cells grew for an extended time in interphase. We used the cell cycle mutant cdc25-22 to arrest cells in interphase, and then measured the N/C ratio (Fig 1D, S1 Table). Eight mutants still exhibited significantly aberrant N/C ratios during interphase arrest; these eight mutants carried deletions of the caf1, crf1, cut8, dss1, nem1, mlo3, spo7 and trm112 genes, and included the four gene deletions noted above with the strongest phenotypes in normal exponentially growing cells (Fig 1B). In addition to an increased N/C ratio, two of the candidate mutants, nem1Δ and spo7Δ, exhibited nuclear shape defects (Fig 1C). Strikingly, this unbiased screen independently identified two components of two complexes, Dss1-Mlo3 and Nem1-Spo7, which are involved in mRNA export from the nucleus and membrane synthesis respectively [13,14].
mRNA export and N/C ratio control
The mRNA export factor Dss1 and the RNA binding protein Mlo3 (orthologous to S. cerevisiae YRA1 and H. sapiens ALYREF) are components of a complex implicated in nuclear mRNA export [13]. To investigate this further we looked at other components of the complex. A third component of the complex, Rae1, targets the Dss1-Mlo3 mRNP to the nuclear pore [13]. The rae1 gene is essential so was not included in our screen of viable strains. To assess the N/C ratio phenotype of cells lacking Rae1 function we used the temperature-sensitive mutant rae1-167. Rae1-167 cells have been reported to accumulate poly(A)+RNA in the nucleus when shifted to the restrictive temperature [15], and we confirmed that poly(A)+RNA accumulates in the nucleus after shift from 25°C to 36°C; this accumulation occurs rapidly, beginning within 15 minutes of temperature shift (S1A Fig). At 25°C, the N/C ratio of this strain was similar to that of wild type (0.080), but after incubation at 36°C for 4 hours the N/C ratio increased by more than 50% to 0.125–0.135, values greater than those of any of the viable gene deletion strains identified in our screen (Fig 2A and 2B, S2 Table). Given this more extreme effect we focused our studies on rae1-167. A N/C ratio increase was detectable within 1 hour of the temperature shift, and became maximal an hour later (Fig 2C). This N/C ratio increase was caused by an increased nuclear growth rate, which was approximately twice that required to maintain a constant N/C ratio (Fig 2D). To determine whether this increase in the N/C ratio caused by inhibiting mRNA export required continued RNA synthesis, we treated cells with 15 μg/ml thiolutin which completely blocks RNA synthesis in wild type cells [16,17]. Although thiolutin treatment did not affect the N/C ratio of wild type cells, the treatment suppressed the increase of N/C ratio observed in rae1-167 cells following temperature shift (Fig 2A and 2B, S2 Table). These results indicate that defects in mRNA export can change the N/C ratio, and that this change requires continued RNA synthesis.
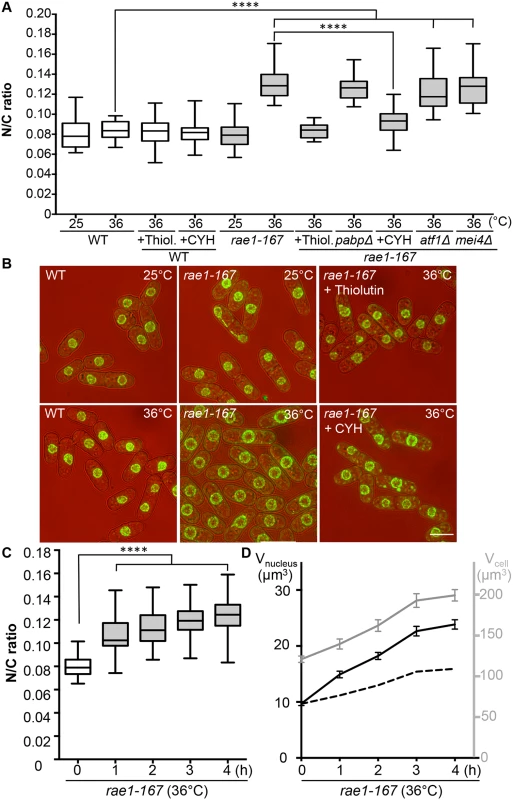
Protein transport and N/C ratio control
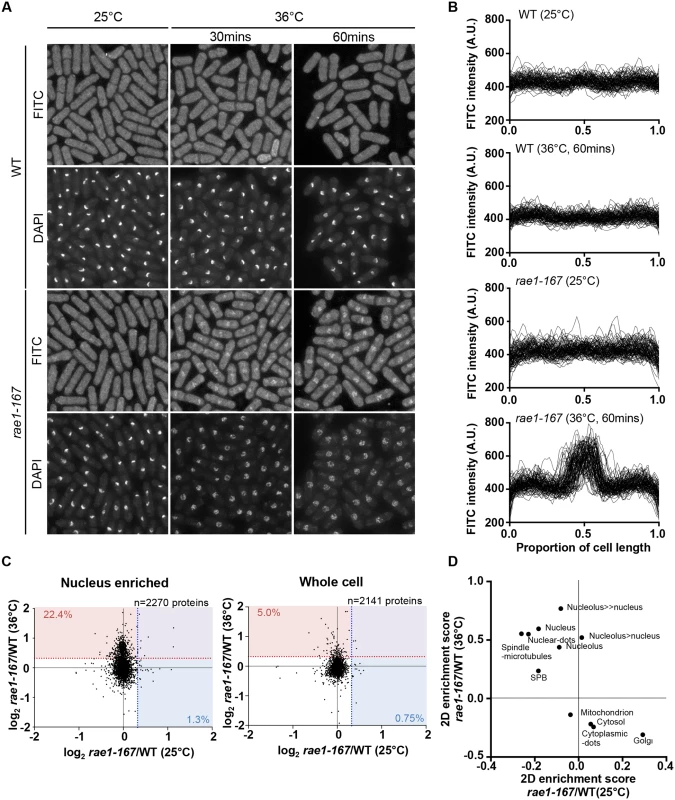
In addition to nuclear poly(A)+RNA accumulation, at 36°C rae1-167 cells exhibit nuclear accumulation of poly(A)-binding protein (Pabp) which shuttles between the nucleus and the cytoplasm in wild type cells (S1B Fig) [18]. Pabp-GFP accumulated in the nucleus within 1 hour of temperature shift in 100% of rae1-167 cells (S1C and S1D Fig). However, deletion of the pabp gene did not significantly suppress the high N/C ratio of rae1-167 cells, so accumulation of Pabp is not sufficient to cause the N/C ratio increase (Fig 2A, S2 Table). To assess whether proteins more generally accumulated in the nuclei of rae1-167 cells, we used the fluorescent protein-staining dye fluorescein isothiocyanate (FITC). DAPI staining was used to identify nuclei, and nuclei of rae1-167 cells appear to show reduced compaction of chromatin following shift to the restrictive temperature (Fig 3A). Proteins were uniformly distributed throughout nuclei and cytoplasms of both wild type and rae1-167 cells at 25°C. Within 30 minutes of shift to 36°C, rae1-167 cells exhibited significant nuclear protein accumulation, in contrast to temperature shifted wild type cells which still had a uniform protein distribution between the nucleus and the cytoplasm (Fig 3A and 3B). This result suggested protein accumulation following mRNA export inhibition also contributes to the N/C ratio increase observed, consistent with previous observations in which prolonged inhibition of nuclear export of proteins by Leptomycin B (LMB), an inhibitor of exportin Crm1 [19], increased the N/C ratio [5]. To investigate this possibility further we tested whether the N/C ratio increase following mRNA export inhibition requires continued protein synthesis by treating cells with 100 μg/ml cycloheximide (CYH) which inhibits protein synthesis [20]. Although CYH treatment did not affect the N/C ratio of wild type cells, the treatment largely suppressed the high N/C ratio observed in rae1-167 cells following shift to 36°C (Fig 2A and 2B, S2 Table). This result indicates that continued protein synthesis is required for the N/C ratio increase in these cells.
Characterisation of nuclear content in rae1-167 cells
We next tested whether it was general bulk protein and mRNA accumulation or accumulation of a smaller number of specific proteins and mRNAs which was taking place during nuclear enlargement. We characterised the protein and mRNA content of rae1-167 nuclei at the level of individual proteins and mRNAs using SILAC mass spectrometry and microarray analysis respectively.
To identify proteins accumulated in rae1-167 nuclei with an enlarged N/C ratio, we compared the protein content of wild type and rae1-167 nuclei. We enriched for nuclei to allow detection of low abundance nuclear proteins that may not be detected in whole cell samples. We confirmed that nuclear enriched samples were enriched for nuclear-localised and depleted for cytosol-localised proteins (P<0.02) (S3 Table). When we compared the protein content of nuclear enriched samples, there was an increase in a subset of proteins in rae1-167 relative to wild type at the restrictive temperature, that was not observed at the permissive temperature. 22.4% of proteins detected at 36°C (509/2270 proteins detected), in contrast to only 1.3% at 25°C (30/2270 proteins detected), were present in the rae1-167 nucleus enriched sample at a level at least 25% higher than in wild type (Fig 3C). Proteins found at this level at 36°C and not at 25°C are listed in S4 Table. To examine what classes of proteins were enriched in rae1-167 nuclei we carried out 2D enrichment analysis [21]. The proteins enriched in rae1-167 nuclei relative to wild type nuclei at 36°C and not at 25°C showed significant enrichment of proteins reported to be localised to the nucleus and subnuclear structures in S. pombe [19] (Fig 3D). Enrichment of specific gene ontology (GO) categories is shown in S2 Fig; nucleic acid binding proteins were enriched in proteins increased in rae1-167 nuclei relative to wild type nuclei at 36°C and not at 25°C. Therefore, our analysis suggests that bulk accumulation of many different proteins localised to the nucleus is taking place in rae1-167 nuclei when the N/C ratio is enlarged, rather than accumulation of a few specific proteins.
We next investigated the steady-state levels of individual mRNA transcripts by microarray analysis to assess whether the accumulation observed is of a few specific mRNAs or bulk accumulation of many mRNAs. Levels of 888 mRNAs were increased at least 2-fold in rae1-167 cells at 36°C relative to 25°C (S5 Table). These included mRNA transcripts of 106 common environmental stress response (CESR) genes [22] and 64 meiotic genes [23]. To examine whether increased expression of either of these specific groups of genes causes the N/C ratio enlargement observed in rae1-167 cells, we used the transcription factor mutants, atf1Δ and mei4Δ, which are respectively defective in induction of most CESR genes and of the middle meiotic genes [22,24]. Deletion of neither atf1 nor mei4 suppressed the high N/C ratio of rae1-167 cells (Fig 2A, S2 Table), indicating that specific transcription-driven increases of CESR gene or middle meiotic gene mRNAs are not the cause of the increased N/C ratio of rae1-167 cells. It is possible that retaining mRNAs in the nucleus could affect their stability; in this situation increased levels of these specific groups of mRNAs could be causative of the nuclear size increase because atf1Δ and mei4Δ mutants would be unlikely to affect their steady state level. Taken together, our mass spectrometry and microarray analyses indicate that general bulk accumulation of a large number of proteins and mRNAs occurs in rae1-167 cells when an N/C ratio increase is observed, and that many of these proteins are normally localised in the nucleus.
Membrane synthesis and N/C ratio control
Two more enlarged N/C ratio candidates identified in our screen were nem1 and spo7. These encode the catalytic and regulatory subunits of the Nem1-Spo7 phosphatase complex, responsible for dephosphorylation and activation of Ned1, a lipin family phosphatidic acid phosphatase [14,25]. In addition to the nuclear size phenotype in these mutants, nuclear shape deformation suggestive of nuclear envelope overproliferation was also observed [26]. These observations suggest that correct regulation of lipid metabolism leading to changes in membrane synthesis plays a role in nuclear size control, and we hypothesised that the increased N/C ratio of nem1 and spo7 deletion mutant cells may be caused by inappropriate nuclear envelope expansion. If this is the case, then inhibition of fatty acid synthesis, which is required for membrane growth [25,26,27], should suppress the increased N/C ratio observed in these mutant cells. To test this, we used a temperature-sensitive mutant cut6-621, which is defective in acetyl-CoA carboxylase and impaired in fatty acid metabolism [28]. The cut6-621 mutant blocked the increase in nuclear size of nem1Δ cells and suppressed the nuclear shape change (Fig 4A and 4B). There was no effect of cut6-621 on the N/C ratio of nem1+ cells. These results indicate that nem1 deletion leads to overproduction of phospholipid by inactivation of Ned1, causing inappropriate expansion of the nuclear envelope resulting in a N/C ratio increase. We next investigated whether new membrane synthesis is required for nuclear enlargement in the rae1-167 mutant, by combining the rae1-167 and cut6-621 mutations and measuring the N/C ratio. The N/C ratio of the double mutant is significantly lower than that of the rae1-167 mutant following shift to 36°C (Fig 4C and 4D, S2 Table), indicating that new membrane synthesis is required for the nuclear size increase observed in rae1-167 cells.
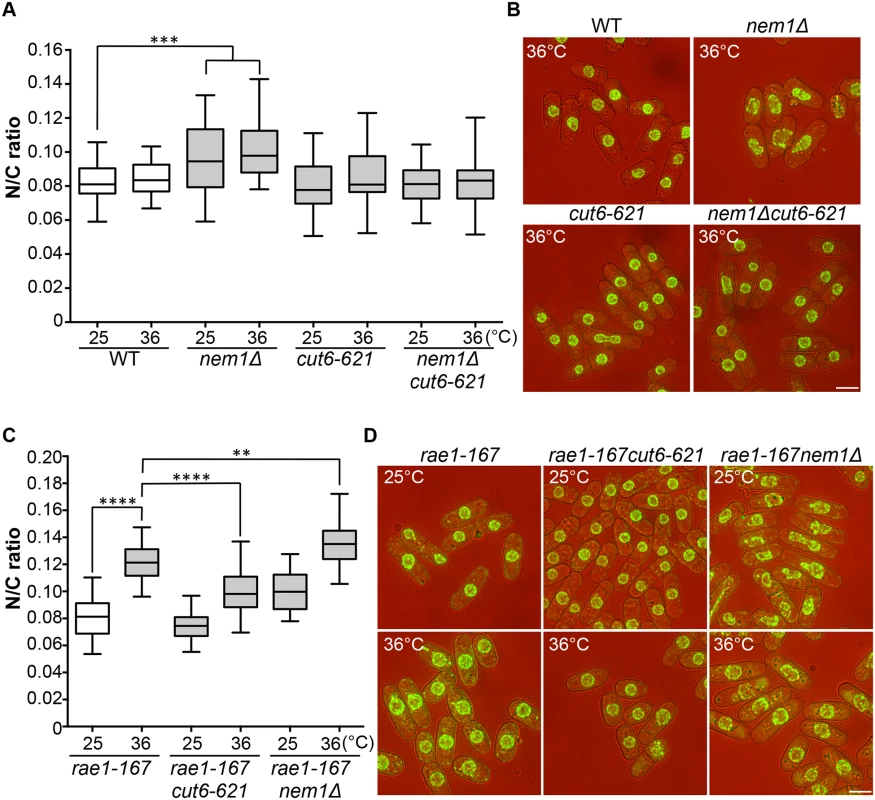
Combining the rae1-167 mRNA export mutant with the nem1Δ membrane synthesis regulation mutant led to a further N/C ratio increase, greater than that observed in either single mutant (Fig 4C and 4D). This suggests that two distinct processes are implicated in nuclear size control, membrane synthesis and nucleocytoplasmic transport. The shape phenotype of the nem1Δ single mutant was also suppressed by the rae1-167 mutation (Fig 4D).
Discussion
Our screen of viable fission yeast gene deletion strains identified 8 genes that when deleted lead to an increased N/C ratio in both exponentially growing and interphase arrested cells. Significantly, among these 8 were 4 genes encoding two components of a complex involved in nuclear mRNA export (dss1 and mlo3) and two components of a complex involved in lipid metabolism (nem1 and spo7). We did not find any gene deletions with a N/C ratio smaller than wild type, suggesting that reduced nuclear size might be deleterious, resulting in lethality. A screen of the diploid heterozygous gene deletion collection [11] could be expected to have less severe effects and may reveal genes that have decreased N/C ratios. It is possible that the rapid growth of unicellular fission yeast cells may require rapid ribosome biogenesis, which results in a limited range of N/C ratio perturbations being viable. A screen of essential genes may identify more severe phenotypes. The four genes, mlo3, caf1, dss1 and trm112, with the strongest deletion phenotypes increased the N/C ratio by 25%, an increase that was also observed in enlarged mutant cells blocked in interphase. All four genes are thought to be involved in RNA metabolism, and two of them, dss1 and mlo3, encode protein components of a complex required for nuclear mRNA export. We examined a third component of this complex, Rae1, which associates with the nuclear pore. A temperature-sensitive mutant of this essential gene, rae1-167, showed a 50% N/C ratio increase at the restrictive temperature, an increase greater than that seen in dss1Δ or mlo3Δ cells. The 50% N/C ratio increase was the result of a doubling of nuclear growth rate, and required continued RNA and protein synthesis. Both the mRNA and protein content of the enlarged nucleus increased. Mass spectrometry and microarray analyses showed that bulk nuclear accumulation of a large number of different proteins and mRNAs, rather than accumulation of a few specific proteins and mRNAs was taking place. This suggests that general accumulation of nuclear content contributes to the N/C ratio increase observed, though it is also possible that one, or a subset, of the many accumulated proteins and mRNAs may more specifically effect the N/C ratio increase.
Membrane growth is required for the nuclear size increase of rae1-167 cells. The cut6-621 mutant impairs nuclear membrane growth and also prevents the rae1-167 nuclear size increase. The importance of membrane growth for nuclear size was also shown by our identification of two lipid metabolic genes, nem1 and spo7, encoding proteins that form a phosphatase complex responsible for activation of Ned1, a lipin family phosphatidic acid phosphatase [14]. Inactivation of Ned1 by either nem1Δ or spo7Δ induced nuclear envelope expansion, resulting in aberrant nuclear shapes and an increased N/C ratio. These changes were observed in cells blocked in interphase and so are not due to aberrant mitosis. These phenotypes, like the N/C ratio increase of rae1-167 cells, were suppressed by inhibition of nuclear membrane growth by the cut6-621 mutation. These observations are consistent with observation that a phosphomimetic mutant of Ned1 exhibits a 34% increase in nuclear surface area in interphase [14]. The rae1-167nem1Δ double mutant exhibited a N/C ratio increase greater than that of either single mutant, suggesting that two distinct processes are important for nuclear size control, both bulk nucleocytoplasmic transport and nuclear membrane growth.
In addition to the components of the Dss1-Mlo3 and Nem1-Spo7 complexes, our systematic screen identified four further gene deletion strains exhibiting enlarged N/C ratios. These carried deletions in caf1, cut8, crf1 and trm112. Caf1 encodes a deadenylase of the CCR4-NOT complex. The multifunctional CCR4-NOT complex has been implicated in many different areas of gene expression, both nuclear and cytosolic. These include regulation of histone modification, regulation of transcription initiation and elongation, nuclear poly(A)-RNA degradation, mRNA export, cytosolic poly(A)-RNA decay and protein turnover [29, 30]. It is possible that disruption of one or a few of these roles of the CCR4-NOT complex by caf1 deletion could lead to nuclear poly(A)-RNA accumulation and therefore effect nuclear size increase by the same mechanisms as those in play in rae1-167 cells. However, due to the diverse roles this complex plays, a separate mechanism is also possible. It is possible that cut8 deletion may cause nuclear size increase by similar mechanisms to those in rae1-167 cells as it encodes a nuclear proteasome tethering factor [31] so could affect nuclear protein levels. Cut8 mutant cells have also been reported to accumulate poly(A)-RNA in the nucleus at the restrictive temperature suggesting a possible role in mRNA export [32]. Although we have focused on the mRNA export role of Dss1 in this study because another component of the Dss1-Mlo3 complex was also identified by our screen, it is of note that Dss1, like Cut8, has been implicated in proteasome function [33]. Crf1 is predicted to encode a TOR-responsive transcriptional corepressor, and deletion of its S. cerevisiae orthologue perturbs repression of ribosomal protein gene transcription [34]. The S. cerevisiae orthologue of trm112 regulates methylation of tRNAs, rRNAs, and translation factors, and is required for synthesis of both 40S and 60S ribosomal subunits [35]. As ribosome biogenesis is dominant in rapidly growing yeast cells and involves both nuclear import of ribosomal proteins and export of ribosomal subunits, it is possible that perturbing the biogenesis process, for example by crf1 or trm112 deletion, could lead to the accumulation of dysfunctional ribosomes or their constituents in the nucleus thus influencing nuclear size. Therefore, the remaining candidates from our screen all encode proteins that might have roles in regulating overall RNA and protein levels, and so impact the RNA and protein content of the nucleus.
Nucleocytoplasmic transport has also been implicated in nuclear size control in metazoa. The study of nuclear assembly in Xenopus egg extracts discussed in the Introduction, and subsequent studies in Xenopus egg extracts and mammalian cells have implicated the transport factors Impα2 and Ntf2 and the import of lamins in determination of nuclear size [9,10,36]. Our data indicates that there are roles for other components involved in nucleocytoplasmic transport and also for nuclear envelope growth in nuclear size homeostasis. Our in vivo study of fission yeast cells in steady state growth has revealed a role for nucleocytoplasmic transport of mRNAs and proteins in interphase nuclear size control, which is dependent on continued RNA and protein synthesis.
Our work has shown that both the accumulation of nuclear content and membrane synthesis, as well as the linkage between these two processes, must be considered when proposing potential models of nuclear size control. Outlined below are two examples of the types of mechanisms for maintenance of the N/C ratio that take account of these considerations. In one, overall cytoplasmic content determines how much protein and RNA is imported into the nucleus, and as a cell grows the resulting increase in nuclear content stimulates new membrane growth, enlarging the nucleus in balance with the cytoplasm. Nuclear content could promote nuclear envelope expansion in one of two ways: increased bulk RNA and protein import could put pressure on the nuclear membrane altering its tension and inducing its expansion [37], or the increased bulk RNA and protein import could lead to increased import of one or a group of specific RNAs and proteins that bring about nuclear envelope expansion. In a second model we suggest that overall cytoplasmic content determines the growth rate of the nuclear membrane, perhaps operating through global cellular membrane growth being controlled by cell size, with a certain proportion of total cellular membrane being delivered to the nucleus. The increased nuclear surface area would result in the incorporation of more nuclear pore complexes, resulting in increased import of protein and RNA, increasing nuclear size in balance with the cytoplasm. Both mechanisms implicate nucleocytoplasmic transport and nuclear membrane growth in interphase nuclear size control, and so are compatible with the data presented here and by others elsewhere. However, in the first model it is the accumulation of nuclear content that is the initial driver of nuclear growth, whilst in the second it is growth of the nuclear membrane. Obviously these are only examples of possible mechanisms.
We conclude that appropriately regulated nucleocytoplasmic transport and nuclear membrane growth are central to nuclear size control. This may be relevant to disease states given that abnormal nuclear size and shape phenotypes are observed in many diseases [38,39] but the role of nuclear size in the pathology of these diseases remains unclear. The genetic identification of processes involved in nuclear size control in fission yeast provides a tractable system in which to investigate this control, and contributes to our understanding of how membrane-bound organelles regulate their overall growth and size.
Materials and methods
Yeast general methods
Strains used are listed in S6 Table. Gene tagging was performed by PCR and homologous recombination [40]. S. pombe media and methods as described previously [41]. For mass spectrometry, cells were grown in SILAC media; heavy labelled samples were grown for >8 generations in media supplemented with heavy arginine (L-Arginine:HCL (U13C6, 99%)) and heavy lysine (L-Lysine:2HCl (U13C6, 99%)) (Cambridge Isotope Laboratories Inc.). All other strains were grown in YE4S. SILAC media used was EMM (6 mM ammonium chloride) supplemented with 0.25 mg/ml leucine, 0.15 mg/ml uridine, 0.04 mg/ml arginine and 0.03 mg/ml lysine [42]. YE4S used was Yeast extract (Difco) supplemented with adenine, leucine, uracil and histidine at 225 mg/l.
Screen for N/C ratio mutants
The 3-stage screen was carried out as described above. Firstly, deletion mutants were incubated at 25°C for 12 to 20h in 300 μl of YE4S in 96-well plates then inoculated onto YE4S agar plates containing DiOC6 at 10 μg/ml (Life technologies) using a pin tool (V & P Scientific, Inc) and incubated at 25°C for 12 to 20h. The N/C ratio of each mutant strain was estimated by comparison with the wild type strain growing in the same plate using a Zeiss Axioskop 40 microscope. 102 of the 2,969 deletion mutants failed to grow on plates so were excluded from the screen. 366 mutant strains were selected for a secondary visual screen in liquid medium. Cells were collected from individual exponentially growing cultures of the 366 candidate mutants, stained with DiOC6 and visually screened to estimate the N/C ratio. The 97 strains selected for the tertiary screen were tagged with the nuclear envelope marker protein Cut11-GFP and the nuclear and cellular volumes measured to assess the N/C ratio [5]. Images were analysed using ImageJ (NIH) as previously described [5]. Nuclear volume of shape mutants was calculated using ImageJ.
Statistical analysis
Unpaired Student’s t tests were used in Figs 1B and 1D, 2A and 2C, 4A and 4C to test statistical significance in pairwise comparisons.
Microscopy and image analysis
For visual screening, mutant cells were observed using a Zeiss Axioskop 40 microscope equipped with a 63x/1.4 NA objective and an AxioCam MRm camera. For protein and poly(A)+RNA localisation and N/C ratio measurement, cells were imaged using a DeltaVision Elite microscope (Applied Precision) comprised of an Olympus IX71 wide-field inverted fluorescence microscope, an Olympus Plan APO 60x oil, 1.42 NA objective, and a Photometrics CoolSNAP HQ2 camera (Roper Scientific). Images were captured in 0.3 or 0.4 μm z-sections over 5 μm and deconvolved using SoftWorx (Applied Precision). Projections of Cut11-GFP images were combined with bright field images. FITC intensity was measured using ImageJ (NIH).
FITC staining
5 ml of exponentially growing cells were fixed with 70% ethanol at 4°C for at least 30 mins, washed with phosphate buffered saline (PBS) then resuspended in PBS containing 1 μg/ml fluorescein isothiocyanate (FITC) (invitrogen). 4,6-diamidino-2phenylindole (DAPI) was used to stain DNA.
In situ hybridisation
The in situ hybridisation method used was described previously [43]. Oligo (dT)50 3’-end labeled with cy3 was used as the hybridisation probe. DAPI was used to stain DNA.
Microarrays
RNA was isolated by acid-phenol extraction and purified using RNeasy (Qiagen). Biotinylated cRNA was hybridised onto GeneChip Yeast Genome 2.0 arrays (Affymetrix) which were scanned with a GeneChip Scanner 3000 and analysed with GCOS v1.4 (Affymetrix) using default analysis settings and global scaling normalisation [44]. NCBI GEO accession number: GSE81666.
Mass spectrometry
Exponentially growing cells were incubated at 36°C or 25°C (as indicated) for 1 hour. Wild type (heavy labeled (H)) and rae1-167 (light labeled (L)) were mixed 1:1 by optical density (inverse labels also mixed). Whole cell and nuclear enriched protein samples were extracted.
For in-gel digestion each SILAC sample was loaded onto a NuPAGE Bis-Tris Protein Gel, 1.0 mm, 10-well (Thermo Fisher), and allowed to migrate through the gel before being stained with coomassie blue. Polyacrylamide gel slices were prepared for mass spectrometric analysis using the Janus liquid handling system (Perkin-Elmer). Each lane was excised into eight equally sized protein gel pieces, destained with 50% acetonitrile + 50 mM ammonium bicarbonate, reduced with 10 mM DTT, and alkylated with 55 mM iodoacetamide. After alkylation, the proteins were digested with 6 ng/μl trypsin overnight at 37°C. The resulting peptides were extracted in 2% formic acid/1% acetonitrile.
Samples were analysed by LC-MS/MS. An LTQ-Orbitrap Velos coupled to an UltiMate 3000 HPLC system for on-line liquid chromatographic separation was used for data acquisition. The extracted peptides were separated over a 70 min gradient elution (75 μm × 50 cm C18 column) with collision-induced dissociation (CID) selected as the activation method.
MaxQuant 1.3.0.5 was used for data processing and quantification. Default MaxQuant parameters were used with the following adjustments: Lys6 and Arg6 were the heavy labels, ‘Filter labelled amino acids’ was deselected, re-quantify was selected with the instruction to keep low-scoring versions of identified peptides within parameter groups and match between runs was selected. Data was searched against a UniProt extracted S. pombe FASTA file amended to include common contaminants. Normalised H/L ratios were used for analysis in Perseus 1.4.0.2. Average log2 rae1-167/WT ratios were calculated, data was annotated with ORFeome localisation data [19] and Gene Ontology terms (default Perseus GO annotation lists), and 2D enrichment analysis was carried out [21] (Benjamini-Hochberg FDR truncation threshold: 0.02). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE [45] partner repository with the dataset identifier PXD004530.
Protein extraction for mass spectrometry
Whole cell samples were produced by quenching cells by adding ice cold 100% (w/v) trichloroacetic acid to a final concentration of 10%. Cells were incubated on ice for at least 20 minutes, washed in ice-cold acetone and stored at -80°C. Pellets were washed and resuspended in lysis buffer (8 M urea, 50 mM ammonium bicarbonate, 5 mM EDTA and cOmplete Mini EDTA-free protease inhibitor cocktail (Roche)). 0.4 mm diameter acid washed glass beads (Sigma) were added and samples beaten to break cells (FastPrep120). Cell debris was pelleted and supernatant protein extracts stored at -80°C.
Nuclear enriched samples were produced using a protocol based on [46] and Experiment 18 [47]. Cells were harvested from 1 L cultures by centrifugation, washed in S buffer (1.4 M sorbitol, 40 mM HEPES and 0.5 mM MgCl2 at pH 6.5), resuspended in S buffer + 10 mM β-mercaptoethanol + 1 mM phenylmethanesulfonyl fluoride (PMSF) and incubated at 32°C for 10 minutes. Cells were harvested by centrifugation and the pellet was resuspended in S buffer + 1 mM PMSF containing 20 mg/gram cell pellet Zymolyase 100T (Amsbio) and incubated at 32°C until cell wall digestion was complete (confirmed by SDS lysis). Remaining steps were performed on ice. Cells were pelleted and washed four times in S buffer then resuspended in 20 ml F buffer (18% Ficoll 400 (w/v), 20 mM PIPES and 0.5 mM MgCl2) + 1 mM PMSF and lysed using a dounce homogeniser. The lysate was layered on 20 ml GF buffer (7% Ficoll (w/v), 20% glycerol, 20 mM PIPES and 0.5 mM MgCl2) and centrifuged at 20,000 g for 30 minutes. The pellet was resuspended in 20 ml F buffer and centrifuged at 3,000 g for 15 minutes. The supernatant was centrifuged at 20,000 g for 25 minutes. Nuclear enriched pellets were resuspended in 250 μl 2X Laemmli buffer (100 mM TRIS (pH 6.8), 4% SDS, 20% glycerol and 0.2 M dithiothreitol), heated to 99°C for 10 minutes and centrifuged. Supernatants were harvested and stored at -80°C.
Nuclear enrichment by this protocol was confirmed by preparation of whole cell and nuclear enriched samples of both wild type and rae1-167 cells grown at 36°C (heavy and light labelled). Heavy labelled nuclear enriched sample was mixed 1:1 by protein concentration (DC Protein Assay (Bio-Rad)) with light labelled whole cell extract for each strain (inverse label mixes also produced and analysed). Samples were analysed by SILAC mass spectrometry as described. Average log2 nuclear enriched/whole cell ratios were calculated, data was annotated with ORFeome localisation data [19] and 2D enrichment analysis was carried out [21]. Benjamini-Hochberg FDR was used for truncation, threshold value 0.02.
Supporting Information
Zdroje
1. Meer GV, Voelker DR, and Feigenson GW. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008; 9: 112–124. doi: 10.1038/nrm2330 18216768
2. Wilson EB. The Karyoplasmic Ratio. In The Cell in Development and Heredity (New York: The Macmillan Company); 1925. pp. 727–733.
3. Gregory T. Genome size evolution in animals. In The Evolution of the Genome, Gregory T., ed. (London: Elsevier Academic Press); 2005. pp. 4–87.
4. Jorgensen P, Edgington NP, Schneider BL, Rupes I, Tyers M, and Futcher B. The size of the nucleus increases as yeast cells grow. Mol. Biol. Cell 2007; 18: 3523–3532. doi: 10.1091/mbc.E06-10-0973 17596521
5. Neumann FR, and Nurse P. Nuclear size control in fission yeast. J. Cell Biol. 2007; 179: 593–600. doi: 10.1083/jcb.200708054 17998401
6. Hara Y, and Merten CA. Dynein-based accumulation of membranes regulates nuclear expansion in Xenopus laevis egg extracts. Dev. Cell 2015; 33: 562–575. doi: 10.1016/j.devcel.2015.04.016 26004509
7. Harris H. The reactivation of the red cell nucleus. J. Cell Sci. 1967; 2: 23–32. 6031005
8. Gurdon JB. Injected nuclei in frog oocytes: fate, enlargement, and chromatin dispersal. J. Embryol. Exp. Morph. 1976; 36: 523–540. 1010977
9. Levy DL, and Heald R. Nuclear size is regulated by importin α and Ntf2 in Xenopus. Cell 2010; 143: 288–298. doi: 10.1016/j.cell.2010.09.012 20946986
10. Jevtić P, Edens LJ, Li X, Nguyen T, Chen P, and Levy DL. Concentration-dependent effects of nuclear lamins on nuclear size in Xenopus and Mammalin cells. J. Biol. Chem. 2015; 290: 27557–27571. doi: 10.1074/jbc.M115.673798 26429910
11. Kim DU, Hayles J, Kim D, Wood V, Park HO, Won M, Yoo HS, Duhig T, Nam M, Palmer G, et al. Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe. Nat. Biotechnol. 2010; 28: 617–623. doi: 10.1038/nbt.1628 20473289
12. Koning AJ, Lum PY, Williams JM, and Wright R. DiOC6 staining reveals organelle structure and dynamics in living yeast cells. Cell Motil. Cytoskeleton 1993; 25: 111–128. doi: 10.1002/cm.970250202 7686821
13. Thakurta AG, Gopal G, Yoon JH, Kozak L, and Dhar R. Homolog of BRCA2-interacting Dss1p and Uap56p link Mlo3p and Rae1p for mRNA export in fission yeast. EMBO J. 2005; 24: 2512–2523. doi: 10.1038/sj.emboj.7600713 15990877
14. Makarova M, Gu Y, Chen JS, Beckley JR, Gould KL, and Oliferenko S. Temporal regulation of lipin activity diverged to account for differences in mitotic programs. Curr. Biol. 2016; 26: 237–243. doi: 10.1016/j.cub.2015.11.061 26774782
15. Brown JA, Bharathi A, Ghosh A, Whalen W, Fitzgerald E, and Dhar R. A mutation in the Schizosaccharomyces pombe rae1 gene causes defects in poly(A)+ RNA export and in the cytoskeleton. J. Biol. Chem. 1995; 270: 7411–7419. 7706287
16. Mendell JT, Medghalchi SM, Lake RG, Noensie EN, and Dietz HC. Novel Upf2p orthologues suggest a functional link between translation initiation and nonsense surveillance complexes. Mol. Cell. Biol. 2000; 20: 8944–8957. 11073994
17. Grigull J, Mnaimneh S, Pootoolal J, Robinson MD, and Hughes TR. Genome-wide analysis of mRNA stability using transcription inhibitors and microarrays reveals posttranscriptional control of ribosome biogenesis factors. Mol. Cell. Biol. 2004; 24: 5534–5547. doi: 10.1128/MCB.24.12.5534-5547.2004 15169913
18. Thakurta AG, Yoon JH, and Dhar R. Schizosaccharomyces pombe spPABP, a homologue of Saccharomyces cerevisiae Pab1p, is a non-essential, shuttling protein that facilitates mRNA export. Yeast 2002; 19: 803–810. doi: 10.1002/yea.876 12112233
19. Matsuyama A, Arai R, Yashiroda Y, Shirai A, Kamata A, Sekido S, Kobayashi Y, Hashimoto A, Hamamoto M, Hiraoka Y. et al. ORFeome cloning and global analysis of protein localization in the fission yeast Schizosaccharomyces pombe. Nat. Biotechnol. 2006; 24: 841–847. doi: 10.1038/nbt1222 16823372
20. Polanshek MM. Effects of heat shock and cycloheximide on growth and division of the fission yeast, Schizosaccharomyces pombe. J. Cell Sci. 1977; 23: 1–23. 893531
21. Cox J, and Mann M. 1D and 2D annotation enrichment: a statistical method integrating quantitative proteomics with complementary high-throughput data. BMC Bioinformatics 2012; 13(Suppl 16); S12.
22. Chen D, Toone WM, Mata J, Lyne R, Burns G, Kivinen K, Brazma A, Jones N, and Bähler J. Global transcriptional responses of fission yeast to environmental stress. Mol. Biol. Cell 2003; 14: 214–229. doi: 10.1091/mbc.E02-08-0499 12529438
23. Mata J, Lyne R, Burns G, and Bähler J. The transcriptional program of meiosis and sporulation in fission yeast. Nat. Genet. 2002; 32: 143–147. doi: 10.1038/ng951 12161753
24. Mata J, Wilbrey A, and Bähler J. Transcriptional regulatory network for sexual differentiation in fission yeast. Genome Biol. 2007; 8 (R217).
25. Santos-Rosa H, Leung J, Grimsey N, Peak-Chew S, and Siniossoglou S. The yeast lipin Smp2 couples phospholipid biosynthesis to nuclear membrane growth. EMBO J. 2005; 24: 1931–1941. doi: 10.1038/sj.emboj.7600672 15889145
26. Tange Y, Hirata A, and Niwa O. An evolutionarily conserved fission yeast protein, Ned1, implicated in normal nuclear morphology and chromosome stability, interacts with Dis3, Pim1/RCC1 and an essential nucleoporin. J. Cell Sci. 2002; 115: 4375–4385. 12376568
27. Yam C, He Y, Zhang D, Chiam KH, and Oliferenko S. Divergent strategies for controlling the nuclear membrane satisfy geometric constraints during nuclear division. Curr. Biol. 2011; 21: 1314–1319. doi: 10.1016/j.cub.2011.06.052 21802294
28. Saitoh S, Takahashi K, Nabeshima K, Yamashita Y, Nakaseko Y, Hirata A, and Yanagida M. Aberrant mitosis in fission yeast mutants defective in fatty acid synthetase and acetyl CoA carboxylase. J. Cell Biol. 1996; 134: 949–961. 8769419
29. Jonstrup AT, Andersen KR, Van LB, and Brodersen DE. The 1.4-Å crystal structure of the S. pombe Pop2 deadenylase subunit unveils the configuration of an active enzyme. Nucleic Acid Res. 2007; 35: 3153–3164. doi: 10.1093/nar/gkm178 17452359
30. Miller JE and Reese JC. Ccr4-Not complex: the control freak of eukaryotic cells. Crit. Rev. Biochem. Mol. Biol. 2012; 47:315–333. doi: 10.3109/10409238.2012.667214 22416820
31. Takeda K, and Yanagida M. Regulation of nuclear proteasome by Rho6/Ubc2 through ubiquitination and destruction of the sensor and anchor Cut8. Cell 2005; 122: 393–405. doi: 10.1016/j.cell.2005.05.023 16096059
32. Ideue T, Azad AK, Yoshida J, Matsusaka T, Yanagida M, Ohshima Y and Tani T. The nucleolus is involved in mRNA export from the nucleus in fission yeast. J. Cell Sci. 2004; 117: 2887–2895. doi: 10.1242/jcs.01155 15161942
33. Jossé L, Harley ME, Pires IM and Hughes DA. Fission yeast Dss1 associates with the proteasome and is required for efficient ubiquitin-dependent proteolysis. Biochem. J. 2006; 393:303–309. doi: 10.1042/BJ20051238 16149916
34. Martin DE, Soulard A, and Hall MN. TOR regulates ribosomal protein gene expression via PKA and the forkhead transcription factor FHL1. Cell 2004; 119: 969–979. doi: 10.1016/j.cell.2004.11.047 15620355
35. Sardana R, and Johnson AW. The methyltransferase adaptor protein Trm112 is involved in biogenesis of both ribosomal subunits. Mol. Biol. Cell 2012; 23: 4313–4322. doi: 10.1091/mbc.E12-05-0370 22956767
36. Vuković LD, Jevtić P, Zhang Z, Stohr BA, and Levy DL. Nuclear size is sensitive to NTF2 protein levels in a manner dependent on Ran binding. J. Cell. Sci. 2016; 129: 1115–1127. doi: 10.1242/jcs.181263 26823604
37. Raucher D, and Sheetz MP. Characteristics of a membrane reservoir buffering membrane tension. Biophys. J. 1999; 77: 1992–2002. doi: 10.1016/S0006-3495(99)77040-2 10512819
38. Webster M, Witkin KL, and Cohen-Fix O. Sizing up the nucleus: nuclear shape, size and nuclear envelope assembly. J. Cell Sci. 2009; 122: 1477–1486. doi: 10.1242/jcs.037333 19420234
39. Zink D, Fischer AH, and Nickerson JA. Nuclear structure in cancer cells. Nat. Rev. Cancer 2004; 9: 677–687.
40. Bähler J, Wu JQ, Longtine MS, Shah NG, McKenzie A 3rd, Steever AB, Wach A, Philippsen P, and Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 1998; 14: 943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y 9717240
41. Moreno S, Klar A, and Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991; 194: 795–823. 2005825
42. Bicho CC, de Lima Alves F, Chen ZA, Rappsilber J, and Sawin KE. A genetic engineering solution to the “arginine conversion problem” in stable isotope labeling by amino acids in cell culture (SILAC). Mol. Cell Proteomics 2010; 9: 1567–1577. doi: 10.1074/mcp.M110.000208 20460254
43. Amberg DC, Goldstein AL, and Cole CN. Isolation and characterization of RAT1: an essential gene of Saccharomyces cerevisiae required for the efficient nucleocytoplasmic trafficking of mRNA. Genes Dev. 1992; 6: 1173–1189. 1628825
44. Wu H, Zheng X, Araki Y, Sahara H, Takagi H and Shimoi H. Global gene expression analysis of yeast cells during sake brewing. Appl. Environ. Microbiol. 2006; 72: 7353–7358. doi: 10.1128/AEM.01097-06 16997994
45. Vizcaíno JA, Csordas A, del-Toro N, Dianes JA, Griss J, Lavidas I, Mayer G, Perez-Riverol Y, Reisinger F, Ternent T, et al. 2016 update of the PRIDE database and related tools. Nucleic Acids Res. 2016; 44(D1): D447–D456. doi: 10.1093/nar/gkv1145 26527722
46. Shimizu M, Roth SY, Szent-Gyorgyi C, and Simpson RT. Nucleosomes are positioned with base pair precision adjacent to the alpha 2 operator in Saccharomyces cerevisiae. EMBO J. 1991; 10: 3033–3041. 1915278
47. Alfa C, Fantes P, Hyams J, McLeod M, and Warbrick E. Experiments with fission yeast: A laboratory course manual: Cold Spring Harbor Lab. Press; 1993.
Štítky
Genetika Reprodukční medicínaČlánek vyšel v časopise
PLOS Genetics
2017 Číslo 5
Nejčtenější v tomto čísle
- A systematic genomic screen implicates nucleocytoplasmic transport and membrane growth in nuclear size control
- The fog of war: How network buffering protects plants’ defense secrets from pathogens
- Epistatic interaction between the lipase-encoding genes and causes liposarcoma in mice
