-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaThe social genome: Current findings and implications for the study of human genetics
article has not abstract
Published in the journal: . PLoS Genet 13(3): e32767. doi:10.1371/journal.pgen.1006615
Category: Perspective
doi: https://doi.org/10.1371/journal.pgen.1006615Summary
article has not abstract
Human health and diseases are influenced by genes and environments [1]. Recent advances in measures of genetic influences have led to calls for parallel advancements in methods to measure environments [2,3]. The article by Baud et al. [4] in the January 2017 issue suggests one provocative path forward: measure the genomes of proximate individuals.
“Social genetic effects” (SGEs, Baud et al. [4]) arise when an organism’s phenotype is influenced by the genetic makeup of that organism’s social environment. To put it another way, SGEs occur when the genotype of organism A influences the phenotype of organism B [5,6] (Fig 1). Previous SGE research focused on related individuals, especially mothers and offspring [7]. Recent work has explored SGEs in unrelated individuals, with a focus on a narrow range of phenotypes, e.g., sexual display behaviors [8] and body size [9]. The study by Baud et al. [4] suggests SGEs are more pervasive. To the extent results obtained from cage-dwelling mice generalize to free-living humans, findings suggest social genotypes are important environmental parameters.
Fig. 1. Social Genetic Effects (SGEs), social genetic correlation, and social epistasis. 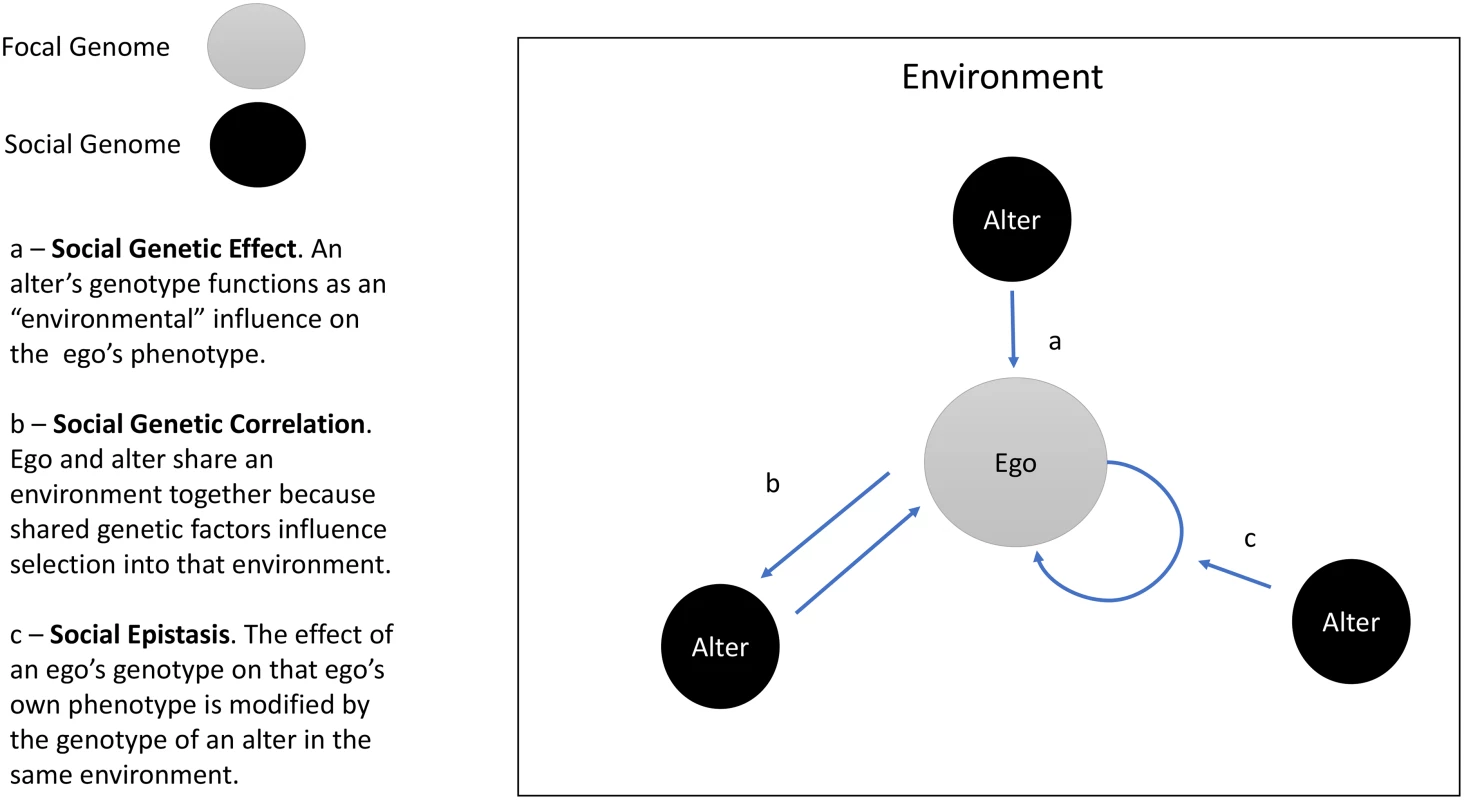
The figure illustrates three social genetic processes: SGEs, social genetic correlation, and social epistasis. The circle labeled “ego” represents the focal individual in an analysis. The circles labeled “alter” represent other individuals within the ego’s social environment. The arrows depict social genetic processes. The present study
Baud et al. conducted experiments with cage-dwelling mice to examine the effects of genetic composition of animals’ social environments on psychosocial and physiological phenotypes. Two separate designs evaluated these SGEs. The first design paired inbred C57BL/6J (B6) and DBA/2J (D2) mice as cage-mates under varying social genetic conditions: genetically homogenous (B6/B6 or D2/D2) and genetically heterogeneous (B6/D2). Analysis compared psychosocial and physiological phenotypes between strains and across social genetic conditions. Alongside phenotypic differences between strains, phenotypes also varied depending on social genetic conditions. Differences were primarily found in psychosocial phenotypes—measures of stress and anxiety—but also in wound healing. Pathway analysis of blood gene expression corroborated phenotypic evidence; SGEs on phenotype were reflected in SGEs on patterns of gene expression.
In the second design, the authors examined a large outbred mouse population (n> 2,000) in which mice were housed 3–6 to a cage. Using a combination of directly measured genetic data and pedigree information, analysis decomposed variance into direct genetic effects (the effects of a mouse’s own genes) and SGEs. This analysis identified SGEs for more than one third of 117 psychological and physiologic phenotypes at the p < 0.05 threshold. SGEs accounted for as much as 29% of phenotypic variance but in general were more modest (5% on average among those traits with p < 0.05 SGE).
In a further analysis, the authors attempted to evaluate whether SGEs might confound estimates of direct genetic effects. They report that cage-mates in the outbred-mouse study tended to be more genetically similar as compared to random pairs of mice. Failing to account for this social genetic structure in the data resulted in inflated estimates of heritability for several traits.
In sum, the study by Baud et al. suggests SGEs (i) are pervasive, affecting many phenotypes; (ii) can be pronounced, contributing as much to phenotypic variance in some cases as direct genetic effects; and (iii) are nonignorable, as they may lead to bias in other estimates if not taken into account.
Implications for research in humans
These experiments with mice highlight opportunities and challenges for social genetic research in humans. One opportunity is to investigate social genotypes as environmental measures. There is already human research investigating social phenotypic effects, e.g., the social “contagion” of obesity (“Are your friends making you fat?”) [10]. But ascertaining causality for social phenotypic influence is challenging [11,12]. Using genetic measures of the social environment to conduct a social version of Mendelian randomization analysis [13] may provide stronger grounds for causal inference.
Baud et al.’s findings further suggest that social genetic factors influencing variation in a given phenotype may be diverse. Thus, genetics previously linked with a particular phenotype may not be the only genetics of interest when considering social genetic effects on that phenotype. Analysis of pleiotropy, e.g., [14,15], may provide a helpful guide in devising more inclusive assays of the social genome.
In addition to direct effects of the social environment/genome, synergies between social and personal genetics are possible. Specifically, social genotyping could be used to study interactions between a person’s genes and the genes of socially proximate individuals. As Baud et al. note, specific genotypes may have different consequences for an organism’s phenotype depending on the prevalence of that genotype in the social environment (Fig 1, arrow b). Such “social epistasis” may be synergistic, with increasing prevalence of genotypes similar to one’s own amplifying genetic effects. This might be expected in settings where the social environment sets norms for behaviors (e.g., in cases like obesity or educational achievement). Other settings could produce antagonistic interactions, in which increasing prevalence of similar genotypes diminishes or reverses a genetic effect.
These opportunities exist alongside challenges. A primary issue is the extent to which social genotypes are independently determined. Individuals who share traits may be more likely to sort into social units together, a phenomenon called homophily [16,17]. Sorting is also observable at the level of individual genetic loci and polygenic predisposition to certain traits [18,19]. Thus, while SGEs may shape an individual’s phenotype or modify the phenotypic effects of that individual’s genes, reverse causation is also possible; i.e., an individual’s phenotype and/or genotype may shape the genetic composition of their social environment (Fig 1, arrow c). Relatedly, human social relationships are nested within larger social structures [20]. Recent findings in humans and monkeys showing genetic influence on position within society [21] and influence of social position on genomic function [22] suggest accounting for the structural context of social relationship will be important. A final issue is the role of social proximity in conditioning SGEs. In contrast to the mice in Baud et al.’s study, for whom the only social relationship was co-housing, human social relationships span a range of proximities (i.e., spouses as compared to Facebook friends). As human social theory suggests a role for weaker ties in some scenarios [23], research is needed to establish the range of designs in which social genotyping may prove informative.
Zdroje
1. Polderman TJ, Benyamin B, De Leeuw CA, Sullivan PF, Van Bochoven A, Visscher PM, et al. Meta-analysis of the heritability of human traits based on fifty years of twin studies. Nat Genet. 2015;47 : 702–709. doi: 10.1038/ng.3285 25985137
2. Khoury MJ, Wacholder S. Invited Commentary: From Genome-Wide Association Studies to Gene-Environment-Wide Interaction Studies-025EFChallenges and Opportunities. Am J Epidemiol. 2009;169 : 227–230. doi: 10.1093/aje/kwn351 19022826
3. Boardman JD, Daw J, Freese J. Defining the environment in gene–environment research: lessons from social epidemiology. Am J Public Health. 2013;103: S64–S72. doi: 10.2105/AJPH.2013.301355 23927514
4. Baud A, Mulligan M, Casale F, Ingels J, Bohl C, Callebert J, et al. Genetic variation in the social environment contributes to health and disease. PLoS Genet 13(1): e1006498. doi: 10.1371/journal.pgen.1006498 28121987
5. Wolf JB, Brodie ED III, Cheverud JM, Moore AJ, Wade MJ. Evolutionary consequences of indirect genetic effects. Trends Ecol Evol. 1998;13 : 64–69. 21238202
6. Moore AJ, Brodie ED III, Wolf JB. Interacting phenotypes and the evolutionary process: I. Direct and indirect genetic effects of social interactions. Evolution. 1997;51 : 1352–1362.
7. Champagne FA, Meaney MJ. Stress during gestation alters postpartum maternal care and the development of the offspring in a rodent model. Biol Psychiatry. 2006;59 : 1227–1235. doi: 10.1016/j.biopsych.2005.10.016 16457784
8. Petfield D, Chenoweth SF, Rundle HD, Blows MW. Genetic variance in female condition predicts indirect genetic variance in male sexual display traits. Proc Natl Acad Sci. 2005;102 : 6045–6050. doi: 10.1073/pnas.0409378102 15840726
9. Bergsma R, Kanis E, Knol EF, Bijma P. The contribution of social effects to heritable variation in finishing traits of domestic pigs (Sus scrofa). Genetics. 2008;178 : 1559–1570. doi: 10.1534/genetics.107.084236 18245326
10. Christakis NA, Fowler JH. The Spread of Obesity in a Large Social Network over 32 Years. N Engl J Med. 2007;357 : 370–379. doi: 10.1056/NEJMsa066082 17652652
11. Cohen-Cole E, Fletcher JM. Is obesity contagious? Social networks vs. environmental factors in the obesity epidemic. J Health Econ. 2008;27 : 1382–1387. doi: 10.1016/j.jhealeco.2008.04.005 18571258
12. VanderWeele TJ. Sensitivity Analysis for Contagion Effects in Social Networks. Sociol Methods Res. 2011;40 : 240–255. doi: 10.1177/0049124111404821 25580037
13. Lawlor DA, Harbord RM, Sterne JA, Timpson N, Davey Smith G. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med. 2008;27 : 1133–1163. doi: 10.1002/sim.3034 17886233
14. Bulik-Sullivan B, Finucane HK, Anttila V, Gusev A, Day FR, Loh P-R, et al. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47 : 1236–1241. doi: 10.1038/ng.3406 26414676
15. Barban N, Jansen R, de Vlaming R, Vaez A, Mandemakers JJ, Tropf FC, et al. Genome-wide analysis identifies 12 loci influencing human reproductive behavior. Nat Genet. 2016;48 : 1462–1472. doi: 10.1038/ng.3698 27798627
16. McPherson M, Smith-Lovin L, Cook JM. Birds of a feather: Homophily in social networks. Annu Rev Sociol. 2001; 415–444.
17. Centola D. The Spread of Behavior in an Online Social Network Experiment. Science. 2010;329 : 1194–1197. doi: 10.1126/science.1185231 20813952
18. Conley D, Laidley T, Belsky D, Fletcher J, Boardman J, Domingue B. Assortative mating and differential fertility by phenotype and genotype across the 20th century. Proc Natl Acad Sci. 2016; 24 : 6647–6652.
19. Domingue BW, Fletcher J, Conley D, Boardman JD. Genetic and educational assortative mating among US adults. Proc Natl Acad Sci. 2014;111 : 7996–8000. doi: 10.1073/pnas.1321426111 24843128
20. House JS, Landis KR, Umberson D. Social relationships and health. Science. 1988;241 : 540–545. 3399889
21. Belsky DW, Moffitt TE, Corcoran DL, Domingue B, Harrington H, Hogan S, et al. The Genetics of Success: How Single-Nucleotide Polymorphisms Associated With Educational Attainment Relate to Life-Course Development. Psychol Sci. 2016;27 : 957–972. doi: 10.1177/0956797616643070 27251486
22. Snyder-Mackler N, Sanz J, Kohn JN, Brinkworth JF, Morrow S, Shaver AO, et al. Social status alters immune regulation and response to infection in macaques. Science. 2016;354 : 1041–1045. doi: 10.1126/science.aah3580 27885030
23. Granovetter MS. The strength of weak ties. Am J Sociol. 1973; 1360–1380.
Štítky
Genetika Reprodukční medicína
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2017 Číslo 3
-
Všechny články tohoto čísla
- 2016 Reviewer and Editorial Board Thank You
- The social genome: Current findings and implications for the study of human genetics
- A rare loss-of-function mutation reduces blood eosinophil counts and protects from asthma
- A variant in the gene in a dog with ichthyosis
- Excess of genomic defects in a woolly mammoth on Wrangel island
- CUZD1 is a critical mediator of the JAK/STAT5 signaling pathway that controls mammary gland development during pregnancy
- Fishing for adaptive epistasis using mitonuclear interactions
- Comparison of independent screens on differentially vulnerable motor neurons reveals alpha-synuclein as a common modifier in motor neuron diseases
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- CUZD1 is a critical mediator of the JAK/STAT5 signaling pathway that controls mammary gland development during pregnancy
- A rare loss-of-function mutation reduces blood eosinophil counts and protects from asthma
- A variant in the gene in a dog with ichthyosis
- Fishing for adaptive epistasis using mitonuclear interactions
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



