-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaSuggestions from Geroscience for the Genetics of Age-Related Diseases
article has not abstract
Published in the journal: . PLoS Genet 12(11): e32767. doi:10.1371/journal.pgen.1006399
Category: Perspective
doi: https://doi.org/10.1371/journal.pgen.1006399Summary
article has not abstract
Background
The relationship between aging and major age-related diseases, such as cardiovascular diseases (CVDs), Alzheimer disease (AD), type 2 diabetes (T2D), and cancer and the genetic contribution to both phenomena are important questions in biomedicine. Over the past few decades, each disease has been studied separately in hundreds of genome-wide association studies (GWAS) involving increasing numbers of patients and SNPs, generating results that can explain only in part the genetics of the traits of interest. On several occasions, the results obtained in one population have not been replicated in others, and the clinical application of these results is questionable.
The new field of “geroscience” [1] proposes a conceptual framework that could lead to more effective approaches for studying the genetics of age-related diseases, starting from the basic observation that their main risk factor is age and aging. Geroscience stresses that the basic molecular and cellular mechanisms underpinning aging and its related pathologies are much more interconnected than previously thought on the basis of purely clinical classifications, and largely overlap. This enables us to study and combat the diseases of the elderly all together, rather than one by one [2].
The Article by Kulminski et al.
The paper by Kulminski et al. [3] provides evidence for how fruitful this approach can be. The authors started by analysing the genetic predisposition to risks of major age-related diseases and mortality, taking advantage of GWAS data generated during the Atherosclerosis Risk in Communities Study (ARCS), and focused on two new promising SNPs (rs222826, rs 222827) on band 2q.22. Using a candidate gene approach, these two SNPs were applied to data from ARCS and from two other studies (Framingham Heart Study and Health and Retirement Study). The combination of advanced statistics and the uniqueness of the datasets, including longitudinal follow-up, allowed them: i) to address the inherent complexity of the genetics of age-related diseases, largely not shaped by natural selection and hidden by age-related heterogeneity and pleiotropic effects of genetic variants [4]; and ii) to explore the causal inferences from selected endophenotypes (body mass index, total and high density lipoproteins). Accordingly, the authors were able to validate risk loci buried in well studied datasets. Notably, in accordance with geroscience, the loci resulted in risks for different age-related diseases, supporting the links between them, and suggesting that this genomic region likely contains elements that play a role in the aging process. In the replication process, the authors noticed that some of the highlighted associations failed to be replicated. Analyzing these apparently contradictory results and profiting from the wealth of available data within these datasets, they found that the association with endophenotypes such as body mass index (BMI) is sensitive to the birth cohort effect, which can be assumed as a rough but informative proxy of environment. This result is in agreement with the intuitive yet neglected idea that genes do not act in isolation, and that observed phenotypes are always the result of gene–environment interactions. This consideration is particularly true for the genetics of age-related diseases, as a given variant interacts with environmental conditions, continuously changing and exerting different selective pressure according to birth cohort (Fig 1). In the last century, pervasive changes in anthropological environments led to significant epidemiological changes. The revolution in hygiene awareness, a major contributor to the unprecedented increase of life expectancy, and the concomitant emergence of an obesogenic environment (easy access to nutrient-rich food; reduced physical activity) exemplify changes that promoted the epidemiological explosion of obesity, metabolic disorders, and eventually of major age-associated diseases. Thus, subsequent generations were exposed to quite different environmental conditions and pressures during the last century, and it is easy to predict that the risk/protective effects of specific alleles changed accordingly. In their causal inference analysis, Kulminski et al. [3] noticed that the correlation of a given allele with a risky endophenotype is also sensitive to chronological age. This result is in line with the antagonistic pleiotropy theory suggesting that a given allele can play different roles at different ages and fits with the remodeling theory of aging [5], according to which the body of each individual undergoes a different or unexpected lifelong process of adaptation to the age-related accumulation of molecular and cellular damages and the consequent functional decline.
Fig. 1. Demography and the genetics of age-related diseases. 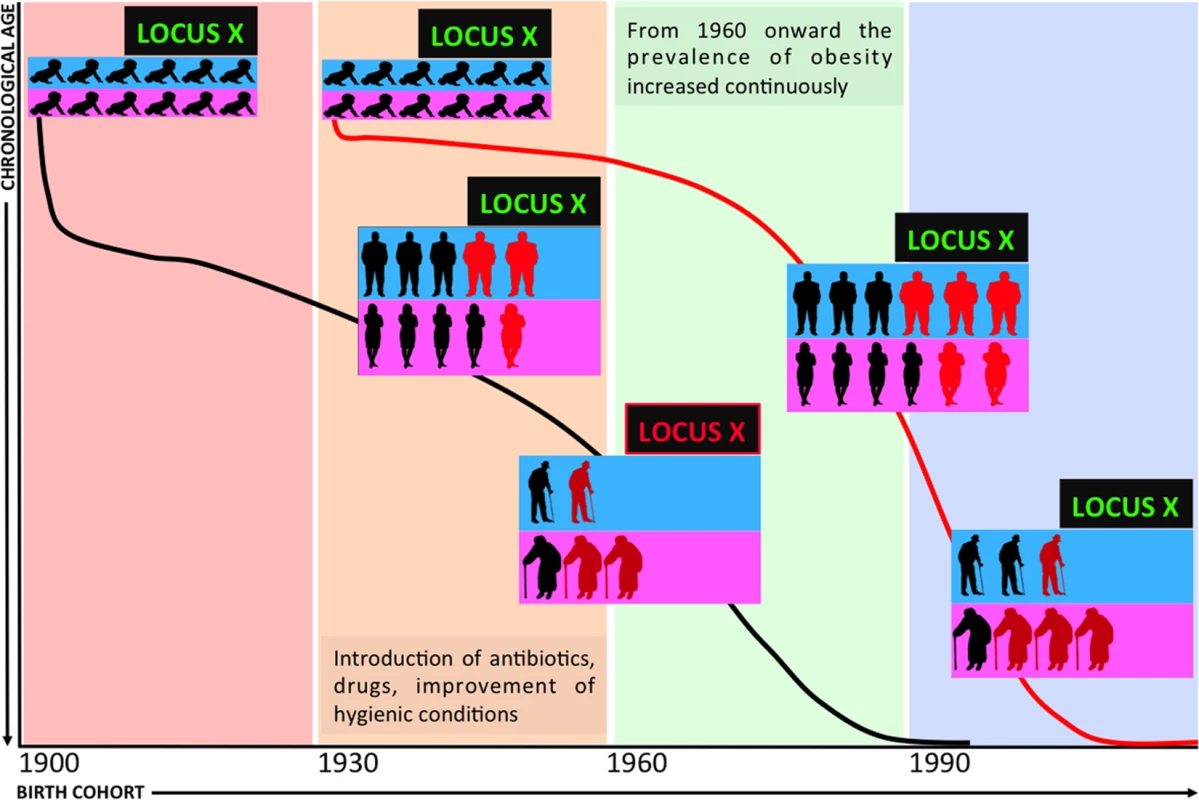
A schematic representation of the effect of birth cohort and chronological age on the genetic risk of age-related diseases is depicted. The interaction between demographic, environmental, and aging factors allows a genetic risk to emerge (LOCUS green becomes LOCUS X red) only in the presence of a unique combination of chronological age and birth cohort, due to the combined effect of changes in the environmental pressures and the physiopathological remodelling that occurs with age. Overall, chronological age and birth cohort are central and independent variables that should be carefully considered in studies on the genetics of major age-related diseases. Disregarding such basic demographic variables in cohorts heterogeneous for age and date of birth confounds analysis and results and can contribute to the difficulty in replicating results across different populations [4]. Such difficulties clearly emerge when different cohorts and datasets are put together, including subjects of different ancestry [6], in order to increase statistical power.
Future Directions
The paper by Kulminski et al. [3] shows how complex the study of the genetics of age-related diseases is in a globalized and changing world. Some people think that a concerted effort to generate whole genome sequences will solve existing problems. This represents a simplistic (and expensive) approach and, on the basis of our experience with GWAS, may have little explanatory or predictive power. The geroscience concept as demonstrated by the work in Kulminski et al., suggests an alternative way forward in which seemingly different phenotypes could have a shared underlying genetic architecture. Important covariates may also be shared, including environment (nutrition, lifestyle, activity, population genetics), sex (since the aging trajectories of men and women are different), and epistatic interactions (including not only the nuclear genome but mitochondrial and microbial genomes) [7–8].
In addition to new computational approaches and efforts, new phenotype models, such as centenarians and their families, could prove extremely useful [9–10] in solving some of the riddles of aging. Time will tell.
Zdroje
1. Kennedy BK, Berger SL, Brunet A, Campisi J, Cuervo AM, Epel ES, et al. Geroscience: linking aging to chronic disease. Cell. 2014 Nov 6;159(4):709–13. doi: 10.1016/j.cell.2014.10.039 25417146
2. Longo VD, Antebi A, Bartke A, Barzilai N, Brown-Borg HM, Caruso C, et al. Interventions to Slow Aging in Humans: Are We Ready? Aging Cell. 2015 Aug;14(4):497–510. doi: 10.1111/acel.12338 25902704
3. Kulminski A, He L, Culminskaya I, Loika Y, Kernogitski Y, Arbeev KG, et al. Pleiotropic Associations of Allelic Variants 1 in a 2q22 Region with Risks of Major Human Diseases and Mortality. PLoS Genet. In press.
4. Yashin AI, De Benedictis G, Vaupel JW, Tan Q, Andreev KF, Iachine IA, et al. Genes, demography, and life span: the contribution of demographic data in genetic studies on aging and longevity. Am J Hum Genet. 1999 Oct;65(4):1178–93. doi: 10.1086/302572 10486337
5. Franceschi C, Valensin S, Bonafè M, Paolisso G, Yashin AI, Monti D, et al. The network and the remodeling theories of aging: historical background and new perspectives. Exp Gerontol. 2000 Sep;35(6–7):879–96. 11053678
6. Sazzini M, Gnecchi Ruscone GA, Giuliani C, Sarno S, Quagliariello A, De Fanti et al. Complex interplay between neutral and adaptive evolution shaped differential genomic background and disease susceptibility along the Italian peninsula. Sci Rep. 2016 Sep 1;6 : 32513. doi: 10.1038/srep32513 27582244
7. Garagnani P, Pirazzini C, Giuliani C, Candela M, Brigidi P, Sevini F, et al. The three genetics (nuclear DNA, mitochondrial DNA, and gut microbiome) of longevity in humans considered as metaorganisms. Biomed Res Int. 2014;2014 : 560340. doi: 10.1155/2014/560340 24868529
8. Corella D, Ordovás JM. Aging and cardiovascular diseases: the role of gene-diet interactions. Ageing Res Rev. 2014 Nov;18 : 53–73. doi: 10.1016/j.arr.2014.08.002 25159268
9. Garagnani P, Giuliani C, Pirazzini C, Olivieri F, Bacalini MG, Ostan R, et al. Centenarians as super-controls to assess the biological relevance of genetic risk factors for common age-related diseases: a proof of principle on type 2 diabetes. Aging (Albany NY). 2013 May;5(5):373–85.
10. Fortney K, Dobriban E, Garagnani P, Pirazzini C, Monti D, Mari D, et al. Genome-Wide Scan Informed by Age-Related Disease Identifies Loci for Exceptional Human Longevity. PLoS Genet. 2015 Dec 17;11(12):e1005728. doi: 10.1371/journal.pgen.1005728 26677855
Štítky
Genetika Reprodukční medicína
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2016 Číslo 11
-
Všechny články tohoto čísla
- Suggestions from Geroscience for the Genetics of Age-Related Diseases
- The Mighty Fruit Fly Moves into Outbred Genetics
- The Rise of FXR1: Escaping Cellular Senescence in Head and Neck Squamous Cell Carcinoma
- A Helping Hand: RNA-Binding Proteins Guide Gene-Binding Choices by Cohesin Complexes
- The Genetic Architecture of Quantitative Traits Cannot Be Inferred from Variance Component Analysis
- Ancient Out-of-Africa Mitochondrial DNA Variants Associate with Distinct Mitochondrial Gene Expression Patterns
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Suggestions from Geroscience for the Genetics of Age-Related Diseases
- The Mighty Fruit Fly Moves into Outbred Genetics
- Ancient Out-of-Africa Mitochondrial DNA Variants Associate with Distinct Mitochondrial Gene Expression Patterns
- A Helping Hand: RNA-Binding Proteins Guide Gene-Binding Choices by Cohesin Complexes
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



