-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaSCA8 CAG/CTG Expansions, a Tale of Two ities: A Unique or Common Case?
article has not abstract
Published in the journal: . PLoS Genet 5(8): e32767. doi:10.1371/journal.pgen.1000593
Category: Perspective
doi: https://doi.org/10.1371/journal.pgen.1000593Summary
article has not abstract
A dozen dominant genetic disorders are caused by aberrant expansions of CAG or CTG trinucleotide repeats. As these repeats are complementary sequences, the expansions are incurred by both strands. However, it is the genic or transcribed strand that has defined the CAG and CTG repeat diseases. CAG repeat–associated diseases represent a group of neurological disorders, including Huntington's disease, dentatorubral pallidoluysian atrophy, spinal and bulbar muscular atrophy, and several spinocerebellar ataxias (SCA1–3, 6, 7, and 17), which are all caused by an expansion of CAG repeats in the coding region of genes with otherwise no other common features. The resulting mutant, toxic proteins bear a polyglutamine expansion (polyQ), which is sufficient to trigger a neuronal pathology [1]. The propensity of polyQ-expanded proteins to progressively misfold and aggregate appears to be doubly deleterious. First, it impairs normal functions or turn-over of host proteins. Second, polyQ aggregates alter neuronal physiology by activating stress responses or recruiting essential components of the cellular machinery [2]. In contrast, the CTG repeat expansion responsible for myotonic dystrophy type 1 (DM1), a multi-systemic disease affecting mainly the muscle but also the brain, is located in the 3′-untranslated region of the DMPK gene. The toxic entity is the CUG-expansion transcript, which adopts secondary structures and forms RNA foci, thereby sequestering RNA-binding proteins implicated in pre-mRNA splicing [3]. Muscleblind-like (MBNL) proteins are splicing factors whose recruitment to RNA foci leads to misregulated splicing of tissue-specific targets. In addition, CELF proteins, including CUG binding protein 1 (CUGBP1), are up-regulated in DM1 tissues. Both events contribute to the reversal from adult to fetal or developmental splicing patterns [4]. Thus, so far, it has been widely accepted that toxic RNA gain-of-function and toxic protein gain-of-function mechanisms underlie the distinction between CTG and CAG repeat–associated diseases [5]. Two articles on spinocerebellar ataxia type 8 (SCA8) published by the group of Laura Ranum, first in 2006 [6] and now in this issue of PLoS Genetics [7], suggest that this view is simplistic, deserving of attention and revision.
Since its discovery, SCA8 has been a curiosity among triplet repeat disorders. It was the first dominant SCA apparently not caused by a CAG expansion–encoding polyglutamine tract, as the uncovered mutation was a CTG repeat expansion in a transcribed gene (ATXN8OS) lacking open reading frames (ORFs) [8]. In 2006, Ranum and colleagues [6] designed transgenic mice carrying a bacterial artificial chromosome (BAC) encompassing the entire human SCA8 locus, with a normal or expanded CTG allele. Characterization of the model revealed bidirectional expression of both CUG - and CAG-expansion transcripts. Unexpectedly, the authors found that the CAG transcript was translated into a nearly pure polyQ protein that formed typical neuronal nuclear inclusions, the specific hallmark of polyQ diseases. The data suggested, therefore, that the pathology resulted, in part, from a toxic protein gain-of-function mechanism and raised the possibility that SCA8 could be caused by both toxic protein and toxic RNA gain-of-functions.
Daughters et al. now present a body of evidence suggesting that the toxic RNA pathomechanism substantially contributes to SCA8 pathology [7]. The authors show that the CUG-repeat transcripts expressed from the SCA8 locus form RNA foci in specific neurons in SCA8 patient and mouse brains. In addition, MBNL1 protein colocalizes with these RNA foci. Furthermore, the authors provide evidence for a genetic interaction between SCA8 and Mbnl1 gene loci by showing that loss of the Mbnl1 gene in SCA8 BACexp mice increases the motor phenotype. These results extend to mice earlier observations in flies showing that SCA8 CUG transcript expression caused toxicity that can be modulated by mutation in muscle-blind or three other genes encoding RNA-binding proteins [9]. Importantly, using cross-linking and immunoprecipitation (CLIP) analysis with an anti-CUGBP1 antibody, Ranum and colleagues identify a specific CUGBP1 RNA target, the GABA-A transporter 4 (GAT4) transcript, which shows up-regulation and a shift in alternative splicing favoring exon 7 insertion in SCA8 mouse and patient brains, resembling expression features detected in the fetal cortex. Interestingly, these changes are specific to SCA8 patients, as GAT4 expression and splicing pattern are normal in DM1 brain. Finally, up-regulation of the GAT4 gene product, which is primarily detected in the granular cell layer of the cerebellar cortex in the SCA8 mouse, fits well with the increased physiological response of granular cells to mossy fiber inputs. The authors hypothesize that transcript-skipping exon 7 is targeted for nonsense-mediated decay, and that shifting in exon 7 insertion in SCA8 patients underlies GAT4 up-regulation. Together, these data indicate that the CUG-expansion transcript expressed at the SCA8 locus is indeed toxic.
Thus, both studies from Ranum's group provide the proof of concept that toxic RNA and toxic protein gain-of-function mechanisms can, together, be involved in SCA8, although the relative contribution of each mechanism remains to be determined. The next obvious question to address is whether bidirectional expression of CAG - and CUG-expansion transcripts is a general feature of CAG/CTG diseases and whether the combined toxic effects of RNA and protein gains-of-function could underlie the pathomechanisms involved in these diseases and account for their complexities. Bidirectional expression at the DM1 locus has been reported, but the antisense transcript does not appear to be translated into a polyQ protein [10]. More puzzling is the case of Huntington's disease–like 2 (HDL2), a dominant inherited disease closely resembling Huntington's disease and caused by CAG/CTG repeat expansion in the JPH3 gene [11]. While expression of CUG-expansion JPH3 transcripts forming toxic RNA foci was demonstrated, the presence of polyQ aggregates was highly suggested by immunodetection using an antibody specifically detecting polyQ expansions, i.e., the 1C2 antibody [12],[13]. If it happens that expression of antisense CUG-expansion transcripts is common in polyQ disorders, undoubtedly this will open new routes of investigation of their pathomechanism. Specifically, such a discovery could help to solve the issue of tissue selectivity, which remains puzzling. Indeed, the pattern of affected tissues correlates neither with the pattern of expression of CAG-expansion transcripts nor with the pattern of polyQ neuronal inclusions. In addition, degenerating neurons differ between polyQ diseases. For instance, the striatum preferentially degenerates in Huntington's disease, while the cerebellum is mostly affected in SCAs. The possible existence of antisense CUG-expansion transcripts could underlie some aspects of tissue selectivity. Toxic RNA effects could add to disease pathomechanism complexity. The relative contribution of both mechanisms (toxic RNA versus toxic protein) could also be tissue - or cell-specific, depending on the relative expression level of CAG - and CUG-expansion transcripts. Finally, data obtained from SCA3 Drosophila models suggested that CAG-expansion transcripts could also be toxic [14]. In that case, the toxicity threshold appeared to exceed 200 CAG repeats, which represent very rare disease alleles in polyQ disorders. One should, however, keep in mind that such high repeat numbers might be reached in some brain regions as a consequence of somatic repeat instability, as is illustrated in the case of Huntington's disease [15]. However, while toxic CUG diseases are typically thought to be associated with large expansions, in the SCA8 patients and mice studied by Ranum, the expansions were relatively short—within the range of many polyQ diseases. Thus, the potential connection between the RNA world and polyQ diseases now has to be clarified.
Fig. 1. Both toxic-protein (polyQ) and toxic-RNA gain-of-functions might underlie SCA8 pathogenesis. 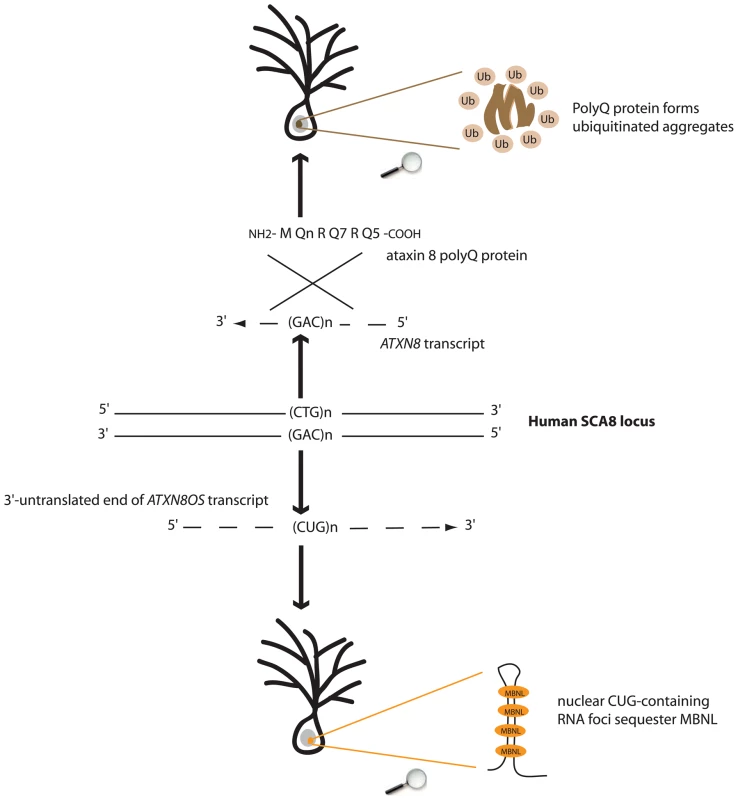
Bidirectional expression of CAG- and CUG-transcripts at the human SCA8 locus results in production of a polyQ protein forming ubiquitinated aggregates and accumulation of RNA foci sequestering MBNL splicing factor.
Zdroje
1. OrdwayJM
Tallaksen-GreeneS
GutekunstCA
BernsteinEM
CearleyJA
1997 Ectopically expressed CAG repeats cause intranuclear inclusions and a progressive late onset neurological phenotype in the mouse. Cell 91 753 763
2. WilliamsAJ
PaulsonHL
2008 Polyglutamine neurodegeneration: protein misfolding revisited. Trends Neurosci 31 521 528
3. O'RourkeJR
SwansonMS
2009 Mechanisms of RNA-mediated disease. J Biol Chem 284 7419 7423
4. RanumLP
CooperTA
2006 RNA-Mediated Neuromuscular Disorders. Annu Rev Neurosci 29 259 277
5. GatchelJR
ZoghbiHY
2006 Diseases of unstable repeat expansion: mechanisms and common principles. Nat Rev Genet 6 743 555
6. MoseleyML
ZuT
IkedaY
GaoW
MosemillerAK
2006 Bidirectional expression of CUG and CAG expansion transcripts and intranuclear polyglutamine inclusions in spinocerebellar ataxia type 8. Nat Genet 38 758 769
7. DaughtersRS
TuttleDL
GaoW
IkedaY
MoseleyML
2009 RNA gain-of-function in spinocerebellar ataxia type 8. PLoS Genet 5(8) e1000600 doi:10.1371/journal.pgen.1000600
8. KoobMD
MoseleyML
SchutLJ
BenzowKA
BirdTD
1999 An untranslated CTG expansion causes a novel form of spinocerebellar ataxia (SCA8). Nat Genet 21 379 384
9. MutsuddiM
MarshallCM
BenzowKA
KoobMD
RebayI
2004 The spinocerebellar ataxia 8 noncoding RNA causes neurodegeneration and associates with staufen in Drosophila. Curr Biol 14 302 308
10. ChoDH
ThienesCP
MahoneySE
AnalauE
FilippovaGN
2005 Antisense transcription and heterochromatin at the DM1 CTG repeats are constrained by CTCF. Mol Cell 20 483 489
11. HolmesSE
O'HearnE
RosenblattA
CallahanC
HwangHS
2001 A repeat expansion in the gene encoding junctophilin-3 is associated with Huntington disease-like 2. Nat Genet 29 377 378
12. RudnickiDD
HolmesSE
LinMW
ThorntonCA
RossCA
2007 Huntington's disease–like 2 is associated with CUG repeat-containing RNA foci. Ann Neurol 61 272 282
13. RudnickiDD
PletnikovaO
VonsattelJP
RossCA
MargolisRL
2008 A comparison of huntington disease and huntington disease-like 2 neuropathology. J Neuropathol Exp Neurol 67 366 374
14. LiLB
YuZ
TengX
BoniniNM
2008 RNA toxicity is a component of ataxin-3 degeneration in Drosophila. Nature 453 1107 1111
15. KennedyL
ShelbournePF
2000 Dramatic mutation instability in HD mouse striatum: does polyglutamine load contribute to cell-specific vulnerability in Huntington's disease? Hum Mol Genet 9 2539 2544
Štítky
Genetika Reprodukční medicína
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2009 Číslo 8
Nejčtenější v tomto čísle- Alternative Mechanisms for Tn Transposition
- SCA8 CAG/CTG Expansions, a Tale of Two ities: A Unique or Common Case?
- Forests of the Night: Refugia of Genetic Diversity in Wild Tigers
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



